Abstract
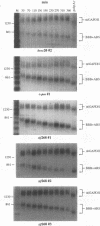
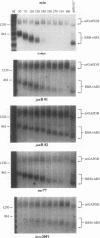
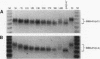
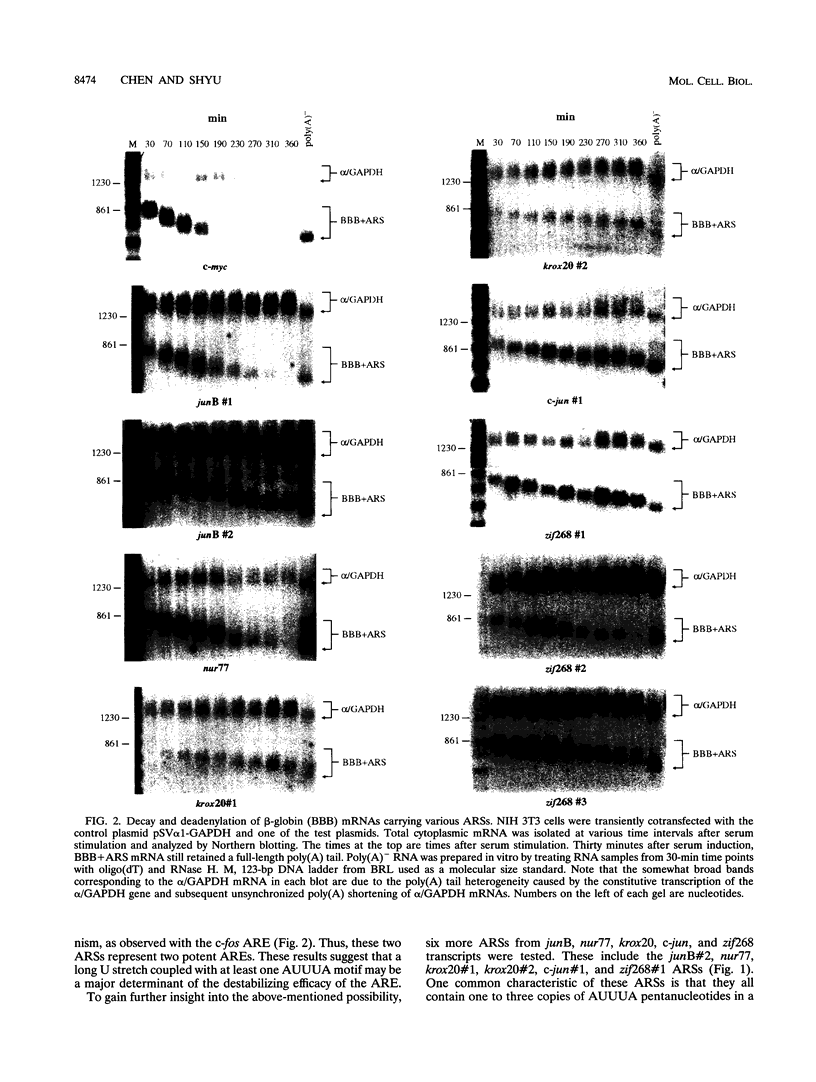
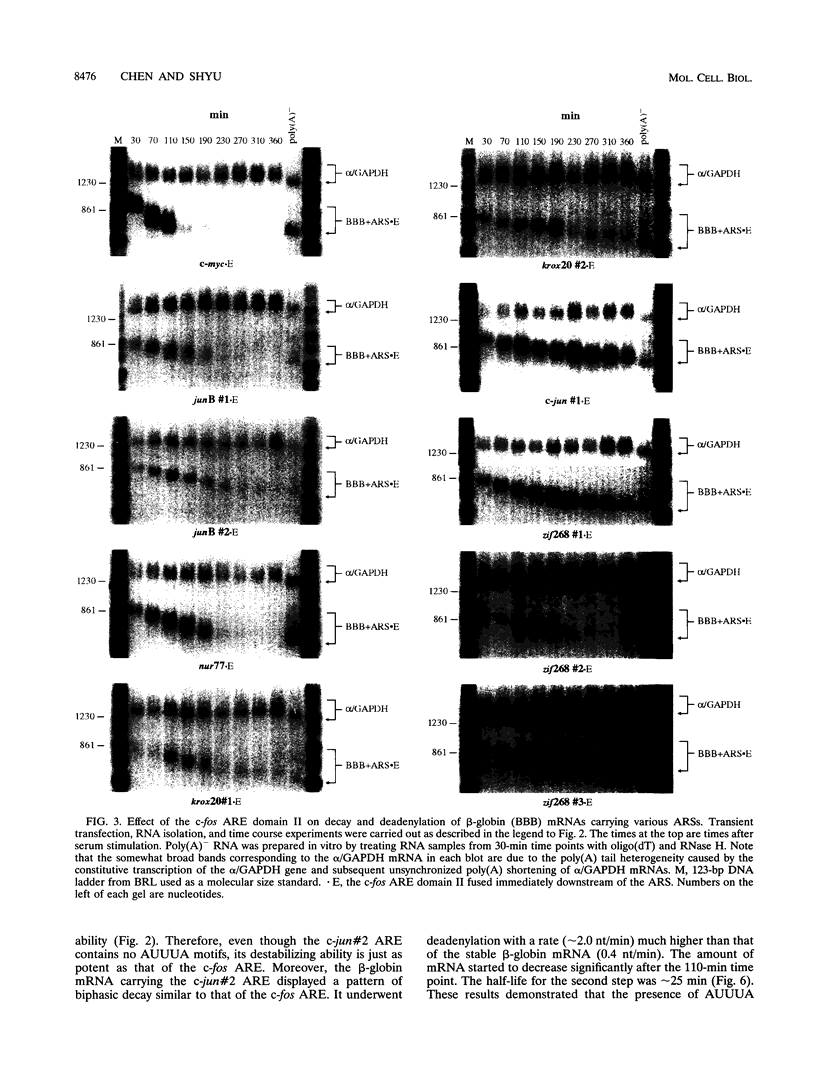
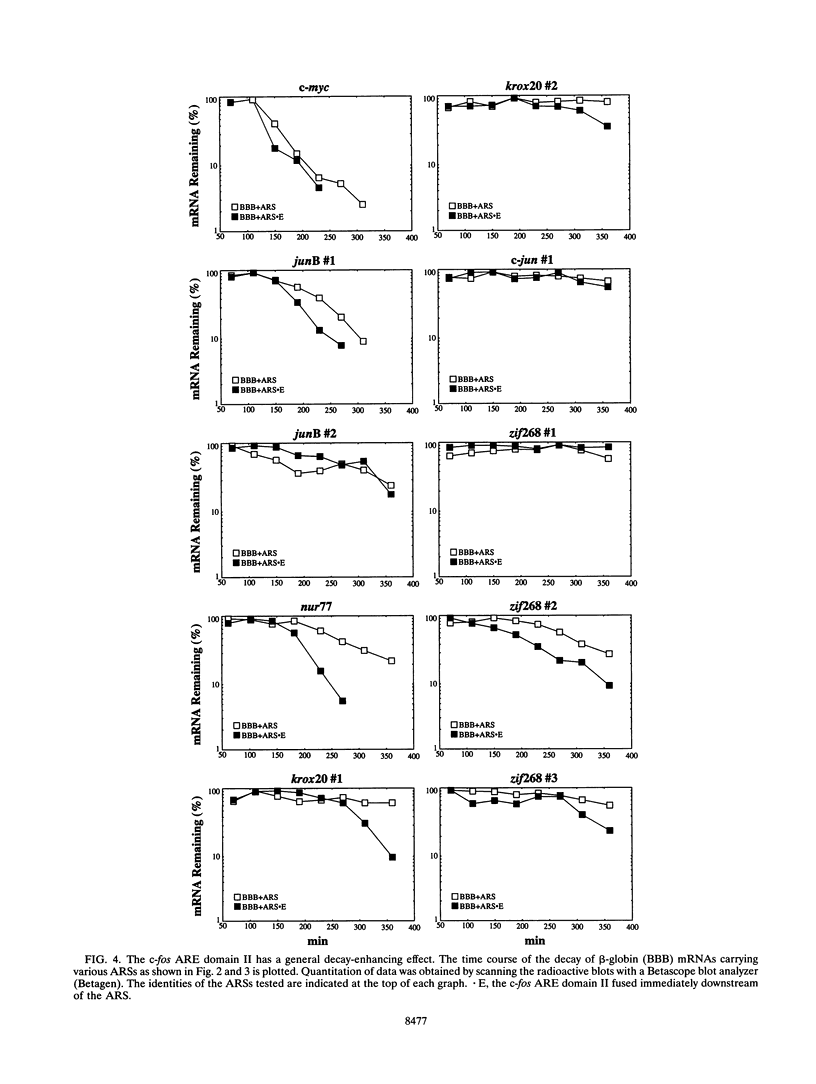
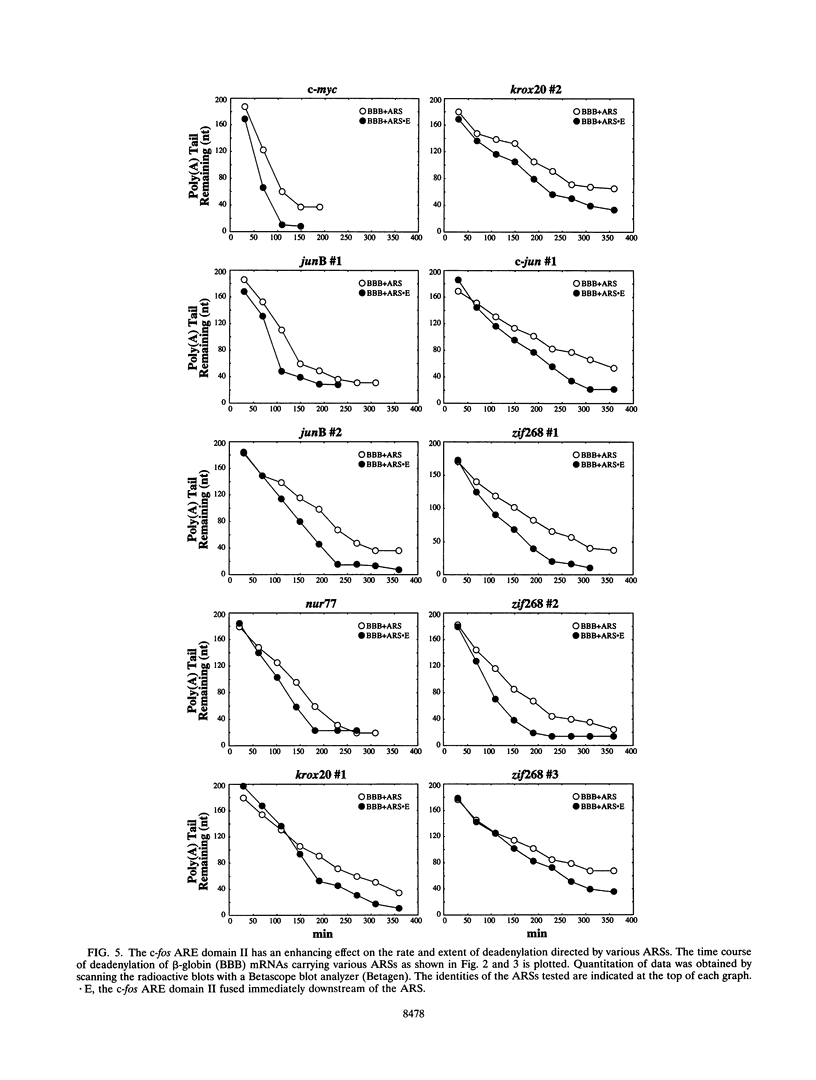
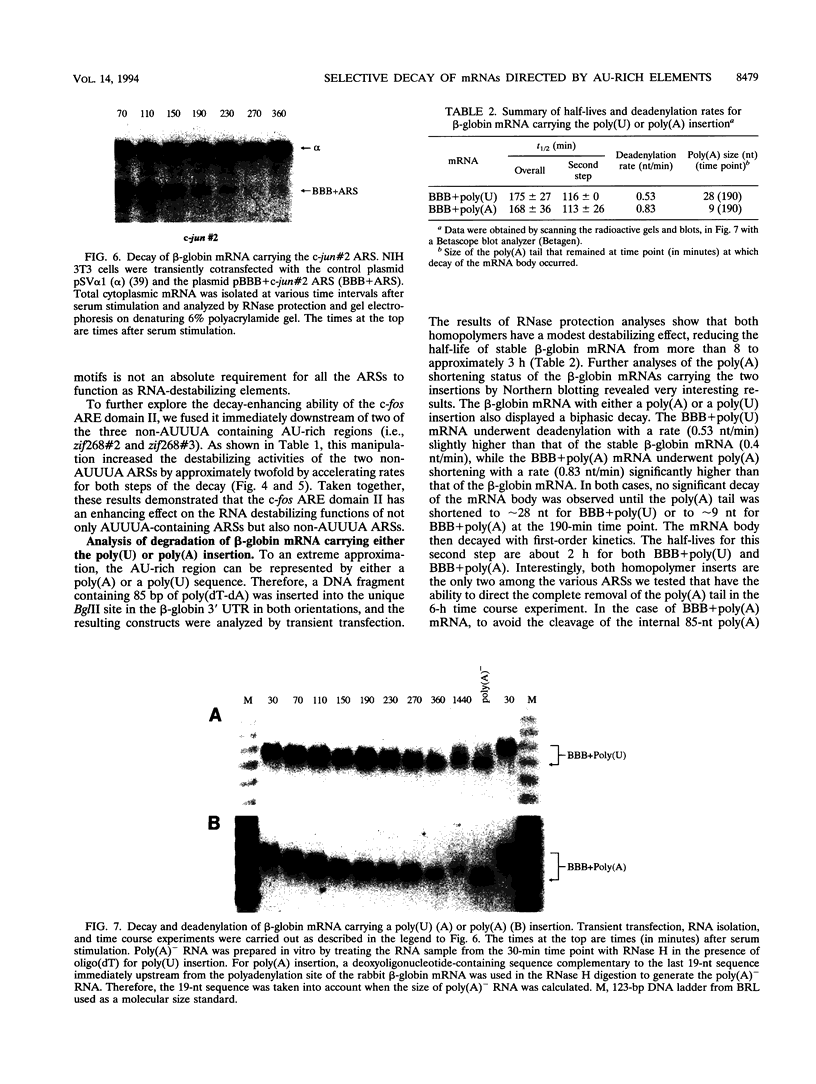
The metabolic lifetime of mRNA can be specified by specific cis-acting elements within mRNA. One type of element is an adenylate- and uridylate-rich element (ARE) found in the 3' untranslated region of many highly unstable mRNAs for mammalian early-response genes (ERGs). Among the better-characterized members of the ERG family are certain genes encoding nuclear transcription factors. Of particular significance was the finding that their mRNAs decay rapidly with kinetics similar to those of c-fos mRNA. Our previous studies of the c-fos ARE-directed mRNA decay have revealed the existence in this ARE of two structurally distinct and functionally interdependent domains, termed domain I and domain II. We proposed that the c-fos ARE-directed decay is a two-step mechanism in which rapid shortening of the poly(A) tail leads to the decay of the mRNA body and further hypothesized that this is a general mechanism by which the ERG AREs mediate rapid mRNA degradation. To test this hypothesis and to further address the generality of the critical structural characteristics within the c-fos ARE, the RNA-destabilizing functions of more than 10 different AU-rich sequences from various nuclear transcription factor mRNAs have been tested. Consistent with the above-mentioned hypothesis is the observation that mRNAs carrying the functional AREs display a biphasic decay, which is characteristic of the proposed two-step mechanism. Our results indicated that the presence of AUUUA pentanucleotides in an AU-rich region does not always guarantee an RNA-destabilizing function for this region. Our results also led to the identification of a novel class of AU-rich destabilizing elements which contains no AUUUA pentanucleotide. The results of sequence comparison and functional tests revealed that a continuous U-rich sequence is a unique feature among the functional AREs. Finally, our experiments further showed that the c-fos ARE domain II has an RNA decay-enhancing ability upon its fusion to heterologous AU-rich regions and defined for the first time an RNA decay-enhancing element, which we termed the RDE element.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aharon T., Schneider R. J. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3' noncoding region is mediated by a cotranslational mechanism. Mol Cell Biol. 1993 Mar;13(3):1971–1980. doi: 10.1128/mcb.13.3.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendral J. M., Sommer D., Macdonald-Bravo H., Burckhardt J., Perera J., Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988 May;8(5):2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P., Allegretto E. A., Okino S. T., Hattori K., Boyle W. J., Hunter T., Karin M. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature. 1988 Mar 10;332(6160):166–171. doi: 10.1038/332166a0. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. AU RNA-binding factors differ in their binding specificities and affinities. J Biol Chem. 1992 Mar 25;267(9):6302–6309. [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3' untranslated region of lymphokine mRNA. Mol Cell Biol. 1991 Jun;11(6):3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnieu A., Piechaczyk M., Marty L., Cuny M., Blanchard J. M., Fort P., Jeanteur P. Sequence determinants of c-myc mRNA turn-over: influence of 3' and 5' non-coding regions. Oncogene Res. 1988 Sep;3(2):155–166. [PubMed] [Google Scholar]
- Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991 May;11(5):2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P., Zerial M., Lemaire P., Almendral J., Bravo R., Charnay P. A gene encoding a protein with zinc fingers is activated during G0/G1 transition in cultured cells. EMBO J. 1988 Jan;7(1):29–35. doi: 10.1002/j.1460-2075.1988.tb02780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Chen T. M., Shyu A. B. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol. 1994 Jan;14(1):416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993 Aug;7(8):1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Decker C. J., Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem Sci. 1994 Aug;19(8):336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Fort P., Marty L., Piechaczyk M., el Sabrouty S., Dani C., Jeanteur P., Blanchard J. M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985 Mar 11;13(5):1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D. R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991 Nov;5(11):2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Herschman H. R. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- Iwai Y., Bickel M., Pluznik D. H., Cohen R. B. Identification of sequences within the murine granulocyte-macrophage colony-stimulating factor mRNA 3'-untranslated region that mediate mRNA stabilization induced by mitogen treatment of EL-4 thymoma cells. J Biol Chem. 1991 Sep 25;266(27):17959–17965. [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987 Dec;7(12):4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeller D. M., Horowitz J. A., Casey J. L., Klausner R. D., Harford J. B. Translation and the stability of mRNAs encoding the transferrin receptor and c-fos. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7778–7782. doi: 10.1073/pnas.88.17.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. F., Nathans D. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1182–1186. doi: 10.1073/pnas.84.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire P., Revelant O., Bravo R., Charnay P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4691–4695. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstein T., June C. H., Ledbetter J. A., Stella G., Thompson C. B. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989 Apr 21;244(4902):339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- Lowell J. E., Rudner D. Z., Sachs A. B. 3'-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992 Nov;6(11):2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'-->3' digestion of the transcript. Genes Dev. 1994 Apr 1;8(7):855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992 Nov;6(11):2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- Munroe D., Jacobson A. mRNA poly(A) tail, a 3' enhancer of translational initiation. Mol Cell Biol. 1990 Jul;10(7):3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N., Laflamme G., Raymond V. 5' untranslated sequences modulate rapid mRNA degradation mediated by 3' AU-rich element in v-/c-fos recombinants. Nucleic Acids Res. 1992 Nov 11;20(21):5753–5762. doi: 10.1093/nar/20.21.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder K., Lau L. F., Nathans D. A gene activated by growth factors is related to the oncogene v-jun. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1487–1491. doi: 10.1073/pnas.85.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993 Aug 13;74(3):413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Savant-Bhonsale S., Cleveland D. W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a > 20S degradation complex. Genes Dev. 1992 Oct;6(10):1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- Schuler G. D., Cole M. D. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell. 1988 Dec 23;55(6):1115–1122. doi: 10.1016/0092-8674(88)90256-5. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Belasco J. G., Greenberg M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991 Feb;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5' element and c-fos 3' sequences. Cell. 1985 Oct;42(3):889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- Whittemore L. A., Maniatis T. Postinduction turnoff of beta-interferon gene expression. Mol Cell Biol. 1990 Apr;10(4):1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T., Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988 Nov 24;336(6197):396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- You Y., Chen C. Y., Shyu A. B. U-rich sequence-binding proteins (URBPs) interacting with a 20-nucleotide U-rich sequence in the 3' untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol Cell Biol. 1992 Jul;12(7):2931–2940. doi: 10.1128/mcb.12.7.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]