Abstract
Our anatomical analysis revealed that a dry maize seed contains four to five embryonic leaves at different developmental stages. Rudimentary kranz structure (KS) is apparent in the first leaf with a substantial density, but its density decreases toward younger leaves. Upon imbibition, leaf expansion occurs rapidly with new KSs initiated from the palisade-like ground meristem cells in the middle of the leaf. In parallel to the anatomical analysis, we obtained the time course transcriptomes for the embryonic leaves in dry and imbibed seeds every 6 h up to hour 72. Over this time course, the embryonic leaves exhibit transcripts of 30,255 genes at a level that can be regarded as “expressed.” In dry seeds, ∼25,500 genes are expressed, showing functional enrichment in transcription, RNA processing, protein synthesis, primary metabolic pathways, and calcium transport. During the 72-h time course, ∼13,900 genes, including 590 transcription factor genes, are differentially expressed. Indeed, by 30 h postimbibition, ∼2,200 genes expressed in dry seeds are already down-regulated, and ∼2,000 are up-regulated. Moreover, the top 1% expressed genes at 54 h or later are very different from those before 30 h, reflecting important developmental and physiological transitions. Interestingly, clusters of genes involved in hormone metabolism, signaling, and responses are differentially expressed at various time points and TF gene expression is also modular and stage specific. Our dataset provides an opportunity for hypothesizing the timing of regulatory actions, particularly in the context of KS development.
Keywords: plant leaf development, plant hormones, gene expression profiling
Maize, a well-studied crop, has been used as a model plant for C4 photosynthesis study, as its leaves possess the kranz structure (KS) for efficient photosynthesis. However, how its leaves develop from seed following imbibition has not been well studied. In particular, it is unclear how KS forms during leaf development. Using the next generation sequencing technology, Li et al. (1) studied the leaf transcriptomes of four regions of 9-d-old third maize leaves: the base, the tip, and two middle regions of the leaf, representing different leaf developmental stages, with the base being the youngest. The data revealed a dynamic transcriptome profile, showing different transcripts enriched in different regions and providing a preliminary view of molecular changes during maize leaf development. However, as the leaf base already exhibits distinct KS, it is not early enough to represent the early leaf development when KS begins to form. Indeed, our anatomical study reveals that KS already exists in a rudimentary form in the first two embryonic leaves of maize dry seeds, and the embryonic leaves develop rapidly after seed imbibition (see below). To correlate the transcriptomic dynamics with the KS development during seed germination, we have obtained the time course transcriptomes of embryonic leaves at every 6 h, starting from dry seeds to hour 72 postimbibition. This set of data provides a clear picture of the transcriptional dynamics of genes for the early leaf development during maize seed germination. Moreover, we use the data to infer the succession of biological processes during this period. Finally, we examine what hormone-related genes and transcriptional regulators are differentially expressed in the embryonic leaves over time, providing a global view of the molecular changes underlying their anatomical and physiological transitions during seed germination.
Results and Discussion
Anatomical Changes During Early Leaf Development.
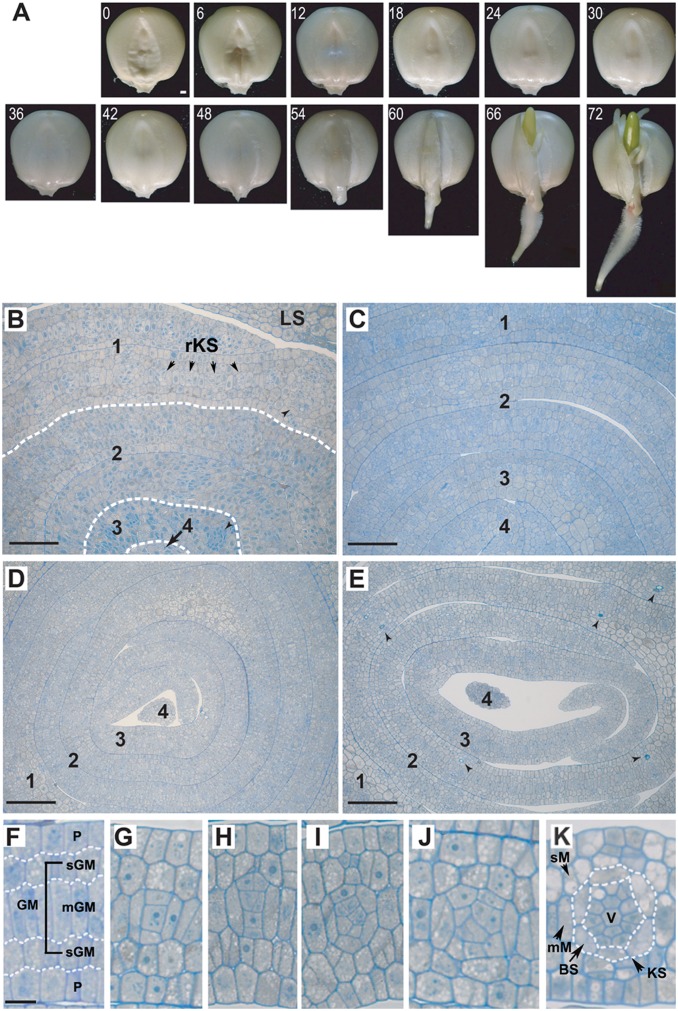
Fig. 1A shows the morphological changes in maize seeds during the first 72 h of imbibition; the tissue swells over time until the appearance of coleoptile at the 72nd hour (T72). A dry maize seed contains four to five embryonic leaves (Fig. 1B). The first leaf already has many well-differentiated rudimentary KS units separated by one to two ground meristem (GM) cells in the middle (median) layer of leaf primodium. The second leaf is less developed, but has recognizable KS units. Upon imbibition, all embryonic leaves expand within 12 h (Fig. 1C). Compared with the first embryonic leaf, the younger leaves contain fewer veins (KS units), frequently separated by more than five palisade-like median ground meristem (mGM) cells. During the next 2 d, the vein density rapidly increases (Fig. 1 D and E).
Fig. 1.
Changes in maize seed morphology and anatomy during germination. (A) Changes in seed morphology from 0 h (dry seed) to 72 h after imbibition. (i) Within the first 36 h: seed expands but no change is seen on the phenotype. (ii) At the 48th hour: primary root breaks the skin. (iii) At the 60th hour: epicotyl is now visible. (iv) At the 66th hour: primary root hairs and lateral roots are now visible. (v) At the 72nd hour: epicotyl becomes green. (B–E) Cross-sections of maize embryonic leaves at (B) 0 h (dry seed; arrows indicate midribs), (C) 12 h, (D) 48 h, and (E) 72 h after imbibition (arrowheads indicate lignified cells in major veins). (F–K) Progressive changes in kranz structure development in embryonic leaves during seed germination. 1–4, first to fourth embryonic leaves; BS, bundle sheath cells; KS, kranz structure (vein + BS); LS, leaf sheath; mGM, palisade-like median ground meristem cells; mM, median layer mesophyll cells; P, protoderm; rKS, rudimentary kranz structure; sGM, round-shaped subprotodermal ground meristem cells; sM, subprotodermal mesophyll cells; V, vein. [Scale bars, (A) 1 mm; (B and C) 50 µm; (D and E) 100 µm; and (F–K) 10 µm.]
The most significant feature of the maize embryonic leaves, except for the main vein regions, is that they consist of five layers of distinct cell types in transverse section (Fig. 1F): two layers of protoderm (P) and three layers of GM cells, with the mGM cell layer located in the middle of the leaf and sandwiched by two layers of round-shaped ground meristem (sGM) cells. The two layers of protoderm form the adaxial and abaxial epidermal cell layers, whereas contiguous mGM cells differentiate into vascular tissue, bundle sheath (BS) cells, and mesophyll (M) cells. Clearly, all KS units of the minor veins arise from the mGM cells, initially with a simple formation of three to four contiguous cells with two cells sandwiching one or two smaller cells. Fig. 1 G–K shows the progressive changes in KS or vein development. The sGM cells next to the protodermal cell layers give rise to the subprotodermal mesophyll (sM) cells (Fig. 1K) separating the veins and the epidermal cells in mature leaves. KS units at different developmental stages can be found in all embryonic leaves, especially in the younger leaves. In initiating new KS units the progenitor mGM cells begin to divide and differentiate into procambium that leads to the formation of various vascular cells of xylem and phloem, whereas the two lateral mGM cells differentiate into bundle sheath (BS) cells (Fig. 1K), likely caused by polarized acquisition of hormones (e.g., auxin, gibberellin, and cytokinin) in these progenitor cells for their division (anticlinal, periclinal, and oblique) and differentiation. Because in mature maize leaves, the veins are normally separated by only two M cells, these M cells may also derive from undifferentiated mGM cells (2). In any event, during the first 72 h of germination the maize embryonic leaves already contain KS units at varying developmental stages, with the most developed ones appearing in the first leaf (e.g., Fig. 1K). Our observation of the initiation and differentiation of minor veins or KS in maize embryonic leaves is consistent with the anatomical study of the base of the third through fifth leaves from 2-wk-old maize plants (2), showing that both main and minor veins have a similar pattern in initiation and development. Also clear is that the two M cells in the middle layer (mM in Fig. 1K) of the maize leaf intervening BSs are ontogenetically more closely related to the BS cells than are the subprotodermal M cells (sM in Fig. 1K).
Transcriptome Profiling of Early Leaf Development.
The RNA samples were taken from the embryonic leaves of dry seeds (denoted T00) and imbibed seeds every 6 h up to T72. The 13 RNA samples were pair-end sequenced with Illumina HiSeq2000 (Table S1). The reads were mapped to the maize genome and the mappable reads were used to estimate the transcription level with the reads per kilobase per million (RPKM) mapped reads measure. The RPKM values were then normalized for comparison between time points.
The current maize annotation contains a “working gene set” (WGS) and a “filtered gene set” (FGS, a subset of WGS). After excluding repetitive elements and pseudogene entries, 63,363 WGS and 39,441 FGS genes remain. To reduce the effect of background transcription, a WGS gene is included in our analysis only if its RPKM value is ≥1 at more than one time point (Dataset S1). With this criterion, 30,255 (48%) WGS genes, including 23,900 (61%) FGS genes, are selected for our analysis (Table S2). For the 2,070 maize transcription factors (TFs) (3), 1,238 genes (60%) are included in our analysis. In the selected gene set, a gene is considered expressed at a time point if its normalized RPKM value is ≥1 at that time point (Dataset S1).
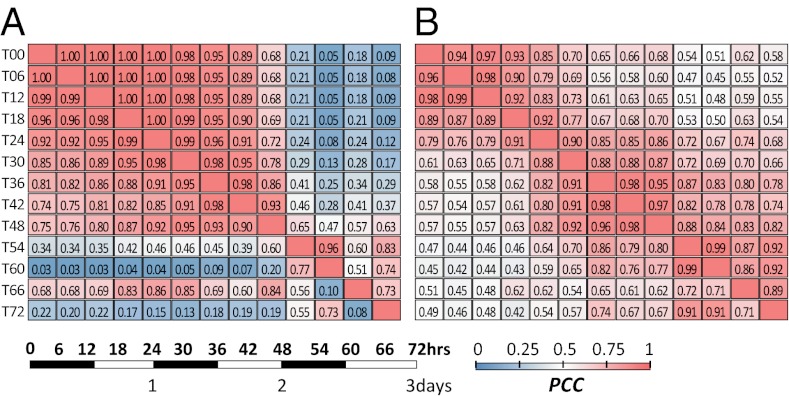
We now ask how similar the expression patterns between time points are. For the collection of all top 1% expressed genes at each time point (693 genes), the correlation (r) in expression level between two consecutive time points decreases very slowly until T42, but then a sharp transition occurs between T48 and T54 (r = 0.65; Fig. 2A, Upper half). This transition is mostly due to the up-regulation of ribosomal protein genes and down-regulation of heat stress and glycolysis genes at T54. Another sharp transition occurs between T60 and T66 (r = 0.51; Fig. 2A, Upper half), due to the up-regulation of heat stress genes and down-regulation of ribosomal protein genes and genes involved in DNA synthesis and glycolysis at T66 (Functional Transition Postimbibition). Some of these changes, such as those attributed to heat stress, may be related to response to sunlight and rising temperature during daytime; T54 was midnight, whereas T66 was noon. At any rate, the top 1% of genes at T54 and later are very different from the top 1% of genes before T30 because all r values are <0.3 (Fig. 2A, Upper half). The bottom 1% (1,415) of genes show a very similar transition pattern to the top 1% but their r values decrease somewhat faster, especially after T48, with the exception of T66 (Fig. 2A, Lower half). The middle 98% (28,152) of genes show a slightly faster decrease in r than the top 1% of genes until T48 but a slower rate of decrease after T48 (Fig. 2B, Upper half). TF genes show a slower rate of decrease in r initially than other genes but a faster rate after T54 (Fig. 2B, Lower half).
Fig. 2.
Expression correlations between time points. (A) Heatmaps of Pearson’s correlation coefficients (PCCs) between time points calculated for the genes at the top 1% expression levels (Upper half) and for the genes at the bottom 1% expression level (Lower half). (B) PCC heatmap for all genes with middle 98% expression level (Upper half) and for TF genes (Lower half). Scale on the Lower Left shows the night (dark) and day (light) cycles.
As mentioned above, an earlier study obtained the transcriptomes of four regions of 9-d-old third maize leaves (1). No significant correlation exists between their transcriptomes and ours, except for the leaf base (the youngest region), and the maximum correlation is only 0.64, which occurs at T60 (Fig. S1). Thus, the base of a 9-d-old maize leaf may not represent the very early leaf development and our time course data provide a unique global view of gene expression over maize early leaf development.
Genes Expressed in the Embryonic Leaves of Dry Seeds.
In dry seeds, transcripts for 20,603 (52%) FGS and 25,401 (40%) WGS genes are already present with RPKM ≥1. These transcripts may jump-start the maize leaf development upon imbibition. To assess the roles of these stored transcripts, we examine MapMan functional categories containing an over- or underrepresented number of expressed genes at T00 (Fig. S2A).
Three types of genes are overrepresented at T00. The first type includes genes involved in transcription, RNA processing, protein synthesis, and primary metabolic pathways, including carbohydrate biosynthesis, the tricarboxylic acid (TCA) cycle, mitochondrial electron transport, and fatty acid, nucleotide, and amino acid biosynthesis. These are likely transcripts stored in the embryonic leaves for meeting the demands for rapid growth upon imbibition. Similarly, cell division genes are overrepresented, potentially contributing to rapid cell proliferation. The second type includes genes involved in calcium transport, likely reflecting the role of calcium in removing the inhibitory effect of abscisic acid (ABA) during germination (4). The third type includes stress genes, whose expression may be caused by dehydration and osmotic stress during seed maturation. Interestingly, genes involved in abiotic stress are overrepresented but those in biotic stress are underrepresented in the quiescent leaves of the dormant seeds.
Some of the above categories are enriched not only in dry seeds but also throughout the time course (Fig. S2A). These categories include primary metabolic pathways, such as the TCA cycle, some components of mitochondrial electron transport, and biosynthesis of amino acids, proteins, and nucleotides, likely for the need of energy and building blocks for early leaf development. On the other hand, genes in some functional categories are underrepresented, including biotic stress, cell wall modification and degradation, and secondary metabolism.
Functional Transition Postimbibition.
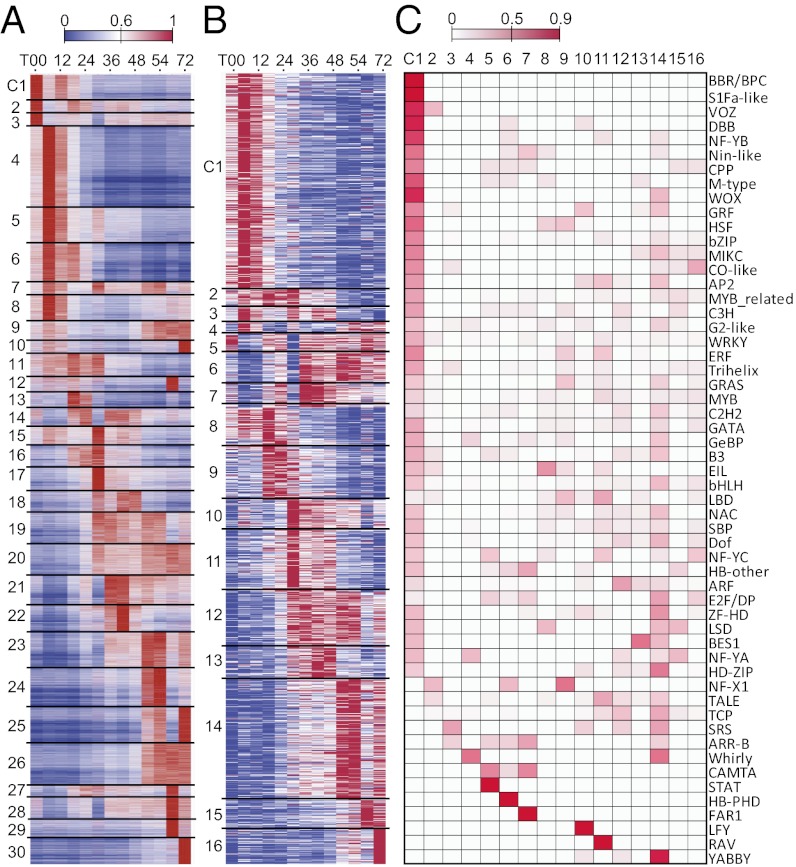
Although some functional categories contain consistently over- or underrepresented numbers of expressed genes over the entire time course, only 852 (2.8%) genes are expressed at similar levels (coefficient of variation <0.2) during the first 72 h of imbibition and 71 of the 852 genes show more than twofold changes in RPKM value during the 72 h. Thus, most genes are up- or down-regulated to various degrees over the 72-h period (Fig. 3A). In addition, some biological processes are enriched in a time-point–specific manner (Fig. S2A). For example, genes involved in cell division and calcium transport are overrepresented until T30 (Fig. S2A). Genes of E3 ligases involved in ubiquitin-mediated protein degradation are particularly prominent at T12–T18, indicating the elevated need for protein turnover. In addition, chromatin structural genes tend to be expressed from T30 to T72. Moreover, many categories have underrepresented numbers of expressed genes in a time-point–specific manner. For example, ethylene signal transduction genes exhibit reduced expression during T54–T72, presumably reflecting a reduced influence of ethylene at this stage.
Fig. 3.
Functional transition and gene expression pattern over the time course. (A) Expression patterns of 30 coexpression modules (C01–C30), ordered according to the time point of their expression peak. For each gene, the value shown is the RPKM value normalized by the maximum value of all RPKM values of the gene in question over all time points. (B) Normalized expression levels of 1,238 TF genes sorted into 16 expression modules (clusters) (C1–16). (C) Proportion of TFs in a family that are members of a cluster.
To assess functional transition due to highly expressed genes, we conducted enrichment tests on genes in the top 10% or 1% (Fig. S2 B and C). Consistent with the above analysis using all genes, categories containing, for example, ATP and protein synthesis or abiotic stress genes are enriched. However, the analysis of the top 10% of genes reveals more categories that are time-point specific. For example, a major class of plant cell wall proteins, hydroxyproline-rich glycoproteins (HRGPs), which are involved in growth and cell wall development, are enriched at T00 and T12. One HRGP (GRMZM2G168651) has been shown to be expressed in maize meristem and vascular tip (5), consistent with the appearance of vascular tissues in embryonic leaves (Fig. 1). Another example is that, at T54, categories containing ribosomal proteins, ATPase transporters, and peroxiredoxins are overrepresented. Peroxiredoxins are targeted to chloroplasts to protect the photosynthetic membrane (6) by scavenging toxic peroxides produced during electron transport. Finally, at T72 genes involved in brassinosteroid (BR) metabolism are overrepresented, suggesting the rapid formation of vascular tissues at this stage, as BR promotes cell expansion and vascular differentiation (7, 8).
Differential Gene Expression Between Time Points.
We identify three types of differentially expressed genes (DEGs) between time points (Table 1). Type 1 DEGs are identified by comparing the transcriptome at a time point against its preceding one, signifying the transition in gene function in 6 h. From T00 to T06, the transcript levels of 229 genes (including 7 TF genes) are decreased, whereas from T12 to T18, 481 genes (including 22 TF genes) are up-regulated. Notably, a sharp increase occurs in down-regulated DEGs from T24 to T30 (1,136 genes, including 28 TF genes) and in up-regulated DEGs from T30 to T36 (1,585 genes, including 50 TF genes). These and other large changes in DEG numbers suggest major developmental or physiological transitions. Interestingly, a group of genes down-regulated at T30 is up-regulated at T36, including RNA regulation of transcription (including many TFs), chromatin remodeling factors, histone acetyltransferases, DNA synthesis/chromatin structure, and cell cycle/division (Fig. S3 A, D, and E).
Table 1.
Numbers of differentially expressed genes and TFs between two time points
| Time (T) | All genes (TF genes): Up-regulated/down-regulated |
||
| T vs. T–6 h | T vs. T00 | New DEGs | |
| T06 | 45 (0)/229 (7) | 45 (0)/229 (7) | 45 (0)/229 (7) |
| T12 | 21 (0)/18 (0) | 51 (2) /81 (0) | 35 (2)/21 (0) |
| T18 | 481 (22)/75 (0) | 466 (35)/43 (0) | 427 (34)/38 (0) |
| T24 | 225 (5)/75 (5) | 1182 (60)/552 (13) | 824 (34)/510 (13) |
| T30 | 136 (16)/1136 (28) | 2035 (152)/2242 (82) | 1142 (96)/1638 (64) |
| T36 | 1585 (50)/189 (9) | 2555 (133)/2423 (81) | 877 (24)/1015 (32) |
| T42 | 20 (0)/32 (0) | 2402 (130)/2404 (88) | 280 (12)/225 (10) |
| T48 | 143 (2)/16 (0) | 2379 (130)/2089 (98) | 305 (9)/119 (15) |
| T54 | 343 (4)/466 (47) | 3700 (148)/2961 (157) | 1351 (46)/686 (50) |
| T60 | 46 (3)/64 (2) | 3980 (171)/3340 (171) | 523 (23)/371 (20) |
| T66 | 718 (17)/705 (17) | 2981 (91)/2602 (140) | 464 (6)/310 (18) |
| T72 | 646 (14)/615 (33) | 3731 (152)/3042 (186) | 424 (20)/223 (20) |
New DEGs, differentially expressed genes not observed until that time point; T-6 h, time point 6 h earlier than time T, that is, the time point preceding T.
Type 2 DEGs are defined by comparisons between a time point and T00, representing genes that are up- and/or down-regulated postimbibition (Table 1 and Fig. S3B); some type 2 DEGs overlap with type 1 DEGs. Many genes become up-regulated 18 h after imbibition and many functional categories are enriched in type 2 DEGs at various time points, including arabinogalactan proteins (AGPs) and cell wall proteins at T18, DNA synthesis/chromatin structure at T24, cell wall cellulose synthesis at T30, fatty acid metabolism at T36, BR signal transduction at T42, the YABBY TF family at T48, and nucleotide metabolism at T54. At T72 genes for nitrate metabolism and chloroplast targeting are up-regulated (Fig. S3B). In Arabidopsis, YABBY members are responsible for specifying the leaf adaxial identity (9). On the other hand, genes in a number of categories are down-regulated such as RNA helicase at T06, late embryogenesis abundant (LEA) proteins at T24, biodegradation of xenobiotics at T30 and T36, ATP synthesis at T36, and many TF families [AP2 (apetala 2)/ERF (ethylene-responsive factor), CO (constans)-like, EIN3 (ethylene insensitive 3)-like and bZIP (basic leucine zipper)] and ubiquitin E3 at T48.
The third type includes genes not differentially expressed until the indicated time points; they are a subset of type 2 DEGs. Type 3 DEGs potentially signify the onset/termination of developmental and/or physiological processes at a particular stage (Fig. S3C). An example is the differential regulation of cell wall proteins (Dataset S1). AGPs and leucine-rich repeat proteins (LRRs) are up-regulated at T18, T30, or T60; cellulose synthases are up-regulated at T18 or T66; cell wall modification proteins are up-regulated at T18, T24, or T54; pectin esterases, which are involved in cell wall modification and breakage, are up-regulated at T24 or T30; and cell wall degradation proteins (e.g., cellulases and pectate lyases) are up-regulated at T24, T30, or T36, which may be important for subsequent cell division (Dataset S1). Differential regulation of genes of the same function/process reveals how distinct components are used at different stages during leaf development.
Overall, the total number of DEGs over the entire time course is estimated to be 13,907 if all pairwise comparisons are included. Thus, close to half (46%) of the 30,255 expressed WGS genes may be regarded as differentially expressed during the 72-h time course. The corresponding proportion for expressed TF genes is 47% (590/1,238).
Coexpression Modules.
Using hierarchical clustering, we classify the expressed WGS genes into 30 coexpression modules, each of which contains genes with highly similar expression patterns (Fig. 3A and Fig. S4A). In the following analysis, we select 20 modules (Fig. S4C) that together contain ∼83% (12,716) of the 15,372 expressed genes with MapMan annotation (Fig. S4 B and D). With some exceptions, the clusters can be sorted according to the time point the peak expression occurs. This clustering analysis allows us to aggregate genes over multiple time points, providing another view of functional transitions along the early maize leaf development.
The 20 selected modules can be roughly divided into early (T00–T12), middle (T18–T48), and late (T54–T72) stages (Fig. 3A and Fig. S4 C and D). The early stage, best represented by modules C01, C04–C06, and C08, coincides with the transition from seed dormancy to germination, which involves extensive physiological changes (10). In C01, the expression level is high at T00 and T12 but becomes lower after T18. C01 includes genes related to lipid metabolism (Fig. 3A and Fig. S4D). These transcripts are likely mRNAs stored for the very early stage of leaf development during germination. In C04–C06, and C08, gene expression peaks at T06 but different modules have distinct profiles after the peak. ABA and ethylene-related genes belong to these modules (Fig. S4 D and E), reflecting the roles of these hormones during early leaf development (11). Genes related to protein degradation and synthesis are also enriched in these modules, suggesting that protein metabolism is essential for the development of embryonic leaves. For example, functional categories related to ubiquitin E3 SCF (Skp1-Cullin-F-box protein) and RING (really interesting new gene) domain proteins are overrepresented in C05 (Fig. S4B), perhaps because several E3 ligases are major components of hormone signaling (12).
The middle stage (T18–T48), represented by C13, C14, C16–C21, and C23, is typified by the up-regulation of auxin and gibberellic acid (GA) related genes, which are involved in cell division and differentiation, and genes in transcriptional regulation (Fig. S4 D and E). This stage coincides with the rapid embryonic leaf and KS development in the first 2 d of imbibition (Fig. 1 C and D). Also, genes in mitochondrial electron transport, ATP synthesis, and glycolysis are overrepresented (Fig. S4D), indicating a heightened energy consumption, presumably taking place following digestion of endosperm. Meanwhile, the elevated transcription levels of transcriptional regulatory genes indicate the onset of multiple regulatory cascades responsible for various aspects of leaf development.
At the late stage (T54–T72), which includes C23–C25, and C30, jasmonate (JA) metabolic genes are up-regulated (Fig. S4E). C30 includes genes involved in photosynthesis, e.g., the RuBisCO (ribulose-1,5-bisphosphate carboxylase oxygenase) small subunit gene. Note that at T66 the entire shoot apex covered by sheath could already be seen outside the seed and became light green (Fig. 1A). Transcripts for photosynthesis genes start to accumulate at T48–T54 and become highly enriched at T72, likely signifying the onset of heightened photosynthetic activities. This is consistent with the analysis of type 1 DEGs, where genes involved in photosynthesis start to be up-regulated at T48 (Fig. S3A).
Developmental Transition of Hormonal Functions.
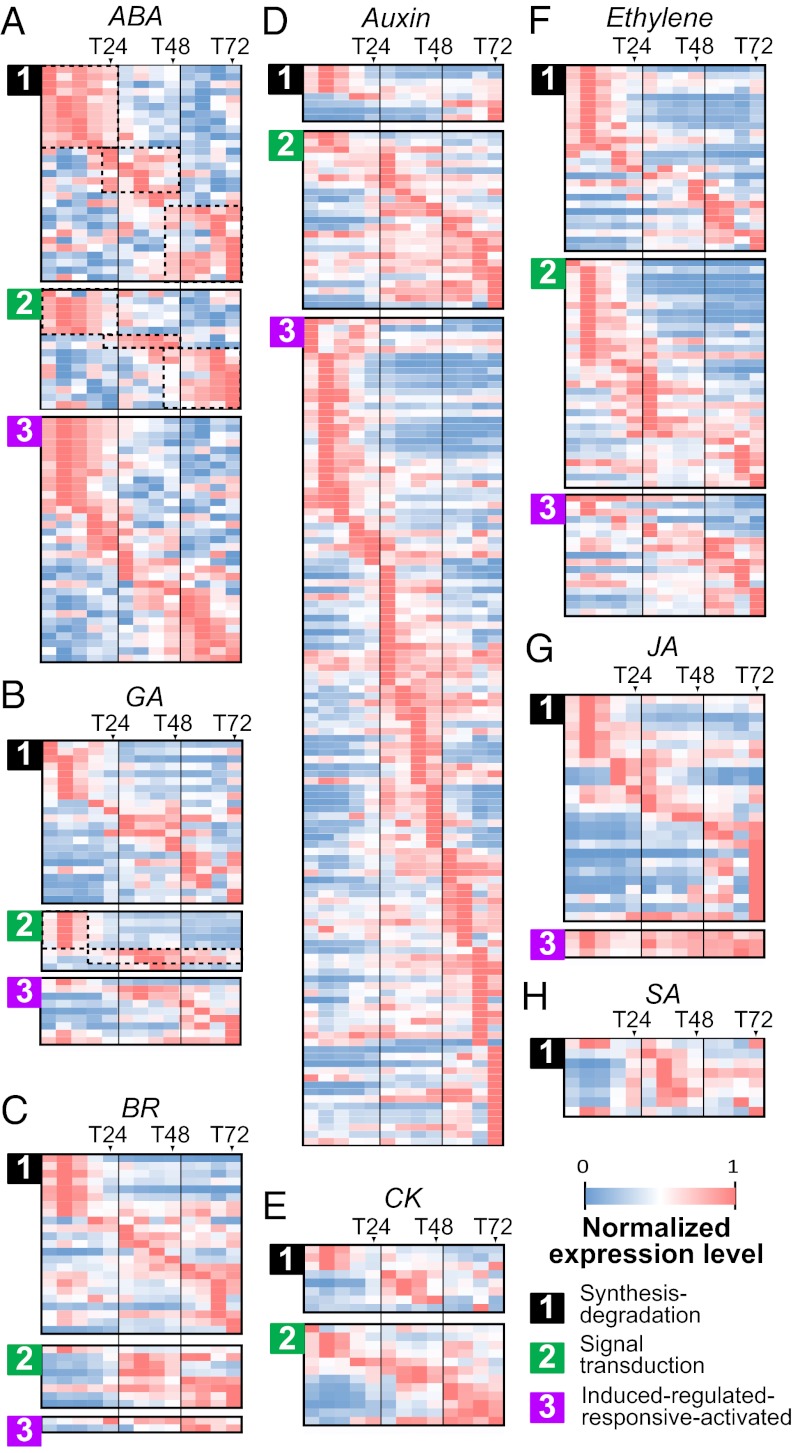
During the first 72 h of leaf development, 431 expressed genes are relevant to the action of eight hormones: ABA, auxin, BR, cytokinin (CK), ethylene, GA, JA, and salicylic acid (SA) (Fig. 4). For each hormone class, genes are classified into three types: (i) synthesis-degradation, (ii) signal transduction, and (iii) induced-regulated-responsive-activated (Fig. 4 and Dataset S2). Most of these hormone-related genes are expressed in a time-point/range–specific manner (Fig. 4). A relatively small number of hormone-related genes have transcripts in the embryonic leaves of dry seeds (T00), including aldehyde oxidase 1 and 4 involved in seed ABA biosynthesis and maize orthologs of GA3 (gibberellin A3) involved in the GA biosynthesis (Dataset S2).
Fig. 4.
Expression patterns of hormone-related genes. Normalized expression levels of genes related to (A) abscisic acid (ABA), (B) gibberellic acid (GA), (C) brassionlide (BR), (D) auxin, (E) cytokinin (CK), (F) ethylene, (G) jasmonic acid (JA), and (H) salicylic acid (SA) are shown. Normalized expression level is calculated as in Fig. 3. For each hormone, its related genes are divided into three functional categories: (i) synthesis-degradation, (ii) signal transduction, and (iii) induced-regulated-responsive-activated.
Many synthesis-degradation (type 1) genes are highly expressed at T06 and continue to express until T12 or T18 (Fig. 4 and Dataset S2). The highly expressed hormone biosynthesis genes include maize orthologs of NCED3 (9-cis-epoxycarotenoid dioxygenase 3) (ABA), GA1 (gibberellin A1), and GA2OX1 (GA 2-oxidase 1) (GA), DET2 (deetiolated2) (BR), and SRG1 (senescence-related gene 1) (ethylene) (Dataset S2), consistent with their important roles in breaking seed dormancy (13). In addition to the early time points, several ABA-, BR-, JA-, and SA-related type 1 genes are also highly expressed at T66 or T72 (Fig. 4 A, C, G, and H). Examples of BR response genes include maize orthologs of Arabidopsis thaliana CYP51G1 (cytochrome P450 51G1) and DWF1 (dwarf 1), which regulate cell elongation, and SMT2 (sterol methyltransferases 2), which regulates vascular differentiation (Dataset S2). For JA synthesis, many lipoxygenase family genes are highly expressed at T72, which may be required for wound response (Dataset S2). Apparently, the surge in expression of these genes in the embryonic leaves at the later stage of germination reflects the active differentiation of vascular tissues and the onset of plant defense mechanisms.
Type 2 genes include those relevant to hormone perception and downstream signaling, e.g., protein kinases and transcriptional regulators. For ABA, GA, BR, and ethylene, there are distinct coexpression modules among the type 2 genes (Fig. 4, type 2). For example, for both type 1 and type 2 ABA genes, there are three distinct modules: from T00 to T24, from T24 to T48, and from T48 to T72 (Fig. 4A, dotted rectangles). The T00–T24 module includes maize VP1 (viviparous1) and the maize ortholog of ABI5 (ABA–insensitive 5), regulating the transition from germination to vegetative growth (Dataset S2), and ABF2 (ABRE-binding factor 2) up-regulated for ABA signaling under water stress (14). The T48–T72 module includes the maize ortholog of LEC2 (leafy cotyledon2) that functions in embryo development (Dataset S2). Type 2 modules also include GA genes (Fig. 4B, type 2). The T00–T12 module includes a GA receptor and a GA signaling repressor, the maize orthologs of GID1c (GA insensitive dwarf 1c) and GAI (GA-insensitive), which are involved in seed germination and stem elongation (Dataset S2). The second type 2 GA module (T18–T72) contains another two maize GAI and SPY (spindly) orthologs with a peak expression at T36–T42 (Dataset S2). SPY is related to suppression of GA response, while promoting CK signaling (15). Thus, T18 may mark a transition point between GA and CK functions, from cell elongation to differentiation, coinciding with the rapid maturation of existing KSs and initiation of new KSs after T18 (Fig. 1C). Another type 2 gene that signifies interactions between hormones is the maize orthologs of the ethylene receptor ETR1 (ethylene receptor1), known to inhibit ABA signaling (Dataset S2). There are seven putative orthologs of CK receptors AHK2 (arabidopsis histidine kinase 2), AHK3, and AHK4, but only one putative ortholog of auxin receptor TIR1 (transport inhibitor response 1) in maize. These AHK genes are important regulators of shoot vascular tissue development in A. thaliana (16) and most of their transcripts start to accumulate after T12. In contrast, TIR1 expression first peaks at T06, followed by a reduction, and peaks again between T30 and T72. Interestingly, the second peak expression coincides with the maize orthologs of BIG (formerly DOC1/TIR3/UMB1/ASA1), PIN1 (pin-formed 1), and PIN4, all of which play important roles in auxin transport and vein patterning (17). These observations are consistent with the vein density increase after T24 (Fig. 1 C and D) and suggest that leaf vein or KS development may be mediated by relocation of the hormones via the action of these receptor and transport genes before T30.
One may expect that the type 3 genes that are induced by the same hormones as those for types 1 and 2 genes would have similar ranges of expression (Fig. 4, type 3). Indeed, two maize orthologs of ABA-responsive genes EM1 (late embryogenesis abundant 1) and EM6 required for buffering water loss during normal seed development (18) exhibit the highest expression levels from T00 to T24 (Dataset S2). Another example is the maize orthologs of ABA-induced HVA22 (HVA22 domain protein), which regulates seed germination by inhibiting GA-mediated vacuolation, are highly expressed at T06, but much lower thereafter (Dataset S2).
Expression Dynamics of TFs.
The 1,238 expressed TF genes are classified into 16 coexpression modules or clusters, which are ordered by their expression peaks (Fig. 3B). Almost all TF genes have relatively narrow windows of expression (Fig. 3B), suggesting regulatory successions that underlie physiological and developmental transitions.
C1, the largest cluster, contains 328 TFs that have high expression levels during the first 24 h, followed by a gradual decrease to a very low level at T72 (Fig. 3B). Most TF families have members in C1 (Fig. 3C), including VP1, which is responsible for the establishment of seed dormancy (19), and Opaque2 (O2) heterodimerizing protein 1, which together with O2 regulates the expression of zein storage protein genes (20). Similar to C1, C8–C11 and C13–C16 have single expression peaks at different time points, indicative of regulatory succession. For example, MWP1 (milkweed pod1), which is in C10, is responsible for adaxial–abaxial leaf surface patterning (21). Because C10 has a peak at T30, the polarity of newly emerged leaves may be established at the second day postimbibition. Another example is Golden 2 (G2), which is involved in maize leaf BS chloroplast development (22). G2 (Zmglk2) transcript is present in dry seeds and is in C14, which peaks at T54, indicating the onset of BS chloroplast development after T48, consistent with the inference from the DEG (Fig. S3 A–C) and cluster (Fig. S3 D and E) analyses. In contrast, the transcript of ZmGlk1, which is responsible for M chloroplast development in maize leaves (23), remains low until T54 (C15). Thus, BS chloroplasts may develop before M chloroplasts. There are other TFs with known functions in these clusters but their roles in early leaf development are not clear. For example, ZFL2 (zea floricaula leafy2) in C10 is known to regulate floral transition and flower development (24) but it is not clear why it is expressed so early postimbibition.
Although the role of most TFs in early leaf development is not known, their orthologs in A. thaliana provide hints on their functions. Take vascular development as an example. Several key A. thaliana regulators have roles in establishing and maintaining procambial and cambial cell populations and for further specification and differentiation into distinct cell types within xylem or phloem (25). For example, A. thaliana BDL (bodenlos), an AUX/IAA family protein, suppresses MP (monopteros), which is important for preprocambial development and procambium differentiation in embryo (26). Expression of BDL ortholog GRMZM2G142768 peaks at T06 and gradually declines thereafter (Fig. S5). In contrast, the expression levels of maize MP orthologs (GRMZM2G034840 and GRMZM2G086949) start arising at T24 when BDL starts to drop. This contrasting pattern of expression suggests that the inhibition for vascular development is released at ∼T24 because BDL is down-regulated by an elevated level of auxin (27). This is consistent with elevated levels of the maize orthologs of TIR1 and PIN1 as discussed above. In addition, the levels of genes involved in vein formation begin to rise at T24 (Fig. S5). Thus, our time course data allows hypotheses to be generated and tested regarding when these TFs may function over the course of early maize leaf development.
Concluding Remarks
Our tissue samples for transcriptomic analysis contained all of the embryonic leaves in the seeds. As the first leaf is the largest, it should have the largest contribution to the transcriptomes. However, as new KS units or veins develop in all of these leaves, our transcriptomes are useful for hypothesizing the regulatory genes involved in KS development. The samples also contained the shoot apical meristem (SAM), but as SAM is small compared with the leaves, its contribution to the transcriptomes may be small. In any event, our transcriptomes reveal a dynamic profile of transcriptional transition in parallel to anatomical transition in maize early leaf development during the first 72 h of seed germination.
Materials and Methods
A full description for Materials and Methods is provided in SI Text.
Plant Growth and Sample Collection.
Seeds of Zea mays cv. White Crystal were imbibed. The embryonic leaves were taken every 6 h for 3 d for transcriptomic analysis. Details for plant growth conditions and anatomical studies are provided in SI Text, Plant Growth Conditions and Sample Collection and SI Text, Anatomical Studies, respectively.
RNA Sequencing and Data Processing.
Total RNA was extracted, purified, quantified, and assessed for quality before Illumina sequencing. The library preparation steps and sequencing details are outlined in SI Text, RNA Extraction and Sequencing. Low quality reads were filtered based on quality scores. Read alignments to the B73 genome as well as gene expression level quantification were carried out following the “alternative protocol B” (28) as detailed in SI Text, Data Processing and Analysis.
Expression Profile Analyses.
Genes were classified into coexpression modules based on the Pearson correlation coefficients of expression profiles between genes using hierarchical clustering (SI Text, Expression Profile Correlation and Clustering). We used the nonparametric method of Tarazona et al. (29) to identify DEGs between two samples (SI Text, Identification of Differentially Expressed Genes). Fisher’s exact tests were conducted to assess functional enrichment based on MapMan annotations (30).
Supplementary Material
Acknowledgments
We thank Jen Sheen, Michael Freeling, and Thomas Okita for comments. This study was supported by Academia Sinica (AS-101-SS-A08 and AS-102-SS-A13) and National Science Council (Taiwan) Fellowship NSC-100-2917-I-564-053 (to S.C.-C.C.) and NSC 99-2321-B-001-041-MY2.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. SRA061194).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301009110/-/DCSupplemental.
References
- 1.Li P, et al. The developmental dynamics of the maize leaf transcriptome. Nat Genet. 2010;42(12):1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- 2.Bosabalidis AM, Evert RF, Russin WA. Ontogeny of the vascular bundles and contiguous tissues in the maize leaf blade. Am J Bot. 1994;81(6):745–752. [Google Scholar]
- 3.Zhang H, et al. PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39(Database issue):D1114–D1117. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey GK, et al. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell. 2004;16(7):1912–1924. doi: 10.1105/tpc.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stiefel V, et al. Expression of a maize cell wall hydroxyproline-rich glycoprotein gene in early leaf and root vascular differentiation. Plant Cell. 1990;2(8):785–793. doi: 10.1105/tpc.2.8.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baier M, Dietz KJ. The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: Its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 1997;12(1):179–190. doi: 10.1046/j.1365-313x.1997.12010179.x. [DOI] [PubMed] [Google Scholar]
- 7.Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 8.Caño-Delgado A, et al. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131(21):5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- 9.Bowman JL, Eshed Y. Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 2000;5(3):110–115. doi: 10.1016/s1360-1385(00)01569-7. [DOI] [PubMed] [Google Scholar]
- 10.Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9(7):1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bentsink L, Koornneef M. Seed dormancy and germination. Arabidopsis Book. 2008;6:e0119. doi: 10.1199/tab.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 13.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida T, et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61(4):672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 15.Greenboim-Wainberg Y, et al. Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;17(1):92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hejátko J, et al. The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell. 2009;21(7):2008–2021. doi: 10.1105/tpc.109.066696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauer M, et al. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 2006;20(20):2902–2911. doi: 10.1101/gad.390806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manfre AJ, Lanni LM, Marcotte WR., Jr The Arabidopsis group 1 LATE EMBRYOGENESIS ABUNDANT protein ATEM6 is required for normal seed development. Plant Physiol. 2006;140(1):140–149. doi: 10.1104/pp.105.072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarty DR, et al. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66(5):895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- 20.Pysh LD, Aukerman MJ, Schmidt RJ. OHP1: A maize basic domain/leucine zipper protein that interacts with opaque2. Plant Cell. 1993;5(2):227–236. doi: 10.1105/tpc.5.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Candela H, Johnston R, Gerhold A, Foster T, Hake S. The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell. 2008;20(8):2073–2087. doi: 10.1105/tpc.108.059709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall LN, Rossini L, Cribb L, Langdale JA. GOLDEN 2: A novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell. 1998;10(6):925–936. doi: 10.1105/tpc.10.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossini L, Cribb L, Martin DJ, Langdale JA. The maize golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell. 2001;13(5):1231–1244. doi: 10.1105/tpc.13.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bomblies K, Doebley JF. Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics. 2006;172(1):519–531. doi: 10.1534/genetics.105.048595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Sebastian J, Lee JY. Signaling and gene regulatory programs in plant vascular stem cells. Genesis. 2011;49(12):885–904. doi: 10.1002/dvg.20795. [DOI] [PubMed] [Google Scholar]
- 26.Lau S, De Smet I, Kolb M, Meinhardt H, Jürgens G. Auxin triggers a genetic switch. Nat Cell Biol. 2011;13(5):611–615. doi: 10.1038/ncb2212. [DOI] [PubMed] [Google Scholar]
- 27.Weijers D, et al. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell. 2006;10(2):265–270. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21(12):2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thimm O, et al. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37(6):914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.