Abstract
Genetically or epigenetically defined reprogramming is a hallmark of cancer cells. However, a causal association between genome reprogramming and cancer has not yet been conclusively established. In particular, little is known about the mechanisms that underlie metastasis of cancer, and even less is known about the identity of metastasizing cancer cells. In this study, we used a model of conditional expression of oncogenic KrasG12D allele in primary mouse cells to show that reprogramming and dedifferentiation is a fundamental early step in malignant transformation and cancer initiation. Our data indicate that stable expression of activated KrasG12D confers on cells a large degree of phenotypic plasticity that predisposes them to neoplastic transformation and acquisition of stem cell characteristics. We have developed a genetically tractable model system to investigate the origins and evolution of metastatic pancreatic cancer cells. We show that metastatic conversion of KrasG12D-expressing cells that exhibit different degrees of differentiation and malignancy can be reconstructed in cell culture, and that the proto-oncogene c-Myc controls the generation of self-renewing metastatic cancer cells. Collectively, our results support a model wherein non-stem cancer cells have the potential to dedifferentiate and acquire stem cell properties as a direct consequence of oncogene-induced plasticity. Moreover, the disturbance in the normally existing dynamic equilibrium between cancer stem cells and non-stem cancer cells allows the formation of cancer stem cells with high metastatic capacity at any time during cancer progression.
Activating Ras mutations are the most frequent oncogenic alterations in human cancers. The three related genes, Hras, Kras, and Nras, are all widely expressed, engage overlapping signaling pathways, and can each exhibit oncogenic activity. The frequency of Kras mutations is higher compared with other Ras isoforms, as they are present in 90% of pancreatic cancers, 50% of colon and thyroid carcinomas, 30% of non–small-cell lung cancers, and 25% of ovarian cancers (1). The consequence of oncogenic activation of different Ras isoforms has been extensively explored in various in vitro studies and, more recently, by using genetic mouse models with targeted mutations of the respective genes (2–7). An important observation from these studies is that individual Ras proteins induce tumors in a cell context-dependent fashion. The functional specificity of Ras isoforms can be determined by the amount of transgene expression (8), by the specific regulatory networks controlled by each isoform (4–7), and by their differential abilities to render cells permissive to oncogene-driven proliferation (9, 10). The unifying theme underlying these studies is that there exists a permissive cellular context for each particular genetic lesion, and that only certain types of cells are capable of cancer initiation. However, defining the causes of preferential occurrence of cell type-specific and tissue-specific cancerous mutations remains one of the unanswered fundamental questions.
Data derived from mouse models of lung and colon cancer indicate that oncogenic Kras accelerates tumor progression by imposing on cells an immature stem-like state in which differentiation is inhibited (11–13). Compelling evidence also exists for Kras-induced reprogramming of pancreatic acinar cells into ductal intraepithelial neoplasia, a histologically well-defined precursor to pancreatic ductal adenocarcinoma (PDAC) (14–17). Although these findings provide evidence of pathological plasticity associated with the early stages of malignant transformation, it is unclear whether Kras-dependent reprogramming is unique to pancreatic tumors or whether it takes place in a broad range of neoplasms. Moreover, the concept of plasticity itself has been challenged, as it remains unclear why only some, but not all, cells in the adult lung, pancreas, and colon have the capacity to give rise to Kras-driven tumors (10, 12, 18, 19). In this study, we sought to address this question and to determine the role of cellular plasticity in the origin of Kras-mediated transformation. To ascertain that genuine reprogramming occurred, we set benchmark criteria and based our assessments on the accepted definition of reprogramming kinetics of somatic cells (20, 21), i.e., a long latency in the appearance of transformed cells in culture, stochastic nature of the process, direct phenotypic conversion occurring in many cells at once, all in the absence of new discernible mutations.
Results
Transformation of Primary Cells by Oncogenic Ras Reveals Stepwise Process.
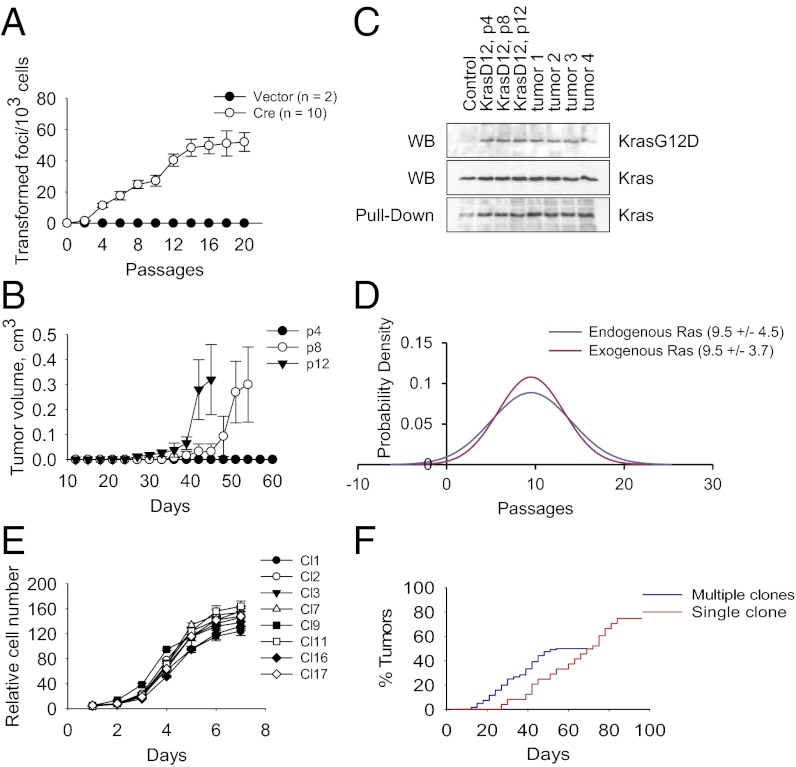
We studied primary fibroblasts derived from LSL KrasG12D p53−/− mice, referred to as LSL KrasG12D p53KO (p53 knockout), that carry a latent point mutant of the Kras2 gene (3). To delete the transcriptional termination cassette (Lox-Stop-Lox) and thus activate expression of the KrasG12D protein, we used a self-excising retroviral vector expressing Cre recombinase (22). The typical efficiency of recombination was >95%, as measured on day 4 postinfection. The cells were passaged over a period of ≥2 mo and monitored for acquisition of a malignant phenotype, defined as the potential to form morphologically transformed foci and generate tumors in nude mice. As expected, activation of KrasG12D expression resulted in rapid and uniform changes in cell proliferation and morphology (Fig. S1). By contrast, the induction of malignant properties showed delayed kinetics, reaching maximum levels at the second month in culture (Fig. 1 A and B). We observed no increase in levels of expression of KrasG12D or activated GTP-bound Kras protein that might underline these time-dependent differences (Fig. 1C). Overexpression of exogenous oncogenic Ras mutants conferred on cells similar time-dependent kinetics. In both cases (endogenous or exogenous KrasG12D), the cells’ acquisition of transformed phenotype closely fit the curve of a normal probability distribution (Fig. 1D), indicating a stochastic process. Thus, genetically identical or nearly identical cells become malignant with different latencies.
Fig. 1.
Transformation of primary cells by oncogenic Ras reveals a stepwise process. (A) Focus formation by LSL KrasG12D p53KO MEFs transduced with vector or Cre-expressing retroviruses. The error bars correspond to SD. (B) Tumor formation by KrasG12D p53KO MEFs (passage 4, 8, or 12) injected s.c. into nude mice. The error bars correspond to SE. (C) Immunoblot analysis of KrasG12D and total Kras expression, or pull-down of GTP-bound active Kras in control LSL KrasG12D p53KO MEFs, KrasG12D p53KO MEFs, or tumors derived from these cells. Passage numbers are indicated. (D) Probability distribution of transformed foci formation by p53KO MEFs expressing endogenous or exogenous KrasG12D. Mean (number of passages) and SD are indicated. (E) Growth curves of clonal KrasG12D p53KO cell lines. (F) Cumulative tumor formation by KrasG12D p53KO clones injected s.c. into nude mice. Multiple and single clone injections are shown.
To confirm these results, we followed transformation of monoclonal cell populations. To this end, early-passage LSL KrasG12D p53KO fibroblasts were infected with Cre-expressing retroviruses and plated at limiting dilutions. Randomly picked clones were expanded for 10 passages (1 mo) before being used for further experiments. All tested clones exhibited similar levels of proliferation (Fig. 1E) and KrasG12D expression (Fig. S1). When injected into nude mice, all clones produced tumors (Fig. 1F), demonstrating that most, if not all, KrasG12D p53KO cells or their clonal derivatives possess tumorigenic potential. However, the efficiency of tumor formation differed, as we obtained an average of 0.5 tumors per injection site. The latency of tumor formation also varied between 18 and 52 d (Fig. 1F). Moreover, groups of mice injected with cells from the same individual clone developed tumors with variable latencies (Fig. 1F). Thus, we conclude that KrasG12D expression per se is not sufficient to render p53-null cells fully tumorigenic. Instead, this process can be modeled with a stochastic distribution, in which occurrences of tumor formation are randomly distributed in time.
Phenotypic Conversion Is a Required Step in Ras-Induced Tumorigenesis.
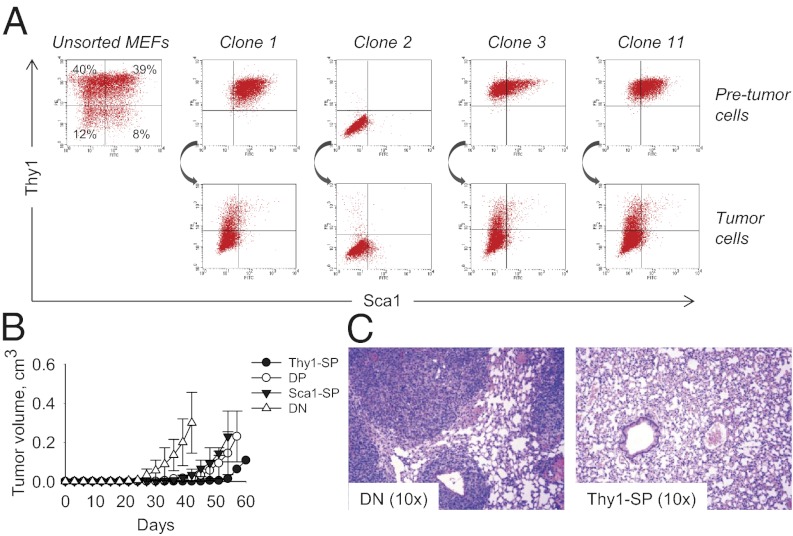
By using cell population statistics and functional complementation analysis, we were unable to find evidence that additional mutations or added genomic instability are required to confer the full malignant phenotype upon KrasG12D p53KO cells (Fig. S2). We thus set out to determine whether the acquisition of malignant properties depends upon the cell type in which KrasG12D is expressed. Primary MEFs, regardless of their p53 genotype, can be FACS-sorted into distinct populations based on the expression of surface markers Thy1 and Sca1 (23, 24). The Thy1-positive subpopulations [Thy1-single positive (SP) and Thy1/Sca1-double positive (DP)] represent the majority of cells (>70%; Fig. 2A and Fig. S3) and are composed of more differentiated cells, whereas Thy1-negative subpopulations [Sca1-SP and Thy1/Sca1 double negative (DN)] contain less mature cells (23, 24). FACS data demonstrated that the Thy1/Sca1 surface phenotypes of clonal KrasG12D cell lines were stable over extended periods of time. Moreover, tumors produced by DN cells were phenotypically identical to the injected parental cells (Fig. 2A). By contrast, all tumors arising from Thy1-positive clones also showed a large amount (as much as 96%) of DN cells (Fig. 2A). To rule out the possibility of cross-contamination, we generated secondary clones. However, when injected into nude mice, these secondary clonal populations again gave rise to tumors containing a large proportion of DN cells (Table S1), suggesting a causal relationship between the presence of these cells and cancer. To prove conclusively the link between DN phenotype and malignancy, we FACS-sorted early-passage KrasG12D p53KO MEFs into Thy1-SP, DP, Sca1-SP, and DN subpopulations, and then injected these cells into nude mice (Fig. 2B). Indeed, all tumors generated by the Thy1-SP, DP, and Sca1-SP subsets were composed, in large part, of DN cells (Fig. S3). Moreover, when injected s.c., DN cells developed tumors more rapidly than other populations (Fig. 2B). When injected into the tail vein, DN cells invaded the periinjection site and generated prominent lung metastases, in contrast to Thy1-positive cells, which failed to do so (Fig. 2C). These data indicate that KrasG12D-expressing DN cells are enriched for tumor-initiating cells. Furthermore, other cell populations adopt the phenotypic characteristics of DN cells before becoming tumorigenic.
Fig. 2.
Phenotypic conversion is a required step in Ras-induced tumorigenesis. (A) FACS analysis of Thy1 and Sca1 expression by total unsorted KrasG12D p53KO MEFs, clonal KrasG12D p53KO cell lines, and the respective tumors. (B) Tumor formation in nude mice by 104 FACS-sorted Thy1-SP, DP, Sca1-SP, and DN KrasG12D p53KO MEFs. (C) Lung metastatic foci in nude mice injected with primary KrasG12D p53KO MEFs. Representative images of lungs from mice injected with 104 FACS-sorted DN and Thy1-SP cells are shown.
Myc Promotes Phenotypic Conversion of KrasG12D-Expressing Cells.
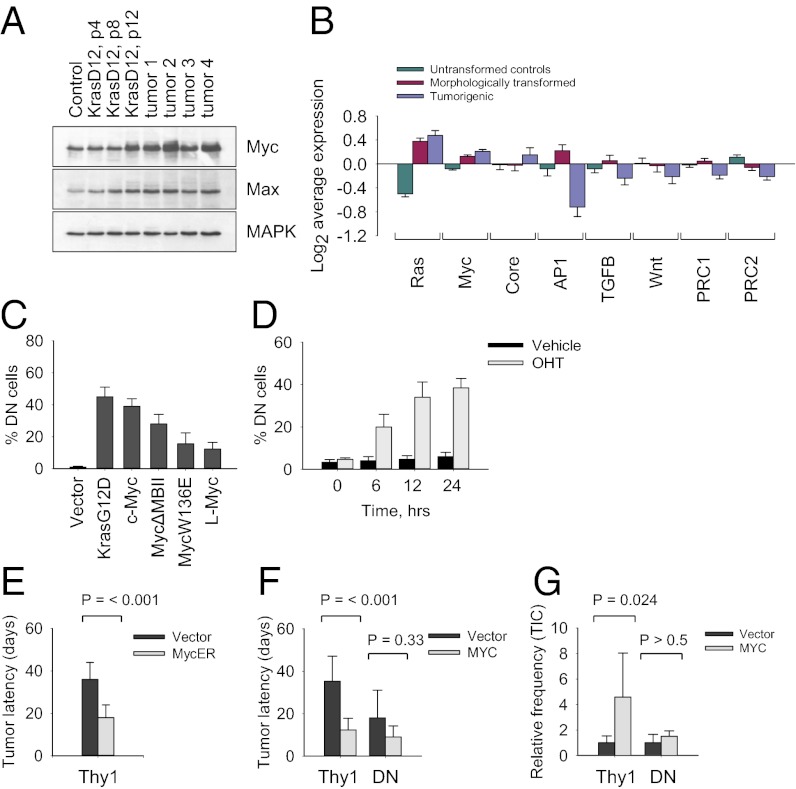
To understand the mechanism of phenotypic switching, we compared gene expression patterns of LSL KrasG12D p53KO MEFs (inactive allele, untransformed controls), early-passage KrasG12D p53KO MEFs (morphologically transformed nontumorigenic cells), and the respective tumor-derived cell lines. We identified ∼320 genes whose expression was changed more than fourfold in the tumorigenic cells (Fig. S4). Functional classification showed that the differentially expressed genes belong to the categories of metabolism, signal transduction, cell adhesion, cytoskeleton organization, and transcriptional regulation (Fig. S4). Of these, an activated Myc expression module (targets of Myc and Myc-interacting proteins) and a repressed polycomb repressive complex (PRC) module (involving PRC1 and PRC2) have been suggested as signatures that are common to reprogrammed cells and cancer cells (25). We identified 46 transcription factors whose expression is significantly changed in tumor cells (Fig. S4). Although Ras transformation did not cause activation of the Myc gene at the transcriptional level, Western blot analysis revealed that Myc protein levels were markedly elevated in tumor cells, implying a posttranslational mechanism of stabilization (Fig. 3A). Gene set enrichment analysis revealed high average activities for the Ras and Myc gene modules (common target genes within each regulatory pathway) in morphologically transformed and tumorigenic cells (Fig. 3B), whereas AP1, Stat3, TGF-β, Wnt, and PRC modules were repressed in tumor cells (Fig. 3B), further indicating that oncogenic transformation of p53-deficient cells is a stepwise process.
Fig. 3.
Myc promotes phenotypic conversion of KrasG12D-expressing cells. (A) Western blot analysis of untransformed and tumor-derived KrasG12D p53KO cells. Passage numbers are indicated. (B) Activity of gene regulatory modules in KrasG12D p53KO cells. (C) Phenotypic conversion of Thy1-positive KrasG12D p53KO cells transduced with the indicated retroviruses. (D) Phenotypic conversion of Thy1-positive KrasG12D p53KO cells transduced with MycER and treated with 1 μM hydroxytamoxifen (OHT) for the indicated periods of time. (E) Relative latencies of tumor formation in nude mice by Thy1-positive KrasG12D p53KO cells transduced with the indicated retroviruses. Cells were treated with 1 μM hydroxytamoxifen for 6 h. (F) Relative latencies of tumor formation in nude mice by Thy1-positive and DN KrasG12D p53KO cells transduced with vector alone or Myc-expressing retroviruses. (G) Relative frequencies of tumor-initiating cells (TICs) in Thy1-positive and DN KrasG12D p53KO cells transduced with the indicated retroviruses.
To identify transcription factors whose expression promotes phenotypic conversion of KrasG12D p53KO cells, we analyzed Thy1-positive and DN cell lines transduced with the identified Ras-inducible genes. Ectopic Myc and Ras, alone or in combination with each other, demonstrated the strongest ability to produce a rapid conversion of Thy1-positive to DN cells (Fig. 3C). We noted that overexpressed KrasG12D enhances Myc protein stability (Fig. S5). Moreover, ectopic Ras and Myc had additive effect with regard to conversion of Thy1-positive to DN cells (Table S2). Through the use of a hormone-inducible form of Myc [c-Myc-estrogen receptor fusion (MycER)], we estimated that the de novo generation of DN cells occurs as early as 6 h postinduction and continues for as long as 24 h (Fig. 3D). Transient excess of Myc activity, provided by the activated MycER, rendered Thy1-positive cells more tumorigenic (Fig. 3E). Furthermore, after becoming malignant, the MycER cells could be propagated for >50 generations and stayed tumorigenic even though Myc was no longer induced. On the contrary, L-Myc as well as c-Myc DNA-binding mutants (W136E and ΔMBII), all of which have low oncogenic potential, also induced partial reprogramming of Thy1-positive cells (Fig. 3C). This suggests that the promotion of phenotypic conversion by Myc is partly independent of its transforming activity, consistent with previous reports (26). A quantitative assessment of rate-limiting steps in Ras-induced tumorigenesis confirmed that activation of Myc is required for transformation of Thy1-positive cells. Thus, Myc-transduced Thy1-positive cells, but not DN cells, gave rise to tumors with an increased penetrance and decreased latency compared with controls (Fig. 3 F and G). By contrast, knockdown of Myc expression by shRNAs impaired tumor development by Thy1-positive and DN cells (Fig. S5). Collectively, these data indicate that Myc functions as a built-in amplifier of oncogenic Ras activity. Moreover, temporal and quantitative fluctuations of Myc levels can prime KrasG12D cells to become malignant (Fig. S4).
Phenotypes of Ras-Transformed Fibroblasts Are Versatile and Changeable.
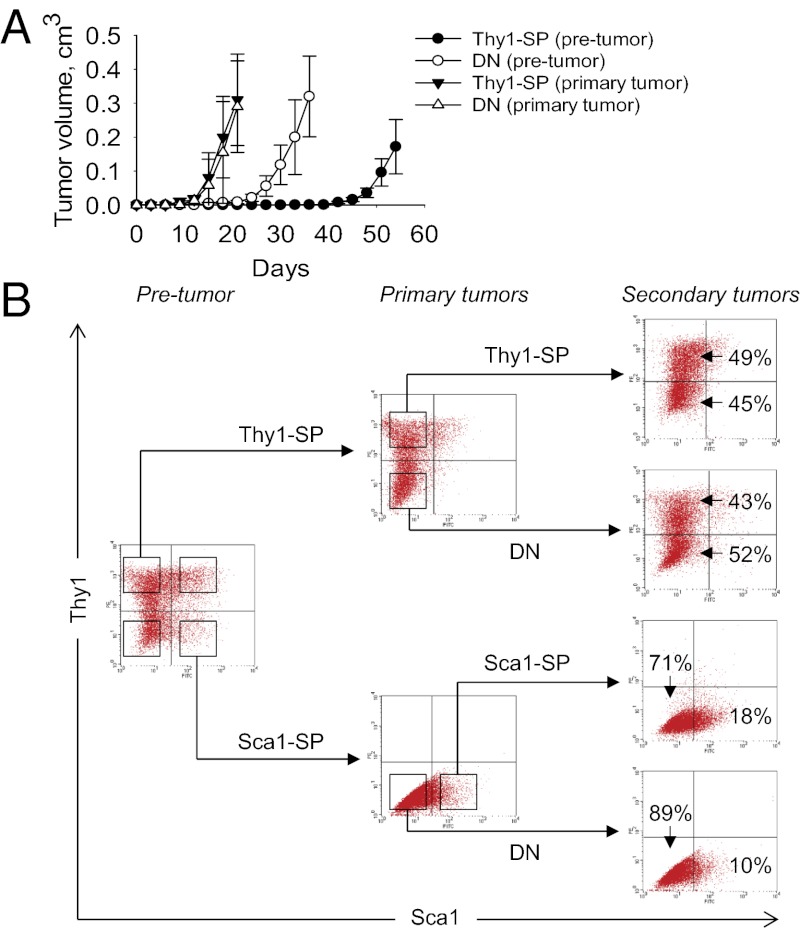
Our results thus far support the notion that non-stem cells have the potential to dedifferentiate and acquire stem cell properties as a direct consequence of oncogene-induced plasticity (27). To test this idea, we FACS-sorted primary tumors derived from Thy1-SP cells and then reinjected the resulting Thy1-SP and DN cell populations into nude mice. In contrast to the variable latencies of primary tumors, whereby DN cells give rise to tumors more rapidly compared with Thy1-SP cells, the corresponding latencies of secondary tumors differed insignificantly, reflecting rapid reacquisition of stem cell characteristic by cancer non-stem cells (Fig. 4A). Regardless of whether a Thy1-SP or DN cell population was used for tumor initiation, all secondary tumors reverted to the original phenotype of primary donor cells (Fig. 4B). Similar results were obtained by using FACS-sorted tumors derived from Sca1-SP cells (Fig. 4B). We thus conclude that KrasG12D-expressing cells acquire malignant properties in a predetermined order, in which their phenotypic conversion precedes the appearance of the transformed phenotype. However, the newly formed tumorigenic cells retain features characteristic of their origin, which favors their differentiation along lineages related to the parental cell.
Fig. 4.
The phenotypes of Kras-transformed fibroblasts are versatile and changeable. (A) Tumor formation in nude mice by 104 FACS-sorted premalignant and tumor-derived Thy1-SP and DN subpopulations of KrasG12D p53KO fibroblasts. (B) FACS analysis of Thy1 and Sca1 expression by premalignant KrasG12D p53KO MEFs and tumors.
Identification of Tumorigenic Cells in KrasG12D-Induced Pancreatic Cancer.
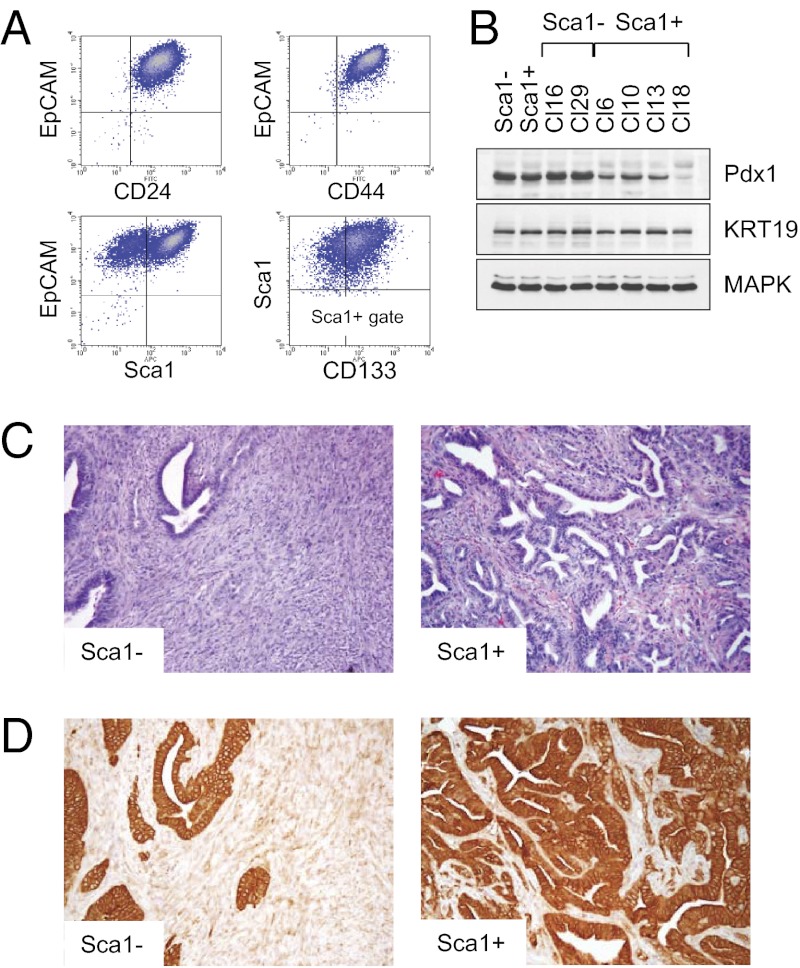
We used a similar strategy to study pathogenesis of pancreatic cancer, a genetically complex and heterogeneous disease. To this end, we have developed conditions for generation of pancreatic cells with cancer stem cell (CSC) properties. The cells were derived from LSL KrasG12D p53KO mice with Nestin-driven GFP expression (28) to mark pancreatic exocrine progenitor cells (29, 30). To activate expression of the KrasG12D protein, we used the self-excision Cre/LoxP strategy described earlier. FACS analysis of the purified pancreatic KrasG12D p53KO cells showed uniform expression of surface markers EpCAM, CD24, and CD44 (Fig. 5A), consistent with the stem cell phenotype in pancreatic cancer cells (31). Nearly 80% of the cells were positive in Sca1 staining (Fig. 5A), indicating their relation to pancreatic duct (30). The Sca1+ fraction was further divided by the expression of CD133, a marker of immature ductal cells (Fig. 5A), whereas GFP could be detected in ∼30% of presumptive exocrine progenitor cells (30, 32). A similar distribution was found in pancreatic cells derived from KrasG12D p53+/+ and KrasG12D p53+/− mice (Fig. S6). When isolated by FACS, the original Sca1+ cells retained their parental phenotype after ˃2 wk in culture, whereas Sca1− cells gave rise to both Sca1− and Sca1+ subsets (Fig. S6). Both Sca1− and Sca1+ subpopulations expressed Pdx1, the earliest marker of pancreatic epithelium, and cytokeratin 19, which is commonly associated with pancreatic ductal epithelial cells (Fig. 5B).
Fig. 5.
Identification of tumorigenic cells in KrasG12D-induced pancreatic cancer. (A) FACS analysis of the purified KrasG12D p53KO pancreatic epithelial cells using the indicated antibodies. (B) Western blot analysis of Sca1− and Sca1+ subsets of primary and clonal KrasG12D p53KO pancreatic epithelial cells. (C) Representative images of tumors from mice injected with Sca1− and Sca1+ subsets of KrasG12D p53KO pancreatic cells. Sarcomatoid carcinoma (Left) and adenocarcinoma (Right) are shown. (D) Immunohistochemical analysis of Sca1− and Sca1+ tumors. Expression of KRT19 in Sca1+ PDAC and low to absent expression in Sca1− tumors is shown.
To determine their tumorigenic potential, FACS-sorted Sca1+ and Sca1− cells were injected s.c. into nude mice. Whereas the two cell populations were both tumorigenic, they gave rise to morphologically distinct cancer types. Thus, tumors derived from Sca1− cells were composed of undifferentiated areas with sarcomatoid or anaplastic features (Fig. 5 C and D). By contrast, the histology of tumors derived from Sca1+ cells exhibited features of differentiated ductal adenocarcinoma with focal areas of poorly differentiated carcinoma (Fig. 5 C and D). Tumors with the undifferentiated areas were morphologically identical to those previously observed in KrasG12D mice nullizygous for p53 or p16Ink4a/p19Arf (16, 33–35). In humans, anaplastic pancreatic cancer is a relatively rare but universally fatal neoplasm. Thus, Sca1− and Sca1+ cells give rise to different histological subtypes of PDAC.
Metastatic Potential of Pancreatic Cancer Cells.
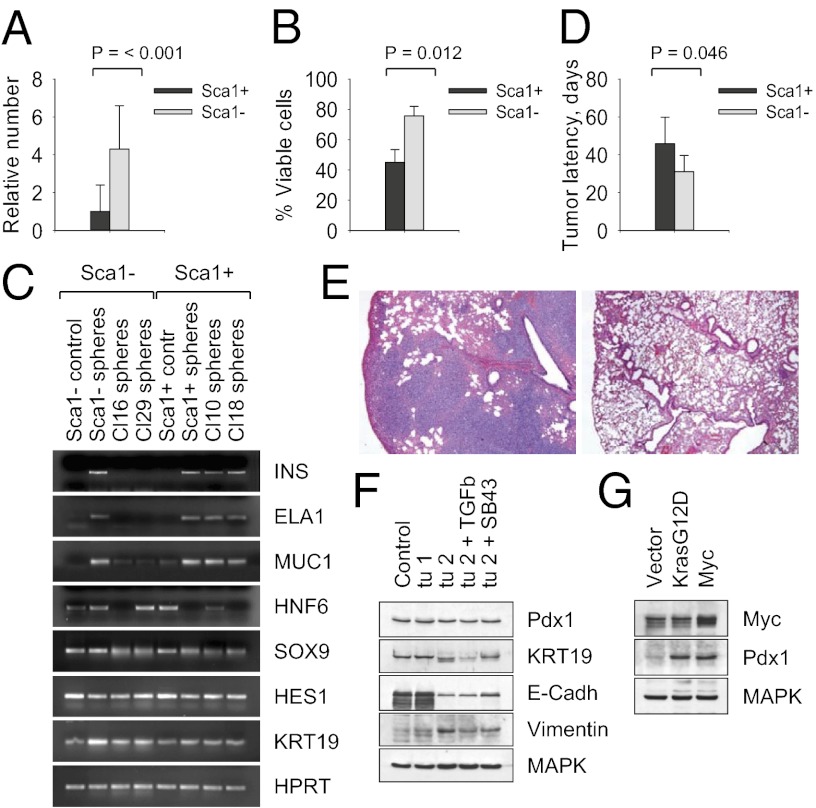
To characterize the cellular precursors of pancreatic cancer in more detail, we generated >50 clonal populations of Sca1+ and Sca1− KrasG12D p53KO cells. Although the clonal cell lines exhibited distinctive phenotypes, we found no differences in levels of KasG12D expression or Erk1/2 phosphorylation (Fig. S6). Moreover, Sca1+ and Sca1− cells formed self-renewing “pancreatospheres” in suspension culture. However, although Sca1− cells showed increased tumor sphere growth (Fig. 6A) and lower levels of anoikis (Fig. 6B), Sca1+ cells displayed a higher capacity to develop into cells that expressed insulin, elastase1, and mucin1, characteristic of islet, acinar, or ductal cell differentiation, respectively (Fig. 6C). By contrast, Sca1− cells were blocked at a less mature stage, as evidenced by the expression of transcription factors Sox9, HNF6, and Hes1 (Fig. 6C). When injected s.c. into nude mice, Sca1− cells developed tumors more rapidly compared with Sca1+ clones (Fig. 6D). When injected into the tail vein, Sca1− cells generated prominent multifocal lung metastases (Fig. 6E). Moreover, Sca1− tumor cells have lost the epithelial features, such as expression of E-cadherin, and acquired mesenchymal characteristics, such as expression of vimentin (Fig. 6F), whereas a large proportion of Sca1+ cell lines (≥20%) did not produce tumors in mice. These data reveal that the Sca1− subset of KrasG12D p53KO cells has substantial metastatic potential, recapitulating highly aggressive human PDAC.
Fig. 6.
Metastatic potential of pancreatic cancer cells. (A) Tumor sphere formation by Sca1+ and Sca1− KrasG12D p53KO cells. (B) Cell viability after 7 d in suspension culture. (C) RT-PCR analysis of Sca1+ and Sca1− subsets of primary and clonal KrasG12D p53KO pancreatic epithelial cells grown as cell monolayers (contr) or tumor spheres. HPRT is a control for equal loading. (D) Latencies of tumor induction in nude mice by Sca1+ and Sca1− KrasG12D p53KO pancreatic cells. (E) Lung metastatic foci in nude mice injected with 104 KrasG12D p53KO Sca1− cells (Left) but not with Sca1+ cells (Right). (F) Western blot analysis of tumors derived from total unsorted (Contr), Sca1+ (tumor 1), or Sca1− (tumor 2) KrasG12D p53KO pancreatic cells maintained for 2 d in the presence of 5 ng/mL TGF-β or 1 µM SB431542. (G) Western blot analysis of Pdx1 expression by clonal Sca1+ cells transduced with vector alone, KrasG12D−, or Myc-expressing retroviruses.
Myc Controls Generation of Self-Renewing Metastatic Pancreatic Cancer Cells.
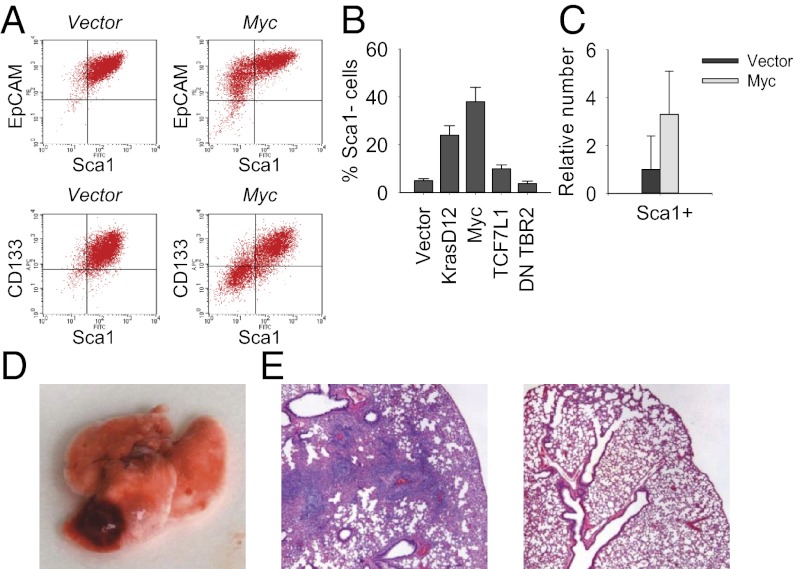
Current evidence indicates that there is genetic heterogeneity among metastasis-initiating cells in human pancreatic cancer, and that seeding metastasis may involve amplification of the Myc gene as an early and critical event (36). Consistent with this, ectopic expression of Myc in Sca1+ cell lines induced robust expression of Pdx1 (Fig. 6G) and converted them rapidly into Sca1-negative cells (Fig. 7 A and B). Moreover, this novel population of pancreatic cells exhibited an increased ability to form tumor spheres when grown in suspension cultures (Fig. 7C) and was capable of generating metastases in mice (Fig. 7 D and E). We observed higher mRNA expression of epithelial-to-mesenchymal transition-associated genes such as Lef1 (component of the Wnt signaling pathway), Twist1, Snail1, Snail2, and Zeb2 (negative feedback regulators of NF-κB) in clonal Sca1-negative KrasG12D p53KO cells (Fig. S6). The induction of metastatic phenotype in Myc-transduced Sca1+ cells also correlated with high expression of Lef1, Twist1, Twist2, and Zeb2 (Fig. S6). Moreover, cell lines transduced with Myc were more similar to embryonic pancreas than adult pancreas in their expression of Sox9, HNF6, Hes1, and Nestin (Fig. S6). By contrast, the disruption of TGF-β signaling via ectopic expression of dominant-negative form of TGF-β receptor 2 failed to produce similar effects (Fig. S6), consistent with the dual role of TGF-β in the progression of pancreatic cancer. Thus, tumors with intact TGF-β signaling frequently exhibit epithelial-to-mesenchymal transition and metastasize, whereas tumors null for TGFBR2 or Smad4 tend to retain a differentiated histopathology with increased expression of epithelial markers (35, 37). In line with this, pharmacologic inhibition of TGF-β signaling in Sca1-negative tumor cells induced expression of epithelial differentiation markers E-cadherin and keratin 19 (Fig. 6F). Collectively, these data implicate Myc in the control of self-renewal and lineage commitment of metastatic pancreatic cancer cells.
Fig. 7.
Myc controls the generation of self-renewing metastatic pancreatic cancer cells. (A) FACS analysis of vector- or Myc-transduced clonal Sca1+ KrasG12D p53KO pancreatic epithelial cells by using the indicated antibodies. (B) Phenotypic conversion of Sca1+ KrasG12D p53KO cells transduced with ectopic KrasG12D, Myc, TCF7L1, or dominant-negative TGFBR2. (C) Tumor sphere formation by vector- or Myc-transduced Sca1+ KrasG12D p53KO cells. (D) Lung metastatic foci in nude mice injected with Myc-transduced Sca1+ KrasG12D p53KO cells. (E) Representative images of lungs from mice injected with Myc-transduced (Left) or vector-transduced Sca1+ cells (Right).
Discussion
Recent evidence of pathological plasticity associated with the early stages of Ras-mediated transformation (12, 14–17) has prompted us to ascertain whether genome reprogramming plays a causal role in cancer development and what is the mechanism involved. Here, we used a well-defined system of primary rodent cells to show that endogenous expression of oncogenic KrasG12D provides a comprehensive view of tumor evolution similar to that seen in human invasive and metastatic disease. We show that, regardless of the origin of a cancer cell (primary MEFs or pancreatic epithelial cells), the initiation and maintenance of the transformed state is a complex process, comprising separable stages. Although the initial stage of transformation, i.e., induction of morphological changes, appears to be rapid and uniform, the acquisition of malignant properties by KrasG12D-expressing cells bears all the characteristics of a time-dependent stochastic process. Our data imply that the evolutionary relationship between premalignant and malignant p53-null cells expressing oncogenic Ras represents the outcome of alterations that accumulate over time rather than one single event. This in turn implies that, although the presence of KrasG12D mutation renders p53-null cells susceptible to transformation, other cooperatively acting genetic or epigenetic switches are needed to convert the cells to malignancy. Our results suggest that oncogene-induced reprogramming is an important early step in acquisition of malignancy and cancer initiation. We also demonstrate that the conversion of precancerous to cancerous cells is determined by oncogenic Ras-induced transcription factors, most notably Myc.
Although Ras and Myc are well known to cooperate to induce tumorigenesis, their potential involvement in cancer invasion and metastasis is not well understood. One of the strongest correlations between presence of Kras mutation and cancer is found in PDACs, in which Kras is mutated in 80% to 95% of cases, and is the most frequent mutation among all cancers (14–17). Although cancer metastasis is the most common cause of death in patients with cancer and is generally considered to be a multistep processes driven by the progressive accumulation of mutations (38), recent evidence suggests that some cells in early-stage pancreatic tumors are predisposed to metastatic spread (39, 40). In support of this hypothesis, we have identified two major populations of pancreatic progenitor cells responsible for KrasG12D-induced tumorigenesis in mice, one highly predisposed, and the other less prone, to metastatic dissemination. We demonstrate that metastatic conversion of pancreatic cells exhibiting different degrees of differentiation and malignancy can be reconstructed in cell culture, and Myc controls the generation of self-renewing metastatic cancer cells. These results have important implications for cancer treatment. According to the CSC theory, cancer is caused by a minority population of self-renewing tumor-initiating cells (also called CSCs), and only eradication of this cell population will lead to a cure (41, 42). However, our results, together with those from previous studies, suggest that there exists a dynamic equilibrium between CSCs and non-stem cancer cells (NSCCs), given that an appropriate mutation that enables reprogramming is present (27, 43, 44). Moreover, these CSCs and NSCCs can be sorted and functionally identified. The NSCCs can also, in appropriate conditions, give rise to CSCs in vivo. In the present proof-of-concept study, we demonstrated conclusively that KrasG12D acts by reprogramming more differentiated cell populations into less differentiated cells poised to become malignant. Considering the fact that there is usually an extended latency period before malignant cells appear, our data support a model in which cells of different degrees of differentiation give rise to tumor cells with different kinetics, reflecting the requirement of more genetic or epigenetic changes for transformation of more differentiated cells. We find that, in contrast to the variable latencies of primary tumors, the latencies of secondary tumors differ insignificantly, reflecting rapid reacquisition of stem cell characteristics by NSCCs. Moreover, stochastic disturbances in the normally existing equilibrium between CSCs and NSCCs allow the formation of CSCs with high metastatic capacity at any time during cancer progression. Thus, therapy approaches that target only CSCs may not be sufficient, because the NSCCs that remain may give rise to CSCs and reinitiate tumorigenesis.
The early detection of cancer-specific genetic and epigenetic changes holds potential for improved cancer diagnostics and pretreatment risk stratification of patients with cancer (45). It should be emphasized that, although persistent alterations in gene expression can be induced by Ras or Myc (46–48), no single subset of downstream target genes was found to account for their prooncogenic activity. Expression of oncogenic Ras induces a highly pleiotropic response in vitro and in vivo (2–7). Likewise, Myc acts as a universal amplifier of the existing gene expression programs, increasing output (49–51). An additional potentially related mechanism involves Myc-mediated regulation of PRC-target genes (47, 48). A striking finding in the present study is that reprogramming induced by oncogenic KrasG12D and Myc is rapid, does not require cell division, and is implemented with high efficiency. Of particular importance is the observation that Myc controls the generation of self-renewing metastatic cancer cells. A future challenge will be to understand the mechanistic underpinnings of the process. The critical question that needs to be answered is whether oncogene-induced reprogramming can be a valid therapeutic target.
Materials and Methods
Primary fibroblasts were derived from day 13–14 embryos using standard techniques. All animal studies were approved by the Institutional Animal Care and Use Committee at the State University of New York at Stony Brook. Molecular, histological, microarray, and other analyses were performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by New York State Stem Cell Science Contract N08T-040 (to U.M.M. and O.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219592110/-/DCSupplemental.
References
- 1.Karnoub AE, Weinberg RA. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerra C, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4(2):111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 3.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5(4):375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, et al. Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc Natl Acad Sci USA. 2009;106(19):7979–7984. doi: 10.1073/pnas.0900343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuhmacher AJ, et al. A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J Clin Invest. 2008;118(6):2169–2179. doi: 10.1172/JCI34385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood. 2011;117(6):2022–2032. doi: 10.1182/blood-2010-04-280750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, et al. Endogenous oncogenic Nras mutation initiates hematopoietic malignancies in a dose- and cell type-dependent manner. Blood. 2011;118(2):368–379. doi: 10.1182/blood-2010-12-326058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkisian CJ, et al. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9(5):493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 9.Collado M, et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436(7051):642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 10.Guerra C, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19(6):728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim CF, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121(6):823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 12.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winslow MM, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473(7345):101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11(3):291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 15.De La O JP, et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci USA. 2008;105(48):18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16(5):379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris JP, 4th, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 2010;120(2):508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, et al. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc Natl Acad Sci USA. 2012;109(13):4910–4915. doi: 10.1073/pnas.1112499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez-Mancera PA, Guerra C, Barbacid M, Tuveson DA. What we have learned about pancreatic cancer from mouse models. Gastroenterology. 2012;142(5):1079–1092. doi: 10.1053/j.gastro.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Hanna J, Carey BW, Jaenisch R. Reprogramming of somatic cell identity. Cold Spring Harb Symp Quant Biol. 2008;73:147–155. doi: 10.1101/sqb.2008.73.025. [DOI] [PubMed] [Google Scholar]
- 21.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: Facts, hypotheses, unresolved issues. Cell. 2010;143(4):508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8(1):233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 23.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460(7259):1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nemajerova A, Kim SY, Petrenko O, Moll UM. Two-factor reprogramming of somatic cells to pluripotent stem cells reveals partial functional redundancy of Sox2 and Klf4. Cell Death Differ. 2012;19(8):1268–1276. doi: 10.1038/cdd.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci USA. 2010;107(32):14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA. 2011;108(19):7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mignone JL, Kukekov V, Chiang AS, Steindler D, Enikolopov G. Neural stem and progenitor cells in nestin-GFP transgenic mice. J Comp Neurol. 2004;469(3):311–324. doi: 10.1002/cne.10964. [DOI] [PubMed] [Google Scholar]
- 29.Means AL, et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132(16):3767–3776. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 30.Rovira M, et al. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci USA. 2010;107(1):75–80. doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 32.Oshima Y, et al. Isolation of mouse pancreatic ductal progenitor cells expressing CD133 and c-Met by flow cytometric cell sorting. Gastroenterology. 2007;132(2):720–732. doi: 10.1053/j.gastro.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 33.Aguirre AJ, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardeesy N, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103(15):5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ijichi H, et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20(22):3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467(7319):1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bardeesy N, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20(22):3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haeno H, et al. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell. 2012;148(1-2):362–375. doi: 10.1016/j.cell.2011.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1-2):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 42.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324(5935):1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 44.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: The time is now. Crit Rev Oncol Hematol. 2008;68(1):1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449(7165):1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodliffe JM, Wieschaus E, Cole MD. Polycomb mediates Myc autorepression and its transcriptional control of many loci in Drosophila. Genes Dev. 2005;19(24):2941–2946. doi: 10.1101/gad.1352305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neri F, et al. Myc regulates the transcription of the PRC2 gene to control the expression of developmental genes in embryonic stem cells. Mol Cell Biol. 2012;32(4):840–851. doi: 10.1128/MCB.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cole MD, Cowling VH. Transcription-independent functions of MYC: Regulation of translation and DNA replication. Nat Rev Mol Cell Biol. 2008;9(10):810–815. doi: 10.1038/nrm2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.