Abstract
Superoxide dismutase–1 (SOD1) is a ubiquitous, Cu and Zn binding, free-radical defense enzyme whose misfolding and aggregation play a potential key role in amyotrophic lateral sclerosis, an invariably fatal neurodegenerative disease. Over 150 mutations in SOD1 have been identified with a familial form of the disease, but it is presently not clear what unifying features, if any, these mutants share to make them pathogenic. Here, we develop several unique computational assays for probing the thermo-mechanical properties of both ALS-associated and rationally designed SOD1 variants. Allosteric interaction-free energies between residues and metals are calculated, and a series of atomic force microscopy experiments are simulated with variable tether positions to quantify mechanical rigidity “fingerprints” for SOD1 variants. Mechanical fingerprinting studies of a series of C-terminally truncated mutants, along with an analysis of equilibrium dynamic fluctuations while varying native constraints, potential energy change upon mutation, frustratometer analysis, and analysis of the coupling between local frustration and metal binding interactions for a glycine scan of 90 residues together, reveal that the apo protein is internally frustrated, that these internal stresses are partially relieved by mutation but at the expense of metal-binding affinity, and that the frustration of a residue is directly related to its role in binding metals. This evidence points to apo SOD1 as a strained intermediate with “self-allostery” for high metal-binding affinity. Thus, the prerequisites for the function of SOD1 as an antioxidant compete with apo state thermo-mechanical stability, increasing the susceptibility of the protein to misfold in the apo state.
Keywords: protein misfolding, frustrated contacts, cooperativity, allosteric communication
Allosteric regulation canonically involves the modulation of a protein’s affinity for a given ligand A through the binding of a separate ligand B to a distinct spatial location on the protein. The modulatory binding site at the distinct location is referred to as the allosteric site, and the interaction between the allosteric site and the putative agonist binding site is referred to as an allosteric interaction. Early models of allostery were used to explain ligand saturation curves for hemoglobin in terms of subunit interactions that would induce binding cooperativity (1, 2). Cooperativity may be quantified through the nonadditivity in the binding energies of ligand and allosteric effector (3). More recently, allostery has been thought to be a more generic property present even in single-domain proteins (4). In this context, positive cooperativity may be modulated through frustrated intermediates (5), which may enhance the conformational changes observed upon ligand binding for allosteric proteins (6). In a single-domain protein, the notion of an allosteric effector can be generalized to intrinsic protein side chains that can enhance protein function or functionally important motion at the expense of native stability—a kind of “self-allostery.” Some allosteric activation mechanisms involve mediation of conformational switches by nonnative intraprotein interactions (7), an effect predicted by energy landscape approaches wherein barriers between conformational states are buffed to lower energies (8). The potential for novel allosteric regulators may vastly broaden candidate targets for drug discovery (9). The internal frustration required for cooperative allosteric function may have deleterious consequences, however, if protein stability is sufficiently penalized in intermediate states to enhance the propensity for misfolding and subsequent aberrant oligomerization, processes known to be involved in neurodegenerative disease (10). Here we show that the ALS-associated protein Cu, Zn superoxide dismutase–1 (SOD1) is embroiled in such a conflict between stability and function.
SOD1 is a homo-dimeric antioxidant enzyme of 32 kDa, wherein each monomer contains 153 amino acids, binds one Cu and one Zn ion, and consists largely of an eight-stranded greek key β barrel with two large, functionally important loops (11–13). Loop VII or the electrostatic loop (ESL, residues 121–142) enhances the enzymatic activity of the protein by inducing an electrostatic funnel toward a redox active site centered on the Cu ion (14). Loop IV or the Zn-binding loop (ZBL, residues 49–83) contains histidines H63, H71, and H80 as well as D83, which coordinate the Zn ion and, along with a disulfide bond between C57 and C146, enforce concomitant tertiary structure in the protein. The Cu, on the other hand, is coordinated by H46, H48, and H120 and the bridging histidine H63 between the Cu and Zn, which are located primarily in the Ig-like core of the protein.
Sporadic ALS (SALS) inclusions are immunoreactive to misfolding-specific SOD1 antibodies (15, 16). Such misfolded aggregates may be initiated from locally (rather than globally) unfolded states that become accessible, for example, via thermal fluctuations or rare events (17); for example, near-native aggregates or aggregation precursors were found for an obligate monomeric SOD1 variant (18) and for familial ALS (FALS) mutants S134N (19, 20) and E,E(SS) H46R (19). The above studies have motivated the present computational study, which focuses on the native and near-native thermo-mechanical properties of mutant, WT, and posttranslationally modified SOD1.
Results
Cumulative Distributions of Work Values Can Discriminate SOD1 Mutants and Premature Variants.
Mechanical probes were used by simulating tethers on the residue closest to the center of mass of the SOD1 variant, and on various residues on the protein surface. The experimental analog to such an in silico approach would require multiple atomic force microscopy (AFM) or optical trap assays involving numerous residue pairs about the protein surface as tethering points. This is difficult and time-consuming to achieve in practice, which thus provides an opportunity for the present simulation approaches. All proteins in this study were taken in the monomeric form—many of them, such as E,E(SH) SOD1, G127X, and G85R, are either naturally monomeric or have significantly reduced dimer stability. The ALS-associated truncation mutant G127X contains a frameshift insertion, which results in six nonnative amino acids following Gly127, after which a truncation sequence terminates the protein 20 residues short of the putative C terminus (21, 22). Here post-translational modifications (PTMs) refer to processes involved in the in vivo maturation of SOD1, including disulfide bond formation and metallation by Cu and Zn.
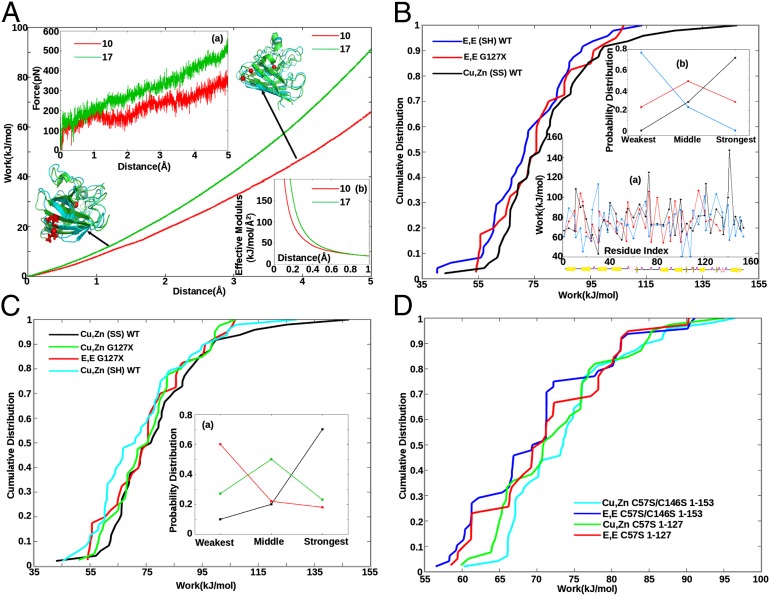
The simulated force-extension profile may be used to obtain the work required to pull a given residue to a given distance (Fig. 1A). In this assay, we found that large effective stiffness moduli were observed for small perturbing distances (Fig. 1 A, b). These were attributable primarily to side chain–side chain “docking” interactions in the native structure (SI Appendix, Fig. S2). The work to pull a given residue to a distance sufficient to constitute an anomalously large fluctuation (e.g., 5 Å) can be calculated as a function of sequence index for a given SOD1 variant, resulting in a characteristic mechanical profile or “mechanical fingerprint” for that protein (Fig. 1 B, a). The work values at 5 Å do not correlate with root mean square fluctuation (RMSF) values of the corresponding residues (23). A representative subset of 48 residues was obtained as described in the SI Appendix. We found that such a profile was independent of how the protein was initially constructed before equilibration—i.e., from what Protein Data Bank (PDB) structure—but was clearly different between WT SOD1 and mutants such as G127X, as well as other PTM variants such as E,E(SH) SOD1 (SI Appendix, Fig. S4). Table S1 in the SI Appendix gives the correlation coefficients between variants. A given PTM globally modulates the mechanical profile, inducing both local and nonlocal changes in stability that can be both destabilizing in some regions and stabilizing in others. A detailed study of the mechanical consequences of mutations or PTMs—e.g., by studying the long-range communication through interaction networks in the mutant vs. WT protein (e.g., refs. 24–27) is an interesting topic of future research.
Fig. 1.
Force (A, a), work (A), and effective modulus (A, b) as a function of extension. Tethers are placed at the Cα atom closest to the center of mass of the SOD1 monomer (H46), and the Cα atom of either residues G10 (red) or I17 (green), in separate pulling assays. Ribbon representations of the protein are also shown; the tethering residue is shown in licorice rendering (in red) and the center Cα as a red sphere. The initial equilibrated (at 0 Å, green ribbon) and final (at 5 Å, blue ribbon) structures are aligned to each other by minimizing RMSD. (B, a) Work profiles of Cu,Zn(SS) WT (black), E,E(SH) WT (blue), and E,E G127X SOD1 (red) vs. sequence index. Secondary structure schematic is shown underneath. (B) Cumulative distributions of the work values in B, a. E,E G127X is more stable than full-length E,E (SH) SOD1 (P = 9e-7). (B, b) Fraction of the 48 incidences that each variant had either the weakest, strongest, or middle work value—e.g., E,E(SH) SOD1 is weakest 80% of the time and is never the strongest variant. (C) Cumulative work distributions for Cu,Zn (SS) WT (black), Cu,Zn G127X (green), E,E G127X (red), and Cu,Zn (SH) WT (cyan). Cu,Zn G127X is destabilized with respect to full-length Cu,Zn (SS) WT (P = 6.2e-8). (C, a) Same analysis as B, b for the variants Cu,Zn(SS) WT, Cu,Zn G127X, and E,E G127X . (D) Cumulative distributions for serine mutant SOD1 variants demonstrate that C-terminal truncation stabilizes the apo form but destabilizes the holo form (Results).
We also found that the mechanical work profile was nearly independent of whether an explicit or implicit solvent model was used in the simulations, even though native basin fluctuations (RMSF) showed significant scatter in comparing implicit and explicit solvent models (SI Appendix, Fig. S5). Equilibrium fluctuations are much more sensitive to solvent models than the large-scale perturbations we consider here. On the other hand, a structure-based Gō model does only a modest job of capturing the mechanical profile and is poor in particular for residues where electrostatic interactions play a significant role in stability (SI Appendix, Fig. S6). Interestingly, the default energy scale of 1 kJ/mol for each contact or dihedral interaction used in current Gō models (28) captures the overall energy scale of the work profile quite well for SOD1, although it must likely be modified for other proteins such as thermophilic proteins.
In support of the thermodynamic relevance of a SOD1 variant’s mechanical fingerprint, we found that the nonequilibrium work values obtained by pulling a residue to 5 Å strongly correlate with the equilibrium-free energy change for the same process, as calculated using the weighted histogram analysis method (WHAM) method (r = 0.96; SI Appendix, Fig. S7). Thus, the work profiles obtained are an accurate measure of the corresponding local thermodynamic stability profile, up to a scaling factor.
Comparing the mechanical fingerprints of E,E(SH) SOD1 and that of G127X (Fig. 1 B, a), we see that many discrepancies between SOD1 variants and WT are difficult to disentangle solely from the work profile, or even from histograms of the work values (SI Appendix, Fig. S3). However, the mechanical discrepancies emerged quite naturally from the cumulative distribution of work values. Mechanical scans were thus used to construct the cumulative distributions, which then allowed us to distinguish stabilizing energetics in various forms of PTM SOD1—e.g., between E,E(SH), G127X, and Cu,Zn(SS) SOD1 in Fig. 1B. Not all residues needed to be sampled to obtain the cumulative distribution; we found convergence to within ∼1 kJ/mol after a sample size of about 40 residues (SI Appendix, Fig. S8). This does not mean the mechanical profile obtained from 48 residues is representative of the stability in the corresponding regions of the protein: the mechanical work values were essentially uncorrelated for amino acids about three residues apart.
Comparison of the cumulative work distributions shows that the ALS-associated mutant Cu,Zn(SS) A4V is slightly more mechanically malleable than Cu,Zn(SS) WT, particularly in the weaker regions of the protein (SI Appendix, Fig. S13). However, the mechanical weakening due to mutation is substantially larger in the E,E(SS) state and largest in the E,E(SH) state (green curves in SI Appendix, Fig. S13). Experimental measurements of melting temperature also have shown increased susceptibility to mutation in the E,E(SH) state for A4V (29). We also note from SI Appendix, Fig. S13 that disulfide reduction in the WT apo state, E,E(SS) → E,E(SH), actually increases the rigidity of the native basin (23), even though the net thermodynamic stability is decreased, primarily due to increased unfolded entropy (30).
Lack of Posttranslational Modifications Mechanically Destabilizes SOD1, However the Truncation Mutant G127X stabilizes the Apo, Disulfide-Reduced Protein.
The cumulative distribution of E,E(SH) lies to the left of that for Cu,Zn(SS) SOD1 in Fig. 1B for all rank-ordered work values, illustrating that that variant is more malleable and thus more susceptible to perturbing forces that might induce the conformational changes accompanying misfolding. Likewise, Cu,Zn(SH) is destabilized with respect to Cu,Zn(SS) in Fig. 1C, and the other PTM variants in SI Appendix, Fig. S14 show that lack of PTMs reduces the mechanical rigidity.
On the other hand, the E,E G127X truncation mutant, which lacks both metals, a disulfide bond, and part of the sequence, shows increased mechanical stability over E,E(SH) WT (P = 9e-7, Fig. 1B). Comparing the profiles, E,E(SH) is most commonly the weakest, Cu,Zn(SS) is most commonly the strongest, and G127X is most commonly in the middle (Fig. 1 B, b). Apparently the C-terminal region of the protein mechanically stresses the remainder of the protein when PTMs are absent, reducing its mechanical stability. However, a metastable holo variant of G127X containing metals in their putative positions is less mechanically stable than holo WT (P = 6.2e-8, Fig. 1C). In the full-length holo protein, native stabilizing interactions are more apt to be minimally frustrated, and thus C-terminal truncation induces softening of the native structure. The metallation of G127X only marginally stabilizes it (P = 6.7e-3, Fig. 1 C, a), while metallation of WT protein results in substantial mechanical stabilization (SI Appendix, Fig. S14). G127X lacks C146, so Cu,Zn(SH) WT may be a more appropriate comparison than Cu,Zn(SS) WT. Cu,Zn(SH) SOD1 is less stable than either Cu,Zn G127X or E,E G127X (Fig. 1C), however Cu,Zn(SH) contains a protononated Cysteine C146, which is destabilizing and not present in G127X .
To deconvolute the effects of protonated Cysteines, we examined a set of four serine mutant proteins: Cu,Zn C57S/C146S 1–153 (full-length), E,E C57S/C146S 1–153, Cu,Zn C57S 1–127 (truncated at residue 127), and E,E C57S 1–127. For this system of proteins, Cu,Zn C57S/C146S 1–153 is clearly the most stable over most of the range of work values (Fig. 1D), and Cu,Zn C57S 1–127 is destabilized with respect to it. E,E C57S 1–127 is further destabilized with respect to its holo form, but is more stable than the full-length apo form E,E C57S/C146S 1–153. This demonstrates that C-terminal truncation stabilizes the apo form but destabilizes the holo form. This conclusion is robust to changes in pulling distance (SI Appendix, Fig. S15).
Short-Length C-Terminal Truncation Mechanically Stabilizes the Apo Protein, While Sufficiently Long C-Terminal Truncation Destabilizes It.
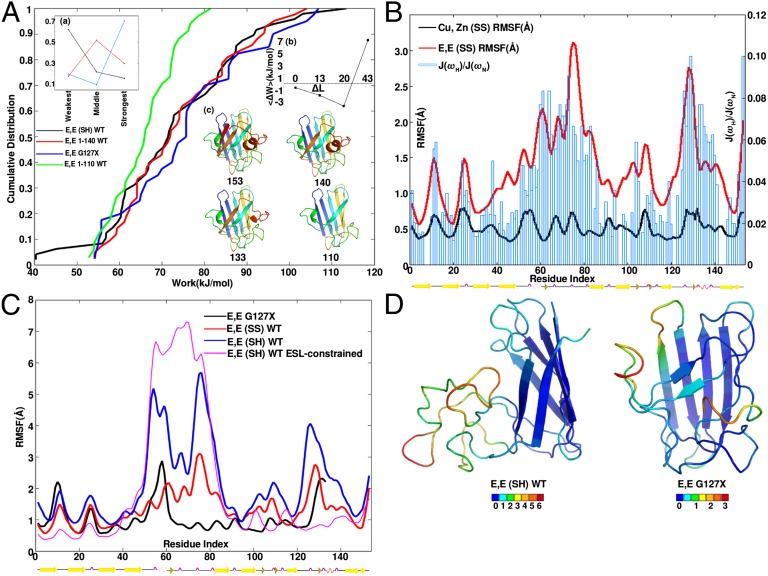
The mechanical properties of two additional truncated constructs, E,E 1–140 (WT sequence) and E,E 1–110, were assayed to investigate the crossover from increased to eventually reduced mechanical stability as the length of truncation is increased (Fig. 2 A, b). All truncation variants are missing the putative disulfide bond. Fig. 2A shows cumulative distributions for the above variants along with E,E(SH) SOD and E,E G127X. The truncation E,E 1–140 mechanically stabilizes E,E(SH) SOD1 (P = 1e-3), and G127X is further stabilized with respect to 1–140 (P = 9e-4). Comparison of the triplets of rank-ordered work values corresponding to E,E(SH), E,E 1–140, and E,E G127X supports this conclusion (Fig. 2 A, a). We have omitted the three least stable work values for all variants in calculating statistical significance; these are outliers for E,E(SH) that would dominate the result toward the conclusion that we have arrived at without their inclusion. Variant E,E 1–110 has significantly compromised mechanical rigidity and may not be thermodynamically stable; the mechanical assay only probes malleability of the native basin.
Fig. 2.
(A) Cumulative distributions of work values for C-terminal–truncated SOD1 variants of variable sequence length show a nonmonotonic trend in mechanical stability. All variants are metal-depleted and have no disulfide bond. Sequences are given in the legend (Results). (A, a) Comparing the cumulative distributions in A, that of the mutant G127X is most commonly the strongest, full-length SOD1 is most commonly the weakest, and 1–140 is most often in the middle. (A, b) Change in work value 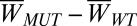 averaged over residues, as a function truncation length, for the SOD1 variants in A. (A, c) Ribbon schematics of the various truncation mutants, colored blue to red from N to C terminus, labeled by C-terminal residue. (B) Simulated native-basin dynamical fluctuations (RMSF) in explicit simple point charge (SPC) solvent, for Cu,Zn(SS) (black) and E,E(SS) SOD1 monomer (red), along with the experimentally measured ratio of spectral density functions J(ωH)/J(ωN) of obligate monomeric E,E(SS) F50E/G51E/E133Q SOD1 (blue bars) (31). Correlation coefficient is r = 0.78. (C) Simulated RMSF for SOD1 variants E,E G127X (black), E,E(SS) (red), E,E(SH) (blue), and E,E(SH) with the ESL constrained to be natively structured (magenta). The presence of native stress is indicated by the increased disorder of the ZBL upon structuring the ESL (Results). (D) Snapshots of typical structures of E,E(SH) and G127X SOD1 from equilibrium simulations, color coded by the mean RMSF for each residue; RMSF increases from blue to red according to the scale bars shown.
averaged over residues, as a function truncation length, for the SOD1 variants in A. (A, c) Ribbon schematics of the various truncation mutants, colored blue to red from N to C terminus, labeled by C-terminal residue. (B) Simulated native-basin dynamical fluctuations (RMSF) in explicit simple point charge (SPC) solvent, for Cu,Zn(SS) (black) and E,E(SS) SOD1 monomer (red), along with the experimentally measured ratio of spectral density functions J(ωH)/J(ωN) of obligate monomeric E,E(SS) F50E/G51E/E133Q SOD1 (blue bars) (31). Correlation coefficient is r = 0.78. (C) Simulated RMSF for SOD1 variants E,E G127X (black), E,E(SS) (red), E,E(SH) (blue), and E,E(SH) with the ESL constrained to be natively structured (magenta). The presence of native stress is indicated by the increased disorder of the ZBL upon structuring the ESL (Results). (D) Snapshots of typical structures of E,E(SH) and G127X SOD1 from equilibrium simulations, color coded by the mean RMSF for each residue; RMSF increases from blue to red according to the scale bars shown.
Simulated Fluctuations Correlate with Experimental Spectral Density Functions for Apo SOD1.
We performed 20 ns equilibrium simulations in explicit solvent (SPC water model) for Cu,Zn(SS) and E,E(SS) SOD1. Native-basin RMSF values show significant scatter between explicit and implicit solvent models compared with mechanical work values (SI Appendix, Fig. S5), so for analysis of equilibrium fluctuations, explicit solvent is used. Fig. 2B plots the resulting equilibrium RMSF for Cu,Zn(SS) and E,E(SS) SOD1; SI Appendix, Fig. S9 also gives the solvent-accessible surface area of backbone amide Nitrogens (SASAN) for the same equilibrium trajectory. The main effect of metal loss is to induce solvent exposed, disordered, and dynamic Zn-binding and ESLs (loops IV and VII). A moderate increase in dynamics is observed for loop VI as well. The preferential increase in dynamics of the ZBL and ESL upon metal loss is consistent with experimental measurements of the ratio of spectral density functions J(ωH)/J(ωN), a measure of dynamics fast compared with the tumbling rate (correlation coefficient r = 0.78). The experimental measurements of spectral density (31) are obtained for a monomeric E,E(SS) SOD1 mutant F50E/G51E/E133Q (32).
Dynamic Fluctuations in C-Terminal Truncation Mutants Reveal Native Frustration in Apo SOD.
Because G127X is more mechanically stable than E,E(SH) SOD1, native basin fluctuations were investigated to see if the extra stability of G127X was recapitulated in equilibrium dynamics. Fig. 2C shows that indeed the RMSF are substantially enhanced in E,E(SH) relative to G127X, in particular in the ZBL and ESL (loops IV and VII). This is true even though β-strand 8, N-terminal to the ESL, constrains E,E(SH) and is absent in G127X. Although more dynamic and mechanically malleable, E,E(SH) is not more solvent-exposed (by SASAN) than G127X, indicating a collapsed, dynamic globule with nonnative interactions (SI Appendix, Fig. S10). As well, E,E(SH) is more dynamic than E,E(SS) SOD1 (Fig. 2C), but less solvent exposed than E,E(SS), particularly in the ZBL and ESL (SI Appendix, Fig. S10). Collapse and nonnative interactions are not hindering the dynamics of the ZBL and ESL; snapshots from simulations for E,E(SH) WT and G127X are shown in Fig. 2D. Similar condensation phenomena are also observed in prion protein (33) and Na+/H+ exchanger regulatory factor (NHERF1) (34). In a large-scale study of 253 proteins across several-fold families (35), RMSF and SASA showed poor correlation (r ≈ 0.35 for backbone carbons).
It is intriguing that the additional constraint of structuring β8 in E,E(SH) SOD1 results in enhanced rather than suppressed disorder in the ZBL. One potential explanation is that because there are more residues in the ESL present in E,E(SH) than in G127X, the ESL forces the ZBL to be more expanded by steric repulsion, and thus more dynamic—i.e., a “polymer brush” effect. Another possible explanation is that the order induced by structuring β8 in E,E(SH) SOD1 induces frustration and consequent strain elsewhere in the protein, resulting in induced disorder in the ZBL. We differentiated these two scenarios by applying native constraints between the Cα atoms in the ESL and the rest of protein, but excluding contacts between the ESL and the ZBL: harmonic springs were applied to all pairs of Cα atoms within 4 Å in the native holo structure that involved contacts either within residues 133–153 or between residues 133–153 and either residues 1–40 or 90–153. This procedure further constrains the ESL and β8 to be natively structured, removing any polymer brush effect, but enhancing any native strain. Consistent with a model involving native frustration, the ZBL loop IV becomes more disordered in E,E(SH) SOD1 upon implementing ESL/β8 native constraints (Fig. 2C).
“Frustratometer” Results and Potential Energy Changes Support Increased Frustration in Apo WT SOD1 with Respect to Apo ALS-Associated Mutants.
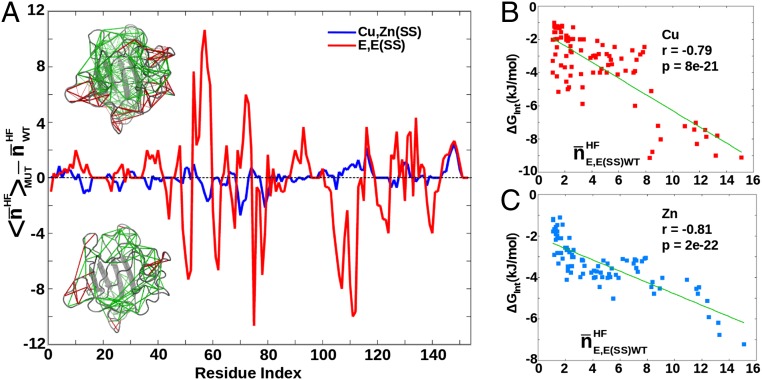
Typically frustrated contacts, according to the “frustratometer” method developed by Wolynes, Ferreiro, and colleagues (5, 36), were found by averaging 50 snapshots from an equilibrium ensemble for WT SOD1, and for each of 22 ALS-associated SOD1 mutants (SI Appendix, Table S3). Both E,E(SS), and Cu,Zn(SS) states were analyzed. Results were averaged over the 22 mutants to yield the mean number of frustrated contacts at a given residue position, for the “average” mutant. Taking the difference of this quantity with that for WT SOD1 gives the mean change in frustration upon mutation, which increased for holo SOD1 by about five total contacts but decreased for apo SOD1 by about 22 total contacts (Fig. 3A). This result again supports a frustrated apo state in SOD1. Direct computation shows on average about 38 more highly frustrated contacts in E,E(SS) SOD1 than in Cu,Zn(SS) SOD1 (SI Appendix, Fig. S17).
Fig. 3.
(A, top cartoon) Frustrated contacts (in red) and unfrustrated contacts (in green) for E,E(SS) WT SOD1. (A, bottom cartoon) Same contacts as top cartoon for the average over 22 ALS E,E(SS) mutants (Results). (A) The mean number of frustrated contacts within a sphere of radius 5 Å centered on each Cα atom is found as a function of residue index. Ensemble averages are taken from 50 snapshots in an equilibrium simulation. This is done for both the Cu,Zn(SS) state and the E,E(SS) state, for both the WT sequence, and for 22 mutant sequences. The 22 mutant sequences are averaged to obtain the ensemble and mutant-averaged number of contacts as a function of residue index i, 〈nHF(i)〉MUT. Plotted is the difference between 〈nHF(i)〉MUT and the corresponding numbers for the WT sequence 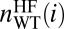 . A positive number would indicate an increase in frustration upon mutation. Holo state is shown in blue and has an average of +5 contacts; apo state is shown in red and has an average of –22 contacts. (B) Interaction-free energy between a residue’s side chain and the Cu ion, plotted as a function of the E,E(SS) ensemble-averaged number of highly frustrated contacts that residue has (r = –0.79, P = 8e-21). (C) Same as in B but for the Zn ion (r = –0.81, P = 2e-22).
. A positive number would indicate an increase in frustration upon mutation. Holo state is shown in blue and has an average of +5 contacts; apo state is shown in red and has an average of –22 contacts. (B) Interaction-free energy between a residue’s side chain and the Cu ion, plotted as a function of the E,E(SS) ensemble-averaged number of highly frustrated contacts that residue has (r = –0.79, P = 8e-21). (C) Same as in B but for the Zn ion (r = –0.81, P = 2e-22).
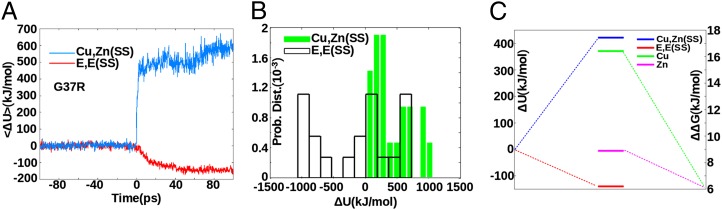
For the same set of mutants, we found that the ensemble-averaged total potential energy in the native state increased upon mutation for the Cu,Zn(SS) protein, but decreased upon mutation in the E,E(SS) protein. This is consistent with the above frustratometer results. The “time-resolved” change in potential energy ΔU upon in silico mutation for a representative mutant (G37R) is shown in Fig. 4A. The distribution of potential energy changes for the 22 ALS-associated mutants listed in SI Appendix, Table S3 is shown in Fig. 4B, which shows potential energy “cost” upon mutation for Cu,Zn(SS) mutants, but a significant shift toward negative stabilizing values for E,E(SS) mutants. The same conclusion is obtained from 30 non-ALS alanine mutants (SI Appendix, Fig. S18). Fig. 4C shows that the net effect of mutation on the potential energy is to increase it on average in the holo state, but to decrease it on average in the apo state, indicating stabilization. Moreover, all ALS mutants facilitate metal release (23); the net effect of mutation on the Cu and Zn binding-free energies is to decrease both of them. Thus, mutations relieve stress in the apo state, while inducing loss of metal binding function. It thus appears that apo SOD1 has evolved to have high affinity for metals at the expense of native stability and increased frustration.
Fig. 4.
(A) Change in potential energy ΔU(t) as a function of in silico time, before and after implementing the mutation G37R. (B) Distribution of the asymptotic potential energy change ΔU(∞) for 22 ALS mutants (Results). (C) Mean potential energy change averaged over mutants, for both the holo state and the apo state, along with the mean difference, WT minus mutants, in both Cu and Zn binding-free energy.
Frustrating Residues in the Apo State Are Allosteric Effectors and Positively Modulate Metal Affinity in Proportion to Their Frustration.
To test the extent to which the residues facilitating metal binding are frustrated, we have calculated the interaction free energy of each WT residue with both Cu and Zn, by considering thermodynamic cycles (3) involving metallation and residue “insertion” from a glycine at the corresponding position (e.g., G4A). The interaction-free energy Gint between each residue side-chain and either Zn or Cu is given (here specifically for residue Ala-4 with Zn) by:
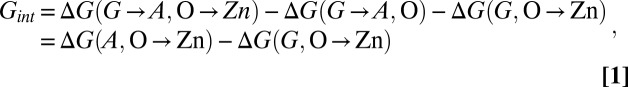 |
where ΔG(A, O→Zn) is the free-energy change of Zn insertion when alanine is present at position 4, and ΔG(G, O→Zn) is the free-energy change of Zn insertion when glycine (no side chain) is present at position 4. The other residues, and Cu interactions, are handled analogously (SI Appendix, Methods).
Fig. 3 B and C plots the above interaction-free energy of a residue with Cu or Zn versus the E,E(SS) ensemble-averaged number of highly frustrated contacts that residue has. Data are obtained for 90 glycine mutants listed in SI Appendix, Table S3, for residues that had at least one highly frustrated contact (SI Appendix). The values of Gint for all 90 mutants listed in SI Appendix, Table S3 are negative, indicating cooperative interactions, wherein the WT residue facilitates binding of the metal. Each of these residues can be thought of as allosteric effectors, positively modulating affinity for either metal. For both Cu and Zn, the larger the degree of frustration in the apo state, the larger the role that residue has in facilitating metal binding. No such trend is seen for Cu,Zn(SS) SOD1 (SI Appendix, Fig. S18). This result strongly supports a model of the apo state as an allosteric intermediate designed for high metal binding affinity at the expense of structural stability.
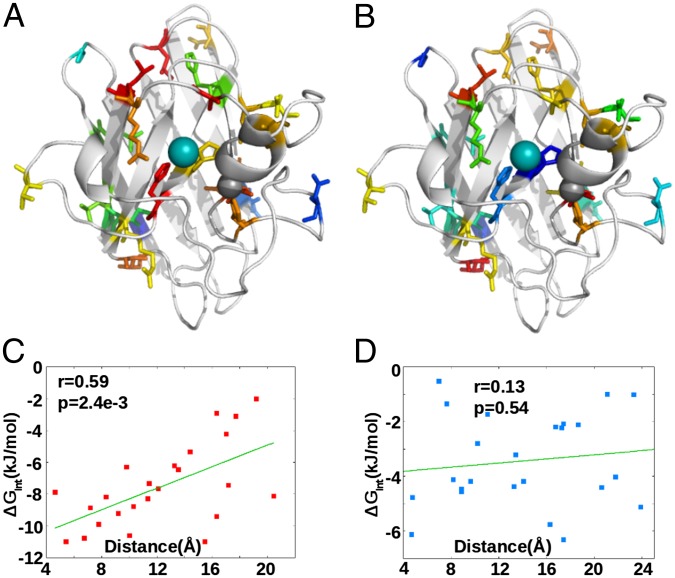
Finally, we test the distance dependence of the interaction-free energy with the metals for the set of 24 ALS-associated mutants in SI Appendix, Table S3. In the case of Cu, the allosteric regulation for metal affinity is significantly correlated with the proximity of the residue to the Cu binding site (Fig. 5 A and C). Interestingly, the allosteric regulation for Zn binding is uncorrelated with distance to the Zn binding site and thus nonlocal. This finding is consistent with experimental results that Zn binding is concomitant with large structural change (partial folding) of the protein (37). Long-range coupling to the Zn-binding region has also been observed in G93A SOD1 (38). The same conclusion is obtained for the 90 frustrated residues given in SI Appendix, Table S3 (SI Appendix, Fig. S19).
Fig. 5.
(A) Residues color-coded by interaction energy with the Cu ion (depicted as a cyan sphere). The extent of interaction is strongest in magnitude for red colored residues and decreases to blue. (B) Same as A for the Zn ion (depicted as a gray sphere). (C) Interaction energy with Cu correlates with the distance of the residue from the Cu ion; residues in close proximity more strongly interact. (D) Interaction energy with Zn does not correlate with distance of the residue to the Zn ion, indicating nonlocal allosteric effects.
Discussion
We have found here a connection between the allosteric design of residues in premature SOD toward high metal-binding affinity and the consequent frustration in the apo state of the protein. A variety of results supported this conclusion. Mechanical profiles were obtained from in silico AFM assays with variable tether positions; in this context it was found that the C-terminal truncation mutant G127X had higher mechanical rigidity than E,E(SH) SOD1, implying the release of internal stresses upon removal of part of the protein. Large truncation lengths eventually destabilized the protein.
The higher malleability of E,E(SH) SOD1 over G127X is recapitulated by larger equilibrium dynamical fluctuations in the native basin. Constraining loop VII (the ESL) to be natively structured only increases fluctuations in loop IV (the ZBL), ruling out polymer brush effects and supporting the native stress hypothesis.
Implementing Wolynes’s “frustratometer” method (5, 36) shows that, perhaps surprisingly, more frustration is present on average in the WT apo state than is present for apo mutants. For the holo state the situation is reversed, however, which is consistent with the notion that mutants facilitate metal release. In fact, every ALS-associated mutant we have studied lowered the affinity for both Cu and Zn (23). We have found here that while these mutants raise the potential energy of the holo state, they tend to lower the potential energy of the apo state and thus stabilize it, consistent with frustratometer results. A general comparative analysis of the decrease in potential energy and frustration for apo ALS-associated mutants, along with the results from their individual mechanical scans, which generally show weakening with respect to local perturbations, is an interesting topic for future work. SI Appendix, Fig. S20 compares the relevant quantities for the ALS mutants A4V and G127X.
Residues in apo SOD1 can be thought of as allosteric effectors for metal binding. By considering the cooperativity in thermodynamic cycles involving mutation to glycine and metal release, we quantified the interaction-free energies between residues in the protein and either Cu or Zn. All interaction energies were negative, indicating positive modulation of metal affinity, Moreover we found that function frustrates stability: the stronger the interaction energy, the more frustrated the residue. For Cu, the strength of the interaction significantly correlates with proximity to the binding site. For Zn, however, there is no correlation with proximity, indicating a nonlocal allosteric mechanism involving propagation of stress release throughout the protein and consistent with the large structural changes accompanying Zn binding.
The above evidence points toward a paradigm wherein sequence evolution toward high metal affinity results in a tradeoff for significant native frustration in the apo state of SOD1. A similar conclusion has been reached from studies of a SOD1 variant with Zn-coordinating ligands H63, H71, H80, and D83 mutated to S, which in the apo form is stabilized with respect to E,E WT SOD1 (39). In this context, the C-terminal truncation in G127X can be seen as an allosteric inhibition mechanism to Zn binding, in that frustration is relieved in the apo native state, but Zn-binding function is lost. A similar scenario is observed for select mutants of subtilisin, a serine protease whose function is regulated by Ca2+ binding. In this protein, the mutation M50F preferentially stabilizes apo subtilisin relative to the holo form, while weakening calcium binding and promoting inactivation in the holo form (40).
Native frustration in apo SOD1 as a result of allosteric cooperativity in metal binding has potential consequences for the misfolding of SOD1. The premature protein, or a protein that perhaps due to an external agent has lost its metals, would show decreased thermal stability relative to one that had not undergone sequence evolution for high metal affinity. In this sense, the tight binding of Zn and Cu essential for enzymatic function of the mature protein as an antioxidant puts the premature form in additional peril for misfolding.
Materials and Methods
A full description of the methods is given in the SI Appendix. Missense and truncation mutants of SOD1, both ALS-associated and rationally designed, were equilibrated and used for mechanical force, dynamic fluctuation, frustratometer, potential energy, and WHAM metal affinity assays. Rationally designed truncation and missense mutants studied here include C57S/C146S, C57S 1–127, and WT sequences 1–110 and 1–140. Frustration and metal-binding allostery assays used either 22 and 24 ALS-associated mutants, respectively, or 90 glycine mutants (SI Appendix, Table S3). Mechanical profiles are obtained after 20 ns pre-equilibration from steered molecular dynamics (MD) simulations (tether speed 2.5 mm/s) in Generalized Born surface area (GBSA) solvent with optimized potentials for liquid simulations using local second-order Møller-Plesset perturbation theory (OPLS-aa/L) force field parameters. Robustness checks are shown in SI Appendix, Fig. S15. Monte Carlo methods yield the statistical significance (error ≈ 2.7 kJ/mol, SI Appendix, Fig. S21). Fluctuation analysis used SPC explicit solvent. Metal binding-free energies are found from WHAM including postrelaxation and validation by thermodynamic cycles. Frustration calculations include protein conformations and protein–protein contacts only; that is, metals are implicit in determining protein conformation but metal–protein interactions are not explicitly included. Frustrated contacts were calculated using the frustratometer server http://lfp.qb.fcen.uba.ar/embnet/.
Supplementary Material
Acknowledgments
We thank Neil Cashman, Will Guest, Ali Mohazab, Eric Mills, Paul Whitford, and Stephen Toope for helpful and supportive discussions. We acknowledge funding from PrioNet Canada, Natural Sciences and Engineering Research Council funding to defray page charge costs, and we acknowledge computational support from the WestGrid high-performance computing consortium.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216597110/-/DCSupplemental.
References
- 1.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 2.Koshland DE, Jr, Némethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 3.Weber G. Energetics of ligand binding to proteins. Adv Protein Chem. 1975;29:1–83. doi: 10.1016/s0065-3233(08)60410-6. [DOI] [PubMed] [Google Scholar]
- 4.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57(3):433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 5.Ferreiro DU, Hegler JA, Komives EA, Wolynes PG. On the role of frustration in the energy landscapes of allosteric proteins. Proc Natl Acad Sci USA. 2011;108(9):3499–3503. doi: 10.1073/pnas.1018980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daily MD, Gray JJ. Local motions in a benchmark of allosteric proteins. Proteins. 2007;67(2):385–399. doi: 10.1002/prot.21300. [DOI] [PubMed] [Google Scholar]
- 7.Gardino AK, et al. Transient non-native hydrogen bonds promote activation of a signaling protein. Cell. 2009;139(6):1109–1118. doi: 10.1016/j.cell.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plotkin SS, Wolynes PG. Buffed energy landscapes: Another solution to the kinetic paradoxes of protein folding. Proc Natl Acad Sci USA. 2003;100(8):4417–4422. doi: 10.1073/pnas.0330720100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christopoulos A. Allosteric binding sites on cell-surface receptors: Novel targets for drug discovery. Nat Rev Drug Discov. 2002;1(3):198–210. [Google Scholar]
- 10.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 11.Tainer JA, Getzoff ED, Beem KM, Richardson JS, Richardson DC. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J Mol Biol. 1982;160(2):181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- 12.Bertini I, Manganl S, Viezzoli MS. In: Structure and Properties of Copper-Zinc Superoxide Dismutases. Adv Inorg Chem. Sykes A, editor. Vol 45. San Diego: Academic; 1998. pp. 127–250. [Google Scholar]
- 13.Valentine JS, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 14.Getzoff ED, et al. Faster superoxide dismutase mutants designed by enhancing electrostatic guidance. Nature. 1992;358(6384):347–351. doi: 10.1038/358347a0. [DOI] [PubMed] [Google Scholar]
- 15.Bosco DA, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13(11):1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg K, et al. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS ONE. 2010;5(7):e11552. doi: 10.1371/journal.pone.0011552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5(1):15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 18.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: Parallels to precursors in amyloid disease. Proc Natl Acad Sci USA. 2006;103(27):10218–10223. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elam JS, et al. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat Struct Biol. 2003;10(6):461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 20.Banci L, et al. Fully metallated S134N Cu,Zn-superoxide dismutase displays abnormal mobility and intermolecular contacts in solution. J Biol Chem. 2005;280(43):35815–35821. doi: 10.1074/jbc.M506637200. [DOI] [PubMed] [Google Scholar]
- 21.Jonsson PA, et al. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127(Pt 1):73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 22.Grad LI, et al. Intermolecular transmission of superoxide dismutase 1 misfolding in living cells. Proc Natl Acad Sci USA. 2011;108(39):16398–16403. doi: 10.1073/pnas.1102645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das A, Plotkin SS. Mechanical probes of SOD1 predict systematic trends in metal and dimer affinity of ALS-associated mutants. J Mol Biol. 2013 doi: 10.1016/j.jmb.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc Natl Acad Sci USA. 2006;103(9):3147–3152. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potter SZ, et al. Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J Am Chem Soc. 2007;129(15):4575–4583. doi: 10.1021/ja066690+. [DOI] [PubMed] [Google Scholar]
- 26.Schuyler AD, Carlson HA, Feldman EL. Computational methods for identifying a layered allosteric regulatory mechanism for ALS-causing mutations of Cu-Zn superoxide dismutase 1. Proteins. 2011;79(2):417–427. doi: 10.1002/prot.22892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards SA, Wagner J, Gräter F. Dynamic prestress in a globular protein. PLOS Comput Biol. 2012;8(5):e1002509. doi: 10.1371/journal.pcbi.1002509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitford PC, et al. An all-atom structure-based potential for proteins: Bridging minimal models with all-atom empirical forcefields. Proteins. 2009;75(2):430–441. doi: 10.1002/prot.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furukawa Y, O’Halloran TV. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J Biol Chem. 2005;280(17):17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 30.Hörnberg A, Logan DT, Marklund SL, Oliveberg M. The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn superoxide dismutase. J Mol Biol. 2007;365(2):333–342. doi: 10.1016/j.jmb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 31.Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. Solution structure of Apo Cu,Zn superoxide dismutase: Role of metal ions in protein folding. Biochemistry. 2003;42(32):9543–9553. doi: 10.1021/bi034324m. [DOI] [PubMed] [Google Scholar]
- 32.Bertini I, Piccioli M, Viezzoli MS, Chiu CY, Mullenbach GT. A spectroscopic characterization of a monomeric analog of copper, zinc superoxide dismutase. Eur Biophys J. 1994;23(3):167–176. doi: 10.1007/BF01007608. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Guest W, Huang A, Plotkin SS, Cashman NR. Immunological mimicry of PrPC-PrPSc interactions: Antibody-induced PrP misfolding. Protein Eng Des Sel. 2009;22(8):523–529. doi: 10.1093/protein/gzp038. [DOI] [PubMed] [Google Scholar]
- 34.Cheng H, et al. Autoinhibitory interactions between the PDZ2 and C-terminal domains in the scaffolding protein NHERF1. Structure. 2009;17(5):660–669. doi: 10.1016/j.str.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson NC, Daggett V. Dynameomics: Large-scale assessment of native protein flexibility. Protein Sci. 2008;17(12):2038–2050. doi: 10.1110/ps.037473.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenik M, et al. Protein frustratometer: a tool to localize energetic frustration in protein molecules. Nucleic Acids Res. 2012;40:W348–W351. doi: 10.1093/nar/gks447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts BR, et al. Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS. J Mol Biol. 2007;373(4):877–890. doi: 10.1016/j.jmb.2007.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Museth AK, Brorsson AC, Lundqvist M, Tibell LA, Jonsson BH. The ALS-associated mutation G93A in human copper-zinc superoxide dismutase selectively destabilizes the remote metal binding region. Biochemistry. 2009;48(37):8817–8829. doi: 10.1021/bi900703v. [DOI] [PubMed] [Google Scholar]
- 39.Nordlund A, et al. Functional features cause misfolding of the ALS-provoking enzyme SOD1. Proc Natl Acad Sci USA. 2009;106(24):9667–9672. doi: 10.1073/pnas.0812046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryan PN. Protein engineering of subtilisin. Biochim Biophys Acta. 2000;1543(2):203–222. doi: 10.1016/s0167-4838(00)00235-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.