Abstract
Mechanical stress plays a key role in many genomic processes, such as DNA replication and transcription. The ability to predict the response of double-stranded (ds) DNA to tension is a cornerstone of understanding DNA mechanics. It is widely appreciated that torsionally relaxed dsDNA exhibits a structural transition at forces of ∼65 pN, known as overstretching, whereby the contour length of the molecule increases by ∼70%. Despite extensive investigation, the structural changes occurring in DNA during overstretching are still generating considerable debate. Three mechanisms have been proposed to account for the increase in DNA contour length during overstretching: strand unpeeling, localized base-pair breaking (yielding melting bubbles), and formation of S-DNA (strand unwinding, while base pairing is maintained). Here we show, using a combination of fluorescence microscopy and optical tweezers, that all three structures can exist, uniting the often contradictory dogmas of DNA overstretching. We visualize and distinguish strand unpeeling and melting-bubble formation using an appropriate combination of fluorescently labeled proteins, whereas remaining B-form DNA is accounted for by using specific fluorescent molecular markers. Regions of S-DNA are associated with domains where fluorescent probes do not bind. We demonstrate that the balance between the three structures of overstretched DNA is governed by both DNA topology and local DNA stability. These findings enhance our knowledge of DNA mechanics and stability, which are of fundamental importance to understanding how proteins modify the physical state of DNA.
Keywords: DNA structure, base-pair stability, DNA melting, single molecules, optical trapping
Many cellular processes, such as DNA repair, gene regulation, and genome compaction, induce mechanical stress (1–3). More than a decade ago, single-molecule studies revealed that torsionally unconstrained double-stranded (ds) DNA exhibits a structural transition at tensile forces of ∼65 pN. Here, the DNA molecule gains ∼70% in contour length (Lc), over a narrow force range, a process known as overstretching (4, 5). Such forces are thought to occur in the cell, for example during sister chromatid separation (6). Detailed knowledge of the microscopic change in DNA structure during overstretching is therefore crucial for understanding DNA–protein interactions at the molecular level.
The molecular structure of torsionally unconstrained dsDNA during overstretching is still the subject of intense debate. Three mechanisms have been proposed to account for the sudden increase in dsDNA contour length at the overstretching transition: strand unpeeling, localized base-pair breaking, and formation of S-DNA (4, 5, 7–16). In the unpeeling model, one strand retracts from its complementary strand via base-pair breaking. This progressively converts dsDNA into two single-stranded (ss) DNA tracts, only one of which is still under tension (10). In the second model, the two complementary tracts of dsDNA both remain under tension, but localized pockets of denatured DNA (ssDNA) are created throughout the molecule (9). We refer to these finite ssDNA domains as “melting bubbles” due to their similarity with thermal denaturation of dsDNA (17). In the S-DNA model, the base-pairing integrity is preserved, and overstretching is accommodated instead by cooperative strand unwinding (8, 15, 18).
Studies combining optical trapping and fluorescence microscopy visualized directly that, under relatively low salt concentrations (5–150 mM NaCl), dsDNA can overstretch via unpeeling from free ends and nicks in the phosphate backbone (10). This finding developed and supported other studies that identified base-pair breaking during dsDNA overstretching (9, 12, 15). The energetics of this unpeeling process were quantified and discussed in the framework of an equilibrium model governed solely by the elastic properties of ssDNA or dsDNA and the underlying sequence (14, 15).
Strand unpeeling, however, must initiate from a discontinuity in the phosphate backbone (either a nick or a free DNA end), which is generally only present transiently inside a cell. Experiments on dsDNA molecules that were each linked with all four strand ends to optically trapped microspheres revealed that replication protein A (RPA, a ssDNA binding protein) can bind to such DNA molecules during overstretching (10). This observation indicates that, in the absence of free ends or nicks in the phosphate backbone, the base-pairing integrity can still be disrupted (at least for salt concentrations <150 mM NaCl). However, in these cases, strand unpeeling was prohibited by applying torsional constrain, preventing the molecule from releasing energy purely by unwinding. The resulting energy cost increases the overstretching transition from 65 pN to 110 pN (10, 18).
To prohibit unpeeling, yet still avoid torsional constraint, Paik and Perkins (13) developed a topologically closed (i.e., no free ends or nicks) but rotationally unconstrained dsDNA molecule. This construct maintains an overstretching transition at ∼65 pN, proving that strand unpeeling is not a requirement for DNA overstretching. This observation substantiates several other reports that found that torsionally unconstrained dsDNA can overstretch via two distinct modes: strand unpeeling and an alternative mechanism (12, 15, 16, 19). The latter is associated with a surprisingly small negative entropy change and is favored when intramolecular electrostatic repulsion is reduced by appropriate salt concentration and temperature (15, 16, 19). In consensus with theoretical studies (8, 11), it has been argued that this additional mechanism of DNA overstretching is consistent with the formation of S-DNA (4, 8, 11, 12, 19). However, as Zhang et al. (16) revealed, the nature of DNA overstretching in the absence of unpeeling is complex; the competition between melting-bubble and S-DNA formation is unclear. Additionally, structural transitions associated with overstretching can also be sequence-dependent, with S-DNA being favored in GC-rich sequences (19).
In an effort to provide an approach that embraces all essential aspects of DNA overstretching (temperature, ionic strength, and DNA topology and sequence), this article, together with a companion study by Zhang et al. (20), reports a unique single-molecule approach to the problem. Using optical tweezers and fluorescence microscopy, the current article reveals that melting-bubble and S-DNA formation can both compete with strand unpeeling during overstretching. We demonstrate that the balance between the three mechanisms is determined by DNA topology as well as the local DNA stability. Zhang et al. (20) use single-molecule calorimetric methods to reach an identical conclusion. The results of these two investigations are mutually supportive.
Results
At Low Ionic Strength (50 mM NaCl), Melting Bubbles Form When Unpeeling Is Prohibited.
We first sought to discover the mechanism of overstretching in the case where no discontinuities (i.e., free ends or nicks) exist in the phosphate backbone, and thus unpeeling is prohibited. To this end, we used a topologically closed, but torsionally unconstrained, dsDNA construct, similar to that reported recently by Paik and Perkins (13). In our case, a modified lambda phage (λ) DNA molecule (∼48,500 bp) was bound between two optically trapped beads via a single biotin embedded in a five-nucleotide loop on one end of the molecule and biotins attached to the complementary strands of the other end of the molecule, as illustrated schematically in Fig. 1A Upper. Force-extension measurements on such a construct, obtained at 50 mM NaCl, yielded two distinct sets of behaviors. In one subset of experiments (∼65% of cases), reported in Fig. 1B, significant hysteresis in force was observed during DNA overstretching and subsequent retraction of the end-to-end distance. The force–distance profile (Fig. 1B) is highly structured during retraction, reminiscent of the stick-slip dynamics of strand unpeeling (14). This suggests that such molecules (designed to be topologically closed) in fact exhibit nicks in the backbone, which serve as nucleation sites for strand unpeeling (Fig. 1B Upper) (13). For the other subset of experiments, however, there was no measureable hysteresis in force during DNA overstretching and subsequent end-to-end retraction (Fig. 1A). This is consistent with reports by Paik and Perkins (13), and we thus ascribe such molecules to topologically closed DNA constructs.
Fig. 1.
At low ionic strength (50 mM NaCl), topologically closed DNA overstretches via formation of melting bubbles. (A) Schematic depiction of topologically closed λ-DNA (Upper), along with the corresponding force-distance curves [Lower, recorded with a pulling/relaxing speed of ∼3 μm/s for extension and retraction of the relative DNA length (L/Lc)]. Gray spheres represent optically trapped beads; orange spheres depict biotin-streptavidin linkages between the DNA molecule and the trapped beads. (B) Schematic illustration (Upper) of the same construct shown in A, but with nicks in the DNA backbone. The corresponding force-distance curves are also shown (Lower, pulling/relaxing speed ∼3 μm/s). (C) Corresponding fluorescence images of EGFP-RPA and Sytox for the construct shown in A. (D) Analogous fluorescence images to those displayed in C, but obtained with a nicked DNA construct similar to that illustrated in B. Strand unpeeling is highlighted by dashed orange arrows. Force-distance curves in A and B were recorded in the absence of fluorescent probes.
We next aimed to determine the ratio of ssDNA to dsDNA for both cases reported above, using fluorescence microscopy. To this end, we used fluorescently labeled (EGFP) RPA to identify regions of ssDNA (21), and the fluorescent DNA binding agent Sytox (22) to probe B-form dsDNA. Sytox was used as a B-DNA probe because this dye has a contrasting excitation–emission spectrum to that of EGFP-RPA (thus allowing dual-color fluorescence measurements). We confirmed that these fluorescent probes do not perturb the structure or thermodynamics of overstretched DNA significantly under our experimental conditions (Figs. S1–S4 and SI Text).
Fig. 1C displays fluorescence images of EGFP-RPA and Sytox, respectively, obtained from experiments on topologically closed DNA that was extended 50% into its overstretching plateau. From the panels in Fig. 1C it is clear that both RPA and Sytox bind in a disperse fashion throughout the DNA molecule. Because the DNA construct is topologically closed, the presence of RPA (and thus ssDNA) can only be explained by localized domains where base pairing is broken (i.e., melting bubbles). The phosphate backbones, however, remain under tension. For comparison, Fig. 1D displays analogous fluorescence images derived from a DNA construct deduced to possess nicks in the backbone. In this case, the entire DNA molecule is accounted for by just two domains each of ssDNA and dsDNA, with contiguous binding of RPA and Sytox, respectively. This behavior is akin to that observed for strand unpeeling (10). Indeed, as the orange arrows in Fig. 1D highlight, application of gentle buffer flow (∼1 μm/s) allows the unpeeled tracts of ssDNA to be identified.
Melting Bubbles Show a Strong Preference for AT-Rich Domains.
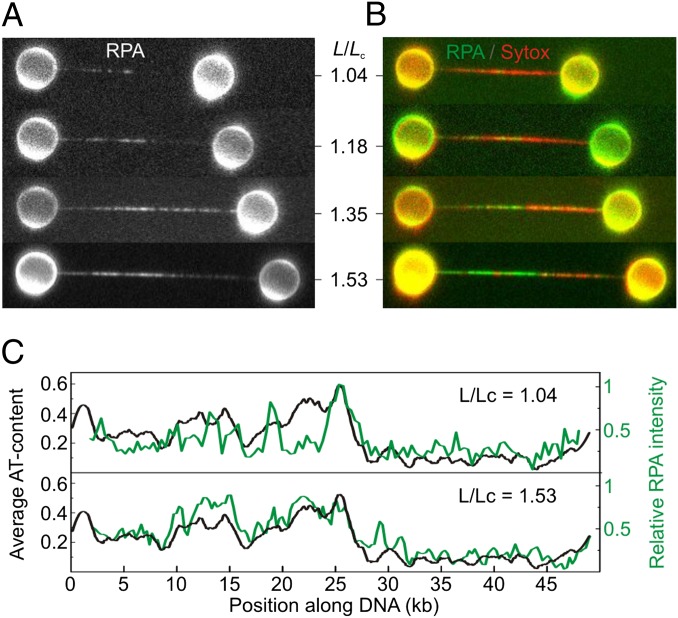
Having demonstrated that topologically closed (but torsionally relaxed) dsDNA can overstretch via localized base-pair melting, we next tracked the evolution of melting bubbles as a function of DNA extension and base-pair sequence. In these experiments, the DNA molecule was extended to various stages of overstretching (defined by the relative DNA extension, 1 < L/Lc > 1.7), and then exposed to RPA and Sytox sequentially. A new DNA molecule was used for each successive extension to exclude the role of prebound RPA or Sytox on the structural response of DNA to applied tension. Fig. 2A displays fluorescence images of EGFP-RPA bound to overstretched DNA, obtained for increasing L/Lc. The composite fluorescence images in Fig. 2B show that Sytox binds to the majority of domains that are not bound already by RPA; under these relatively low salt concentrations (50 mM NaCl), melting bubbles continue to form as the DNA molecule is extended to lengths close to the end of the overstretching process.
Fig. 2.
At low ionic strength (50 mM NaCl), melting bubbles nucleate in AT-rich regions. (A) Selection of fluorescence images of EGFP-RPA binding to topologically closed λ-DNA molecules as a function of relative DNA extension (L/Lc). A different DNA molecule was used for each extension. (B) Composite fluorescence images displaying the binding of EGFP-RPA (in green) and Sytox (in red). DNA molecules shown here are the same as those in A. (C) Variation in fluorescence intensity of EGFP-RPA along the DNA molecule (in green) for L/Lc = 1.04 and 1.53, respectively. Black traces display the AT-content of λ-DNA (in 100-bp bins) and are oriented in the direction that best matches the RPA fluorescence intensity (SI Text, Correlating Melting-Bubble Formation with AT-Rich DNA). These, and 10 further examples, are also shown in Fig. S5.
The sequence of λ-DNA is nonpalindromic and the molecule can be oriented in the optical traps in either direction. Fig. S5 compares RPA binding to overstretched λ-DNA for 12 different molecules with random alignment. Two examples from Fig. S5 are given in Fig. 2C. Strikingly, in every case, there is a strong correlation between the relative fluorescence intensity of EGFP-RPA (as a function of DNA length) and the AT-content of the DNA molecule (binned over 100 bp) when aligned in one direction. Such a correlation suggests that melting bubbles occur preferentially in AT-rich regions (refer to Figs. S5 and S6 for more details).
At High Ionic Strength (150 mM NaCl plus 20 mM MgCl2), Melting-Bubble Formation is Disfavored.
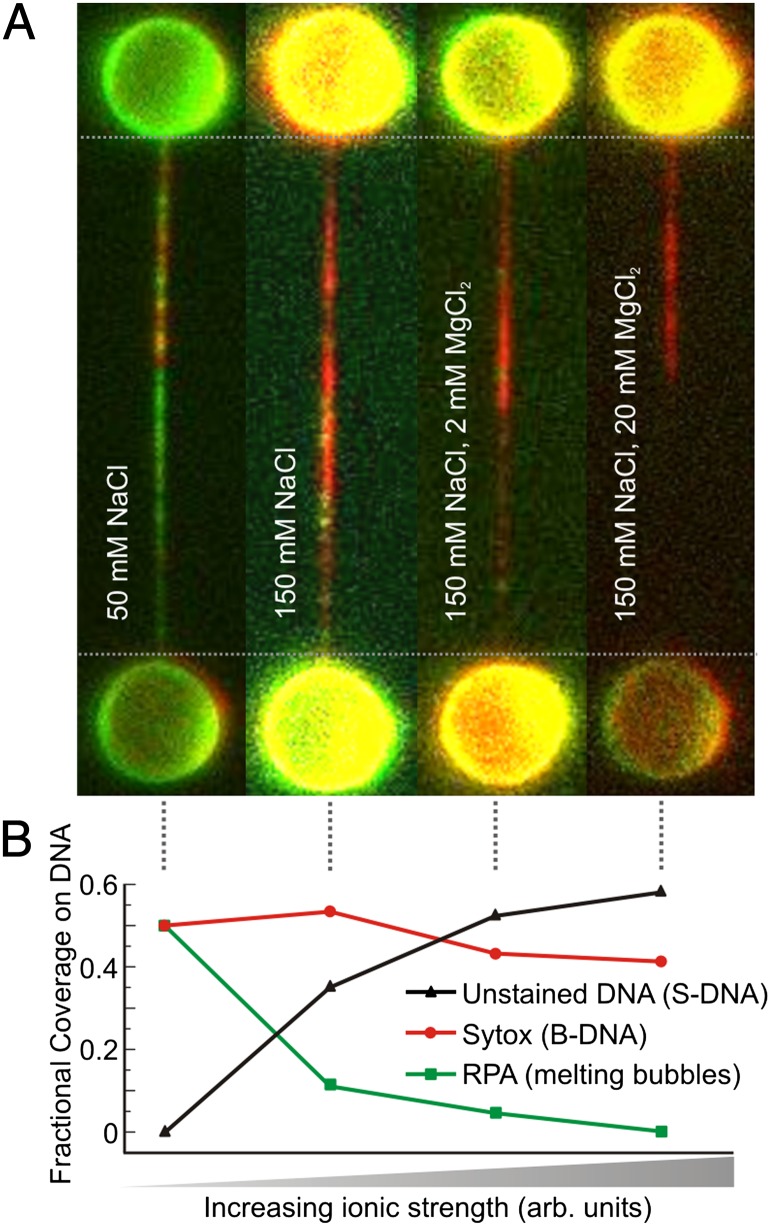
It is well established that thermally induced DNA melting depends strongly on solvent ionic strength (23). Dissolved ions shield the negative charges of DNA (2e− per base pair) and thus decrease electrostatic repulsion between the two DNA strands, stabilizing the double helix. To examine the effect of electrostatic shielding on tension-induced DNA melting, we measured the fraction of ssDNA (using EGFP-RPA) and B-form dsDNA (using Sytox) as a function of salt concentration. Figs. 2 A and B revealed that, at 50 mM NaCl and for L/Lc ∼1.35, there are melting bubbles dispersed throughout the DNA molecule. Fig. 3A shows composite fluorescence images of DNA at L/Lc ∼1.35 exposed sequentially to EGFP-RPA and Sytox, as a function of both mono- and divalent salt concentration, obtained with the same procedure as for Fig. 2B. The fractional coverage of EGFP-RPA, Sytox, and unstained DNA as a function of ionic strength is summarized in Fig. 3B. From these data, it is immediately apparent that increasing the concentration of both NaCl and MgCl2 substantially reduces the fraction of overstretched DNA that is labeled by EGFP-RPA, until there is almost no binding at high ionic strength (150 mM NaCl plus 20 mM MgCl2). We investigated the binding rate of RPA on ssDNA as a function of salt concentration (Fig. S2). There was no appreciable difference, satisfying us that any change in RPA binding to overstretched DNA is not due to the ionic strength per se, but rather to the fundamental structure of the molecule. Moreover, many of those regions that are not stained by RPA are also not bound by Sytox. The findings indicate that as the ionic strength increases either (i) the average size of melting bubbles reduces, eventually becoming smaller than the footprint of RPA (30 nucleotides), and thus unable to be bound by the protein, or (ii) a different structural form of overstretched DNA exists that cannot be stained by either RPA or Sytox.
Fig. 3.
Observable melting bubbles disappear with increasing salt concentration. (A) Selection of composite fluorescence images from sequential binding of EGFP-RPA (green) and Sytox (red) to topologically closed λ-DNA molecules as a function of salt concentration. In each case, a separate DNA molecule was extended midway into its overstretching plateau (L/Lc = 1.35) and exposed to (i) 50 mM NaCl, (ii) 150 mM NaCl, (iii) 150 mM NaCl plus 2 mM MgCl2, and (iv) 150 mM NaCl plus 20 mM MgCl2. (B) Fractional coverage of RPA (revealing melting bubbles), Sytox (labeling B-DNA), and unstained DNA for the four images reported in A. Each data point has an associated error of ≤10% (SI Text, Estimating the Fraction of DNA Bound by Fluorescent Probes). Note that the salt concentration used in the far left image of A is the same as that used in Figs. 1 and 2.
We argue that, as the ionic strength increases, the formation of melting bubbles too small to be detected by RPA binding is unlikely for the following two reasons. First, it is expected that, for fully overstretched DNA, some melting bubbles (<30 bp in size) would coalesce, due to their density at high L/Lc and their known preference for AT-rich regions (Fig. 2C). This would result in bubbles >30 bp, which could then be bound by RPA. However, at 150 mM NaCl plus 20 mM MgCl2, little RPA binding is observed at all, even when incubating the overstretched DNA molecule in RPA for up to 7 min (Fig. S3C). Second, if increasing salt concentration simply reduces the size of melting bubbles to <30 bp, it is anticipated that Sytox binding should become more disperse throughout the molecule to accommodate the many discrete melting bubbles. However, as appreciated from Fig. 3A, the opposite is true: Sytox binding becomes more contiguous as the ionic strength increases.
Based on the above arguments, we conclude that the formation of melting bubbles becomes less likely as the salt concentration increases, until it is nearly negligible at 150 mM NaCl plus 20 mM MgCl2. Instead, an alternative structure of overstretched DNA is increasingly favored as the ionic strength increases; this particular form of DNA very likely exhibits base-pairing interactions (revealed by the absence of RPA binding), although the base-stacking distance and helical pitch must change to accommodate the length increase (resulting in no Sytox binding). These observations are consistent with the formation of the putative DNA structure known as S-DNA (8, 11, 15, 16, 19).
In Topologically Open DNA, Unpeeling Is Suppressed with Increasing Ionic Strength.
The findings reported above demonstrate that when unpeeling is prohibited using a topologically closed DNA construct overstretching proceeds via two competing pathways: formation of melting bubbles and S-DNA. The latter dominates as electrostatic shielding increases. We now consider the effect of electrostatic shielding in the case where discontinuities (free ends or nicks) are present in the phosphate backbone and thus a third mechanism, strand unpeeling, is also possible.
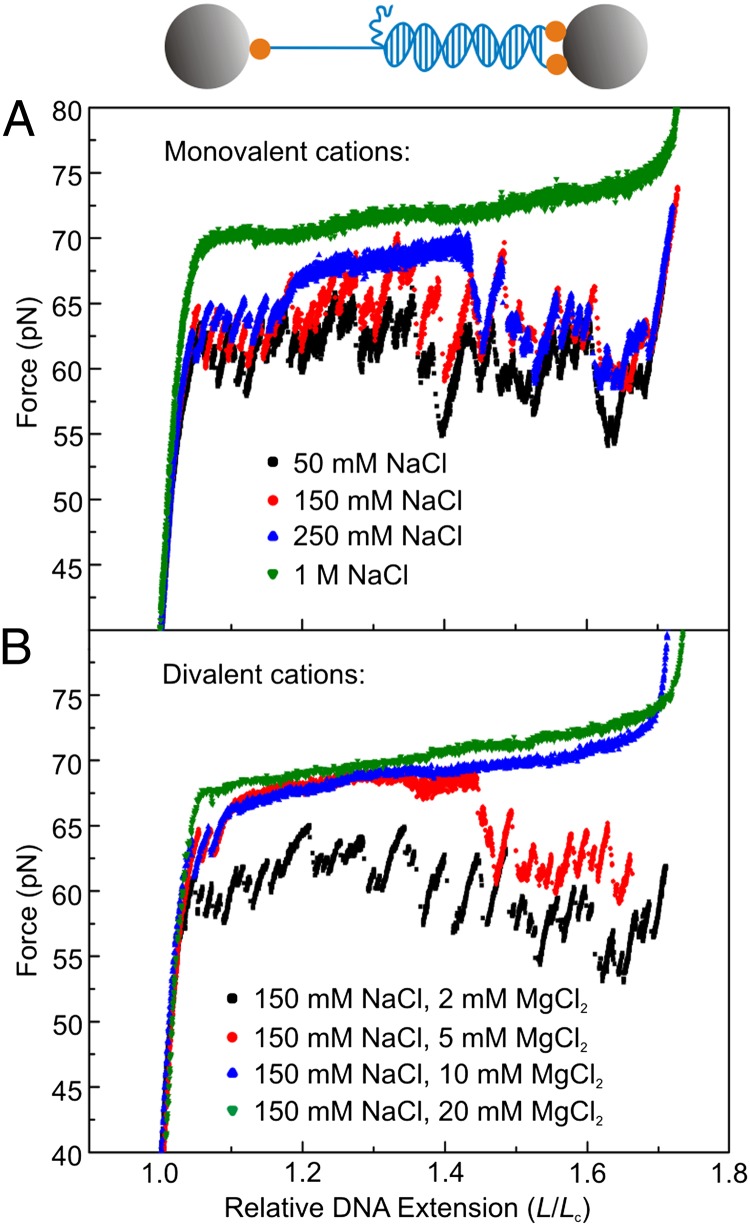
We performed DNA force-extension measurements over a wide range of mono- and divalent salt concentrations, in the absence of fluorescence markers. For these experiments, we used a DNA construct that has one unconstrained DNA end (Fig. 4A Upper), such that unpeeling can only initiate from one end of the molecule and can progress in only one direction (14). This allows us to follow the location of the unpeeling front in the DNA sequence with high resolution during force-extension measurements, which is essential for distinguishing potential effects of ionic strength from those of DNA sequence. Force-extension measurements with this DNA construct at increasing monovalent salt concentrations (Fig. 4A) reveal that at lower ionic strength (≤250 mM NaCl) the overstretching plateau is highly structured with “saw-tooth-like” features. This behavior is characteristic of strand unpeeling via stick-slip dynamics (14). However, at higher monovalent salt concentrations (≥500 mM NaCl), the overstretching plateau in the force-extension curve changes its characteristics: there is little or no resolvable structure. Fig. 4B reveals that the overstretching mechanics depend even more critically on divalent salt concentration, because evidence of stick-slip dynamics is absent for MgCl2 concentrations above only ∼10 mM, together with 150 mM NaCl.
Fig. 4.
Increasing ionic strength suppresses strand unpeeling in topologically open DNA during overstretching. (A) Force-extension measurements of DNA molecules with identical sequence at different monovalent salt concentrations, ranging from 50 mM to 1 M NaCl. Inset: Schematic of the DNA construct used, containing only one unconstrained DNA end. Gray spheres represent optically trapped beads; orange spheres represent biotin-streptavidin linkages between the DNA molecule and the trapped beads. (B) Divalent salt titration of the force-extension behavior of DNA molecules with the same sequence as in Fig. 4A. Monovalent salt concentration: 150 mM NaCl. The divalent salt was increased from 2 mM to 20 mM MgCl2. All data in this figure were obtained using a 8,350-kb DNA fragment (14). Compared with experiments on λ-DNA, the presence of a single melting front, the more heterogeneous sequence, and the slower stretching speed used (10 nm/s) allow for identification of stick-slip dynamics that are not resolved in Fig. 1B (recorded with λ-DNA).
These findings indicate that as the ionic strength increases the probability for unpeeling (with observable stick-slip dynamics in force-extension measurements) decreases. Such a conclusion is consistent with the formation of melting bubbles or S-DNA: in both cases, the respective structural changes can nucleate and progress at many locations throughout the DNA molecule.
At High Ionic Strength (150 mM NaCl plus 20 mM MgCl2) S-DNA Is Favored in Topologically Open DNA.
The force-extension curves in Fig. 4 reveal that, under conditions of intermediate salt concentration (e.g., 250 mM NaCl), the mechanism of overstretching can switch suddenly from unpeeling (with characteristic stick-slip dynamics) to the formation of either melting bubbles or S-DNA (with no structure in the overstretching plateau). To explain these switching events, we considered the local GC-content of switching locations (over a binning window of 40 bp). From the histogram in Fig. S7 (focused on the first ∼4,000 bp of the DNA molecule), it is evident that unpeeling arrest occurs most likely at DNA regions with high GC-content and thus high base-pair stability. In topologically closed dsDNA, it was shown that during overstretching, S-DNA is favored over melting bubbles at higher ionic strength (Fig. 3) due to increased base-pair stability. This leads us to suspect that, in the case of topologically open dsDNA, S-DNA is formed during overstretching at the expense of strand unpeeling (particularly in GC-rich regions).
To test this hypothesis, we estimated the ratio of ssDNA to dsDNA under high-salt conditions of 150 mM NaCl plus 20 mM MgCl2. Topologically open λ-DNA was exposed to EGFP-RPA and Sytox, sequentially, and fluorescence images were recorded for both. Fig. 5A presents dual-color images from this experiment; for each image, a separate DNA molecule is probed at increasing L/Lc. From this figure, it is clear that as each DNA molecule is overstretched further (e.g., L/Lc > 1.4), considerable domains evolve that are labeled by neither RPA nor Sytox. The fractional coverage of RPA, Sytox, and unstained DNA as a function of L/Lc is summarized in Fig. 5B. The unlabeled domains must arise from either small melting bubbles (<30 bp, unable to be bound by RPA) or S-DNA. As discussed earlier, small melting bubbles would be expected to coalesce when L/Lc > 1.5, yielding bubbles >30 bp. This was not found to be the case here. Thus, we deduce that in topologically open DNA, the domains that are not bound by any fluorescent probe are associated with S-DNA. Our studies on topologically closed DNA (Fig. 3A) revealed that, under the same conditions of 150 mM NaCl plus 20 mM MgCl2, S-DNA is also strongly favored over the formation of melting bubbles. As the ionic strength increases, S-DNA is therefore increasingly dominant in both topologically open and closed DNA when overstretched.
Fig. 5.
At high ionic strength (150 mM NaCl plus 20 mM MgCl2), S-DNA formation is favored over strand unpeeling in topologically open DNA. (A) Selection of composite fluorescence images from sequential binding of EGFP-RPA (green) and Sytox (red) to λ-DNA (with one free end on each side of the molecule) as a function of relative DNA extension (L/Lc). Data obtained at 150 mM NaCl plus 20 mM MgCl2. The white circles indicate regions of RPA binding. (B) Fractional coverage of RPA, Sytox, and unstained regions on the DNA molecule for each end-to-end distance reported in A. Each data point has an associated error of ≤10% (SI Text, Estimating the Fraction of DNA Bound by Fluorescent Probes). (C) Comparison of fluorescence images from sequential binding of EGFP-RPA and Sytox recorded under low-salt conditions (50 mM NaCl), illustrating domains of both unpeeling and melting bubbles in λ-DNA with a single nick in the phosphate backbone. Images obtained under mild buffer flow (∼1 μm/s); the orange circle denotes the unpeeled tract of ssDNA bound by RPA.
Despite the dominance of S-DNA when overstretching at high ionic strength (150 mM NaCl plus 20 mM MgCl2), we still identified small regions of ssDNA when using topologically open λ-DNA. These are highlighted by the white circles in Fig. 5A. The fractional coverage of ssDNA (determined by EGFP-RPA binding, Fig. 5B) was relatively constant, at ∼10–15%, independent of L/Lc. We found that these small domains of RPA binding were usually terminated by loose tracts of ssDNA (free from tension) that could be labeled by mitochondrial single-stranded binding protein (mtSSB) (Fig. S8). This finding indicates that the small regions of ssDNA in Fig. 5A are associated with, primarily, strand unpeeling (10).
In topologically open λ-DNA, strand unpeeling can thus always occur to some extent, although S-DNA is increasingly favored as the ionic strength increases. This does not preclude that melting bubbles can form in topologically open DNA as well. Both strand unpeeling and melting bubbles involve base-pair breaking, which is more likely in AT-rich domains. At the lowest salt concentrations tested (50 mM NaCl), and when several nicks are present in the DNA backbone, strand unpeeling is generally favored over melting-bubble formation (e.g., Fig. 1D). However, for <150 mM NaCl, when a single nick exists in a domain with high GC-content, we sometimes observe both unpeeling and the formation of melting bubbles. To illustrate this, consider Fig. 5C, which displays fluorescence images for an overstretched DNA molecule (L/Lc ∼1.35) exposed sequentially to EGFP-RPA and Sytox. These images were obtained with gentle buffer flow (∼1 μm/s). Using the structural investigation reported in this article, we can recognize clearly an unpeeling domain, characterized by both contiguous RPA binding and no Sytox binding. The limit of the unpeeling domain is identified by a loose tract of ssDNA, exposed by the buffer flow. The remainder of the DNA molecule is composed of an intermittent and noncontiguous array of RPA and Sytox, characteristic of melting bubbles dispersed among regions of B-DNA (similar to that shown in Fig. 1C).
Discussion
Our unique experimental approach combining optical tweezers with fluorescence microscopy has shed light on the mechanics of DNA overstretching. We have shown that overstretching can proceed via three fundamentally different mechanisms: (i) strand unpeeling from discontinuities in the DNA backbone; (ii) formation of internal melting bubbles; and (iii) creation of S-DNA, involving strand unwinding but retention of base pairing. As discussed below, the balance between the three mechanisms can be determined by DNA topology, DNA sequence, and ionic strength. Zhang et al. (20) further show that melting bubbles are favored over formation of S-DNA as the temperature is increased. The different mechanisms of overstretching compete over a broad range of solvent conditions, including those occurring in the cell. These findings confirm and unify the varying and competing interpretations of DNA overstretching (4, 8, 12, 13, 15, 19, 20, 23, 24).
DNA topology has a defining role in the mechanism of overstretching. When the DNA construct is topologically closed (but torsionally unconstrained), which prohibits unpeeling, overstretching involves a competition between melting-bubble and S-DNA formation (Fig. 3). However, when there are free ends or nicks in the phosphate backbone, unpeeling and melting-bubble and S-DNA formation can all occur during DNA overstretching (Fig. 5). Unpeeling and melting-bubble formation both involve base-pair breaking, yielding regions of ssDNA. These mechanisms show strong sequence dependence, reflecting the difference in base-pair stability between AT and GC pairs. At higher ionic strengths, unpeeling is suppressed in DNA regions with high GC-content (Fig. 4 and Fig. S7). This is consistent with Bosaeus et al. (19), who report that, in topologically open DNA (at 1 M NaCl), base-pair breaking and S-DNA formation are favored during overstretching in AT- and GC-rich DNA, respectively. Further, we confirm here that melting bubbles develop preferentially in AT-rich domains (Fig. 2C).
In addition to DNA topology and sequence, the mechanism of overstretching is highly sensitive to ionic strength, as highlighted in Figs. 3 and 4. At the lowest salt concentrations investigated (50 mM NaCl), overstretching proceeds only via mechanisms that involve base-pair breaking: unpeeling and melting-bubble formation. Unpeeling is forbidden in topologically closed DNA but becomes increasingly favored over melting-bubble formation as the number of discontinuities (nicks and free ends) increases. At the other extreme, S-DNA is the primary structure of overstretched DNA at the highest salt concentrations used (>500 mM NaCl or >150 mM NaCl plus 10 mM MgCl2). In between those salt concentrations, unpeeling and melting-bubble and S-DNA formation can all occur during overstretching.
There is a direct correlation between the ionic strength and the degree of base-pair breaking during overstretching. As the ionic strength increases, there is greater electrostatic shielding within the DNA molecule, and therefore base-pair breaking is suppressed, rendering S-DNA more favorable than strand unpeeling or formation of melting bubbles. This conclusion is consistent with a number of recent studies (15, 16, 19). Strikingly, we find that mono- and divalent salt act differently, the latter more pronounced than expected on the basis of valence (e.g., Fig. 4). This chimes with the observation that divalent cations, such as Mg2+, specifically condense to the phosphate backbone (24). Divalent cations tend to be fully hydrated, causing the interaction with dsDNA to be more sequence-specific. Water molecules of the solvation shell can interact specifically with the bases by accepting or donating hydrogen bonds. This has been shown to result in an enhanced affinity of Mg2+ for DNA with an increased GC-content (25), supporting our observation that the probability of base-pair breaking is more sensitive to Mg2+ than to Na+.
Our findings, obtained with and without the use of fluorescent probes, are complementary to calorimetric studies by Zhang et al. (20) published in parallel with this work. For clarity, we highlight two specific differences between these studies. First, different types of transitions may be observed at the same salt concentrations in the two studies. This is largely attributed to the fact that the DNA constructs used in each case have different lengths (typically 48.5 kb and 7.25 kb, respectively), with different local distributions of GC-percentage along the molecule. There will thus be differences in local base-pair stability between the two DNA constructs, which could affect the relative fraction of S-DNA at a given salt concentration. Additionally, in our case, the probability of DNA melting might be enhanced by local heating effects arising from the optical traps. The second difference is that, in contrast to our study (Fig. 1A), Zhang et al. (20) report that melting-bubble formation is a hysteretic transition and that the resulting overstretched DNA is significantly shorter than S-DNA. This difference can be explained if, under the lowest salt conditions considered in our study (50 mM NaCl), the overstretched DNA contains a mixture of S-DNA and small melting bubbles. The latter may yield a nonhysteretic transition and similar extension to that of S-DNA. For more details, refer to SI Text, Biochemical Procedures and Analyses.
Nevertheless, despite these differences, the conclusions reached from our respective studies are the same; together they offer clear experimental evidence for the existence of both melting bubbles and S-DNA in overstretched DNA. The observation of melting bubbles during overstretching mirrors reports of melting bubbles in dsDNA in response to both thermal stress (26) and applied torque (27). Meanwhile, the dominance of S-DNA in overstretched DNA under high ionic strength (>500 mM NaCl or >150 mM NaCl plus 10 mM MgCl2) validates the model put forward by several groups in recent years (8, 11, 15, 16, 19). Our findings provide a clear answer to the long-running debate over the structure of overstretched DNA and expand our knowledge of DNA mechanics, which is of fundamental importance to understanding how proteins modify the physical state of DNA.
Materials and Methods
The experimental procedure for recording two-color fluorescence measurements to inspect the overstretched DNA structure was as follows: (i) overstretch the dsDNA construct; (ii) move the overstretched molecule into a buffer containing EGFP-RPA and excite, then detect EGFP fluorescence; and (iii) transfer the overstretched molecule into a buffer containing Sytox and excite, then detect Sytox fluorescence. The visualization of fluorescently labeled RPA, mtSSB, and Sytox was performed using a combined fluorescence and dual-beam optical trapping instrument, which has been described in detail before (10, 28). RPA was fluorescently labeled as discussed previously (29). In all experiments reported in this article, the concentrations of RPA, mtSSB, and Sytox used were 3, 60, and 2 nM, respectively, unless stated otherwise. For each fluorescent probe, the incubation time for the DNA was 20, 5, and 10 s, respectively. Unless stated otherwise, all fluorescence images are snapshots (1-s exposure) recorded at the end of this incubation period. All experiments were performed in a 10 mM Tris buffer at pH 7.6–7.8 and at 22 ± 2 °C (excluding the global heating of the buffer caused by the optical traps, estimated at ≤5 °C by measuring the temperature of the buffer using a calibrated infrared camera). Biochemical protocols for generating the three different DNA constructs used are discussed in SI Text, Biochemical Procedures and Analyses.
Supplementary Material
Acknowledgments
The authors thank Jie Yan, Xinghua Zhang, Hu Chen, and Andreas Biebricher for discussions and Geraldine Farge and Sandrine D’Haene for biochemical assistance. This work is part of the research program of the Stichting voor Fundamenteel Onderzoek der Materie, which is financially supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) (G.J.L.W. and E.J.G.P.). G.J.L.W. acknowledges the European Research Council for a starting grant. G.J.L.W. and E.J.G.P. are recipients of Vici grants from the NWO. G.A.K. thanks the European Molecular Biology Organisation for a long-term postdoctoral fellowship. P.G. is supported by ATLAS, a European Commission-funded Marie Curie early-stage training network. U.B. thanks the Royal Netherlands Academy of Arts and Sciences for a visiting professor grant. M.M. is supported by the Fondation ARC pour la Recherche sur le Cancer and by the Agence Nationale de la Recherche.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. V.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213676110/-/DCSupplemental.
References
- 1.Dame RT, Noom MC, Wuite GJL. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444(7117):387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 2.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438(7067):460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanner NA, et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15(2):170–176. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271(5250):795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 5.Cluzel P, et al. DNA: An extensible molecule. Science. 1996;271(5250):792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 6.Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97(2):542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix 1. Thermodynamic analysis. Biophys J. 2001;80(2):882–893. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocco S, Yan J, Léger JF, Chatenay D, Marko JF. Overstretching and force-driven strand separation of double-helix DNA. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70(1 Pt 1):011910. doi: 10.1103/PhysRevE.70.011910. [DOI] [PubMed] [Google Scholar]
- 9.Shokri L, McCauley MJ, Rouzina I, Williams MC. DNA overstretching in the presence of glyoxal: Structural evidence of force-induced DNA melting. Biophys J. 2008;95(3):1248–1255. doi: 10.1529/biophysj.108.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Mameren J, et al. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc Natl Acad Sci USA. 2009;106(43):18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitelam S, Geissler PL, Pronk S. Microscopic implications of S-DNA. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82(2 Pt 1):021907. doi: 10.1103/PhysRevE.82.021907. [DOI] [PubMed] [Google Scholar]
- 12.Fu H, Chen H, Marko JF, Yan J. Two distinct overstretched DNA states. Nucleic Acids Res. 2010;38(16):5594–5600. doi: 10.1093/nar/gkq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paik DH, Perkins TT. Overstretching DNA at 65 pN does not require peeling from free ends or nicks. J Am Chem Soc. 2011;133(10):3219–3221. doi: 10.1021/ja108952v. [DOI] [PubMed] [Google Scholar]
- 14.Gross P, et al. Quantifying how DNA stretches, melts and changes twist under tension. Nat Phys. 2011;7:731–736. [Google Scholar]
- 15.Fu H, et al. Transition dynamics and selection of the distinct S-DNA and strand unpeeling modes of double helix overstretching. Nucleic Acids Res. 2011;39(8):3473–3481. doi: 10.1093/nar/gkq1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Chen H, Fu H, Doyle PS, Yan J. Two distinct overstretched DNA structures revealed by single-molecule thermodynamics measurements. Proc Natl Acad Sci USA. 2012;109(21):8103–8108. doi: 10.1073/pnas.1109824109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inman RB. Denaturation maps of the left and right sides of the lambda DNA molecule determined by electron microscopy. J Mol Biol. 1967;28(1):103–116. doi: 10.1016/s0022-2836(67)80081-0. [DOI] [PubMed] [Google Scholar]
- 18.Léger JF, et al. Structural transitions of a twisted and stretched DNA molecule. Phys Rev Lett. 1999;83:1066. [Google Scholar]
- 19.Bosaeus N, et al. Tension induces a base-paired overstretched DNA conformation. Proc Natl Acad Sci USA. 2012;109(38):15179–15184. doi: 10.1073/pnas.1213172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, et al. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry. Proc Natl Acad Sci USA. 2013;110:3865–3870. doi: 10.1073/pnas.1213740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385(6612):176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 22.Yan X, et al. Development of a mechanism-based, DNA staining protocol using SYTOX orange nucleic acid stain and DNA fragment sizing flow cytometry. Anal Biochem. 2000;286(1):138–148. doi: 10.1006/abio.2000.4789. [DOI] [PubMed] [Google Scholar]
- 23.SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 24.Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nat Struct Biol. 1999;6(4):346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 25.Chiu TK, Dickerson RE. 1 A crystal structures of B-DNA reveal sequence-specific binding and groove-specific bending of DNA by magnesium and calcium. J Mol Biol. 2000;301(4):915–945. doi: 10.1006/jmbi.2000.4012. [DOI] [PubMed] [Google Scholar]
- 26.Guéron M, Kochoyan M, Leroy JL. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987;328(6125):89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- 27.De Vlaminck I, et al. Torsional regulation of hRPA-induced unwinding of double-stranded DNA. Nucleic Acids Res. 2010;38(12):4133–4142. doi: 10.1093/nar/gkq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross P, Farge G, Peterman EJG, Wuite GJL. Combining optical tweezers, single-molecule fluorescence microscopy, and microfluidics for studies of DNA-protein interactions. Methods Enzymol. 2010;475:427–453. doi: 10.1016/S0076-6879(10)75017-5. [DOI] [PubMed] [Google Scholar]
- 29.Modesti M. Fluorescent labeling of proteins. Methods Mol Biol. 2011;783:101–120. doi: 10.1007/978-1-61779-282-3_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.