Significance
Most proteins that reside in the bacterial outer membrane are β sheets that fold into a unique cylindrical structure known as a “β barrel.” Here we describe significant insights into the function of the Bam complex, a protein machine that catalyzes the insertion of β barrel proteins into the membrane by an unknown mechanism. By analyzing the assembly of autotransporters, a specialized family of outer membrane proteins, we found that the function of the Bam complex can be divided into an initial substrate binding stage and a subsequent insertion stage that is surprisingly sensitive to structural distortions in client proteins.
Keywords: membrane protein assembly, outer membrane proteins, protein translocation
Abstract
Autotransporters are bacterial virulence factors that contain an N-terminal extracellular (“passenger”) domain and a C-terminal β barrel (“β”) domain that anchors the protein to the outer membrane. The β domain is required for passenger domain secretion, but its exact role in autotransporter biogenesis is unclear. Here we describe insights into the function of the β domain that emerged from an analysis of mutations in the Escherichia coli O157:H7 autotransporter EspP. We found that the G1066A and G1081D mutations slightly distort the structure of the β domain and delay the initiation of passenger domain translocation. Site-specific photocrosslinking experiments revealed that the mutations slow the insertion of the β domain into the outer membrane, but do not delay the binding of the β domain to the factor that mediates the insertion reaction (the Bam complex). Our results demonstrate that the β domain does not simply target the passenger domain to the outer membrane, but promotes translocation when it reaches a specific stage of assembly. Furthermore, our results provide evidence that the Bam complex catalyzes the membrane integration of β barrel proteins in a multistep process that can be perturbed by minor structural defects in client proteins.
Autotransporters are a very large superfamily of virulence factors produced by Proteobacteria and Chlamydia that consist of an N-terminal extracellular domain (passenger domain) and a C-terminal β barrel domain (β domain) that anchors the protein to the outer membrane (OM) (1). Passenger domains range in size from ∼20 kDa to over 400 kDa and have been shown to mediate a variety of different virulence functions (2). Following their translocation across the OM, many passenger domains are released from the cell surface by a proteolytic cleavage. Experimental and in silico studies have suggested that virtually all passenger domains form a β-helical structure, despite the fact that their primary amino acid sequence is poorly conserved (3–6). β domains are generally ∼30 kDa in size, and although they also display considerable sequence diversity, they can all be identified as members of the pfam03797 (smart00869) family of protein domains. Several divergent β domains have been crystallized and have been shown to form nearly superimposable 12-stranded β barrels that are traversed by an α-helical segment (7–10). The α-helical segment generally extends into the extracellular space and links the passenger domain to the β domain. In a few cases, however, the passenger domain is released in an intrabarrel cleavage reaction that leaves a small α-helical segment inside the barrel (11). Available evidence suggests that the incorporation of the α-helical segment into the β domain pore occurs in the periplasm (where the β domain appears to undergo considerable folding) and is required for the integration of the β domain into the OM (12).
Although there is general agreement that the passenger domain is translocated across the OM in a C- to N-terminal direction (13, 14), the mechanism of translocation has been hotly debated. Early experiments in which the β domain was deleted showed that it plays an essential role in translocation and led to the proposal that it forms a channel through which the covalently linked passenger domain is secreted (15). Recently, however, several findings have challenged the “autotransporter” hypothesis. Crystallographic analysis has shown that the pore formed by the β domain is ∼10 Å in diameter and therefore only wide enough to accommodate a completely unfolded polypeptide in a hairpin conformation or a single polypeptide in an α-helical conformation. Molecular dynamics simulations have confirmed that the β domain is extremely stable and unlikely to expand spontaneously (16, 17). Nevertheless, both native and modified passenger domains that have acquired tertiary structure in the periplasm are secreted efficiently by the autotransporter pathway (18, 19). Furthermore, the observation that the peptide inside the β domain is in an α-helical conformation at an early stage of translocation is incompatible with passenger domain translocation through the β domain pore (20). Finally, crosslinking experiments (13) have shown that during its transit across the OM, the passenger domain interacts with BamA, a component of a complex that binds to β barrel proteins and facilitates their integration into the OM by an unknown mechanism (21–24). Interestingly, members of the BamA superfamily produced by bacteria and chloroplasts have been shown to mediate protein translocation reactions (25). In addition to BamA, which consists of a β barrel domain and five periplasmic POTRA (polypeptide transport associated) domains, the Bam complex contains four lipoproteins called BamB, BamC, BamD, and BamE.
An analysis of the interactions between cellular factors and the β domain of a model autotransporter produced by Escherichia coli O157:H7 called EspP has recently led to a new model in which the translocation of the passenger domain and the assembly of the β domain are interconnected (26). EspP is a member of the SPATE (serine protease autotransporters of Enterobacteraceae) family of autotransporters whose passenger domains are released in an intrabarrel cleavage reaction (11). Site-specific photocrosslinking experiments showed that the EspP β domain interacts with the periplasmic chaperone Skp and components of the Bam complex in a temporally and spatially regulated fashion in vivo. Skp is a homotrimer that resembles a jellyfish with long, flexible α-helical tentacles that form a large central cavity (27, 28). The results revealed that although the entire β domain initially interacts with Skp, discrete regions of the polypeptide subsequently interact with BamA, BamB, and BamD. The data suggest the existence of an assembly intermediate in which the EspP β domain is effectively surrounded by components of the Bam complex. Interestingly, the results also suggested that the passenger domain is not only normally secreted and cleaved before the completion of β domain assembly, but that the completion of β domain assembly is strictly dependent on the completion of passenger domain translocation. To account for these results and other recent observations on autotransporter biogenesis, it was proposed that the passenger domain is secreted through a channel comprised of an incompletely closed β domain, BamA, and possibly other factors that have not yet been identified (26).
Although these results provide insight into the later stages of autotransporter assembly, they do not address the mechanism by which the translocation of the passenger domain across the OM is initiated. One possibility is that once the β domain captures the appropriate α-helical peptide in the periplasm, it simply serves as a targeting signal that guides the passenger domain to the OM. In that case, it is likely that the initiation of passenger domain translocation would be closely coupled to the binding of the β domain to the Bam complex. Alternatively, the initiation of translocation might depend on the completion of an additional step in β domain assembly that occurs after the β domain docks onto the Bam complex. Here we describe an analysis of several EspP β domain mutants that strongly supports the latter hypothesis. We found that the mutation of two highly conserved residues, G1066 and G1081, perturbs the stability of the β domain and delays the initiation of passenger domain translocation. In vivo site-specific photocrosslinking and other experiments showed that the mutations delay both the exposure of the passenger domain on the cell surface after the β domain binds to the Bam complex and the integration of the β domain into the lipid bilayer. By uncoupling the initiation of translocation from the interaction of the β domain with the Bam complex, our results imply that the β domain must undergo a transition after it reaches the OM before translocation can begin. Moreover, our results suggest that the Bam complex facilitates the integration of β barrel proteins into the OM in a reaction that can be divided into discrete stages.
Results
Mutation of Two Conserved Residues in the EspP β Domain Delays the Initiation of Passenger Domain Translocation.
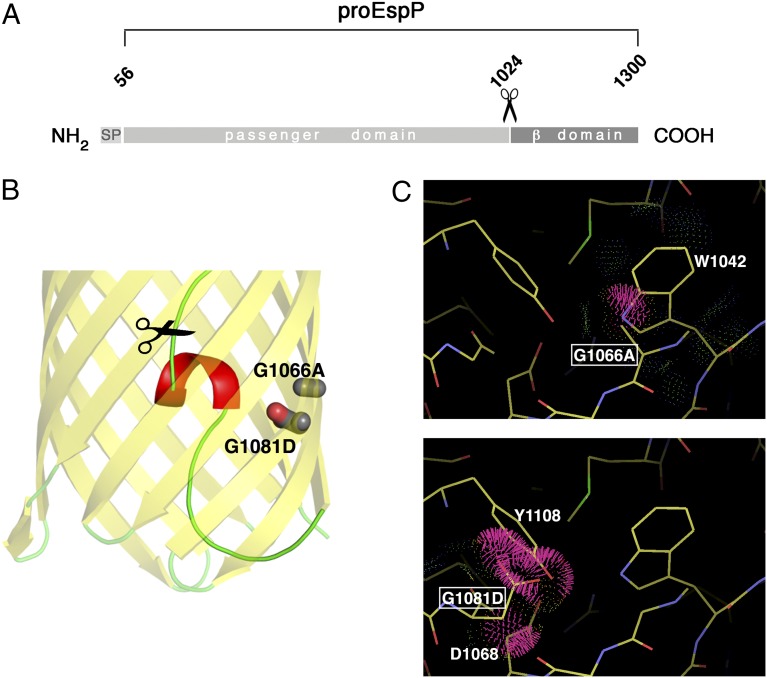
Although autotransporter β domains do not contain any invariant residues, a multiple sequence alignment of 100 divergent β domains revealed the presence of 10 highly conserved residues, three of which are glycines (Fig. S1). Two of the conserved glycine residues (G1066 and G1081) reside in the second and third β strands of the EspP β domain (residues 1024–1300) (Fig. 1 A and B). Based on the crystal structure of the EspP β domain (8), the side chain of any amino acid larger than glycine at position 1066 (including the methyl group of an alanine residue) is predicted to project into the lumen of the β barrel and clash sterically with the side chain of a second highly conserved residue, W1042 (Fig. 1 B and C, and Fig. S1). Similarly, amino acid side chains at position 1081 are predicted to clash sterically with the side chains of another highly conserved residue (Y1108) and a moderately conserved residue (D1068) inside the β barrel (Fig. 1 B and C, and Fig. S1).
Fig. 1.
Mutation of residues 1066 and 1081 creates potential steric clashes inside the EspP β barrel. (A) Illustration of EspP showing the signal peptide (SP) (residues 1–55), the passenger domain (residues 56–1023), and the β domain (residues 1024–1300). ProEspP is the precursor form of the protein that is observed before the proteolytic release of the passenger domain from the β domain. (B) Modeling of the G1066A and G1081D mutations based on the crystal structure of the EspP β domain (8). The passenger domain cleavage site is shown. (C) Steric clashes created by the introduction of the G1066A (Upper) and G1081D (Lower) mutations were predicted in Coot using Molprobity tools (41).
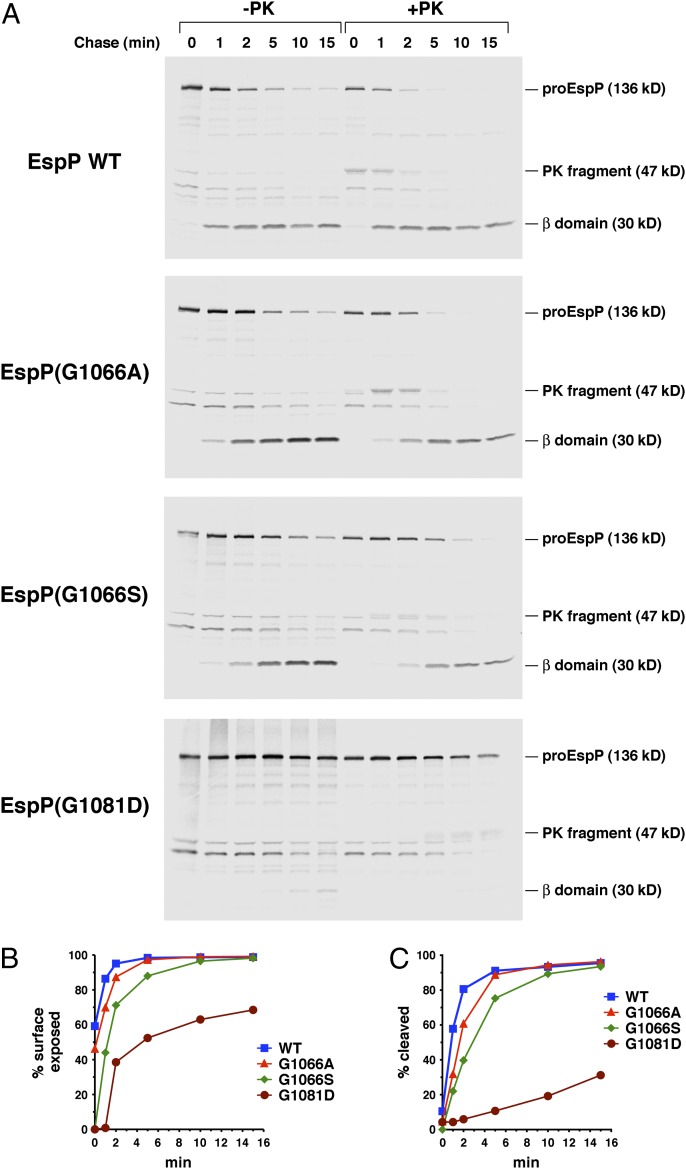
We performed a heat modifiability assay to determine the effect of mutations at positions 1066 and 1081 on the stability of the EspP β domain. This assay is based on the observation that native β barrel proteins are often resistant to SDS denaturation and migrate relatively rapidly on SDS gels unless they are heated to a high temperature (29). E. coli strain AD202 (MC4100 ompT::kan) transformed with a plasmid encoding wild-type EspP under the control of the trc promoter (pRLS5) or a derivative of pRLS5 encoding a mutant EspP protein was grown in minimal medium. After isopropyl-β-D-thiogalactopyranoside (IPTG) was added to induce expression of the plasmid-borne gene, cells were lysed by sonication. Cell extracts were then heated in SDS/PAGE sample buffer and the cleaved EspP β domain was detected by Western blot using an antibody generated against a C-terminal peptide. Consistent with previous results (8), the wild-type β domain was resistant to SDS-denaturation when heated up to 70 °C (Fig. 2, first blot). A fraction of the β domain molecules were denatured at 75 °C, and complete denaturation was observed at 90 °C. Even the very mild G1066A mutation, however, reduced the stability of the β domain. Some of the mutant molecules were denatured at 65 °C, and complete denaturation was observed at 75 °C (Fig. 2, second blot). The slightly stronger G1066S mutation and a glycine-to-aspartate mutation at position 1081 (G1081D) had a more deleterious effect and led to the complete denaturation of the β domain even at 25 °C (Fig. 2, blots 3 and 4). These results show that mutations at positions 1066 and 1081 distort the structure of the EspP β domain and reduce its overall stability.
Fig. 2.
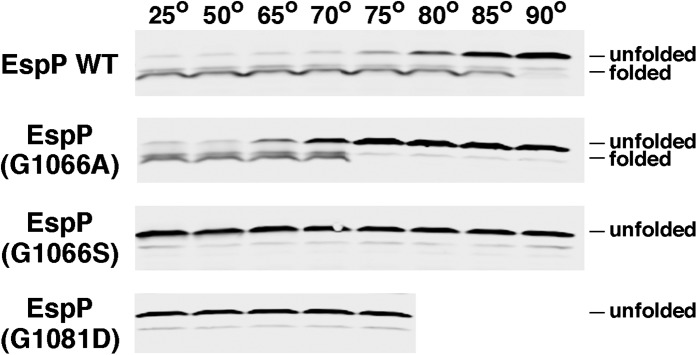
Mutation of residues 1066 and 1081 reduces the stability of the EspP β domain. AD202 transformed with pRLS5 (Ptrc-espP) or a pRLS5 derivative encoding the indicated EspP mutant were harvested after the addition of IPTG. Cell membranes were isolated and heated at the indicated temperature before proteins were resolved by SDS/PAGE. The cleaved EspP β domain was detected by Western blot using an antiserum generated against a C-terminal peptide.
We next wished to determine whether the mutation of G1066 and G1081 affects EspP biogenesis. To this end AD202 cells transformed with pRLS5 or a mutated version of pRLS5 were grown in minimal medium at 37 °C and espP expression was induced by the addition of IPTG. Cells were then subjected to pulse-chase labeling and divided into two equal portions, one of which was treated with proteinase K (PK). Finally, samples were subjected to immunoprecipitation with the anti-EspP C-terminal antiserum and resolved by SDS/PAGE. We assessed the exposure of the passenger domain on the cell surface by quantitating the fraction of proEspP (the precursor form of the protein that contains covalently linked passenger and β domains) that was sensitive to PK digestion. We also assessed the cleavage of the protein into discrete passenger and β domain fragments by determining the ratio of free β domain to proEspP. This proteolytic maturation step has been shown to require the completion of passenger domain translocation (13, 20). As previously reported (20), nearly 90% of the wild-type passenger domain was exposed within the first minute (Fig. 3A, Top gel; Fig. 3B, squares). Because cleavage of the passenger domain lagged behind cell surface exposure by about 1 min (Fig. 3A, Top gel, and Fig. 3C), an ∼47 kDa C-terminal PK fragment could be observed at early time points (Fig. 3A, Top gel, lanes 7–8). This fragment contains the β domain plus a PK-resistant ∼17-kDa passenger domain segment and is derived from an intermediate in which the passenger domain is partially or completely secreted but not yet cleaved (20).
Fig. 3.
Mutation of EspP residues 1066 and 1081 delays the initiation of passenger domain translocation. (A) AD202 transformed with pRLS5 (Ptrc-espP) or a pRLS5 derivative encoding the indicated EspP mutant were subjected to pulse-chase labeling after the addition of IPTG. Half of the cells were treated with PK, and immunoprecipitations were peformed use the C-terminal anti-EspP antiserum. The percent of the passenger domain that was surface exposed or released by proteolytic cleavage in A is shown in B and C.
Interestingly, the mutations all delayed the initiation of passenger domain translocation. The G1066A and G1066S mutations slowed the cell-surface exposure of the passenger domain by ∼1 min and ∼2 min, respectively (Fig. 3A, gels 2 and 3, and Fig. 3B, triangles and diamonds). The observation that the mutations did not clearly increase the lag in the cleavage of the passenger domain (Fig. 3C) strongly suggests that they do not directly affect either the processivity of translocation or the cleavage reaction. Consistent with this conclusion, G1066 is located far from the cleavage site and the mutations do not appear to impinge on any residues that play a role in the cleavage reaction (8, 30) (Fig. 1 B and C). In contrast, the G1081D mutation strongly delayed passenger domain translocation, and even after 15 min less than two-thirds of the passenger domain molecules were exposed on the cell surface (Fig. 3A, Bottom gel, and Fig. 3B, circles). The G1081D mutation clearly impaired passenger domain cleavage more drastically than cell surface exposure (Fig. 3C) and therefore must exert an independent effect on proteolytic processing. Indeed, an aspartate at position 1081 might be expected to produce this effect because it clashes with D1068, a residue that is required for efficient cleavage (11). When the G1081D mutation was introduced into EspPΔ5, a derivative that lacks the entire passenger domain except 26 C-terminal residues that remain embedded inside the β domain, proteolytic maturation was more rapid (Fig. S2). This finding confirmed that the delay in the translocation of the full-length passenger domain in turn reduced the rate of cleavage. Furthermore, the observation that the G1066A and G1081D mutations also delayed the secretion of the 116 residue passenger domain fragment associated with an EspP deletion mutant designated EspP*Δ1 (12) (Fig. S3) corroborated the conclusion that they affect an early stage of the translocation reaction.
Although the mutations described above affect both the structure of the EspP β barrel and the initiation of passenger domain translocation, it is important to note that mutations that produce stability defects in heat modifiability assays do not always exert the same effect on protein biogenesis. The D1120N mutation, for example, causes EspP*Δ1 to migrate as an unfolded polypeptide on SDS/PAGE at all temperatures. Although the mutation abolishes passenger domain cleavage, it has no discernible effect on passenger domain translocation (11).
G1066A and G1081D Mutations Uncouple Passenger Domain Translocation from the Targeting of the EspP β Domain to the Bam Complex.
We next wished to determine the mechanism by which mutations at positions 1066 and 1081 delay the initiation of passenger domain translocation. The results of the heat modifiability assay suggested that the mutations might impede the folding of the β domain, thereby slowing its release from periplasmic chaperones and its subsequent targeting to the OM. Alternatively, the mutations might affect an assembly step that is both downstream of the binding of the β domain to the Bam complex and necessary for the onset of passenger domain translocation. We combined pulse-chase labeling with site-specific photocrosslinking to distinguish between these two possibilities. As shown previously, this approach provides temporal and spatial information about EspP biogenesis in vivo by tracking interactions between specific segments of the protein and cellular factors (26). The method involves the coexpression of an amber suppressor tRNA and an amino acyl-tRNA synthetase derived from Methanococcus jannaschii to incorporate the photoactivable amino acid analog p-benzoyl-L-phenylalanine (Bpa) at amber codons engineered into a protein of interest (31). Because of its small size, Bpa can be crosslinked only to proteins that are within 4 Å of the polypeptide backbone. AD202 transformed with two plasmids that encode an EspP amber mutant and the amber suppression system (pDULEBpa) (32) were grown at 37 °C. IPTG was added to induce espP expression, and cells were subjected to pulse-chase labeling. Two identical samples were removed from radiolabeled cultures at various time points, and one sample was UV irradiated. Immunoprecipitations were conducted using antisera that recognize the C-terminal EspP peptide and specific cellular proteins to identify crosslinking products.
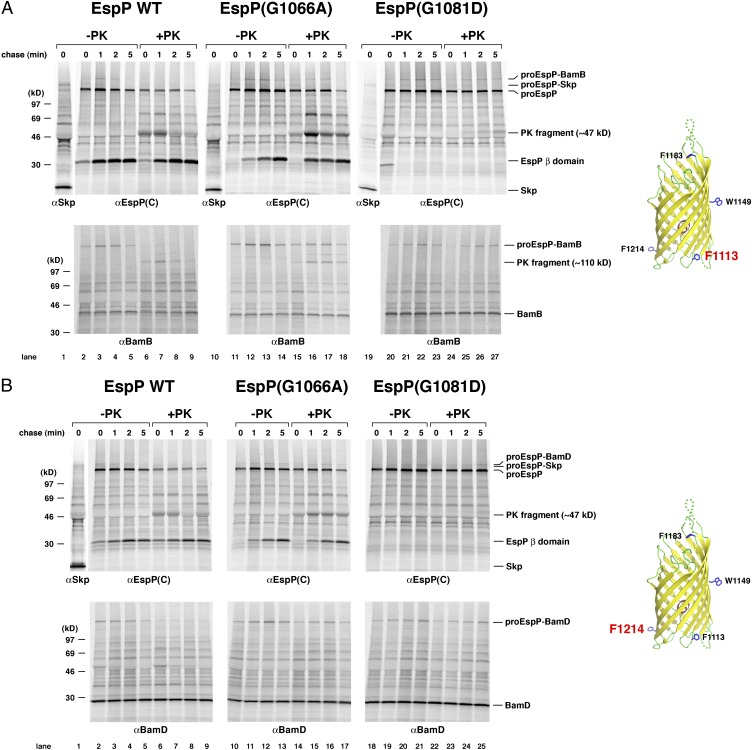
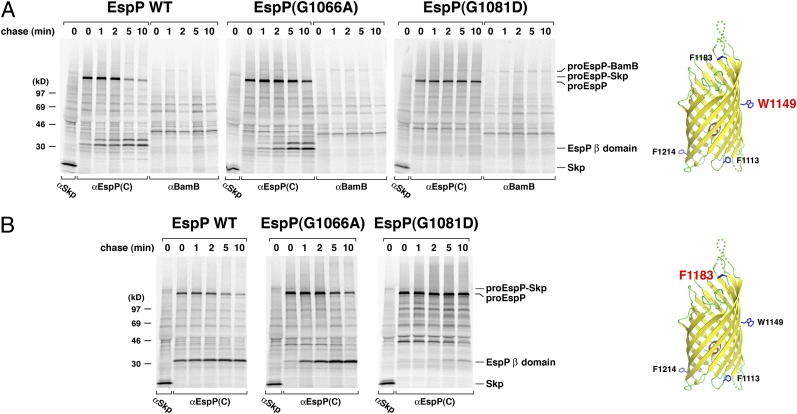
Consistent with previous results (26), we found that residues in the wild-type EspP β domain interact initially with the periplasmic chaperone Skp and subsequently with the Bam complex. When Bpa was incorporated at residue 1113, which is located on the periplasmic side of the fully folded β domain, an ∼155-kDa polypeptide that was immunoprecipitated by both anti-EspP and anti-Skp antisera was observed only in UV-irradiated samples at 0- and 1-min time points (Fig. 4A, lanes 1–3, and Fig. S4). An ∼155-kDa crosslinking product was not observed in a Δskp strain (Fig. S5). Based on its size, this polypeptide must correspond to the pro form of EspP (∼135 kDa) crosslinked to Skp (∼17 kDa). A larger proEspP-BamB crosslinking product was also observed but peaked later, presumably because the EspP β domain interacted with the Bam complex after it was released from Skp (Fig. 4A, lanes 2–5). A similar pattern was generated when the crosslinker was incorporated at residue 1214, which is located ∼120° away from residue 1113 on the periplasmic side of the β barrel, except that proEspP was crosslinked to BamD instead of BamB (Fig. 4B, lanes 1–5, and Fig. S4). As also previously reported (26), residues 1149 and 1183, which are located in the middle of the β barrel and in an extracellular loop, respectively, were crosslinked to Skp at early time points. Residue 1149 was also crosslinked weakly to BamB, but residue 1183 was not crosslinked to any other cellular factors (Fig. 5, Left). The spatially restricted crosslinking of discrete regions of the EspP β domain to specific Bam complex subunits strongly suggests that the β domain is uniquely oriented during its interaction with the Bam complex.
Fig. 4.
Mutation of EspP residues 1066 and 1081 uncouples passenger domain translocation from the binding of the β domain to the Bam complex. AD202 transformed with pDULEBpa and pRI22 (Plac-espP) or a pRI22 derivative encoding EspP(G1066A) or EspP(G1081D) with an amber mutation at residue 1113 (A) or 1214 (B) were subjected to pulse-chase labeling after the addition of IPTG. One aliquot of cells was UV-irradiated and an equal aliquot was untreated, and PK was added to half of each sample. Immunoprecipitations were subsequently conducted using the indicated antisera. Only the UV-irradiated samples are shown; untreated samples are shown in Fig. S4.
Fig. 5.
Crosslinking of wild-type and mutant EspP midbarrel and extracellular loop residues to Skp and BamB. AD202 transformed with pDULEBpa and pRI22 or a pRI22 derivative encoding EspP(G1066A) or EspP(G1081D) with an amber mutation at residue 1149 (A) or 1183 (B) were subjected to pulse-chase labeling after the addition of IPTG. One aliquot of cells was UV-irradiated and an equal aliquot was untreated. Immunoprecipitations were subsequently conducted using the indicated antisera. Only the UV-irradiated samples are shown.
To determine the disposition of the EspP passenger domain when the wild-type β domain was bound to the Bam complex, we treated half of the UV-irradiated cells with PK. If the passenger domain was exposed on the cell surface, then the proEspP-BamB and proEspP-BamD crosslinking products should be sensitive to the protease. Indeed the EspP(1113AMB)-BamB crosslinking product was nearly completely cleaved (Fig. 4A, lanes 6–9). A major PK fragment that migrated at ∼110 kDa was immunoprecipitated by both anti-EspP and anti-BamB antisera and presumably contained the C-terminal ∼47 kDa of EspP plus BamB (∼41 kDa). The EspP(1214AMB)-BamD crosslinking product was similarly almost completely sensitive to PK digestion (Fig. 4B, lanes 6–9). In contrast, EspP(1113AMB)-Skp and EspP(1214AMB)-Skp crosslinking products were resistant to PK digestion. This observation is consistent with the view that Skp interacts stably with the β domain only before EspP is targeted to the OM. In addition, the protease resistance of the proEspP-Skp crosslinking products demonstrates that the OM remained intact during the protease treatment. Taken together, the results show that there is normally a very close temporal link between the interaction of the β domain with the Bam complex and the initiation of passenger domain secretion.
Additional crosslinking experiments strongly suggested that the G1066A and G1081D mutations do not affect either the interaction of the β domain with Skp or the targeting of EspP to the Bam complex. Crosslinks between the mutant β domains and Skp were observed when Bpa was introduced at positions 1113, 1149, 1183, and 1214 (Fig. 4, lanes 10–14 and 19–23, and Fig. 5, Center and Right). Like wild-type EspP, the mutant proteins dissociated from Skp rapidly. We did not see a significant difference in either the time course of the interaction between Skp and wild-type EspP, EspP(G1066A), or EspP(G1081D) or in the level of the crosslinking product. In fact, we did not see any indication that the mutations prolonged or enhanced Skp binding even in experiments in which the EspP-Skp crosslinking product was especially prominent (Fig. S6). Crosslinks between the mutant β domains and Bam complex subunits appeared at the same time as crosslinks between the wild-type β domain and Bam complex subunits and followed the same pattern of spatial restriction (Fig. 4, lanes 10–14 and 19–23, and Fig. 5). The results suggest that the mutations do not grossly impair β domain folding. Consistent with this conclusion, neither mutation appeared to significantly affect the incorporation of an α-helical segment into the lumen of the EspP*Δ1 β barrel (12) (Fig. S3). Crosslinks between the mutant β domains and the Bam complex persisted longer than the equivalent crosslinks involving the wild-type β domain. For example, almost none of the EspP(1113AMB)-BamB crosslinking product remained at 5 min, but the EspP(G1066A/1113AMB)-BamB product was still very prominent at the same time point (Fig. 4A, compare lanes 2–5 and 11–14). The greater persistence of the crosslinking products paralleled the longer half-life of proEspP and corroborates the conclusion that the Bam complex dissociates from the β domain only after the passenger domain is cleaved (26).
Interestingly, treatment of the UV-irradiated samples with PK yielded strong evidence that the mutations delay the initiation of passenger domain translocation by impairing a step in EspP biogenesis that occurs after the β domain is targeted to the Bam complex. Unlike the EspP(1113AMB)-BamB and EspP(1214AMB)-BamD crosslinking products, a significant fraction of the EspP(G1066A/1113AMB)-BamB and EspP(G1066A/1214AMB)-BamD crosslinking products was resistant to PK digestion (Fig. 4A, lanes 15–18 and Fig. 4B, lanes 14–17). Consistent with the observation that the G1081D mutation delays the initiation of translocation more profoundly, the EspP(G1081D/1113AMB)-BamB and EspP(G1081D/1214AMB)-BamD crosslinking products appeared to be even more resistant to cleavage (Fig. 4A, lanes 24–27 and Fig. 4B, lanes 22–25). Indeed it was difficult to detect the ∼110-kDa PK fragment that normally results from the cleavage of the crosslinking product formed between residue 1113 and BamB. The results imply that the mutations at least transiently trap a hitherto unidentified assembly intermediate in which the β domain is bound to the Bam complex but the passenger domain is located entirely in the periplasmic space, and is therefore resistant to PK cleavage. Presumably this intermediate accumulates because a posttargeting assembly step that is required for the onset of passenger domain translocation occurs more slowly.
We next wished to rule out the possibility that the delay between the targeting of the β domain to the OM and the initiation of passenger domain translocation was an off-pathway phenomenon caused by the mutations. Because the incubation of cells at low temperature has proven to be useful to slow down autotransporter biogenesis and to capture assembly intermediates (12), we incorporated Bpa into wild-type EspP at 25 °C and subjected cells to pulse-chase labeling and UV irradiation, as described above. Crosslinks were formed between both residues 1113 and 1214 and Skp, between residue 1113 and BamB, and between residue 1214 and BamD (Fig. S7, lanes 1–4). As expected, the disappearance of the EspP-Skp crosslinking products and the concomitant appearance of the EspP-BamB and EspP-BamD crosslinking products was slower than at 37 °C. Interestingly, when cells were treated with PK, the EspP(1113AMB)-BamB and EspP(1214AMB)-BamD crosslinking products were partially resistant to cleavage (Fig. S7, lanes 5–8). These results demonstrate that the initiation of passenger domain secretion can be uncoupled from the targeting of the β domain to the OM not only by introducing specific mutations into the EspP β domain, but also by slowing the overall rate of assembly.
G1066A Mutation Reveals the Pretranslocation State of the Passenger Domain.
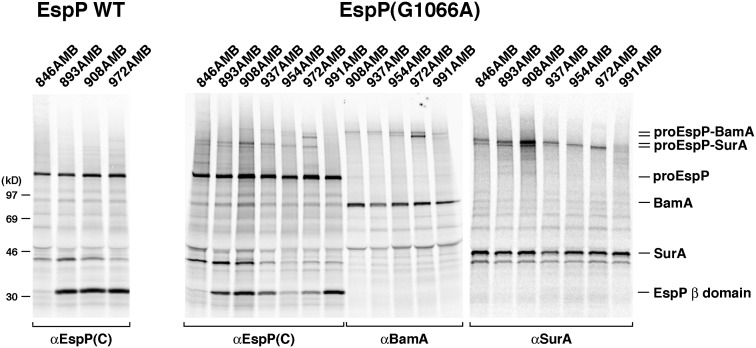
Crosslinking experiments in which Bpa was incorporated into the passenger domain of EspP(G1066A) provided further evidence that the mutation delays the initiation of passenger domain translocation and yielded insight into the status of the passenger domain before translocation begins. Amber mutations were introduced into wild-type EspP or the G1066A mutant at one of seven positions between residues 846 and 991. These residues are located in the first segment that is transported across the OM; passenger domain residues ∼996–1023 appear to be embedded inside the EspP β domain before the initiation of translocation and remain in place until the passenger domain is cleaved (12, 20, 30). Based on the finding that intermolecular interactions can be detected when passenger domain secretion stalls (13, 20), we hypothesized that residues in this segment of EspP(G1066A) could be crosslinked to cellular factors during the delay in the initiation of translocation. AD202 transformed with pDULE and a plasmid encoding one of the amber mutants were pulse-labeled, subjected to a 1-min chase, and UV-irradiated. Consistent with previous results, no crosslinks between the wild-type passenger domain and other proteins were seen (Fig. 6, Left). In contrast, all of the EspP(G1066A) residues were clearly crosslinked to cellular factors (Fig. 6, Right). Although weak crosslinking to BamA was detected (except when Bpa was introduced at residue 972), much stronger crosslinking to the periplasmic chaperone SurA was observed. As in previous site-specific photocrosslinking experiments, some of the crosslinking products migrated as doublets that presumably reflect the formation of bonds between the photoprobe and distinct sites in the target protein (26, 33).
Fig. 6.
Esp(G1066A) C-terminal passenger domain residues interact with SurA. AD202 transformed with pDULEBpa and pRI22 or a pRI22 derivative encoding EspP(G1066A) with an amber mutation at the indicated residue were pulse-labeled and subjected to a 1-min chase after the addition of IPTG. One aliquot of cells was UV-irradiated and an equal aliquot was untreated. Immunoprecipitations were subsequently conducted using anti-EspP C-terminal, anti-BamA, and anti-SurA antisera. Only the UV-irradiated samples are shown.
These results are striking for two reasons. First, they imply that the G1066A mutation delays EspP biogenesis at a stage when residue 972 (which is only ∼25 amino acids away from the first residue that emerges from the top of the β barrel) is located in the periplasm. This localization would only be possible if the passenger domain is in a hairpin conformation and no more than a very small loop is exposed on the cell surface. Second, the crosslinking pattern implies that the G1066A mutation traps an assembly intermediate that is very different from those observed after translocation has already been initiated. In the latter case, crosslinks to SurA are only observed when Bpa is introduced at least ∼80 residues away from the stall point; more proximal residues are crosslinked exclusively to BamA (13, 20) (Fig. S8). The data strongly suggest that whereas a long stretch of the passenger domain interacts with BamA after translocation is initiated, the passenger domain is in a state in which it is not yet engaged by BamA when EspP assembly is delayed by the G1066A mutation.
EspP Point Mutations Delay the Integration of the β Domain into the OM.
The location of G1066 and G1081 in the β domain strongly suggested that the mutation of these residues delays the initiation of passenger domain translocation by interfering with β domain assembly. In one scenario, the mutations might exert their effect at least in part by slowing the integration of the β domain into the OM. Alternatively, the mutations might simply interfere with a postintegration conformational change in the β domain that is required for the passage of the passenger domain into the extracellular space. To distinguish between these two possibilities, we deleted the EspP passenger domain and examined the effect of the mutations on the membrane integration of the free β domain. Initially we found that the β domain is translocated inefficiently through the Sec machinery, presumably because its N terminus is slightly basic (34). To overcome this problem, we used a construct (β′) in which an aspartate residue was inserted between the signal peptide and the β domain.
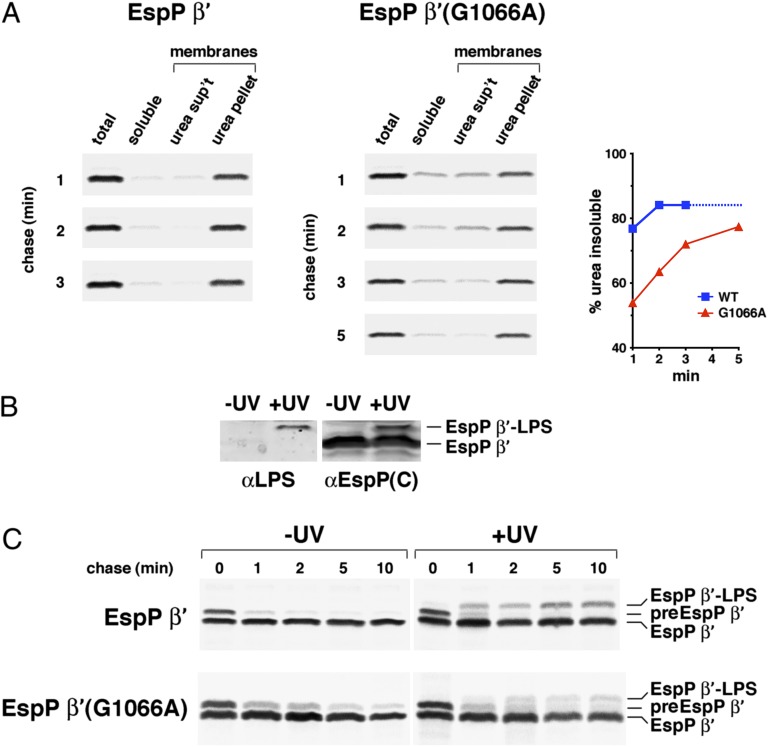
The results provided clear evidence that the point mutations delay the insertion of the β domain into the OM and supported the first hypothesis. In one type of experiment, AD202 transformed with a plasmid that produces wild-type β′ or β′ containing the G1066A or G1081D mutation were subjected to pulse-chase labeling and cells were fractionated into membrane and soluble components. The membrane fractions were then treated with urea to remove any proteins that were not fully integrated into the lipid bilayer. Most of the wild-type β′ protein was associated with the membrane fraction and was resistant to urea extraction by 1 min (Fig. 7A). In contrast, EspP β′ (G1066A) did not reach the same level of resistance to urea extraction until ∼5 min. Despite a partial (and highly reproducible) loss of EspP β′(G1081D) during the fractionation procedure, the finding that a significant amount of protein was urea extractable even after 10 min indicated that the mutation impairs membrane integration even more severely (Fig. S9). A second set of experiments was based on the observation that residue 1149, which is located in the middle of the EspP β barrel, can be crosslinked to LPS when that residue reaches the outer leaflet of the OM (26). As illustrated in Fig. 7B, when cells producing EspP β′(1149AMB) were irradiated and analyzed by Western blot, a ∼33-kDa band was detected by both anti-EspP and anti-LPS antisera. To determine the rate at which residue 1149 is localized to the outer leaflet of the OM, cells were subjected to pulse-chase labeling before UV irradiation and immunoprecipitations were performed using the anti-EspP C-terminal antiserum. Crosslinking of the wild-type β′ protein to LPS peaked by ∼1 min (Fig. 7C). The G1066A mutation appeared to delay the interaction between residue 1149 and LPS, however, because a strong crosslinking signal was not detected until 2 min. Presumably because of a slight shift in the position of residue 1149 relative to the lipid bilayer, significant crosslinking between the mutant protein and LPS was not observed.
Fig. 7.
The G1066A mutation delays the membrane integration of the EspP β domain. (A) AD202 transformed with pJH61 (Ptrc-espP β′) or a pJH61 derivative encoding EspP β′(G1066A) were subjected to pulse-chase labeling after the addition of IPTG. Cells were fractionated and equivalent amounts of each fraction were used for immunoprecipitations with an anti-EspP C-terminal antiserum. The percent of the protein recovered during the fractionation procedure that was isolated in the membrane fraction and that was resistant to urea extraction at each time point is shown. We assume that the percent of wild-type EspP β′ that was urea insoluble did not increase after 3 min (dotted line). (B) AD202 transformed with pDULEBpa and pJH61 were incubated with IPTG. Cell membrane fractions were then analyzed by Western blot using anti-LPS and anti-EspP C-terminal antisera. (C) AD202 transformed with pDULEBpa and pJH61 or a pJH61derivative encoding EspP β′(G1066A) were subjected to pulse-chase labeling after the addition of IPTG. One aliquot of cells was UV-irradiated and an equal aliquot was untreated, and immunoprecipitations were conducted using an anti-EspP C-terminal antiserum.
Discussion
In this article we describe evidence for an additional step in the biogenesis of autotransporter proteins. Previously it was unclear whether the β domain is essential for passenger domain secretion simply because it acts as a targeting signal that directs the passenger domain to the transport machinery in the OM or because it plays a more direct role in the secretion reaction. Our characterization of EspP β domain mutants fortuitously helped to distinguish between these two possibilities. We found that the mutation of two highly conserved glycine residues, G1066 and G1081, delayed the initiation of passenger domain translocation. By following the time course of assembly of the mutant proteins in living cells using a site-specific photocrosslinking approach, we showed that the mutations transiently trap the protein at a stage during which the β domain is bound to the Bam complex but the passenger domain has not yet emerged on the cell surface. We also observed the same assembly intermediate when the biogenesis of wild-type EspP was deliberately slowed down. Interestingly, although previous studies showed that EspP passenger domains located close to the OM interact with BamA when passenger domain translocation stalls (13), our results indicate that the equivalent residues interact primarily with SurA at this stage of assembly. By uncoupling the binding of the EspP β domain to the Bam complex from the initiation of passenger domain translocation, our results demonstrate that the targeting of the passenger domain to the OM is not sufficient to trigger the transport reaction and imply that translocation is dependent on a subsequent event. The data indicate that the mutations distort the structure of the β domain only modestly, but nevertheless delay its integration into the lipid bilayer of the OM. Taken together, our results suggest a scenario in which the β domain must undergo further assembly after binding to the Bam complex (perhaps by moving into the lipid bilayer or a specific position relative to the lipid bilayer) before translocation can begin.
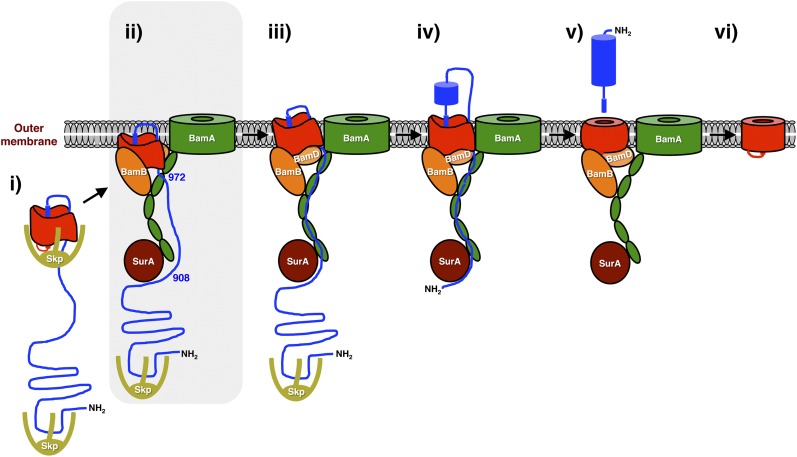
Based on the results described here, we can now add more details to the model for autotransporter biogenesis that we recently proposed (26). Previous work suggests that the EspP β domain begins to fold in the periplasm and incorporates a peptide that spans the passenger domain–β domain junction into a protected environment (12). We propose that the incorporation of a second peptide derived from the C terminus of the passenger domain creates a small hairpin that protrudes from the top of the β barrel (Fig. 8, step i). Subsequently the incompletely folded β domain binds to several Bam complex subunits in a unique orientation but remains associated with the inner leaflet of the OM or is only partially integrated into the lipid bilayer (Fig. 8, step ii). Although the exact status of the β domain is unclear, the finding that the G1066A and G1081D mutants (whose assembly stalls at this stage) appeared to be crosslinked to BamB more efficiently than wild-type EspP when Bpa was introduced at residue 1149 (Fig. 5A) suggests that the middle of the β domain is exposed to the periplasm. At this stage the hairpin that protrudes from the top of the β barrel is not yet exposed on the cell surface and the segment of the passenger domain that emerges from the bottom of the β barrel is close to—but not fully engaged by—the BamA POTRA domains. Progression through the assembly pathway requires a change in the conformation or positioning of the β domain, which in turn exposes the hairpin and “licenses” the initiation of translocation (Fig. 8, step iii). Based on our analysis of the EspP G1066A and G1081D mutants, the β domain may insert more completely into the lipid bilayer at this stage, but other changes that open the translocation pore might also occur. Our crosslinking results (13, 20) (Fig. S8) suggest that an ∼80-residue segment of the passenger domain then binds to the BamA POTRA domains, which facilitate a stepwise or progressive transfer of the passenger domain through a channel composed of the open EspP β domain, BamA, and possibly other factors. The β domain subsequently undergoes an additional transition after the initiation of translocation (26) (Fig. 8, step iv). Following the completion of translocation the passenger domain is cleaved, the assembly of the β domain is completed, and the β domain dissociates from the Bam complex (Fig. 8, steps v and vi).
Fig. 8.
Model of EspP biogenesis. The EspP β domain undergoes considerable folding in the periplasm and incorporates the C terminus of the passenger domain in a hairpin configuration (i). The protein is then targeted to the OM, where specific regions of the β domain interact with BamA, BamB, and BamD. Previously unpublished results presented here indicate that the hairpin is not exposed on the cell surface at this stage and suggest that although the passenger domain is not bound stably to the BamA periplasmic (POTRA) domains, residues 908 and 972 are in close proximity to SurA and BamA, respectively (ii, shaded). A conformational change in the β domain is subsequently required to “license” the initiation of passenger domain translocation (iii). The passenger domain then interacts with the BamA POTRA domains and moves in a stepwise or processive fashion to a transport channel composed of the β domain, BamA and possibly other proteins. After the initiation of translocation (iv), the β domain undergoes an additional conformational change. Following the completion of translocation, the passenger domain is released in an intrabarrel cleavage reaction (v). Finally, the β domain is released into the lipid bilayer following the completion of its assembly (vi). Many aspects of this model are based on previous studies. For simplicity, BamC and BamE have been omitted. BamD is also largely hidden in step ii.
Although this proposal is consistent with all available data, one possible drawback of the model is that it postulates the existence of an open β domain that persists through several stages of autotransporter biogenesis. This conformation would be unstable and would presumably have to be maintained by chaperones or the Bam complex. In an alternative scenario, the β domain would fold into a closed structure in the periplasm and incorporate only the passenger domain–β domain junction peptide. Assuming that the passenger domain does not pass directly through the hydrophobic environment of the OM, additional OM proteins would subsequently have to be recruited to create a hydrophilic translocation channel. Although no such factors are known, it is interesting to note that when the translocation of the EspP passenger domain stalls, at least one residue near the stall site can be crosslinked to an unidentified cellular protein (13). An OM protein called TamA was also recently suggested to play a role in passenger domain secretion (35).
Our data raise two intriguing questions about the status of autotransporters between the binding of the β domain to the Bam complex and the initiation of passenger domain translocation. The especially strong crosslinking of EspP residues 908 and 972 to SurA and BamA, respectively (Fig. 6), is consistent with a scenario in which the former residue is in close proximity to POTRA domain 1, the segment of BamA to which SurA binds (36), and the latter residue is near the periplasmic side of the β domain (Fig. 8, step ii). The distance between these two residues is also consistent with the observation that ∼80 residues are in proximity to BamA when passenger domain translocation stalls, but more distal residues are close to SurA (13, 20) (Fig. S6). It is not clear, however, why significant crosslinking between residues 937 and 954 and SurA was observed. Although multiple SurA molecules might bind to the passenger domain at this stage, it is also possible that the dynamics of the Bam complex or SurA brings segments of the passenger domain that are close to the OM into proximity to the chaperone. Second, it is unclear why strong crosslinking of residues 1113 and 1214 to BamB and BamD is observed both before and after the initiation of passenger domain translocation if the β domain moves physically through the OM between these two stages. One interesting possibility is that flexible N-terminal tethers enable lipoproteins to remain bound to their client proteins and move significant distances during the assembly process.
Finally, our data provide evidence that the Bam complex facilitates the membrane integration of β barrel proteins in a reaction that can be divided into at least two stages. In the first stage, the Bam complex may simply bind to β barrel proteins. Available evidence indicates that this interaction involves the recognition of a highly conserved C-terminal sequence motif (23). In a subsequent step (or steps), the Bam complex catalyzes the actual movement of client proteins into the lipid bilayer. Surprisingly, our analysis of EspP mutants suggests that even modest defects in the folding or structure of OM proteins can impede their membrane integration. One explanation of the results is that the Bam complex effectively performs a quality-control function and is activated by very specific structural cues from substrates. Perhaps more likely, the Bam complex might lower the free energy of membrane insertion, but the efficiency of insertion is ultimately dictated by the biophysical properties of each substrate. Indeed a recent study showed that although the Sec machinery greatly enhances the partitioning of α-helical transmembrane segments into a lipid bilayer, insertion efficiency is influenced by the nonpolar surface area of individual amino acid side chains (37). Moreover, although the generality of our results remains to be determined, it is noteworthy that the mutation of a conserved glycine residue in the β domain of a trimeric autotransporter also appears to affect its membrane insertion (38). Thus, the efficient assembly of at least closely related classes of OM proteins may depend on the ability of the polypeptide to pack tightly or to remain flexible at specific locations.
Materials and Methods
Bacterial Strains, Growth Conditions, and Reagents.
All experiments were conducted in the E. coli strain AD202 (MC4100 ompT::kan) (39). Unless otherwise noted, cultures were grown at 37 °C in M9 medium containing 0.2% glycerol and all of the L-amino acids, except methionine and cysteine (40 μg/mL). Ampicillin (100 μg/mL) and tetracycline (5 μg/mL) were added as needed. Polyclonal antisera generated against EspP N- and C-terminal peptides have been described previously (40). Polyclonal rabbit antisera were generated against C-terminal peptides of BamA and BamD (CQPFKKYDGDKAEQFQFNIGKT and CQMNAQAEKVAKIIAANSSNT), His-tagged Skp that was purchased from MyBioSource.com (catalog no. MBS203478) and His-tagged SurA that was produced and purified as described previously (24). An antiserum against BamB was obtained from Dan Kahne (Harvard University, Cambridge, MA), and a monoclonal antibody against LPS (HyCult Biotech) was obtained from Tom Silhavy (Princeton University, Princeton, NJ).
Plasmid Construction.
Plasmids pRLS5 and pJH61, which encode full-length espP and espP β′ under the control of the trc promoter, pRI22, which encodes His-tagged espP under the control of the lac promoter, and pDULEBpa have been described previously (13, 18, 32, 40). Missense and amber mutations were introduced into espP by site-directed mutagenesis using the QuikChange II Site-Directed Mutagenesis Kit (Agilent).
Cell Membrane Isolation and Heat Modifiability Assays.
Overnight cultures were washed and diluted into fresh M9 at OD550 = 0.02. When the cultures reached OD550 = 0.2, EspP synthesis was induced by the addition of 10 μM IPTG. After 30 min, cells were collected by centrifugation (2,500 × g, 15 min, 4 °C), resuspended at 10 OD units/mL in PBS, and sonicated. Unbroken cells were pelleted (2,500 × g, 5 min, 4 °C), and 1 mL of each cell extract was centrifuged in a Beckman TLA100.2 rotor (100,000 × g, 30 min, 4 °C). In heat modifiability experiments, the resulting membrane pellet was resuspended in PBS at 40 OD units/mL. Aliquots were then mixed with 2× Laemmli loading buffer and heated to a temperature between 25 °C and 90 °C for 15 min. Proteins were resolved by SDS/PAGE on 8–16% minigels (Life Technologies) and the EspP β domain was detected by Western blot.
Pulse-Chase Labeling and Photocrosslinking.
Cultures were grown as described above and EspP synthesis was induced by the addition of 10 μM IPTG (for experiments in which cells contained pRLS5 or pJH61 derivatives) or 200 μM IPTG (for crosslinking experiments in which cells contained pRl22 derivatives). Pulse-chase labeling and photocrosslinking were performed essentially as previously described (13). In experiments performed at low temperature, cultures were shifted to 25 °C 7 min before pulse labeling. Cells were pipetted over ice, concentrated by centrifugation and resuspended in 1 mL M9 medium. Proteins in all samples were collected by TCA precipitation. In some experiments, resuspended cells were divided in half, and one half was treated with PK as previously described (13) before TCA precipitation. Immunoprecipitations were performed as previously described (12), and proteins were resolved by SDS/PAGE on 8–16% minigels. Percent surface exposure and percent passenger domain cleavage were calculated as previously described (20). To detect crosslinking between EspP β′ and LPS, the radio labeling step was omitted. Cell membranes were isolated as described above and then resuspended in PBS containing 5% Elugent at 20 OD units/mL Insoluble material was then removed by centrifugation (TLA100.2 rotor, 100,000 × g, 30 min, 4 °C) before proteins were TCA precipitated and analyzed by Western blot.
Urea Extractions.
Cultures were subjected to pulse-chase labeling essentially as previously described (13). Cells were then pipetted over ice and collected by centrifugation. Cell pellets were resuspended in TBS (20 mM Tris pH 7.4/150 mM NaCl) at 3 OD units/mL and sonicated. Unbroken cells were removed by centrifugation and a portion of the supernatant (representing the total cell lysate) was removed. The remainder of each sample was centrifuged in a Beckman TLA100.2 rotor (100,000 × g, 30 min, 4 °C). The resulting supernatant was defined as the soluble fraction and the pellet as the membrane fraction. The membranes were resuspended in 150 μL 20 mM Tris pH 7.4/100 mM glycine/6M urea and incubated at 25 °C for 1 h (21). The urea insoluble fraction was then pelleted in a Beckman TLA-100 rotor (150,000 × g, 30 min, 4 °C) and resuspended in TBS. After urea-containing fractions were diluted 1:25 in water, proteins in all samples were collected by TCA precipitation and resolved by SDS/PAGE on 8–16% minigels.
Supplementary Material
Acknowledgments
We thank Travis Barnard for helping to construct Fig. 1 and Yihong Ye for providing valuable comments on the manuscript. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases Intramural Research Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219076110/-/DCSupplemental.
References
- 1.Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: Mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol. 2012;10(3):213–225. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- 2.Henderson IR, Nataro JP. Virulence functions of autotransporter proteins. Infect Immun. 2001;69(3):1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emsley P, Charles IG, Fairweather NF, Isaacs NW. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature. 1996;381(6577):90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- 4.Otto BR, et al. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J Biol Chem. 2005;280(17):17339–17345. doi: 10.1074/jbc.M412885200. [DOI] [PubMed] [Google Scholar]
- 5.Junker M, et al. Pertactin β-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc Natl Acad Sci USA. 2006;103(13):4918–4923. doi: 10.1073/pnas.0507923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangwer KA, et al. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc Natl Acad Sci USA. 2007;104(41):16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oomen CJ, et al. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23(6):1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnard TJ, Dautin N, Lukacik P, Bernstein HD, Buchanan SK. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat Struct Mol Biol. 2007;14(12):1214–1220. doi: 10.1038/nsmb1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Berg B. Crystal structure of a full-length autotransporter. J Mol Biol. 2010;396(3):627–633. doi: 10.1016/j.jmb.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 10.Zhai Y, et al. Autotransporter passenger domain secretion requires a hydrophobic cavity at the extracellular entrance of the β-domain pore. Biochem J. 2011;435(3):577–587. doi: 10.1042/BJ20101548. [DOI] [PubMed] [Google Scholar]
- 11.Dautin N, Barnard TJ, Anderson DE, Bernstein HD. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J. 2007;26(7):1942–1952. doi: 10.1038/sj.emboj.7601638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ieva R, Skillman KM, Bernstein HD. Incorporation of a polypeptide segment into the β-domain pore during the assembly of a bacterial autotransporter. Mol Microbiol. 2008;67(1):188–201. doi: 10.1111/j.1365-2958.2007.06048.x. [DOI] [PubMed] [Google Scholar]
- 13.Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA. 2009;106(45):19120–19125. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol. 2009;71(5):1323–1332. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- 15.Pohlner J, Halter R, Beyreuther K, Meyer TF. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325(6103):458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 16.Khalid S, Sansom MS. Molecular dynamics simulations of a bacterial autotransporter: NalP from Neisseria meningitidis. Mol Membr Biol. 2006;23(6):499–508. doi: 10.1080/09687860600849531. [DOI] [PubMed] [Google Scholar]
- 17.Tian P, Bernstein HD. Molecular basis for the structural stability of an enclosed β-barrel loop. J Mol Biol. 2010;402(2):475–489. doi: 10.1016/j.jmb.2010.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skillman KM, Barnard TJ, Peterson JH, Ghirlando R, Bernstein HD. Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Mol Microbiol. 2005;58(4):945–958. doi: 10.1111/j.1365-2958.2005.04885.x. [DOI] [PubMed] [Google Scholar]
- 19.Saurí A, Ten Hagen-Jongman CM, van Ulsen P, Luirink J. Estimating the size of the active translocation pore of an autotransporter. J Mol Biol. 2012;416(3):335–345. doi: 10.1016/j.jmb.2011.12.047. [DOI] [PubMed] [Google Scholar]
- 20.Peterson JH, Tian P, Ieva R, Dautin N, Bernstein HD. Secretion of a bacterial virulence factor is driven by the folding of a C-terminal segment. Proc Natl Acad Sci USA. 2010;107(41):17739–17744. doi: 10.1073/pnas.1009491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299(5604):262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 22.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121(2):235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Robert V, et al. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4(11):e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328(5980):890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58(5):1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- 26.Ieva R, Tian P, Peterson JH, Bernstein HD. Sequential and spatially restricted interactions of assembly factors with an autotransporter β-domain. Proc Natl Acad Sci USA. 2011;108(31):E383–E391. doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walton TA, Sousa MC. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell. 2004;15(3):367–374. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 28.Korndörfer IP, Dommel MK, Skerra A. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat Struct Mol Biol. 2004;11(10):1015–1020. doi: 10.1038/nsmb828. [DOI] [PubMed] [Google Scholar]
- 29.Freudl R, et al. An outer membrane protein (OmpA) of Escherichia coli K-12 undergoes a conformational change during export. J Biol Chem. 1986;261(24):11355–11361. [PubMed] [Google Scholar]
- 30.Barnard TJ, et al. Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter. J Mol Biol. 2012;415(1):128–142. doi: 10.1016/j.jmb.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci USA. 2002;99(17):11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods. 2005;2(5):377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 33.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94(6):795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 34.Tian P, Bernstein HD. Identification of a post-targeting step required for efficient cotranslational translocation of proteins across the Escherichia coli inner membrane. J Biol Chem. 2009;284(17):11396–11404. doi: 10.1074/jbc.M900375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selkrig J, et al. Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol. 2012;19(5):506–510, S1. doi: 10.1038/nsmb.2261. [DOI] [PubMed] [Google Scholar]
- 36.Bennion D, Charlson ES, Coon E, Misra R. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol. 2010;77(5):1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Öjemalm K, et al. Apolar surface area determines the efficiency of translocon-mediated membrane-protein integration into the endoplasmic reticulum. Proc Natl Acad Sci USA. 2011;108(31):E359–E364. doi: 10.1073/pnas.1100120108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grosskinsky U, et al. A conserved glycine residue of trimeric autotransporter domains plays a key role in Yersinia adhesin A autotransport. J Bacteriol. 2007;189(24):9011–9019. doi: 10.1128/JB.00985-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akiyama Y, Ito K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the ompT protease in vitro. Biochem Biophys Res Commun. 1990;167(2):711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 40.Szabady RL, Peterson JH, Skillman KM, Bernstein HD. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA. 2005;102(1):221–226. doi: 10.1073/pnas.0406055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.