Abstract
Pretreatments such as dilute acid at elevated temperature are effective for the hydrolysis of pentose polymers in hemicellulose and also increase the access of enzymes to cellulose fibers. However, the fermentation of resulting syrups is hindered by minor reaction products such as furfural from pentose dehydration. To mitigate this problem, four genetic traits have been identified that increase furfural tolerance in ethanol-producing Escherichia coli LY180 (strain W derivative): increased expression of fucO, ucpA, or pntAB and deletion of yqhD. Plasmids and integrated strains were used to characterize epistatic interactions among traits and to identify the most effective combinations. Furfural resistance traits were subsequently integrated into the chromosome of LY180 to construct strain XW129 (LY180 ΔyqhD ackA::PyadC′fucO-ucpA) for ethanol. This same combination of traits was also constructed in succinate biocatalysts (Escherichia coli strain C derivatives) and found to increase furfural tolerance. Strains engineered for resistance to furfural were also more resistant to the mixture of inhibitors in hemicellulose hydrolysates, confirming the importance of furfural as an inhibitory component. With resistant biocatalysts, product yields (ethanol and succinate) from hemicellulose syrups were equal to control fermentations in laboratory media without inhibitors. The combination of genetic traits identified for the production of ethanol (strain W derivative) and succinate (strain C derivative) may prove useful for other renewable chemicals from lignocellulosic sugars.
Keywords: lignocellulose, metabolic engineering
The carbohydrate component of lignocellulose represents a potential feedstock for renewable fuels and chemicals (1–3), an alternative to food crops and petroleum. However, the cost-effective use of lignocellulosic sugars in fermentation remains challenging (4, 5). Unlike starch, lignocellulose has been designed by nature to resist deconstruction (2, 6). Crystalline fibers of cellulose are encased in a covalently linked mesh of lignin and hemicellulose. Steam pretreatment with dilute mineral acids is an efficient approach to depolymerize hemicellulose (20–40% of biomass dry weight) into sugars (hemicellulose hydrolysate, primarily xylose) and to increase the access of cellulase enzymes (2, 3, 6). However, side-reaction products (furfural, 5-hydroxymethylfurfural, formate, acetate, and soluble lignin products) are formed during pretreatment that hinder fermentation (7, 8). Furfural (dehydration product of pentose sugars) is widely regarded as one of the most important inhibitors (6–8). The concentration of furfural is correlated with the toxicity of dilute acid hydrolysates (9). Although overliming to pH 10 with Ca(OH)2 can be used to reduce the level of furfural and toxicity, inclusion of this step increases process complexity and costs (9, 10).
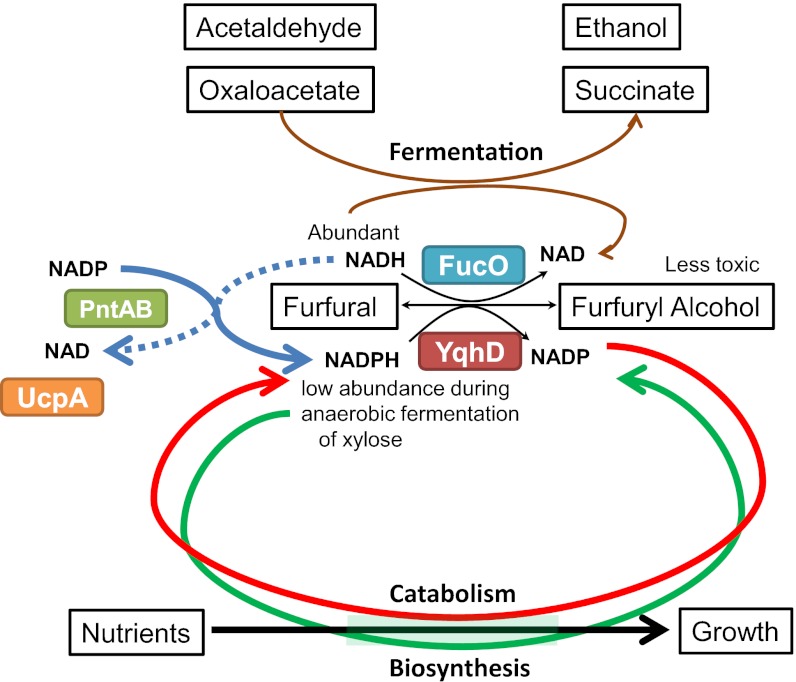
Escherichia coli and yeasts have proven to be excellent biocatalysts for metabolic engineering (11, 12). However, both are inhibited by furan aldehydes (7, 8, 13–15) and both contain NADPH-dependent oxidoreductases that convert furfural and hydroxymethylfurfural (dehydration product of hexose sugars) into less toxic alcohols (15–17). It is this depletion of NADPH by oxidoreductases such as YqhD (low Km for NADPH) that has been proposed as the mechanism for growth inhibition in E. coli (Fig.1) (18–20). Growth resumed only after the complete reduction of furan aldehydes (19). A similar furan aldehyde-induced delay in growth has been reported for fermenting yeasts (14, 15). The NADPH-intensive pathway for sulfate assimilation was identified as an early site affected by furfural (18). Addition of cysteine (18), deletion of yqhD (19) or increased expression of pntAB (transhydrogenase for interconversion of NADH and NADPH) increased tolerance to furan aldehydes (18, 21) (Fig. 1). Furfural tolerance was also increased by overexpression of an NADH-dependent propanediol (and furfural) oxidoreductase (FucO) normally used for fucose metabolism (17) and by overexpression of a cryptic gene (ucpA) adjacent to a sulfur assimilation operon (22) (Fig. 1). However, none of these traits alone fully eliminated the problem of furfural toxicity.
Fig. 1.
Model showing relationships of furfural resistance traits, metabolism, and reducing cofactors. NADPH-linked reduction of furfural by YqhD is proposed to compete with biosynthesis, starving key steps in biosynthesis such as sulfate assimilation (18, 19). Deletion of yqhD or increased expression of pntAB (NADH/NADPH transhydrogenase) mitigated this problem by increasing the availability of NADPH. Overexpression of fucO increased the rate of furfural reduction and used NADH, an abundant cofactor during sugar fermentation (17). The cryptic gene ucpA is required for native furfural tolerance and further increased furfural resistance when overexpressed (22).
Little is known about the interactions among these four genetic traits. In this study, combinations of traits were constructed and tested for furfural tolerance in a derivative of E. coli W (LY180). Optimal combinations identified for ethanol production by LY180 derivatives were subsequently reengineered into strain KJ122, strain C engineered to produce succinate (precursor for solvents and plastics) (17). Resulting strains exhibited an increase in furfural tolerance and an increase in tolerance to toxins in hemicellulose hydrolysates.
Results
Epistatic Interactions Among Four Furfural Resistance Traits in Ethanologenic LY180.
Previous studies have shown that deletion of yqhD and increased expression of fucO, ucpA, or pntAB from plasmids each improved growth of ethanologenic E. coli LY180 in the presence of 10 mM furfural (17–19, 22). Further constructions (SI Text and Table S1) were made to allow a comparison of all combinations of these genetic traits using pTrc99a-based plasmids for expression of target genes (fucO, ucpA, and fucO-ucpA). Three new derivatives of LY180 were constructed for use as host strains: ΔyqhD, adhE::pntAB, and ΔyqhD adhE::pntAB. Integration of pntAB behind the adhE promoter in LY180 provided furfural tolerance equivalent to pTrc99a expressing pntAB (uninduced). Higher levels of pntAB expression with inducer were inhibitory in the absence or presence of furfural (18).
Ethanol production from 100 g/L xylose was complete after 48 h in control cultures lacking furfural (Fig. 2A). Ethanol production at this time point was selected as a comparative measure of tolerance to 15 mM furfural. All individual traits except fucO improved ethanol production in the presence of 15 mM furfural (Fig. 2A). Combinations of two traits (Fig. 2B) were more effective than single traits with two exceptions: (i) ΔyqhD with pntAB integration and (ii) ΔyqhD with the ucpA plasmid (pLOI4856). All binary combinations with fucO were beneficial. Because growth and ethanol production were also inhibited by excess pntAB expression (18), the negative interactions between pntAB (increased NADPH production) and ΔyqhD (reduced NADPH consumption) could result from a similar problem. The poor performance of LY180 ΔyqhD containing the ucpA plasmid suggests that this cryptic gene may be associated with a similar action. Among ternary combinations, the combination of ΔyqhD adhE::pntAB and ucpA plasmid was particularly sensitive to furfural inhibition. Ethanol titer was low (13 g/L) when all four genetic traits were combined, comparable to strains with a single resistance trait (Fig. 2B). The most effective combinations were plasmid expression of fucO-ucpA in a host strain with either ΔyqhD or adhE::pntAB. Both constructs produced close to 30 g/L ethanol after 48 h in medium with 15 mM furfural, about 70% of the ethanol titer in control fermentations without furfural (Fig. 2B).
Fig. 2.
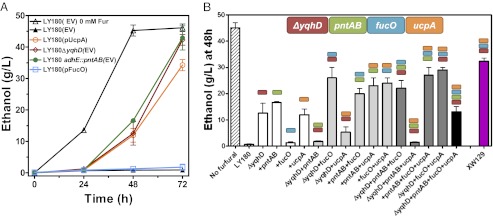
Epistatic interactions of furfural resistance traits during ethanol production. Fermentations were conducted in isopropyl-β-D-1-thiogalactopyranoside mineral salts medium (100 g/L xylose, 0.1 mM IPTG, and 12.5 mg/L ampicillin) with 15 mM furfural. (A) Single furfural-resistant traits. LY180 containing empty vector pTrc99a (EV) was included as a control with and without furfural. LY180 ΔyqhD and LY180 adhE::pntAB also contained an empty vector to reduce differences related to plasmid burden. (B) Comparison of furfural tolerance for ethanol production (48 h). Test strains contain either empty vector or plasmids for expression of fucO, ucpA, or fucO-ucpA. Ethanol titers of parent strain LY180 (hatched bars) were included with or without furfural for comparison. Modified strains contain a single trait (open bars), two traits (light gray bars), three traits (dark gray bars), or four traits (black bar). Strain XW129 (LY180 ΔyqhD ackA::PyadC′fucO-ucpA) was obtained after promoter engineering and chromosomal integration (purple bar). The four-color boxes at the top represent a key to genetic traits. Stacked boxes correspond to traits in each respective strain. Data represent averages of at least two experiments with SDs.
Constructing Plasmid-Free Strains for Ethanol Production (Integration of fucO-ucpA).
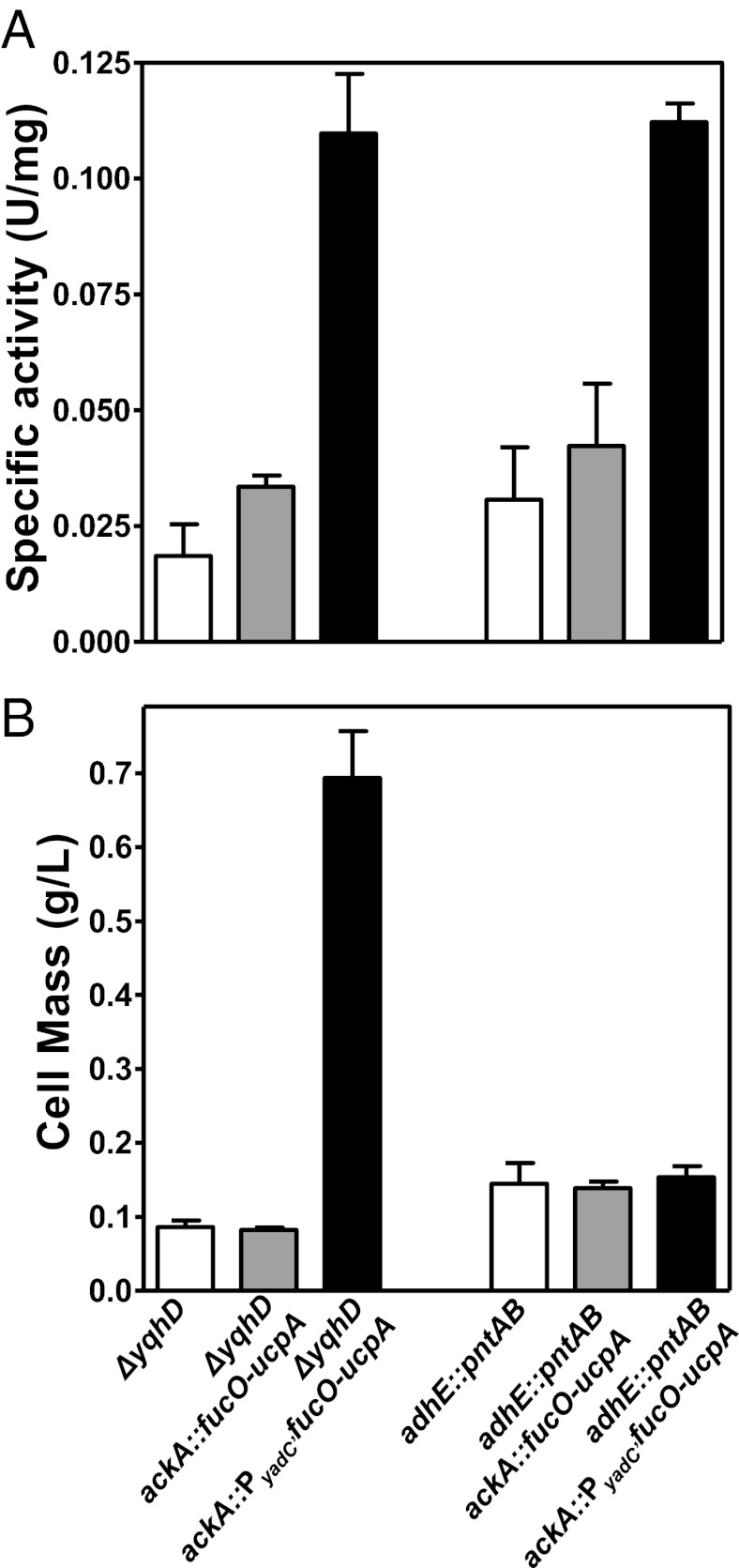
The use of plasmids, antibiotics, and expensive inducers allowed an investigation of gene interactions but is unlikely to provide the desired genetic stability needed for commercial strains. Chromosomal integration of fucO-ucpA behind a strong promoter such as ackA (highly expressed in mRNA arrays) (18, 20, 22) was tested as a replacement for plasmid pTrc fucO-ucpA in LY180 adhE::pntAB and LY180 ΔyqhD. However, FucO activity of the integrated strains was low (Fig. 3A) and furfural tolerance (12.5 mM) was unchanged (Fig. 3B). Integration behind the strong pflB promoter (18, 20, 22, 23) also did not provide sufficient expression of fucO-ucpA for furfural tolerance. Clearly, a more efficient approach was needed.
Fig. 3.
Comparison of in vitro furfural reductase activity and furfural resistance. NADH-linked furfural-dependent reductase activity (A) and furfural tolerance for growth (B) are shown for plasmid-free strains containing predicted optimal combinations of furfural resistance traits. Cell mass was measured from tube cultures (n = 3) grown for 48 h in AM1 minimal media containing 50 g/L xylose with 12.5 mM furfural. Data represent averages of at least two experiments with SDs.
A function-based selection was used to identify a useful promoter. A promoter probe vector was constructed for fucO-ucpA as a derivative of pACYC184 (low copy) with an appropriately engineered upstream BamH1 site (Fig. S1A and SI Text). Random Sau3A1 fragments (E. coli W chromosome) were ligated into this site, and resulting plasmids transformed into LY180 ΔyqhD. After selection for large colonies on furfural (12 mM) plates and further screening, the most effective promoter was identified by sequencing as a 600-bp internal fragment of the E. coli yadC gene, designated PyadC′, in plasmid pLOI5259 (Fig. S1B). With this promoter, constitutive expression of fucO on a low-copy plasmid (pACYC184) was equal to induced expression of fucO from a high-copy plasmid (pTrc99a) (Fig. S1). The nucleotide sequences for this promoter and more details concerning isolation are included in SI Text.
The expression cassette from pLOI5259 (ackA′::PyadC′fucO-ucpA -ackA′) was amplified by PCR (Table S1) and integrated into the chromosomes of LY180 ΔyqhD and LY180 adhE::pntAB by precisely replacing the ackA coding region including 22 bp immediately upstream. Resulting strains were designated XW129 and XW131, respectively. Although both integrated strains produced fourfold to sixfold higher FucO activity than the respective parent strain (Fig. 3A), furfural tolerance was only improved in XW129 (Fig. 3B). It is possible that the higher level of FucO produced with plasmids (0.7 U/mg protein; Fig. S1D) is required to increase tolerance in the adhE::pntAB strain (XW131), where yqhD remains functional. Further studies have focused on the useful combination of genetic traits assembled in XW129 (ΔyqhD and increased expression of fucO-ucpA).
Integration of Traits Restored Ethanol Fermentation in 15 mM Furfural.
Strain XW129(LY180 ΔyqhD ackA::PyadC′fucO-ucpA) was compared with the parent LY180 during batch fermentation in AM1 mineral salts medium (100 g/L xylose) with and without 15 mM furfural (Fig. 4 A and B). In the absence of furfural, ethanol yields for both strains were equal. In the presence of 15 mM furfural, growth and fermentation of LY180 was completely blocked. Only 5 mM furfural was metabolized (reduced to furfuryl alcohol) by LY180 after 72 h. Addition of 15 mM furfural delayed the growth of strain XW129 by 24 h, during which time furfural was fully reduced. However, the time required to complete fermentation was extended by only 6 h. The final ethanol yield for strain XW129 with 15 mM furfural was equal to the control without added furfural, 90% of the theoretical yield. Despite being sixfold lower in FucO activity (Fig. 3A and Fig. S1D), ethanol titers (32 g/L after 48 h) for strain XW129 (LY180 ∆yqhD ackA::PyadC′fucO-ucpA) with integrated fucO-ucpA were equivalent to LY180 ∆yqhD with induced expression of fucO-ucpA from plasmids (Fig. 2B). This suggests that the metabolic burden of plasmid maintenance and producing larger amounts of target protein (FucO, UcpA) may have countered any benefit from the additional activities.
Fig. 4.
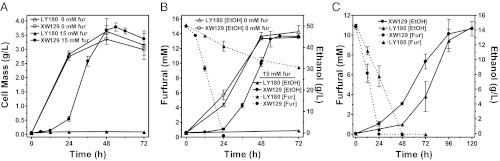
Comparison of batch fermentations for the parent LY180 and the plasmid-free, furfural-resistant strain XW129. Furfural resistance traits in XW129 (LY180 ΔyqhD ackA::PyadC′fucO-ucpA) improved fermentation with furfural in AM1 medium and also improved the fermentation of hemicellulose hydrolysate. For A (cell mass) and B (ethanol and furfural), fermentations were conducted in mineral salt medium AM1 (100 g/L xylose and 15 mM furfural). Control fermentations without furfural were also included. Fermentations (C) were also conducted using hemicellulose hydrolysate containing 36 g/L total sugar, supplemented with components of AM1 medium and 0.5 mM sodium metabisulfite. Data represent averages of at least two experiments with SDs.
Furfural-Resistance Traits also Increased Resistance to Hemicellulose Hydrolysate.
The inhibitory role of furfural in dilute acid hydrolysates of hemicellulose was confirmed in part by a comparison of batch fermentations containing sugarcane bagasse hemicellulose hydrolysate (Fig. 4C). The onset of rapid ethanol production was delayed in hydrolysate, similar to the delay with 15 mM furfural in AM1 medium containing 10% xylose (Fig. 4B). The onset of rapid ethanol production in AM1 medium with furfural and in hydrolysate medium (LY180 and XW129) again coincided with the depletion of furfural. Although total fermentation time in hydrolysate medium and final ethanol titers were similar for both the parent LY180 and the engineered strain XW129, the furfural-resistant XW129 reduced furfural at twice the volumetric rate of LY180. This more rapid reduction of furfural by XW129 shortened the initial delay in ethanol production by 24 h, half that of the parent (Fig. 4C).
Reengineering E. coli KJ122 for Conversion of Hemicellulosic Hydrolysates to Succinate.
Strain LY180 is derived from E. coli KO11, a sequenced strain that has acquired many mutations during laboratory selections for growth in mixed sugars, high sugars, lactate resistance, and other conditions (24–26). It is possible that some of the mutations in KO11 or the heterologous genes encoding ethanol production in this strain may be critical for engineering furfural tolerance and improving resistance to hemicellulose hydrolysate. To address this concern, we have reconstructed the optimal traits for furfural resistance in KJ122, a succinate-producing derivative of E. coli C (27). Initially, strain KJ122 was unable to effectively ferment 100 g/L xylose (Fig. 5A). Mutants with fivefold improvement of succinate titer were readily selected after 40 generations of serial cultivation in xylose AM1 medium. A clone was isolated and designated XW055 with a succinate yield from xylose of 0.9 g/g, equivalent to the yield reported previously for glucose (27).
Fig. 5.
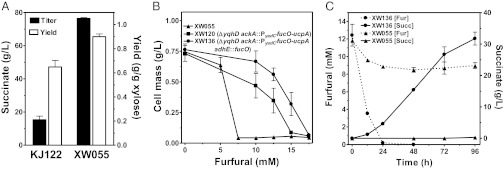
Engineering furfural-resistant derivatives of E. coli C for hemicellulose conversion to succinate. (A) Fermentation titer and yield (96 h) for parent KJ122 and mutant XW055 selected for improved xylose metabolism. Strains were grown in AM1 medium containing 100 g/L xylose as described previously (27) using KOH/K2CO3 to automatically maintain pH 7. Yield was calculated as grams of succinate produced per grams of xylose metabolized. (B) Comparison of furfural tolerance in tube cultures containing AM1 medium (50 g/L xylose, 100 mM MOPS, and 50 mM KHCO3). Strain XW055 was compared with strains XW120 and XW136 containing chromosomally integrated traits for furfural resistance. Cell mass was measured after incubation for 48 h. (C) Fermentation of hemicellulose hydrolysate (components of AM1 medium, 0.5 mM sodium metabisulfite,100 mM potassium bicarbonate, and 36 g/L total sugar). Strain XW136 (XW055 ΔyqhD ackA::PyadC′fucO-ucpA* adhE::fucO) completed the reduction of furfural in 24 h, coincident with the onset of rapid fermentation. Strain XW055 was unable to completely metabolize furfural or ferment sugars in hemicellulose hydrolysate. Data for furfural and succinate are shown by broken lines and solid lines, respectively. All data represent averages of at least two experiments with SDs.
The same genetic tools used to construct furfural tolerance in ethanol-producing biocatalysts were used to engineer XW055 (Fig. S2 and Fig. 5B). As with ethanol biocatalysts, combining a yqhD deletion with integration of pntAB was not helpful (Fig. S2). The most effective combination for succinate production was ΔyqhD and ackA::PyadC′fucO-ucpA, resulting in strain XW120 (Fig. S2 and Fig. 5B). These genetic changes increased the minimal inhibitory concentration of furfural from 7.5 mM (XW055) to 15 mM (XW120). Plasmid derivatives of pTrc99a expressing fucO alone and ucpA alone were tested in XW120. Addition of a fucO plasmid further increased furfural tolerance (Fig. S3). The benefit of this plasmid was supplied by another chromosomal integration, replacing the coding region of adhE with the coding region of fucO to make XW136. The additional expression of fucO from the adhE promoter increased furfural tolerance to 17.5 mM (Fig. 5B).
XW055 and the furfural-resistant XW136 (XW055, ΔyqhD ackA::PyadC′fucO-ucpA adhE::fucO) were compared during batch fermentation using hemicellulose hydrolysate as a source of sugar (Fig. 5C). Hydrolysate medium contained 12 mM furfural and completely inhibited growth and fermentation of the parent. During 96 h of incubation, the parent reduced only 3 mM furfural and was unable to grow or effectively ferment hemicellulose sugars. In contrast, furfural (12 mM) was completely reduced within 24 h by the furfural-resistant strain XW136. With this strain, fermentation of hemicellulose sugars (primarily xylose) into succinate was complete after 96 h with a yield of 0.9 g/g. This succinate yield from hemicellulose sugars was equivalent to that of the parent organism (KJ122) during the fermentation of glucose in AM1 mineral salts medium without furfural (27).
Discussion
Importance of Furfural As an Inhibitor in Hemicellulose Hydrolysate.
Microbial biocatalysts can be used to produce renewable chemicals from lignocellulosic sugars. Large-scale implementation of biobased processes has the potential to replace petroleum for solvents, plastics, and fuels without disrupting food supplies or animal feed. Costs for such processes remain a challenge and can be reduced by developing biocatalysts that are tailored for specific feedstocks. Inhibitors formed during the deconstruction of lignocellulose such as furfural are part of this challenge. Previous studies have shown that furfural was unique in binary combinations of inhibitors, increasing the toxicity of other compounds (soluble lignin products, formate, acetate, etc.) in hemicellulose hydrolysates (13). Our studies demonstrate that removal of furfural is essential before rapid growth and metabolism of sugars by E. coli biocatalysts (Fig. 4 B and C and Fig. 5C). The starting strain for ethanol production, LY180, was more resistant to furfural, as well as hemicellulose hydrolysates, than the starting strain for succinate production, XW055 (Fig. S4, Fig. 4C, and Fig. 5C). However, the same combination of furfural-resistance traits was optimal for furfural tolerance with both strains. Genetic changes that increased furfural tolerance also increased resistance to hemicellulose hydrolysate, establishing the importance of furfural for toxicity and the generality of this approach. Although furfural is not the only inhibitor present in hydrolysate, enzymatic reduction of this compound should allow further studies to identify additional genes that confer resistance to remaining toxins. By developing biocatalysts that are resistant to furfural and other hemicellulose toxins, remaining toxins in hydrolysates can reduce the cost of fermentations by serving as a barrier that prevents the growth of undesirable contaminants.
Epistatic Interactions of Beneficial Traits for Furfural Tolerance.
There is no established strategy to predict the epistatic interactions of target genes, and searching for the optimal combination of beneficial genetic traits is challenging (28). A general model is included to illustrate interactions among the four genetic traits for furfural tolerance (Fig. 1). Energy generation and growth require nutrients, intermediates from carbon catabolism, and balanced oxidation and regeneration of NADPH and NADH. YqhD has a low Km for NADPH that competes effectively with biosynthesis, limiting growth by impeding NADPH-intensive processes such as sulfate assimilation (18). Increasing PntAB transhydrogenase partially restored this imbalance using NADH as a reductant (abundant during fermentation) (18). However, the combination of a yqhD deletion and increased expression of pntAB was more sensitive to furfural inhibition than either alone (Fig. 2B). NADPH-dependent furfural reductase YqhD may play a positive role for furfural tolerance in strains where pntAB expression has been increased. Pyridine nucleotide transhydrogenase activity of PntAB couples proton translocation and makes the reduction of NADP by NADH a costly energy process (29). This increase in energy demand during expression of yqhD and pntAB could reduce fitness, despite potential benefits of reducing furfural to the less toxic alcohol. FucO can serve as a more effective furfural reductase because it uses NADH (abundant during fermentation) as the reductant and does not compete for biosynthetic NADPH. Like pntAB, increased expression of ucpA in a yqhD deletion strain did not further increase furfural tolerance. This epistatic interaction suggests the UcpA-dependent furfural resistance may also involve NADPH availability (Fig. 2B).
Two furfural-resistant strains have been isolated and characterized previously, EMFR9 [selected for furfural tolerance (19)] and MM160 [selected for hydrolysate resistance (17)]. Each contains a mutation that improves furfural tolerance by silencing YqhD using completely different mechanisms, IS10 disruption of adjacent yqhC (transcriptional activator for yqhD) and a nonsense mutation in yqhD, respectively (17, 20). Silencing genes such as yqhD can be caused by a myriad of genetic changes (30). An increase in fitness by gene silencing would be expected to emerge early in populations under growth-based selection. No mutations were found in these strains that increased expression of ucpA, pntAB, or fucO (18–20). Genetic solutions for gain-of-function mutants can be very limited and much less abundant (30, 31). Also, recovery of mutants with increased expression of ucpA and pntAB would be prevented by their negative interactions with yqhD silencing. Very high levels of fucO expression were needed that may require multiple mutations, dramatically limiting recovery without deliberate genetic constructions.
Succinate Fermentation from Lignocellulose Sugars.
Succinic acid is currently produced from petroleum derived maleic anhydride and can serve as a starting material for synthesis of many commodity chemicals used in plastics and solvents (32). Genetically engineered strains of E. coli (33) and native succinate producers such as Actinobacillus succinogenes (34–36) and Anaerobiospirillum succiniciproducens (37) have been tested for lignocellulose conversion to succinate. However, fermentation using these strains required costly additional steps (34), nutrient supplementation (33–37), and mitigation of toxins in hydrolysates by overliming or treating with activated charcoal (33, 36). Reengineering derivatives of KJ122 using known combinations of furfural resistance traits resulted in strain XW136 that now ferments hemicellulose hydrolysates in mineral salts medium without costly detoxification steps (32 g/L succinic acid with a yield of 0.9 g/g sugars; Fig. 5C). The ability to use defined genetic traits for furfural tolerance to improve tolerance to inhibitors in hemicellulose hydrolysates should prove useful as a starting point for many new biocatalysts and products.
Materials and Methods
Strains and Growth Conditions.
Strains used in this study are listed in Table S1. Ethanologenic E. coli LY180 (a derivative of E. coli W; ATCC 9637) and succinate-producing E. coli KJ122 (a derivative of E. coli C; ATCC 8739) were developed previously in our laboratory (19, 27). Strains XW092 (LY180, ΔyqhD), XW103 (LY180, adhE::pntAB), XW109(LY180, ΔyqhD adhE::pntAB), XW115 (LY180, ΔyqhD ackA::fucO-ucpA), XW116 (LY180, adhE::pntAB ackA::fucO-ucpA), XW129 (LY180, ΔyqhD ackA::PyadC′fucO-ucpA), and XW131 (LY180, adhE::pntAB ackA::PyadC′fucO-ucpA) were genetically engineered for furfural tolerance using LY180 as the parent strain. Strain KJ122 (succinate production from glucose) was serially transferred in pH-controlled fermenters (27) at 48-h intervals for ∼40 generations to isolate a mutant with improved xylose fermentation (designated XW055). Strains XW120 (XW055, ΔyqhD ackA::PyadC′fucO-ucpA) and XW136 (XW055, ΔyqhD ackA::PyadC′fucO-ucpA adhE::fucO) were genetically engineered using XW055 as the parent strain. Cultures were grown in low-salt xylose AM1 medium as described previously (38).
Genetic Methods.
Methods for seamless chromosomal deletion, gene replacement, or integration were described previously using Red recombinase technology (12, 27). Plasmids, primers, and construction details are listed in Table S1. The Clone EZ PCR Cloning Kit from GenScript was used for gene replacement on the plasmid. Constructions were made in Luria broth containing 20 g/L xylose or 50 g/L arabinose (inducer for lambda Red recombinase; Gene Bridges) or 100 g/L sucrose (for counter selection of sacB). Antibiotics were added when required. Additional details are included in SI Text and Table S1.
Identification of Promoter for fucO-ucpA Cassette.
A genome-wide promoter library with more than 10,000 clones was constructed in plasmid pLOI4870 (pACYC184 derivative) by ligating Sau3A1 fragments of E. coli genomic DNA into a unique BamH1 site immediately upstream from a promoterless fucO-ucpA cassette (Fig. S1). The library was transformed into LY180 ΔyqhD cells with selection under argon for large colonies on AM1–xylose plates containing 12 mM furfural and 40 mg/L chloramphenicol. Of more than 10,000 transformants, 176 exhibited a large colony phenotype and were further compared using a BioScreen C growth curve analyzer (BioScreen).The most effective clone was identified and designated plasmid pLOI5237 containing a 1,600-bp insert. Subcloning reduced the size of this promoter fragment to 600 bp (pLOI5259). This smaller fragment was identified by sequencing as part of the yadC coding region. The BamH1-furfural resistance cassette in pLOI4870 and pLOI5259 (includes upstream promoter fragment) were bordered by segments of ackA for chromosomal integration. Additional details are provided in SI Text.
NADH-Dependent Furfural Reductase Assay and SDS/PAGE.
The preparation of cell crude lysates and furfural reductase assay were as described previously (17). Soluble protein lysates (15 µg of protein) were also analyzed on 12% (wt/vol) SDS/PAGE gels (Bio-Rad).
Furfural Tolerance in Tube Cultures.
Furfural toxicity was measured in tube cultures (13 mm × 100 mm) as described previously for ethanol strains (17, 22). For succinate strains, tubes contained 4 mL AM1 medium with 50 g/L xylose, 50 mM KHCO3, and 100 mM 3-(N-morpholino)propanesulfonic acid (MOPS) as a buffer. Tubes were inoculated with starting cell density of 44 mg/L. Cell mass was measured at 550 nm after incubation for 48 h (37 °C).
Fermentation of Ethanol or Succinate.
Ethanol fermentations with xylose were carried out as described previously (17, 22), with and without furfural. For succinate production from xylose, seed precultures of strains were grown in sealed culture tubes containing AM1 medium (20 g/L xylose, 50 mM KHCO3, and 100 mM MOPS). After incubation for 16 h, preinocula were diluted into 500-mL fermentation vessels containing 300 mL of AM1 media (100 g/L xylose, 1 mM betaine, and 100 mM KHCO3) at an initial density of 6.6-mg dry cell weight. After 24 h of growth, these seed cultures were used to provide starting inocula for batch fermentations (AM1 medium, 100 g/L xylose, and 100 mM KHCO3). Fermentations were maintained at pH 7.0 by automatic addition of base containing additional CO2 (2.4 M K2CO3 in 1.2 M KOH) as described previously (27). Quantitative analyses of sugars, ethanol, furfural, and succinate were as described previously (17, 27, 39).
Preparation and Fermentation of Hemicellulose Hydrolysates.
Hemicellulose hydrolysate was prepared as described previously (40, 41). Briefly, sugarcane bagasse (Florida Crystals) impregnated with phosphoric acid (0.5% of bagasse dry weight) was steam-treated for 5 min at 190 °C (40, 41). Hemicellulose syrup (hydrolysate) was recovered using a screw press, discarding solids. After removal of fine particulates with a Whatman GF/D glass fiber filter, clarified hydrolysate was stored at 4 °C (pH 2.0). Hydrolysate was adjusted to pH 9.0 (5 M ammonium hydroxide) and stored for 16 h (22 °C) before use in fermentations, declining to pH 7.5. Batch fermentations (300 mL) were conducted in pH-controlled vessels containing 210 mL hemicelluloses hydrolysate supplemented with 0.5 mM sodium metabisulfite, components of AM1 medium (38), and inoculum. Potassium bicarbonate (100 mM) was included for succinate production. Final hydrolysate medium contained 36 g/L total sugar (primarily xylose), furfural 1.2 g/L, hydroxymethylfurfural 0.071 g/L, formic acid 1.1 g/L, and acetic acid 3.2 g/L. Precultures and seed cultures were prepared as described above. After 20 h of incubation, seed cultures were used to provide a starting inoculum of 66 mg for hemicelluloses hydrolysate fermentations producing succinate or 13 mg for ethanol. Fermentations were maintained at pH 7.0 by the automatic addition of base (2.4 M K2CO3 in 1.2 M KOH for succinate or 2 M KOH for ethanol).
Supplementary Material
Acknowledgments
This research was supported by US Department of Agriculture Grant 2011-10006-30358 and by the Myriant Corporation.
Footnotes
Conflict of interest statement: L.O.I. has less than 4% stock ownership in a company currently producing monomers for renewable plastics from edible sugars. This company hopes to use less expensive, wood-derived sugars that do not compete with food in the future.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217958110/-/DCSupplemental.
References
- 1.Carole TM, Pellegrino J, Paster MD. Opportunities in the industrial biobased products industry. Appl Biochem Biotechnol. 2004;115:871–885. doi: 10.1385/abab:115:1-3:0871. [DOI] [PubMed] [Google Scholar]
- 2.Gírio FM, et al. Hemicelluloses for fuel ethanol: A review. Bioresour Technol. 2010;101(13):4775–4800. doi: 10.1016/j.biortech.2010.01.088. [DOI] [PubMed] [Google Scholar]
- 3.Saha BC. Hemicellulose bioconversion. J Ind Microbiol Biotechnol. 2003;30(5):279–291. doi: 10.1007/s10295-003-0049-x. [DOI] [PubMed] [Google Scholar]
- 4.Alvira P, Tomás-Pejó E, Ballesteros M, Negro MJ. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: A review. Bioresour Technol. 2010;101(13):4851–4861. doi: 10.1016/j.biortech.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 5.Harris EE. Wood saccharification. Adv Carbohydr Chem. 1949;4:153–188. [Google Scholar]
- 6.Geddes CC, Nieves IU, Ingram LO. Advances in ethanol production. Curr Opin Biotechnol. 2011;22(3):312–319. doi: 10.1016/j.copbio.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Almeida JR, Bertilsson M, Gorwa-Grauslund MF, Gorsich S, Lidén G. Metabolic effects of furaldehydes and impacts on biotechnological processes. Appl Microbiol Biotechnol. 2009;82(4):625–638. doi: 10.1007/s00253-009-1875-1. [DOI] [PubMed] [Google Scholar]
- 8.Mills TY, Sandoval NR, Gill RT. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol Biofuels. 2009;2:26. doi: 10.1186/1754-6834-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez A, Rodriguez ME, York SW, Preston JF, Ingram LO. Effects of Ca(OH)(2) treatments (“overliming”) on the composition and toxicity of bagasse hemicellulose hydrolysates. Biotechnol Bioeng. 2000;69(5):526–536. doi: 10.1002/1097-0290(20000905)69:5<526::aid-bit7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 10.Martinez A, et al. Detoxification of dilute acid hydrolysates of lignocellulose with lime. Biotechnol Prog. 2001;17(2):287–293. doi: 10.1021/bp0001720. [DOI] [PubMed] [Google Scholar]
- 11.Abbott DA, Zelle RM, Pronk JT, van Maris AJ. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: Current status and challenges. FEMS Yeast Res. 2009;9(8):1123–1136. doi: 10.1111/j.1567-1364.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- 12.Jarboe LR, et al. Metabolic engineering for production of biorenewable fuels and chemicals: Contributions of synthetic biology. J Biomed Biotechnol. 2010;2010:761042. doi: 10.1155/2010/761042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaldivar J, Martinez A, Ingram LO. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng. 1999;65(1):24–33. doi: 10.1002/(sici)1097-0290(19991005)65:1<24::aid-bit4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Taherzadeh MJ, Gustafsson L, Niklasson C, Lidén G. Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2000;53(6):701–708. doi: 10.1007/s002530000328. [DOI] [PubMed] [Google Scholar]
- 15.Liu ZL, Blaschek HP. Biomass conversion inhibitors and in situ detoxification. In: Vertès AA, Blaschek HP, Yukawa H, editors. Biomass to Biofuels: Strategies for Global Industries. West Sussex, United Kingdom: John Wiley and Sons; 2010. pp. 233–259. [Google Scholar]
- 16.Liu ZL, Moon J, Andersh BJ, Slininger PJ, Weber S. Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2008;81(4):743–753. doi: 10.1007/s00253-008-1702-0. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, et al. Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl Environ Microbiol. 2011;77(15):5132–5140. doi: 10.1128/AEM.05008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller EN, et al. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl Environ Microbiol. 2009;75(19):6132–6141. doi: 10.1128/AEM.01187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller EN, et al. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl Environ Microbiol. 2009;75(13):4315–4323. doi: 10.1128/AEM.00567-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner PC, et al. YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance. J Ind Microbiol Biotechnol. 2011;38(3):431–439. doi: 10.1007/s10295-010-0787-5. [DOI] [PubMed] [Google Scholar]
- 21.Miller EN, Turner PC, Jarboe LR, Ingram LO. Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180. Biotechnol Lett. 2010;32(5):661–667. doi: 10.1007/s10529-010-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Miller EN, Yomano LP, Shanmugam KT, Ingram LO. Increased furan tolerance in Escherichia coli due to a cryptic ucpA gene. Appl Environ Microbiol. 2012;78(7):2452–2455. doi: 10.1128/AEM.07783-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawers RG. Differential turnover of the multiple processed transcripts of the Escherichia coli focA-pflB operon. Microbiology. 2006;152(8):2197–2205. doi: 10.1099/mic.0.28951-0. [DOI] [PubMed] [Google Scholar]
- 24.Turner PC, et al. Optical mapping and sequencing of the Escherichia coli KO11 genome reveal extensive chromosomal rearrangements, and multiple tandem copies of the Zymomonas mobilis pdc and adhB genes. J Ind Microbiol Biotechnol. 2012;39(4):629–639. doi: 10.1007/s10295-011-1052-2. [DOI] [PubMed] [Google Scholar]
- 25.Yomano LP, York SW, Zhou S, Shanmugam KT, Ingram LO. Re-engineering Escherichia coli for ethanol production. Biotechnol Lett. 2008;30(12):2097–2103. doi: 10.1007/s10529-008-9821-3. [DOI] [PubMed] [Google Scholar]
- 26.Jarboe LR, Grabar TB, Yomano LP, Shanmugan KT, Ingram LO. Development of ethanologenic bacteria. Adv Biochem Eng Biotechnol. 2007;108:237–261. doi: 10.1007/10_2007_068. [DOI] [PubMed] [Google Scholar]
- 27.Jantama K, et al. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng. 2008;101(5):881–893. doi: 10.1002/bit.22005. [DOI] [PubMed] [Google Scholar]
- 28.Sandoval NR, et al. Strategy for directing combinatorial genome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2012;109(26):10540–10545. doi: 10.1073/pnas.1206299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke DM, Bragg PD. Purification and properties of reconstitutively active nicotinamide nucleotide transhydrogenase of Escherichia coli. Eur J Biochem. 1985;149(3):517–523. doi: 10.1111/j.1432-1033.1985.tb08955.x. [DOI] [PubMed] [Google Scholar]
- 30.Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4(6):457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 31.Nam H, Conrad TM, Lewis NE. The role of cellular objectives and selective pressures in metabolic pathway evolution. Curr Opin Biotechnol. 2011;22(4):595–600. doi: 10.1016/j.copbio.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinlay JB, Vieille C, Zeikus JG. Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol. 2007;76(4):727–740. doi: 10.1007/s00253-007-1057-y. [DOI] [PubMed] [Google Scholar]
- 33.Liu R, et al. Fermentation of xylose to succinate by enhancement of ATP supply in metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2012;94(4):959–968. doi: 10.1007/s00253-012-3896-4. [DOI] [PubMed] [Google Scholar]
- 34.Li Q, et al. Efficient conversion of crop stalk wastes into succinic acid production by Actinobacillus succinogenes. Bioresour Technol. 2010;101(9):3292–3294. doi: 10.1016/j.biortech.2009.12.064. [DOI] [PubMed] [Google Scholar]
- 35.Zheng P, Dong JJ, Sun ZH, Ni Y, Fang L. Fermentative production of succinic acid from straw hydrolysate by Actinobacillus succinogenes. Bioresour Technol. 2009;100(8):2425–2429. doi: 10.1016/j.biortech.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 36.Borges ER, Pereira N., Jr Succinic acid production from sugarcane bagasse hemicellulose hydrolysate by Actinobacillus succinogenes. J Ind Microbiol Biotechnol. 2011;38(8):1001–1011. doi: 10.1007/s10295-010-0874-7. [DOI] [PubMed] [Google Scholar]
- 37.Lee PC, Lee SY, Hong SH, Chang HN, Park SC. Biological conversion of wood hydrolysate to succinic acid by Anaerobiospirillum succiniciproducens. Biotechnol Lett. 2003;25(2):111–114. doi: 10.1023/a:1021907116361. [DOI] [PubMed] [Google Scholar]
- 38.Martinez A, et al. Low salt medium for lactate and ethanol production by recombinant Escherichia coli B. Biotechnol Lett. 2007;29(3):397–404. doi: 10.1007/s10529-006-9252-y. [DOI] [PubMed] [Google Scholar]
- 39.Martinez A, Rodriguez ME, York SW, Preston JF, Ingram LO. Use of UV absorbance To monitor furans in dilute acid hydrolysates of biomass. Biotechnol Prog. 2000;16(4):637–641. doi: 10.1021/bp0000508. [DOI] [PubMed] [Google Scholar]
- 40.Geddes CC, et al. Simplified process for ethanol production from sugarcane bagasse using hydrolysate-resistant Escherichia coli strain MM160. Bioresour Technol. 2011;102(3):2702–2711. doi: 10.1016/j.biortech.2010.10.143. [DOI] [PubMed] [Google Scholar]
- 41.Varga E, Réczey K, Zacchi G. Optimization of steam pretreatment of corn stover to enhance enzymatic digestibility. Appl Biochem Biotechnol. 2004;114:509–523. doi: 10.1385/abab:114:1-3:509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.