Abstract
Ribonucleotide reductase (RNR) catalyzes the conversion of nucleoside diphosphates to deoxynucleoside diphosphates (dNDPs). The Escherichia coli class Ia RNR uses a mechanism of radical propagation by which a cysteine in the active site of the RNR large (α2) subunit is transiently oxidized by a stable tyrosyl radical (Y•) in the RNR small (β2) subunit over a 35-Å pathway of redox-active amino acids: Y122• ↔ [W48?] ↔ Y356 in β2 to Y731 ↔ Y730 ↔ C439 in α2. When 3-aminotyrosine (NH2Y) is incorporated in place of Y730, a long-lived NH2Y730• is generated in α2 in the presence of wild-type (wt)-β2, substrate, and effector. This radical intermediate is chemically and kinetically competent to generate dNDPs. Herein, evidence is presented that NH2Y730• induces formation of a kinetically stable α2β2 complex. Under conditions that generate NH2Y730•, binding between Y730NH2Y-α2 and wt-β2 is 25-fold tighter (Kd = 7 nM) than for wt-α2|wt-β2 and is cooperative. Stopped-flow fluorescence experiments establish that the dissociation rate constant for the Y730NH2Y-α2|wt-β2 interaction is ∼104-fold slower than for the wt subunits (∼60 s−1). EM and small-angle X-ray scattering studies indicate that the stabilized species is a compact globular α2β2, consistent with the structure predicted by Uhlin and Eklund’s docking model [Uhlin U, Eklund H (1994) Nature 370(6490):533–539]. These results present a structural and biochemical characterization of the active RNR complex “trapped” during turnover, and suggest that stabilization of the α2β2 state may be a regulatory mechanism for protecting the catalytic radical and ensuring the fidelity of its reactivity.
Keywords: conformational equilibria, radical transfer, unnatural amino acid
Ribonucleotide reductase (RNR) is the sole enzyme responsible for the conversion of nucleoside diphosphates (NDPs) to 2′-deoxynucleoside diphosphates (dNDPs), providing the cell with the monomeric precursors necessary for DNA replication and repair (1, 2). The class I RNRs are composed of two subunits, α and β; the active form of the prototypical Escherichia coli class Ia enzyme is generally accepted to be α2β2 (3, 4). The α2 subunit houses the active site, where the four NDP substrates (CDP, ADP, GDP, and UDP) are reduced, and two distinct regulatory sites, where allosteric effectors (ATP, dGTP, dTTP, and dATP) bind. The specificity site dictates which of the four substrates is reduced, whereas the activity site binds ATP/dATP and regulates the overall rate of reduction (5). The β2 subunit, an obligate dimer, contains a diferric-tyrosyl radical cofactor (Y122•) that is essential for catalysis. X-ray crystal structures of the individual E. coli β2 and α2 subunits have been solved (6, 7). However, the weak interaction between α2 and β2 (8), the conformational rearrangements induced by nucleotide binding (2, 9, 10), and complicated subunit equilibria (11) have precluded detailed structural characterization of any active RNR complexes. We now report the characterization of an active, kinetically stable α2β2 complex that forms transiently during turnover.
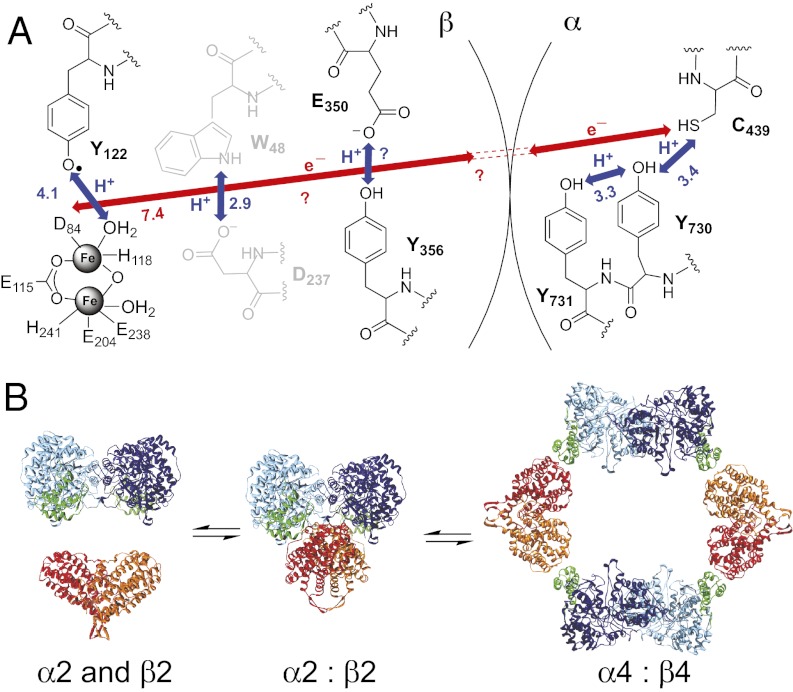
Nearly 2 decades ago, Uhlin and Eklund (7) put forth a docking model for the active E. coli α2β2 complex based on shape complementarity between the structures of the individual subunits. Their model predicted a 35-Å distance between the diferric-Y122• cofactor in β2 and the active site cysteine (C439) in α2, the transient oxidation of which is a prerequisite for nucleotide reduction (1). A radical transfer (RT) pathway of conserved aromatic amino acids was proposed to account for kinetically competent radical propagation over this long distance (7). The thermodynamics of Y oxidation require loss of a proton to accompany loss of an electron, and the more detailed mechanism for proton-coupled electron transfer shown in Fig. 1A has emerged from experiments conducted in our laboratories (12, 13).
Fig. 1.
(A) Model for long-range (∼35 Å), reversible RT in E. coli class Ia RNR. Orthogonal proton-coupled electron transfer (PCET) is proposed to be operative in the β2 subunit, whereas colinear PCET is proposed within the α2 subunit. Y356 has not been observed in any crystal structure of β2; thus, its location relative to the other residues is unknown. W48 and D237 are shown in gray because their participation has not been established experimentally. (B) Three-state model for E. coli class Ia solution equilibrium between free α2 and β2, α2β2, and α4β4. The relative distribution between these populations depends on the concentration of protein and the presence and concentrations of nucleotides in solution.
Evidence for the utilization of an amino acid pathway in long-range RT has been derived from several types of experiments. Initial site-directed mutagenesis studies of the conserved residues (Fig. 1A) supported their importance in nucleotide reduction but provided little insight into the mechanism of RT (14, 15). RNR’s fidelity to a specific redox pathway involving Y356, Y730, and Y731 (Fig. 1A) has become apparent from recent experiments in which these residues have been site-specifically replaced with unnatural Y analogs with modified redox properties (9, 10). For example, incorporation of 3-aminotyrosine (NH2Y) in place of the three transiently oxidized Ys in the RT pathway has been mechanistically informative. NH2Y has a reduction potential ∼190 mV lower than Y at pH 7, and when incorporated at position 356 of β2, position 731 of α2, or position 730 of α2, it generates a chemically and kinetically competent NH2Y• intermediate in the presence of both protein subunits, substrate, and effector. Furthermore, NH2Y RNRs are capable of making dNDPs with 3–12% the activity of wild-type (wt) RNR (9, 10).
The ability to incorporate NH2Y in place of the three pathway Ys and 3-hydroxytyrosine (DOPA) in place of Y356 provided an opportunity to test the validity of the docking model experimentally using pulsed electron-electron double resonance (PELDOR) spectroscopy. This method allows for measurement of the distance between two weakly coupled paramagnetic species, and is applicable to RNR because of its half-site reactivity (i.e., one dNDP must be generated on the first α|β pair before RT is initiated on the second α|β pair). The PELDOR measurements of three diagonal distances between a NH2Y• (or DOPA•) generated under turnover conditions on the first α|β pair and the Y122• remaining on the second α|β pair provided experimental support for the α2β2 docking model (16).
Additional support for the docking model was provided by recent physical biochemical studies of E. coli RNR that demonstrated a solution equilibrium across the three interconverting subunit states: α2 + β2 ↔ α2β2 ↔ α4β4 (11, 17) (Fig. 1B). The relative populations of these states were shown to depend on protein concentration and the identity and concentration of nucleotide effectors. A shape reconstruction of the small-angle X-ray scattering (SAXS) curves obtained at low micromolar protein concentrations in the presence of CDP indicated that the major species was a α2β2 complex resembling Uhlin and Eklund’s model (11). On addition of the negative activity effector dATP, or on increasing the protein concentration, the equilibrium was shifted toward an inactive α4β4 ring, as characterized by SAXS, EM, and X-ray crystallography (11, 18).
We present herein evidence that generation of NH2Y730•, formed in the reaction of Y730NH2Y-α2 with wt-β2, CDP, and ATP, induces the formation of a kinetically stable α2β2 complex of E. coli RNR. Compared with the noncooperative weak interaction(s) between the wt subunits (Kd = 0.18 μM) (8), Y730NH2Y-α2 and wt-β2 interact cooperatively and with increased subunit affinity (Kd = 7 nM), as determined by competitive inhibition and affinity chromatography pull-down assays. Stopped-flow (SF) fluorescence experiments using 6-acetyl-2-dimethylaminonaphthalene–labeled β2 (DAN-β2) indicate that the dissociation rate constant (koff) for the Y730NH2Y-α2|wt-β2 interaction is slowed by ∼104 relative to wt-α2|wt-β2. Characterization of the tightly interacting species by single-particle EM reveals an α2β2 complex resembling the original docking model, whereas SAXS experiments support the conclusion that generation of an NH2Y• shifts the subunit equilibrium relative to the wt enzyme (11) (Fig. 1B) strongly in favor of a globular α2β2 structure. Finally, activity assays indicate that the complex is active or exists in rapid equilibrium with an active state. Together, these data report on a previously unrecognized regulatory function of the E. coli RNR: the generation of a transiently stabilized α2β2 complex to prevent the loss of the catalytic radical during turnover. These results also provide an image of a class I RNR trapped in its catalytically relevant quaternary structure.
Results
Strength of Y730NH2Y-α2|wt-β2 Interaction.
The formation of NH2Y• in the reactions of NH2Y RNRs (Y730NH2Y-α2, Y731NH2Y-α2, or Y356NH2Y-β2) with the second wt subunit, substrate, and allosteric effector has been extensively studied (9, 10). One of the more striking observations is the apparent stability of NH2Y• formed at positions 730, 731, and 356, which persists on the minute time scale. The lifetimes of these on-pathway radicals, compared with the microsecond lifetimes of Y• analogs in solution (19), prompted us to investigate the cause(s) of their increased stability. X-ray crystal structures of Y730NH2Y-α2 and Y731NH2Y-α2 (10) indicated that the mutated pathway residues are superimposable with Y730 and Y731 in the wt structure (7), and are partially solvent-exposed in α2 alone. Similarly, Y356 is located in the C-terminal tail of β and is conformationally disordered and solvent-exposed in the structure of β2 alone (6). Thus, the long lifetimes of NH2Y•s led us to surmise that the radical must be shielded from solvent, a condition that would require enhanced interaction between the subunits.
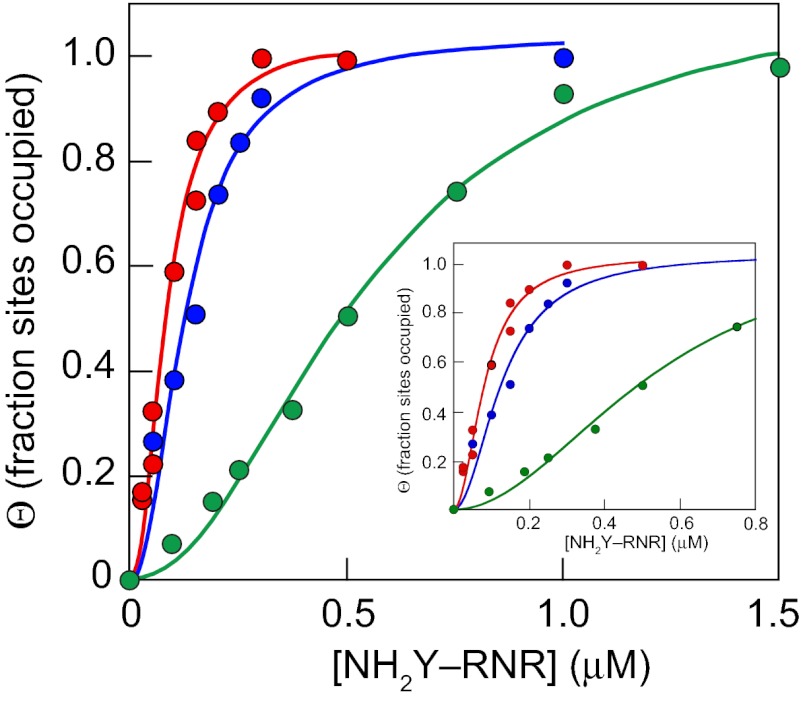
As a first test of this contention, we measured the equilibrium dissociation constant (Kd) between Y730NH2Y and wt-β2 in the presence of CDP and ATP (i.e., turnover conditions); the t1/2 of NH2Y730• formed under these conditions is ∼2.7 min at 25 °C (20). The Kd for Y730NH2Y was determined using a spectrophotometric competitive inhibition assay (8) in which the wt-α2|wt-β2 interaction is disrupted by titrating increasing concentrations of an inhibitor (Y730NH2Y-α2) into the assay mixture. The corresponding decrease in RNR activity is representative of the amount of inhibitor in the complex, and is used to extrapolate a Kd for the Y730NH2Y-α2|wt-β2 interaction. A plot of [Y730NH2Y-α2]bound vs. [Y730NH2Y-α2]free is sigmoidal, rather than the hyperbolic curve observed for the wt-α2|wt-β2 interaction. Analysis of the data by the Hill model for cooperative binding gives Kd = 7 nM for the Y730NH2Y-α2|wt-β2 interaction (Fig. 2, red). This Kd is 26-fold tighter than the wt-α2|wt-β2 (0.18 μM) interaction measured by the same method using the inactive mutant Y122F-β2 as a competitive inhibitor of the wt association (14). A Kd = 8 nM was determined for association between the analogous affinity-tagged protein, His6-Y730NH2Y-α2 (10), and wt-β2, indicating that the N-terminal His6-tag does not disrupt the subunit interactions. The Kds between Y731NH2Y-α2 and wt-β2 and those between Y356NH2Y-β2 and wt-α2 were also determined. In both cases, the binding was cooperative, with Kd = 17 nM and Kd = 0.30 μM, respectively (Fig. 2, blue and green). Thus, the Y731NH2Y-α2|wt-β2 interaction is 10-fold stronger than the corresponding wt interaction. The basis for the weaker Kd between Y356NH2Y-β2 and wt-α2 is not understood at present but may reflect decreased stability of the NH2Y• at this position compared with positions in α2.
Fig. 2.
Kds between NH2Y RNRs and a second wt subunit as determined by the spectrophotometric competitive inhibition assay (8). The following α2|β2 interactions were explored in the presence of CDP and ATP: Y730NH2Y-α2 and wt-β2 (red), Y731NH2Y-α2 and wt-β2 (blue), and Y356NH2Y-β2 and wt-α2 (green). The data shown are the average of two to three replicates, with error between measurements <10%. The data were fit (solid lines) to the Hill equation for cooperative binding. (Inset) Expanded view of the data at low [NH2Y RNR] is shown.
We further hypothesized that the origin of the enhanced interaction between Y730NH2Y-α2 and wt-β2 rests in the ability to generate the stable pathway radical, NH2Y730•, rather than in ground-state structural differences between Y730NH2Y-α2 and wt-α2. To this end, the Kd for the interaction between Y731F/Y730NH2Y-α2 and wt-β2 was also measured, because the additional Y731F mutation prevents NH2Y• formation (21). The data (Fig. S1) reveal a loss of cooperativity and a Kd = 1.14 μM, which is >160-fold weaker than that for the Y730NH2Y-α2 and wt-β2 interaction. This result supports the model that formation of an on-pathway NH2Y• stabilizes the subunit interaction.
A nickel-nitriliotriacetic acid (Ni-NTA) affinity chromatography “pull-down” assay was designed to validate the enhanced interaction between Y730NH2Y-α2 and wt-β2 independently. A 1:1 mixture of His6-Y730NH2Y-α2 and wt-β2 (10 μM) was combined with CDP and ATP under single-turnover conditions (i.e., no reductant) to generate NH2Y730•, and the reaction mixture was incubated for 30 s with Ni-NTA agarose. The resin was washed to remove nonspecifically bound proteins, and bound protein(s) were then eluted with high [imidazole]. The entire procedure was complete within 3 min of initial mixing. The interaction between His6-α2(wt) and wt-β2 was examined under identical conditions. The protein content after each step was analyzed by SDS/PAGE (Fig. S2), and comparative densitometry analysis revealed that twice as much wt-β2 coeluted with His6-Y730NH2Y-α2 as with His6-α2(wt), consistent with a Y730NH2Y-α2|wt-β2 interaction that is stronger than the wt subunit interaction.
Kinetics of Y730NH2Y-α2|wt-β2 Interaction.
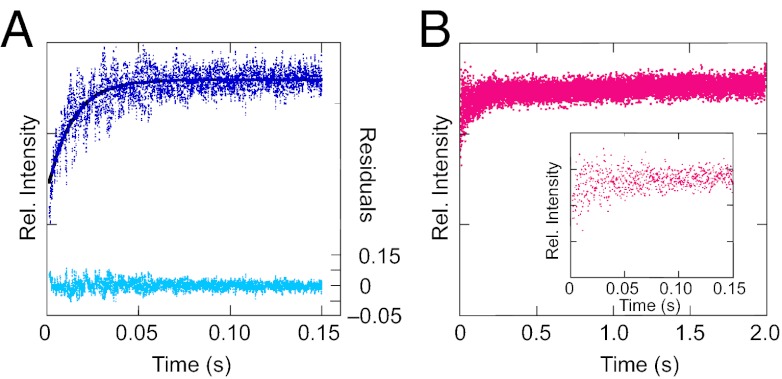
The long t1/2 of NH2Y730• suggested that the Y730NH2Y-α2|wt-β2 interaction is tight and kinetically stable. To investigate this possibility, the koffs for various α2|β2 interactions were measured by SF fluorescence spectroscopy, with the prediction that the koff for Y730NH2Y-α2|wt-β2 would be significantly slower than that for wt-α2|wt-β2. We used DAN-β2(V365C), a mutant β2 labeled with an environmentally sensitive fluorophore at a minimally disruptive position on the C-terminal tail (22), as a competitive inhibitor of β2 to measure koff. Four subunit interactions were measured in the presence of CDP and ATP: wt-α2 with wt-β2 or Y122•-reduced diferric-β2 (met-β2, incapable of initiating RT) and Y730NH2Y-α2 with wt-β2 or met-β2. For each experiment, α2 (1 eq), unlabeled β2 (3 eq), and nucleotides were combined in one syringe and rapidly mixed with an excess of DAN-β2(V365C) (105 eq) in the second syringe, and the total fluorescence at wavelengths >420 nm was monitored. An increase in fluorescence intensity in this regime reports on the displacement of the fluorophore to a more hydrophobic environment, as is predicted to occur when the disordered β-tail binds to α (22). The averaged kinetic traces for the wt-β2|wt-α2 and met-β2|wt-α2 reactions are shown in Fig. 3A and Fig. S3A, respectively. Monoexponential fits to the data give koff = 74 s−1 and koff = 63 s−1, respectively. The similarity between these rate constants indicates that the presence of neither Y122• nor the active site disulfide formed during turnover has a significant impact on koff. The averaged fluorescence trace of an analogous experiment with Y730NH2Y-α2 and met-β2 (Fig. S3B) gave a koff = 38 s−1, with a total fluorescence change similar to that of the wt-α2 reactions. Thus, koff of Y730NH2Y-α2 from met-β2 is approximately twofold slower than with wt-α2, even in the absence of NH2Y730• formation.
Fig. 3.
SF fluorescence measurements of koff for various α|β complexes. The average of 10–20 individual kinetic traces is shown for wt-β2 and wt-α2 (A, blue) and wt-β2 and Y730NH2Y-α2 (B, pink), both with CDP/ATP. (A) Monoexponential fit to the data is indicated by the solid line, and residuals to the fit are shown in cyan. Note the differences in time scales between A and B. (B, Inset) Expanded view of the first 150 ms of the wt-β2|Y730NH2Y-α2 trace.
Finally, koff was measured for Y730NH2Y-α2 and wt-β2 in the presence of CDP and ATP, conditions that produce the long-lived radical. In contrast to the previous experiments, there was no significant increase in fluorescence over the first 150 ms (Fig. 3B, Inset), and the small amplitude change observed over longer times (Fig. 3B, 2 s) constitutes <15% of the total fluorescence change observed over 150 ms in the three previous experiments. This change may reflect interaction of DAN-β2(V365C) with the small fraction (<20%) of unbound Y730NH2Y-α2 in the initial solution (10). Details regarding the quantitative assessment of the amount of Y730NH2Y-α2 interacting with wt-β2 in our experiments are provided in SI Materials and Methods. Because the koff for the Y730NH2Y-α2|wt-β2 interaction was too slow to measure on the SF time scale, steady-state fluorescence spectroscopy was conducted under identical conditions. A slow fluorescence increase was observed over 40 min; no increase was observed on this time scale for the wt-α2|wt-β2 control (Fig. S4). Fitting the data gives a koff on the order of 10−3 s−1, which is in agreement with the decay constant for NH2Y730• (4 × 10−3 s−1) and suggests that the lifetime of the Y730NH2Y-α2|wt-β2 interaction and the pathway radical are correlated. The combined fluorescence experiments establish that the Y730NH2Y-α2|wt-β2 interaction is kinetically stable compared with the wt subunit interaction.
Structure of Y730NH2Y-α2|wt-β2 Complex Characterized by EM.
The strength and kinetic stability of the Y730NH2Y-α2|wt-β2 interaction suggested that the complex might be visualized by EM, potentially providing valuable information about the structure and oligomeric state of the association. Y730NH2Y-α2 was combined with wt-β2 in the presence of CDP and ATP, applied to the EM grid with minimal incubation time relative to the lifetime of the NH2Y730•, and preserved and contrasted with stain. Individual particles were identified in the images, and to avoid reference bias, particles were aligned and classified without the use of an initial model.
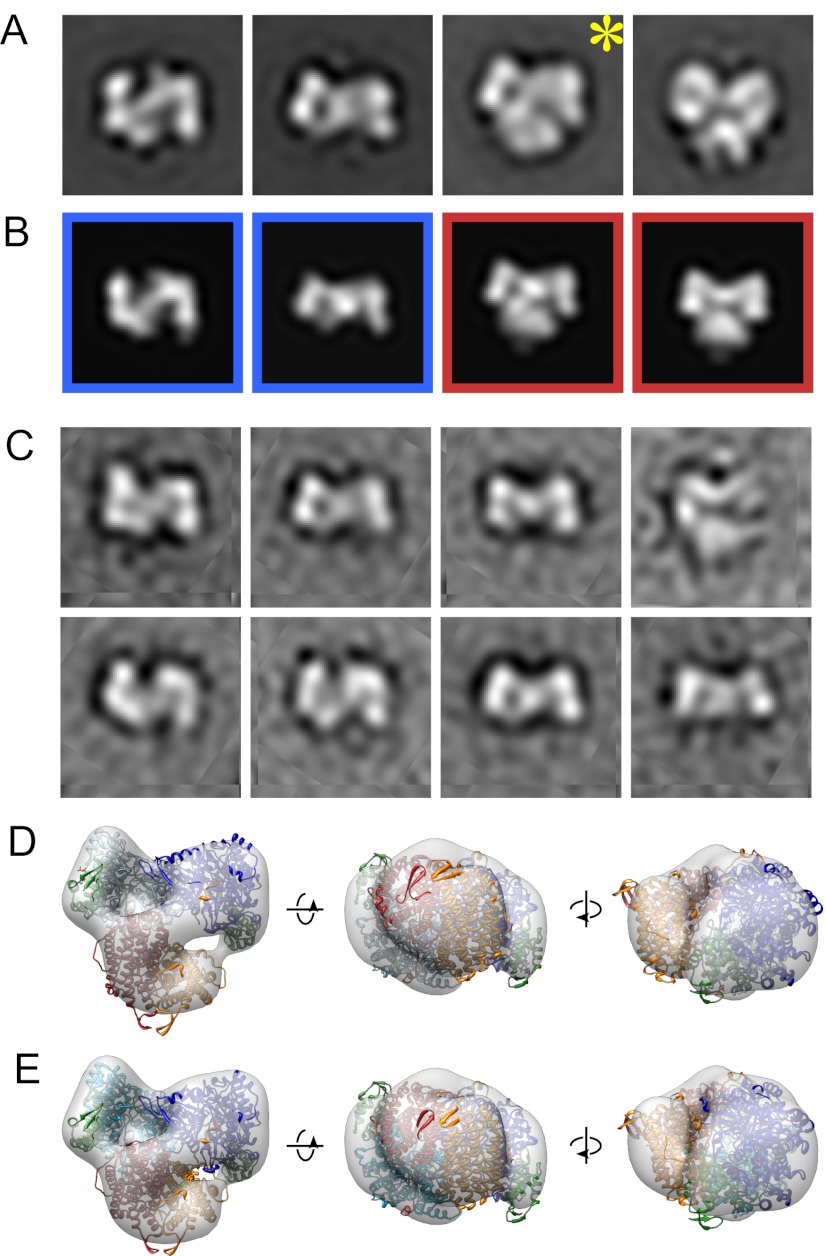
The resulting class averages were compared with projections of an α2 crystal structure (23) or the α2β2 docking model (7) (Fig. 4 A and B and Fig. S5). This comparison suggests that the majority of the averages represent different views of a complex that adopts a subunit arrangement consistent with the α2β2 docking model. Several other averages clearly represent views of the free α2 subunit. To determine whether observation of the α2β2 complexes is dependent on NH2Y730• formation, we prepared a specimen with met-β2 substituted for wt-β2. Strikingly, the averages from this control experiment almost exclusively correspond with dissociated α2 and β2 subunits (Fig. 4C), indicating that NH2Y730• formation is a prerequisite for visualization of the α2β2 complex.
Fig. 4.
Visualization of the NH2Y730•-stabilized α2β2 complex by EM. (A) Class averages of Y730NH2Y-α2 with wt-β2 in the presence of CDP and ATP. (B) Two-dimensional projections created from dTTP/GDP-bound α2 crystal structure [Protein DataBank (PDB) ID code 4R1R] (23) (blue outline) and the α2β2 docking model (7) (red outline) that best correlate with the class averages in A. (C) Class averages of Y730NH2Y-α2 with met-β2 in the presence of CDP and ATP. (D) Three-dimensional reconstruction from tilted views (gray) of the particles in the class marked by the asterisk in A. Structures of α2 and β2 (PDB ID codes 4R1R and 1RIB) iteratively fit into the reconstruction are shown in ribbons (α2 chains in light and dark blue with activity sites in green, β2 chains in red and orange). (E) Three-dimensional reconstruction as in D fit with the α2β2 docking model. Averages and projections are 209 Å wide at the specimen level.
Some α2β2 class averages correspond to orientations in which subunits are not overlapping and can be easily identified. These views offer an opportunity to understand the subunit arrangement in the α2β2 complex better, especially when considered relative to a 3D map calculated from tilted-pair particle images (24). X-ray models of α2 and β2 were separately fit into the 3D EM map (Fig. 4D) and compared with fitting of the α2β2 docking model (7) as a single rigid unit (Fig. 4E). The results from these two approaches agree [within the limitations related to low (∼32 Å) resolution and possible stain-induced deformation of the EM map], providing direct experimental support for the docking model.
Structure of Y730NH2Y-α2|wt-β2 Complex and Its Quaternary Equilibria Characterized by SAXS.
Recent analytical ultracentrifugation and SAXS experiments with wt-α2 and met-β2 support a general model for E. coli RNR in which an α2β2 complex resembling the docking model is an intermediate between the subunit dissociated state and the inhibited α4β4 state (11) (Fig. 1B). In the wt model, a shift in equilibrium toward α4β4 occurs in the presence of increasing protein concentrations or inhibitory concentrations of dATP. The enhanced stability of the Y730NH2Y-α2|wt-β2 interaction should lead to increased abundance of α2β2 relative to the other states. To test this hypothesis and to examine the contribution(s) of RT to the stability of the α2β2 complex, we investigated the distribution of solution species formed by various combinations of α2 (wt and Y730NH2Y), β2 (wt and met), substrate, and effector by SAXS. Data were processed as described previously (11) to yield the scattering intensity, I, vs. q, a function of scattering angle.
Free subunits were first characterized under identical conditions by Guinier and Kratky analyses, as described previously (11). Although a small amount of inherent aggregation was detected in Y730NH2Y-α2, q ranges with well-defined radii of gyration (Rg) could be identified in all samples by automated Guinier analysis (25) (Fig. S6 A–D). This analysis yielded similar extrapolated Rg values for both forms of β2 (30.2 ± 3.7 Å for wt and 31.1 ± 1.4 Å for met), as well as for the two forms of α2 (42.0 ± 4.3 Å for wt and 45.2 ± 5.4 Å for Y730NH2Y). Shape information could be readily acquired from Kratky analysis of the mid-q region of scattering data, which is unaffected by the presence of aggregates (Fig. S7). Scattering from folded, globular species decays as q−4, which gives rise to a pronounced peak when plotted in Kratky representation (Iq2 vs. q), whereas multimodal peaks are indicative of distinct, folded domains in a nonglobular arrangement (26). In the case of wt-β2 and met-β2, monomodal peaks are observed, consistent with the compact monomer arrangement within the β2 dimer. Furthermore, the curves are nearly superimposable, indicating that the radical does not affect the shape of this subunit. By comparison, the Kratky curves of wt-α2 and Y730NH2Y-α2 display peaks with shoulders, consistent with the less globular monomer arrangement in α2 dimers. Importantly, the curves are nearly identical in shape, indicating that the mutation does not change the overall shape or fold of α2.
Interactions between subunits were examined under conditions in which mixtures of α2β2 and α4β4 are expected to form based on previous work with wt-α2 and met-β2 (11), namely, at a high protein concentration (30 μM) in the presence of GDP and dTTP. Guinier and Kratky analyses were again used. Previously, it was shown that the globular α2β2 complex (Rg ∼40 Å) has a monomodal Kratky curve with a peak at q ∼0.04 Å−1, whereas the highly nonglobular α4β4 complex (Rg ∼70 Å) has a multimodal Kratky curve with distinct peaks at q ∼0.024 Å−1 and ∼0.06 Å−1 (11). Mixtures of α2β2 and α4β4 thus lead to apparent Rg between 40 and 71 Å and a smearing of Kratky peaks. Qualitatively, greater definition of multimodal peaks indicates higher levels of α4β4, whereas sharpening of the Kratky curve to a single peak at q ∼0.04 Å−1 indicates higher levels of α2β2.
A reaction mixture representing a preturnover state consisting of 30 μM wt-α2 and met-β2 with dTTP/GDP led to a bimodal Kratky curve and a corresponding Rg of 61.6 ± 0.1 Å, suggestive of a α2β2/α4β4 mixture (Fig. 5A, red) and consistent with previous observations (11). Replacing met-β2 with wt-β2 resulted in a similar Kratky curve (Fig. 5A, blue) with minimal changes to the Rg (60.3 ± 0.2 Å), indicating that the distributions of α2β2 and α4β4 are similar for the pre- and postturnover states. Likewise, a similar Kratky curve and nearly identical Rg (64.3 ± 0.2 Å) were obtained from a reaction mixture consisting of 30 μM Y730NH2Y-α2 and met-β2 with dTTP/GDP (Fig. 5B, red), indicating that Y730NH2Y-α2 is able to make the same intersubunit interactions as wt-α2. Moreover, this result demonstrates that the Y730NH2Y mutation does not significantly perturb the distribution of α2β2 and α4β4 under nonturnover conditions. When Y730NH2Y-α2 was reacted with wt-β2, GDP, and dTTP, however, a dramatic change was observed (Fig. 5B, blue). A single prominent peak was observed at q ∼0.04 Å−1, and the corresponding Rg was much smaller (48.2 ± 5.4 Å), indicating that the α2β2 complex is dominant under turnover conditions. Previous work has shown that substrate is required for formation of NH2Y730• (9, 10). Consistent with this observation, removal of substrate from the Y730NH2Y-α2|wt-β2 reaction mixture led to a less prominent peak (Fig. S8A, red), suggesting that the stability of the α2β2 complex is dependent on RT.
Fig. 5.
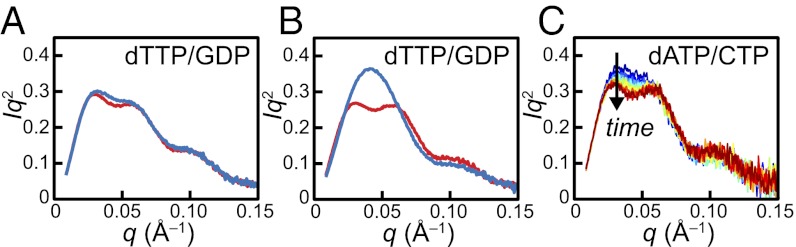
SAXS reveals the stability of Y730NH2Y-α2|wt-β2 complex. (A) Preturnover mixture of 30 μM wt-α2 with met-β2 (red) and postturnover mixture of 30 μM wt-α2 with wt-β2 (blue) in the presence of 0.1 mM dTTP and 1 mM GDP display a bimodal Kratky curve, indicative of the presence of α4β4. (B) Preturnover mixture of 30 μM Y730NH2Y-α2 with met-β2 in the presence of 0.1 mM dTTP and 1 mM GDP (red) also exhibits a bimodal Kratky curve. Initiation of turnover with Y730NH2Y-α2 and wt-β2 leads to a dramatically different Kratky curve (blue) that is consistent with a stable α2β2 complex. (C) Initiating turnover of 30 μM Y730NH2Y-α2 and wt-β2 in the presence of 175 μM dATP and 1 mM CDP leads to a slow change from a largely monomodal (blue) to bimodal (red) Kratky curve, consistent with the slow conversion of α2β2 to α4β4. Time proceeds from 2–22 min in increments of 2 min.
The kinetic stability of the NH2Y730•-induced α2β2 complex was examined under conditions in which wt-α4β4 is maximally favored (11) (i.e., in the presence of dATP). A reaction mixture consisting of 30 μM Y730NH2Y-α2 and wt-β2 was first incubated with CDP (1 mM), conditions that have been shown to generate 70% the amount of NH2Y730• observed with the matched CDP/ATP pair (10). Sufficient dATP to saturate both effector sites (175 μM) was then added to the reaction, and the mixture was monitored every 2 min for a total of 22 min. Initially, a largely monomodal peak was observed in the Kratky curve (Fig. 5C, blue), demonstrating the stability of this α2β2 complex even under strongly inactivating conditions. With time, the Kratky curve slowly becomes more bimodal (Fig. 5C, red), approaching the state observed with wt-α2 and wt-β2 (Fig. S8B). Singular value decomposition analysis indicates that the data can be described as a two-state transition that is not completed over the course of 22 min (Fig. S9), whereas the change in shape of the Kratky curve is consistent with the conversion of α2β2 to α4β4. These results are consistent with very slow dissociation of the α2β2 complex, even under strongly inhibitory conditions.
Catalytic Activity of the Y730NH2Y-α2|wt-β2 Complex.
We next investigated whether the stabilized complex is an active form of the enzyme. A modified RNR activity assay was conducted in which the Y730NH2Y-α2|wt-β2 complex was preformed by addition of His6-Y730NH2Y-α2 to a mixture of wt-β2 (2 eq), CDP, and ATP (mix 1). This solution was briefly aged and then diluted into a second solution containing the components of the standard steady-state RNR assay, including additional β2 (5 eq) and the reducing agents required for multiple turnovers, thioredoxin (TR), thioredoxin reductase (TRR), and NADPH (mix 2) (9). The activity of His6-Y730NH2Y-α2 from the preformed NH2Y-α2|wt-β2 complex was 86 (±10) nmol⋅min−1⋅mg−1. As a positive control, wt-β2 was omitted from mix 1 (i.e., no preformed complex) but included in mix 2. Under these conditions, His6-Y730NH2Y-α2 had a specific activity of 88 (±1) nmol⋅min−1⋅mg−1, consistent with previous reports (10). This result indicates that the Y730NH2Y-α2|wt-β2 complex induced by NH2Y730• formation is either active itself or exists in rapid equilibrium with an active form of the complex. The fluorescence experiments indicate that koff for the complex is slow compared with the steady-state kcat. This observation suggests it is the Y730NH2Y-α2|wt-β2 complex itself that is an active quaternary structure. However, the effect of reductant(s) on the stability of the Y730NH2Y-α2|wt-β2 complex has not been studied and could influence koff in the steady state.
Discussion
The results described herein provide very strong evidence that the reaction of Y730NH2Y-α2, wt-β2, substrate, and effector generates a kinetically stable α2β2 complex and that the stability of this complex originates from the formation of a radical, NH2Y730•, on the RT pathway. Remarkably, the transfer of the equivalent of a single hydrogen atom from NH2Y730 in α2 to Y122• in β2 in active RNR increases the kinetic stability of a 260-kDa complex by a factor of ∼104 relative to wt-α2. Recent studies have indicated that the affinity between E. coli α2 and β2 is increased in the presence of matched substrate/effector pairs and that the binding of substrate/effector to α2 conformationally gates radical initiation in β2, indicating a role for nucleotide-induced conformational triggering of RT in the active α2β2 complex (10, 27). The present studies extend this hypothesis by demonstrating that RT, in turn, transiently induces tight association between the subunits. We rationalize that this enhanced subunit affinity evolved as a regulatory mechanism to prevent quenching of transiently formed Y•s, which would result in RNR inactivation through loss of the catalytic oxidant.
Although the current studies have focused specifically on Y730NH2Y-α2, studies with other RNRs containing unnatural Y analogs suggest that localizing a radical on the pathway is a general mechanism for strengthening and stabilizing α2β2 interactions. For example, we have recently reported the replacement of Y122• with several high-energy radical initiators (NO2Y122• or FnY122•s). Incubation of these mutant β2s with wt-α2, substrate, and effector converts up to 50% of the initial radical at position 122 to a Y356• intermediate (28, 29). The lifetime of the Y356• depends on the identity of the radical initiator at position 122; however, in all cases, the new radical persists on the minute time scale. Recalling that Y356•, located on the C-terminal tail of free β2, would likely be reduced on the microsecond time scale (19), the observation of a long-lived Y356• suggests a comparably long-lived α2β2 association. In retrospect, a similar stabilization of the α2β2 complex likely accounts for the observations with the mechanism-based inhibitor 2′-azido-2′-deoxynucleotide, which reacts in the presence of α2 and β2 to generate a nucleotide-based, nitrogen-centered radical (N•) in the enzyme active site. Fifty percent of the total initial Y• is rapidly lost concomitant with 50% formation of the N• and 90% loss of total enzyme activity, consistent with the half-site reactivity model. The complete loss of Y•, however, occurs only after 30 min, an observation suggestive of very slow subunit dissociation before complete Y• quenching (30).
The characterization of a stable, NH2Y•-induced complex has provided a unique opportunity to visualize the active α2β2 complex “trapped” during catalysis by EM. Although the experiment was challenging due to the kinetic complexity and extent of NH2Y730• formation, class averages and 3D reconstructions reveal a compact, globular structure consistent with the docking model. SAXS experiments provide additional support for the solution stabilization of a Y730NH2Y-α2|wt-β2 complex. These experiments, in combination with the previous PELDOR distances (16) and SAXS data (11), establish the docking model as an accurate representation of the active RNR structure.
In summary, the results presented above provide a visual representation of the active structure of the E. coli class Ia RNR. They also indicate that subtle conformational changes, which are induced by nucleotide binding and long-range RT, regulate RNR by preventing quenching of the catalytic radical at positions along the pathway, a loss that would result in enzyme inactivation. This mechanism is one of many that E. coli RNR uses to ensure the fidelity of its complicated radical chemistry.
Materials and Methods
Kd Measurements.
Kds for the interaction between NH2Y-α2s and wt-β2 or wt-α2 and Y356NH2Y-β2 were determined in assay buffer [50 mM Hepes, 15 mM Mg2+, 1 mM EDTA, (pH 7.6)] at 25 °C by the competitive inhibition assay (8). Noncooperative binding data were fit to the standard Kd equation (31) (Eq. S1); cooperative binding data were fit to the Hill equation (Eq. S5).
Pull-Down Assays.
To a solution of untagged wt-β2 (10 μM) in EDTA-free assay buffer with CDP/ATP at 25 °C was added either His6-α2(wt) or His6-Y730NH2Y-α2 (10 μM). The reaction mixture was combined with Ni-NTA resin in the same buffer, incubated on ice (30 s), and centrifuged, and the supernatant was then removed. The resin was washed twice with EDTA-free assay buffer with 300 mM NaCl and 15 mM imidazole, and it was resuspended in EDTA-free assay buffer with 250 mM imidazole to elute bound protein. The protein in the supernatant at each step was assessed by 8% (wt/vol) SDS/PAGE.
SF Fluorescence Experiments.
DAN-β2(V365C) was prepared according to the protocol reported in the literature (22). SF fluorescence experiments were performed at 25 °C as described previously (32). In all cases, one syringe contained α2 (wt or Y730NH2Y, 0.16 μM), β2 (wt or met, 0.5 μM), CDP (1 mM), and ATP (3 mM) in assay buffer. A second syringe contained DAN-β2(V365C) (17.5 μM) in assay buffer. The contents of the syringes were mixed 1:1, and the fluorescence was monitored at >420 nm.
Structural Characterization by EM.
Y730NH2Y-α2 (1.5 μM), wt-β2 or met-β2 (3 μM), ATP (3 mM), and CDP (1 mM) were combined in assay buffer. The reaction mixture (8 μL) was applied to a carbon-coated EM grid, rinsed with assay buffer/ATP/CDP, and stained with a 2% uranyl acetate solution that also contained 0.2% trehalose. The specimens were imaged at 120 kV on a Tecnai F20 electron microscope (FEI) equipped with an UltraScan 4000 CCD camera (Gatan) using Leginon (33) operated in manual low-dose mode at a magnification of 50,000× with a pixel size of 2.18 Å at the specimen level. For the preparation with wt-β2, 48 pairs of images of the untilted and −55° tilted specimen were collected, and an initial dataset of 26,845 particle pairs was windowed from the CCD frames and processed using SPIDER (34). For the preparation with met-β2, 2,187 particles were windowed from 5 untilted images using SPIDER, and they were aligned and classified using the Iterative Stable Alignment and Clustering program in SPARX (35).
SAXS Analysis.
SAXS data were collected using a Pilatus 100-K detector and an in vacuo 2-mm pathlength quartz capillary flow cell (36) at the Cornell High Energy Synchrotron Source (CHESS) G1 station. Data were processed following previously described protocols (11, 37). The momentum transfer variable is defined as q = 4π/λsinθ, where λ is the X-ray wavelength and 2θ is the scattering angle with respect to the beam. Reaction components were mixed immediately before data acquisition. For each sample, exposures that did not exhibit time-dependent changes were averaged. All samples were in assay buffer with the nucleotide concentrations specified in the figure legends.
Activity Assays.
His6-Y730NH2Y-α2 (5 μM), wt-β2 (10 μM), [3H]-CDP (1 mM, 20,000 cpm/nmol), and ATP (3 mM) in a total volume of 25 μL were mixed in assay buffer at 25 °C, and the resulting solution was aged for 30 s. A fraction of this mixture (20 μL) was then diluted into a mixture (200 μL) containing additional wt-β2 (2.5 μM), [3H]-CDP (1 mM), ATP (3 mM), TR (30 μM), TRR (0.5 μM), and NADPH (1 mM) in assay buffer, and RNR activity was measured (9).
Supplementary Material
Acknowledgments
We thank Marco Jost and Jeremy Setser (Massachusetts Institute of Technology) for assistance in SAXS data collection, Prof. Sol Gruner (Cornell University) for access to wet laboratory space, and Drs. Richard Gillilan and Soeren Nielsen (CHESS) for setting up the flow cell. CHESS is supported by the National Science Foundation and the National Institute of General Medical Sciences via National Science Foundation Award DMR-0936384; the Macromolecular Diffraction Facility at CHESS resource is supported by National Institute of General Medical Sciences Award GM103485. EM was performed at the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health through the National Center for Research Resources P41 program (RR017573). This research was supported by National Science Foundation Graduate Research Fellowship 1122374 (to L.O.) and by National Institutes of Health Grants K99GM100008 (to N.A.), F32GM904862 (to N.A.), GM67167 (to F.J.A.), P30-ES002109 (to C.L.D.), GM47274 (to D.G.N.), and GM29595 (to J.S.). C.L.D. is a Howard Hughes Medical Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220691110/-/DCSupplemental.
References
- 1.Stubbe J, van Der Donk WA. Protein radicals in enzyme catalysis. Chem Rev. 1998;98(2):705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 2.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 3.Brown NC, Reichard P. Ribonucleoside diphosphate reductase. Formation of active and inactive complexes of proteins B1 and B2. J Mol Biol. 1969;46(1):25–38. doi: 10.1016/0022-2836(69)90055-2. [DOI] [PubMed] [Google Scholar]
- 4.Thelander L. Physicochemical characterization of ribonucleoside diphosphate reductase from Escherichia coli. J Biol Chem. 1973;248(13):4591–4601. [PubMed] [Google Scholar]
- 5.Brown NC, Reichard P. Role of effector binding in allosteric control of ribonucleoside diphosphate reductase. J Mol Biol. 1969;46(1):39–55. doi: 10.1016/0022-2836(69)90056-4. [DOI] [PubMed] [Google Scholar]
- 6.Nordlund P, Sjöberg BM, Eklund H. Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature. 1990;345(6276):593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 7.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370(6490):533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 8.Climent I, Sjöberg BM, Huang CY. Carboxyl-terminal peptides as probes for Escherichia coli ribonucleotide reductase subunit interaction: Kinetic analysis of inhibition studies. Biochemistry. 1991;30(21):5164–5171. doi: 10.1021/bi00235a008. [DOI] [PubMed] [Google Scholar]
- 9.Seyedsayamdost MR, Xie J, Chan CT, Schultz PG, Stubbe J. Site-specific insertion of 3-aminotyrosine into subunit α2 of E. coli ribonucleotide reductase: Direct evidence for involvement of Y730 and Y731 in radical propagation. J Am Chem Soc. 2007;129(48):15060–15071. doi: 10.1021/ja076043y. [DOI] [PubMed] [Google Scholar]
- 10.Minnihan EC, Seyedsayamdost MR, Uhlin U, Stubbe J. Kinetics of radical intermediate formation and deoxynucleotide production in 3-aminotyrosine-substituted Escherichia coli ribonucleotide reductases. J Am Chem Soc. 2011;133(24):9430–9440. doi: 10.1021/ja201640n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ando N, et al. Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase. Proc Natl Acad Sci USA. 2011;108(52):21046–21051. doi: 10.1073/pnas.1112715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stubbe J, Nocera DG, Yee CS, Chang MCY. Radical initiation in the class I ribonucleotide reductase: Long-range proton-coupled electron transfer? Chem Rev. 2003;103(6):2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 13.Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Proton-coupled electron transfer: The mechanistic underpinning for radical transport and catalysis in biology. Philos Trans R Soc Lond B Biol Sci. 2006;361(1472):1351–1364. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Climent I, Sjöberg BM, Huang CY. Site-directed mutagenesis and deletion of the carboxyl terminus of Escherichia coli ribonucleotide reductase protein R2. Effects on catalytic activity and subunit interaction. Biochemistry. 1992;31(20):4801–4807. doi: 10.1021/bi00135a009. [DOI] [PubMed] [Google Scholar]
- 15.Ekberg M, Sahlin M, Eriksson M, Sjöberg BM. Two conserved tyrosine residues in protein R1 participate in an intermolecular electron transfer in ribonucleotide reductase. J Biol Chem. 1996;271(34):20655–20659. doi: 10.1074/jbc.271.34.20655. [DOI] [PubMed] [Google Scholar]
- 16.Seyedsayamdost MR, Chan CT, Mugnaini V, Stubbe J, Bennati M. PELDOR spectroscopy with DOPA-β2 and NH2Y-α2s: Distance measurements between residues involved in the radical propagation pathway of E. coli ribonucleotide reductase. J Am Chem Soc. 2007;129(51):15748–15749. doi: 10.1021/ja076459b. [DOI] [PubMed] [Google Scholar]
- 17.Rofougaran R, Crona M, Vodnala M, Sjöberg BM, Hofer A. Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J Biol Chem. 2008;283(51):35310–35318. doi: 10.1074/jbc.M806738200. [DOI] [PubMed] [Google Scholar]
- 18.Zimanyi CM, et al. Tangled up in knots: Structures of inactivated forms of E. coli class Ia ribonucleotide reductase. Structure. 2012;20(8):1374–1383. doi: 10.1016/j.str.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holder PG, Pizano AA, Anderson BL, Stubbe J, Nocera DG. Deciphering radical transport in the large subunit of class I ribonucleotide reductase. J Am Chem Soc. 2012;134(2):1172–1180. doi: 10.1021/ja209016j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seyedsayamdost MR. 2007. Investigation of the mechanism of radical propagation in E. coli ribonucleotide reductase by site-specific incorporation of unnatural amino acids. PhD thesis (Massachusetts Institute of Technology, Cambridge, MA)
- 21.Minnihan EC, Seyedsayamdost MR, Stubbe J. Use of 3-aminotyrosine to examine the pathway dependence of radical propagation in Escherichia coli ribonucleotide reductase. Biochemistry. 2009;48(51):12125–12132. doi: 10.1021/bi901439w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan AQ, Wang Y, Plate L, Stubbe J. Methodology to probe subunit interactions in ribonucleotide reductases. Biochemistry. 2008;47(49):13046–13055. doi: 10.1021/bi8012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eriksson M, et al. Binding of allosteric effectors to ribonucleotide reductase protein R1: Reduction of active-site cysteines promotes substrate binding. Structure. 1997;5(8):1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 24.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146(Pt 2):113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 25.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Cryst. 2006;39(Pt 2):277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mertens HD, Svergun DI. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J Struct Biol. 2010;172(1):128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Ge J, Yu G, Ator MA, Stubbe J. Pre-steady-state and steady-state kinetic analysis of E. coli class I ribonucleotide reductase. Biochemistry. 2003;42(34):10071–10083. doi: 10.1021/bi034374r. [DOI] [PubMed] [Google Scholar]
- 28.Yokoyama K, Uhlin U, Stubbe J. A hot oxidant, 3-NO2Y122 radical, unmasks conformational gating in ribonucleotide reductase. J Am Chem Soc. 2010;132(43):15368–15379. doi: 10.1021/ja1069344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minnihan EC, Young DD, Schultz PG, Stubbe J. Incorporation of fluorotyrosines into ribonucleotide reductase using an evolved, polyspecific aminoacyl-tRNA synthetase. J Am Chem Soc. 2011;133(40):15942–15945. doi: 10.1021/ja207719f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fritscher J, et al. Structure of the nitrogen-centered radical formed during inactivation of E. coli ribonucleotide reductase by 2′-azido-2′-deoxyuridine-5′-diphosphate: Trapping of the 3′-ketonucleotide. J Am Chem Soc. 2005;127(21):7729–7738. doi: 10.1021/ja043111x. [DOI] [PubMed] [Google Scholar]
- 31.Seyedsayamdost MR, Stubbe J. Forward and reverse electron transfer with the Y356DOPA-β2 heterodimer of E. coli ribonucleotide reductase. J Am Chem Soc. 2007;129(8):2226–2227. doi: 10.1021/ja0685607. [DOI] [PubMed] [Google Scholar]
- 32.Reece SY, Lutterman DA, Seyedsayamdost MR, Stubbe J, Nocera DG. Re(bpy)(CO)3CN as a probe of conformational flexibility in a photochemical ribonucleotide reductase. Biochemistry. 2009;48(25):5832–5838. doi: 10.1021/bi9005804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20(2):237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen SS, Møller M, Gillilan RE. High-throughput biological small-angle X-ray scattering with a robotically loaded capillary cell. J Appl Cryst. 2012;45(Pt 2):213–223. doi: 10.1107/S0021889812000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ando N, Chenevier P, Novak M, Tate MW, Gruner SM. High hydrostatic pressure small-angle X-ray scattering cell for protein solution studies featuring diamond windows and disposable sample cells. J Appl Cryst. 2008;41(Pt 1):167–175. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.