Abstract
Here we present an X-ray crystallography structure of the clinically relevant tigecycline antibiotic bound to the 70S ribosome. Our structural and biochemical analysis indicate that the enhanced potency of tigecycline results from a stacking interaction with nucleobase C1054 within the decoding site of the ribosome. Single-molecule fluorescence resonance energy transfer studies reveal that, during decoding, tigecycline inhibits the initial codon recognition step of tRNA accommodation and prevents rescue by the tetracycline-resistance protein TetM.
Tetracyclines are broad-spectrum antibiotic agents that bind to elongating ribosomes and inhibit delivery of the ternary complex elongation factor thermo unstable (EF-Tu)⋅GTP⋅aminoacyl (aa)-tRNA to the ribosomal A site (1). Crystal structures of tetracyclines bound to the 30S subunit identified one common primary binding site that overlaps with the anticodon stem–loop of an A-site–bound tRNA (2–4). The widespread use of tetracyclines in the past has led to an increase in acquired tetracycline-resistance determinants among clinically relevant pathogenic bacteria, limiting the utility of many members of this class. Of the variety of tetracycline-specific resistance mechanisms, efflux and ribosome protection are the most common (5). Ribosome protection is mediated by ribosome protection proteins, with the best characterized being TetO and TetM (6). Ribosome protection proteins bind to tetracycline-stalled translating ribosomes and chase the drug from the ribosome, thus allowing translation to continue. The third generation of tetracycline derivatives, such as tigecycline, display enhanced antimicrobial activity compared with tetracycline, as well as overcoming efflux and ribosome protection mechanisms (7, 8).
Results and Discussion
X-Ray Crystallography Structure of 70S⋅Tigecycline Complex.
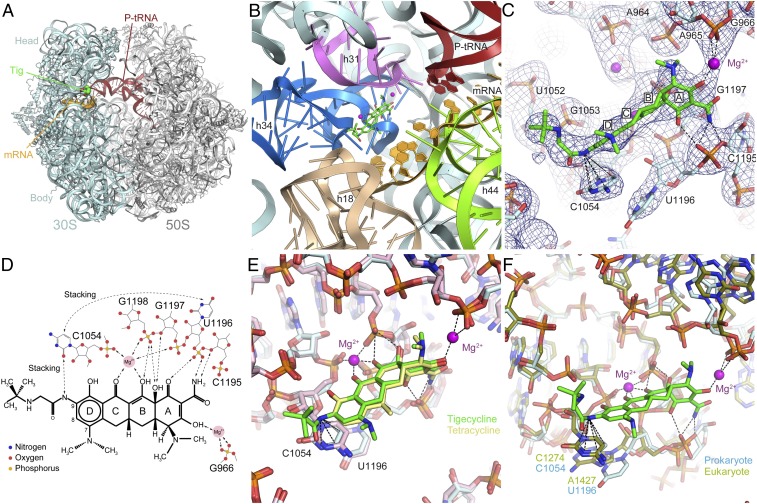
To address the molecular basis for the enhanced properties of tigecycline, we have determined an X-ray crystallography structure of tigecycline bound to a Thermus thermophilus 70S ribosome initiation complex containing P-site tRNAfMet and mRNA at 3.3-Å resolution (Fig. 1A and Table S1). The binding site of tigecycline comprises nucleotides of helix 31 (h31) and helix 34 (h34) of the 16S rRNA located on the head of 30S subunit (Fig. 1B). Electron density for tigecycline was observed only at the primary tetracycline binding site (Fig. S1) and not at any of the previously reported secondary tetracycline binding sites (2, 3) (Fig. S2), even though cocrystallization was performed at similar concentrations (60 µM) of tigecycline as used previously for tetracycline (4–80 µM) (2, 3).
Fig. 1.
X-ray crystal structure of tigecycline on the 70S ribosome. (A) Overview from the A site of the 70S ribosome with tRNAfMet in the P site (red), mRNA (orange) and tigecycline (green) bound. (B) View of the tigecycline binding site showing the rRNA elements in the vicinity of the site. (C) The fully refined electron density map (2Fobs−Fcalc) contoured at 1.2 sigma for the area surrounding the tigecycline binding site. Dashed lines indicate the stacking of the 9-t-butylglycylamido moiety of tigecycline with nucleobase C1054 and the coordination of the additional Mg2+ connecting tigecycline to G966 (h31). (D) Schematic chemical structure of tigecycline showing possible hydrogen bonds and other interactions with Mg2+ ions and bases from 16S rRNA. (E) Comparison of the binding modes of tigecycline (green) and tetracycline (yellow) via superimposition of the 16S rRNA. (F) Comparison of the prokaryotic T. thermophilus and the eukaryotic Saccharomyces cerevisiae tetracycline binding sites by superimposition of h34. Note that the nucleotide equivalents to C1054 and U1196 in S. cerevisiae are C1274 and A1427, the latter of which is slightly shifted, whereas the rest of the binding pocket is nearly identical.
At 3.3-Å resolution, the tigecycline molecule could be unambiguously fit into the electron density (Fig. 1C and Fig. S1), allowing the mode of interaction with the nucleotides of the 16S rRNA to be ascertained (Fig. 1D). The polar edge of the tigecycline molecule, containing many hydrophilic functional groups, interacts directly with the phosphate-oxygen backbone of nucleotides C1195 and U1196 in h34 as well as indirectly with G1197–G1198 and C1054 via a coordinated Mg2+ ion (Fig. 1 D and E), as reported previously for tetracycline (2). In addition, ring A of tigecycline coordinates a second Mg2+ ion to facilitate an indirect interaction with the phosphate-backbone of G966 in h31 (Fig. 1 C–E). We also determined a structure of tetracycline bound to the T. thermophilus 70S ribosome initiation complex containing P-site tRNAfMet and mRNA at 3.45-Å resolution (Table S1). Interestingly, initial cocrystallization for tetracycline was performed by using the same conditions as for tigecycline, i.e., with 60 µM drug and fivefold excess of tRNAfMet vs. ribosomes; however, density for nonspecific binding of tRNAfMet in the A site was observed, rather than tetracycline (Fig. S3). To obtain electron density for tetracycline, it was necessary to perform cocrystallization with higher concentrations of tetracycline (300 µM), coupled with lower excess (1.5-fold) of tRNAfMet vs. ribosomes (Fig. S4). These observations reemphasize the increased affinity of tigecycline vs. tetracycline for the ribosome (7–9), as well as illustrating the increased ability of tigecycline vs. tetracycline to compete with tRNA for binding at the A site. The structure of tetracycline bound to the 70S ribosome also suggests that tetracycline likely coordinates a second Mg2+ ion (Fig. 1E and Fig. S5), which was not suggested previously (2). Moreover, we note that no density was observed for the lower-affinity secondary tetracycline binding sites under our crystallization conditions (2, 3) (Fig. 1E and Fig. S6).
The major difference between tigecycline and tetracycline is the presence of 7-dimethylamido and 9-t-butylglycylamido moieties attached to ring D of tigecycline (Fig. 1D). Whereas the 7-dimethylamido substitution does not establish interactions with the ribosome, the glycyl nitrogen atom of the 9-t-butylglycylamido moiety of tigecycline forms stacking interactions with the π-orbital of nucleobase C1054 of the 16S rRNA (Fig. 1 C–E). Strong electron density for the remainder of the 9-t-butylglycylamido moiety of tigecycline suggests that, despite the lack of any apparent contact with the ribosome, it adopts a very rigid conformation (Fig. 1C), which may contribute to the stacking interaction with C1054. Indeed, binding of tigecycline appears to also enhance stacking between C1054 and U1196 (Fig. 1 C and D), similar to what is seen when tRNA is bound to the A site (10–12). In contrast, these nucleotides appear more flexible and adopt diverse conformations in ribosome structures with an empty A site (Fig. S7). The exclusive use of stacking and backbone interactions, together with the high structural conservation of the tetracycline binding site from bacteria to eukaryotes (13) (Fig. 1F), is consistent with the broad-spectrum activity of tetracyclines (5). Thus, we believe insights gained from the structures of tetracycline/tigecycline on the T. thermophilus ribosome can therefore be transferred to other bacteria. Although tetracycline activity has not been demonstrated against T. thermophilus strains to our knowledge, tetracyclines have been documented to have inhibitory activity against eukaryotic translation in vitro (14) (Fig. S8).
Binding and Inhibitory Properties of Tetracycline Derivatives.
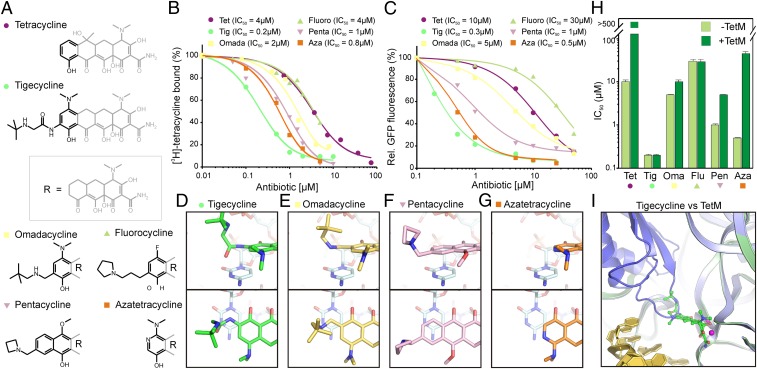
To investigate the contribution of the stacking interaction between the 9-t-butylglycylamido moiety of tigecycline and C1054, we used a series of tetracycline derivatives (Fig. 2A) and compared their ribosome binding (Fig. 2B) and translation inhibitory properties (Fig. 2C). The first two compounds tested, omadacycline (15) and 9-propylpyrrolidyl-7-fluorocyline (16), have amide bond replacements in the 9-position (Fig. 2A). Based on the tigecycline⋅70S structure presented here, these compounds would not be expected to form efficient stacking interactions with C1054 (Fig. 2 D and E). Indeed, omadacycline and 9-propylpyrrolidyl-7-fluorocycline had significantly reduced binding affinities (IC50 of 2 µM and 4 µM, respectively) with respect to tigecycline (IC50 of 0.2 µM), and were comparable to tetracycline (IC50 of 4 µM). Similar trends were also observed for the inhibition of an Escherichia coli in vitro translation system (Fig. 2C), suggesting a strong correlation between binding affinity and translation inhibitory activity. Next, we rationalized that, if stacking with C1054 is important for drug activity, introduction of an additional aromatic ring to generate a pentacycline should allow π-orbital stacking (Fig. 2F) and thus improve the binding and inhibitory properties of the drug. Indeed, the 7-methoxy-10-azetidinomethyl pentacycline (17) exhibited improved binding affinity (IC50 of 1 µM) and translation inhibitory activities (IC50 of 1 µM) compared with tetracycline, although it was less potent than tigecycline (Fig. 2 B and C). Collectively, these data support the hypothesis that stacking interactions with C1054 enhance the binding and inhibitory properties of tetracycline derivatives.
Fig. 2.
Binding and inhibitory properties of tetracycline derivatives. (A) Chemical structures of tetracycline, tigecycline, omadacycline, 9-propylpyrrolidyl-7-fluorocyline (fluorocycline), 7-methoxy-10-azetidinomethyl pentacycline (pentacycline), and 7-dimethylamido 8-azatetracycline (azatetracycline). (B) The inhibitory effect of tetracycline derivatives from A were monitored by using an E. coli in vitro transcription/translation assay monitoring the fluorescence of GFP as a function of antibiotic concentration. (C) The ability of tetracycline compounds from panel A to compete for binding to the E. coli 70S ribosome with [3H]tetracycline was monitored as a function of antibiotic concentration. (D–G) The stacking interaction of the (D) glycyl side chain of tigecycline (green) as observed in the tigecycline⋅70S structure is compared with models for the (E) omadacycline (yellow), (F) pentacycline (pink), and (G) azatetracycline (orange) docked on the 70S ribosome based on the tigecycline⋅70S structure. (H) The ability of the tetracycline derivatives from panel A to overcome TetM-mediated resistance was determined by performing translation inhibition assays as in C in the absence (light green) and presence (dark green) of TetM. The IC50 is presented as a log-scale (in µM). (I) Superimposition of the 70S ribosome structure with TetM (19) (blue) on the 70S structure with tigecycline (green) and mRNA (11) (gold).
Next we investigated the correlation between binding affinity and the ability to overcome TetM-mediated resistance. Here, we included 7-dimethylamido 8-azatetracycline (18), which lacks a C9 substitution (Fig. 2 A and G), yet had a similar binding affinity and translation inhibitory activity (both IC50s of 0.8 µM) as the 7-methoxy-10-azetidinomethyl pentacycline (IC50 of 1 µM; Fig. 2 B and C). To monitor the ability of each compound to overcome TetM-mediated resistance, the inhibitory effects of each compound were tested in an E. coli in vitro translation assay in the presence and absence of TetM (Fig. 2H). As expected (8), TetM dramatically relieved the translation inhibition of tetracycline, whereas TetM had no effect on the IC50 of tigecycline (Fig. 2H). Omadacycline, 9-propylpyrrolidyl-7-fluorocyline, and the 7-methoxy-10-azetidinomethyl pentacycline were all able to overcome TetM-mediated resistance, whereas 7-dimethylamido 8-azatetracycline, despite its increased binding affinity (Fig. 2B), was not able to overcome TetM-mediated resistance, with the IC50 increasing by ∼100 fold in presence of TetM (Fig. 2H). These findings suggest that binding affinity alone is not sufficient to overcome TetM-mediated resistance. Instead, TetM resistance appears to depend more on the presence of bulky substitutions at the 9-position. Indeed, superimposition of the tigecycline⋅70S X-ray structure determined here with a recent structure of TetM bound to the 70S ribosome (19) reveals that the drug and TetM sterically overlap near the domain IV loop of TetM and the 9-t-butylglycylamido moiety of tigecycline (Fig. 2I).
Effect of Tetracyclines and Glycylcyclines on tRNA Selection.
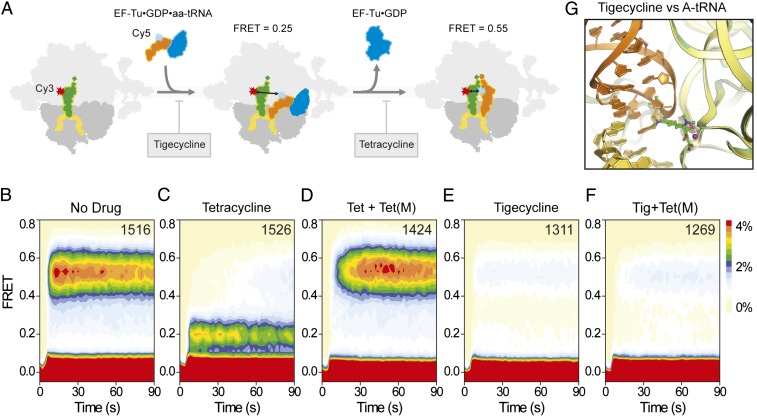
To investigate the interplay among tigecycline, ternary complex EF-Tu⋅GTP⋅aa-tRNA, and TetM on the ribosome, we used single-molecule fluorescence resonance energy transfer (FRET) imaging methods (20). Delivery of ternary complex containing Cy5-labeled Phe-tRNAPhe to 70S ribosomes bound with Cy3-labeled tRNAfMet at the P-site was monitored via FRET between the donor (Cy3) and acceptor (Cy5) fluorophores upon tRNA binding to the A site (Fig. 3A). In the absence of drug, tRNAs rapidly accommodated, resulting in the accumulation of a high (0.55) FRET state, previously shown to correspond to a pretranslocation complex configuration where A- and P-site tRNAs are classically positioned (20) (Fig. 3B). In the presence of tetracycline (40 µM; 10× Kd), aminoacyl-tRNA accommodation was efficiently blocked (21), resulting in repetitive ternary complex binding and release events from a low-(∼0.2) FRET configuration, as anticipated from a defect in the selection process subsequent to codon recognition but before GTP hydrolysis (20, 21) (Fig. 3C). The same experiment performed in the presence of tetracycline and TetM alleviated inhibition, restoring the capacity of aminoacyl-tRNA to rapidly accommodate into the A site, as evidenced by emergence of the high-FRET (∼0.55) state upon ternary complex delivery (Fig. 3D). When ternary complex delivery to the ribosome was monitored in the presence of tigecycline (2 µM; 10× Kd), aminoacyl-tRNA accommodation was again efficiently blocked at the low-FRET state. However, the duration of the transient binding events was relatively short-lived compared with those observed in the presence of tetracycline, consistent with an increased potency of tigecycline in its capacity to inhibit the initial selection process (20, 21) (Fig. 3E and Fig. S9). Strikingly, the addition of TetM did not alleviate the inhibition of ternary complex binding by tigecycline (Fig. 3F), consistent with the ability of tigecycline to overcome TetM-mediated resistance (7, 8) (Fig. 2I).
Fig. 3.
Effect of tetracyclines and glycylcyclines on tRNA selection. (A) Schematic illustrating the delivery of EF-Tu⋅tRNA⋅GTP ternary complex containing cognate Phe-tRNAPhe(Cy5-acp3U47) to 70S E. coli ribosomes containing P-site OH-tRNAfMet(Cy3-s4U8), leading to low (0.2) FRET during initial steps of codon recognition, and high (0.55) FRET upon A-site tRNA accommodation. Tigecycline is more effective than tetracycline at blocking the initial selection process. Tetracycline is nevertheless effective at preventing transitions into the fully accommodated, high-FRET state. (B–F) Single-molecule FRET imaging of aa-tRNA selection performed under direct 532-nm excitation following a 5-min incubation (B) in the absence of drugs or with (C) 40 µM tetracycline, (D) 40 µM tetracycline and 0.1 µM TetM, (E) 2 µM tigecycline, or (F) 2 µM tigecycline and 0.1 µM TetM.
Conclusion
Our findings indicate that the increased potency of tigecycline compared with tetracycline results from the increased affinity of tigecycline for the ribosome (7–9) (Fig. 1B) via stacking interactions of the 9-t-butylglycylamido moiety of tigecycline with C1054 of the 16S rRNA (Figs. 1 C–E and 2D). The presence of 9-t-butylglycylamido moiety of tigecycline, rather than the increased affinity compared with tetracycline, enables tigecycline to overcome TetM-mediated resistance (Fig. 2H). This observation is consistent with direct overlap of the 9-t-butylglycylamido moiety and domain IV loop of TetM (Fig. 2I). Moreover, we show that tigecycline is much more effective at blocking the initial selection process of tRNA entry into the A site (Fig. 3 C and E), in agreement with the steric clash between the drug and the anticodon stem–loop of the A-site tRNA (Fig. 3G).
Materials and Methods
Reagents.
Tetracycline (EMD Biosciences) and tigecycline (Sigma) were purchased from commercial sources, whereas PTK0796, TP767, pentacycline (TP556D), and azacycline (TP120C) were provided by Tetraphase. [3H]Tetracycline was purchased from Perkin-Elmer. Ribosomes from T. thermophilus cells were isolated as described previously (22, 23). Purified native uncharged E. coli tRNAfMet used for crystallographic studies was supplied by Chemical Block. The 30-nt-long mRNA [5′-GGCAAGGAGGUAAAA AUG UAC (A)6-3′] was purchased from Dharmacon (Shine–Dalgarno sequence and initiation codon are underlined). Labeling and charging of tRNAfMet(s4U8) and tRNAPhe(acp3U47) was as previously described by (21, 24, 25). tRNAfMet(s4U8) and tRNAPhe(acp3U47) were purchased from Sigma. Recombinant purified TetM protein was prepared as described previously (26).
Complex Formation and Crystallization.
The ribosomal complexes were formed in 10 mM Tris-acetate, pH 7.0, 40 mM KCl, 7.5 mM magnesium acetate, 0.5 mM DTT, by incubating 70S ribosomes (3 μM) with mRNA, tRNAfMet, and antibiotic (tetracycline or tigecycline) for 30 min at 37 °C. Crystals were grown at 24 °C by sitting-drop vapor diffusion based on the previously described procedure (23). Detailed methods can be found in SI Materials and Methods.
Data Collection, Processing, and Structure Determination.
Data on all complexes were collected at 100 K at the Swiss Light Source, PSI, using the Pilatus 6M detector. A very low dose mode was used and huge redundancy was collected (27). The initial model (from ref. 12, with tRNAs, mRNA, and metal ions removed) was correctly placed within each data set with Phenix (28) by rigid body refinement with each molecule defined as a rigid body. A detailed description is provided in SI Materials and Methods.
Single-Molecule Fluorescence Experiments.
Single-molecule FRET data were acquired by using a prism-based total internal reflection microscope as previously described (25, 29). Detailed materials and methods can be found in SI Materials and Methods.
Binding and Inhibitory Assays.
Inhibitory activity of the tetracycline compounds was assessed in an E. coli-coupled in vitro transcription/translation assay (5 PRIME) by using GFP fluorescence as a readout, as described previously (30, 31). Binding of all tetracycline compounds to 70S ribosomes was examined by using a competition assay as described previously (8). Further details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Michael B. Feldman, James B. Munro, and Peter Geggier for helpful comments; Natalia Demeshkina for help during X-ray data collection; and S. Duclaud for ribosome preparation. This work was supported by the Tri-Institutional Training Program in Computational Biology and Medicine (D.S.T.); National Institute of General Medical Sciences Grant 5R01GM079238-03; Deutsche Forschungsgemeinschaft Grant FOR1805/WI3285/1-2; Human Frontiers of Science Foundation Grant RGY88/2008; European Molecular Biology Organisation (EMBO) Young Investigator Programme (D.N.W.); and French National Research Agency Grants ANR BLAN07-3-190451 (to M.Y.) and ANR-11-BSV8-006 01 (to G.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4G5K, 4G5L, 4G5T, and 4G5U).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216691110/-/DCSupplemental.
References
- 1.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44(6):393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 2.Brodersen DE, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103(7):1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 3.Pioletti M, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20(8):1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray JB, et al. Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site. Chem Biol. 2006;13(2):129–138. doi: 10.1016/j.chembiol.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Chopra I, Roberts M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell SR, Tracz DM, Nierhaus KH, Taylor DE. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob Agents Chemother. 2003;47(12):3675–3681. doi: 10.1128/AAC.47.12.3675-3681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergeron J, et al. Glycylcyclines bind to the high-affinity tetracycline ribosomal binding site and evade Tet(M)- and Tet(O)-mediated ribosomal protection. Antimicrob Agents Chemother. 1996;40(9):2226–2228. doi: 10.1128/aac.40.9.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossman TH, et al. Target- and resistance-based mechanistic studies with TP-434, a novel fluorocycline antibiotic. Antimicrob Agents Chemother. 2012;56(5):2559–2564. doi: 10.1128/AAC.06187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson MW, et al. Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob Agents Chemother. 2006;50(6):2156–2166. doi: 10.1128/AAC.01499-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16(5):528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17(9):1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 12.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484(7393):256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 14.Budkevich TV, El’skaya AV, Nierhaus KH. Features of 80S mammalian ribosome and its subunits. Nucleic Acids Res. 2008;36(14):4736–4744. doi: 10.1093/nar/gkn424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutcliffe JA. Antibiotics in development targeting protein synthesis. Ann N Y Acad Sci. 2011;1241:122–152. doi: 10.1111/j.1749-6632.2011.06323.x. [DOI] [PubMed] [Google Scholar]
- 16. Xiao X, et al. (2011) Antibacterial activity of novel 7,9-disubstituted tetracycline analogs. Posters Presented at the 51st Annual Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC), September 17–20, 2011. Abstract F1-1855. (American Society for Microbiology, Washington) Available at http://tphase.com/files/P1%20F1-1855-7,9-Disubstituted%20Tetracycline%20Analogs.pdf.
- 17.Sun C, et al. Synthesis and antibacterial activity of pentacyclines: A novel class of tetracycline analogs. J Med Chem. 2011;54(11):3704–3731. doi: 10.1021/jm1015395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark RB, et al. 8-Azatetracyclines: Synthesis and evaluation of a novel class of tetracycline antibacterial agents. J Med Chem. 2011;54(5):1511–1528. doi: 10.1021/jm1015389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dönhöfer A, et al. Structural basis for TetM-mediated tetracycline resistance. Proc Natl Acad Sci USA. 2012;109(42):16900–16905. doi: 10.1073/pnas.1208037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geggier P, et al. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol. 2010;399(4):576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11(10):1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 22.Gogia ZV, Yusupov MM, Spirina TN. Structure of Thermus thermophilus ribosomes. Method of isolation and purification of ribosomes. Molekul. Biol. (USSR) 1986;20(2):519–526. [Google Scholar]
- 23.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17(5):555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 24.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101(35):12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25(4):505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikolajka A, et al. Differential effects of thiopeptide and orthosomycin antibiotics on translational GTPases. Chem Biol. 2011;18(5):589–600. doi: 10.1016/j.chembiol.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller M, Wang M, Schulze-Briese C. Optimal fine φ-slicing for single-photon-counting pixel detectors. Acta Crystallogr D Biol Crystallogr. 2012;68(pt 1):42–56. doi: 10.1107/S0907444911049833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96(6):2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starosta AL, et al. Identification of distinct thiopeptide-antibiotic precursor lead compounds using translation machinery assays. Chem Biol. 2009;16(10):1087–1096. doi: 10.1016/j.chembiol.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starosta A, et al. Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem Biol. 2010;17(5):1–10. doi: 10.1016/j.chembiol.2010.04.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.