Summary
Chlamydia pneumoniae is responsible for a high prevalence of respiratory infections worldwide and has been implicated in atherosclerosis. Inflammation is regulated by transcription factor (TF) networks. Yet, the core TF network triggered by chlamydiae remains largely unknown. Primary human coronary artery endothelial cells were mock-infected or infected with C. pneumoniae to generate human transcriptome data throughout the chlamydial developmental cycle. Using systems network analysis, the predominant TF network involved receptor, binding and adhesion, and immune response complexes. Cells transfected with interfering RNA against activator protein-1 (AP-1) members FOS, FOSB, JUN and JUNB had significantly decreased expression and protein levels of inflammatory mediators interleukin (IL)6, IL8, CD38 and tumor necrosis factor compared with controls. These mediators have been shown to be associated with C. pneumoniae disease. Expression of AP-1 components was regulated by MAPK3K8, a MAPK pathway component. Additionally, knockdown of JUN and FOS showed significantly decreased expression of Toll-like receptor (TLR)3 during infection, implicating JUN and FOS in TLR3 regulation. TLR3 stimulation led to elevated IL8. These findings suggest that C. pneumoniae initiates signaling via TLR3 and MAPK that activate AP-1, a known immune activator in other bacteria not previously shown for chlamydiae, triggering inflammation linked to C. pneumoniae disease.
Introduction
Inflammation is a natural biological response to stimuli such as microbial infection. Chronic inflammation, however, can lead to severe human diseases such as atherosclerosis and cancer (Karin et al., 2006). The extent of inflammation is tightly controlled by transcription factors (TFs). For example, the NF-κB complex of TFs mediates inflammation involved in antimicrobial bioprocesses and disease pathogenesis (Pikarsky et al., 2004). Yet, the key TFs that control inflammation initiated by a particular pathogen remain elusive.
Chlamydia pneumoniae is an obligate intracellular bacterial pathogen that is responsible for a high prevalence of upper and lower respiratory tract infections worldwide (Campbell et al., 2004). C. pneumoniae has also been implicated in atherosclerosis based on compelling evidence from in vitro and animal studies in addition to data from human populations (Selzman et al., 2003, Belland et al., 2004, Campbell et al., 2004). Some of the most persuasive evidence comes from the direct initiation or exacerbation of vascular lesions in rabbit and murine models in response to C. pneumoniae infection (Selzman et al., 2003, Belland et al., 2004, Campbell et al., 2004).
Human hosts activate multiple TFs such as AP-1 and NF-κB that mediate inflammation (Dechend et al., 1999, Miller et al., 2000, Huang et al., 2008). The NF-κB complex has been regarded as a major TF complex in the regulation of inflammation caused by C. pneumoniae infection (Dechend et al., 1999). However, recent discoveries reveal that NF-κB is not necessary for the stimulation of inflammation during C. trachomatis infection (Misaghi et al., 2006, Le Negrate et al., 2008). This organism actually stimulates the formation of a protein complex, ChlaDub1, to inhibit NF-κB activation (Misaghi et al., 2006, Le Negrate et al., 2008), suggesting that NF-κB may not be the most important complex controlling inflammation initiated by chlamydiae (Le Negrate et al.), although this has not yet been shown for C. pneumoniae. Nevertheless, with or without NF-κB, the human host activates a variety of inflammatory responses and releases various cytokines to cope with C. pneumoniae infection. For example, IL-8, which contains promoters for multiple TFs including binding sites for AP-1 and NF-κB, is released during C. pneumoniae infection of endothelial cells (Krull et al., 2004) and is linked to inflammatory disease development at this site (Campbell et al., 2004, Krull et al., 2004). This suggests that at least one alternative TF complex other than NF-κB is activated in response to C. pneumoniae.
An obvious candidate is the AP-1 complex, a ubiquitous dimeric protein complex composed of Jun (for example, JUN) and Fos (for example, FOS) subfamilies (Curran et al., 1988). AP-1 is commonly activated during microbial infection with bacteria and viruses (Seo et al., 2004, Xie et al., 2005). In in vitro studies, C. pneumoniae infection of human vascular smooth muscle cells induced both NF-κB and AP-1 (Miller et al., 2000). In the murine model, the product of the immediate early gene c-Fos (FOS), a subunit of AP-1, is activated in the heart during C. pneumoniae infection (Huang et al., 2008).
Similar to NF-κB, AP-1 contains transcriptional regulator binding sites for most inflammatory mediators, and AP-1 can also bind promoters of inflammatory mediators independent of NF-κB during inflammation (Cho et al., 2001, Balasubramanian et al., 2003). Therefore, AP-1 independently mediates the release of inflammatory mediators such as IL8 (Yeo et al., 2004). AP-1 also interacts with other TFs to modulate expression of inflammatory mediators during infection with pathogens such as Group B Streptococcus (Vallejo et al., 2000). AP-1 functions vary depending on the combination of AP-1 complex components and conditions. The FOS and JUN complexes normally function as positive factors in regulating inflammation (Shaulian et al., 2002). However, the functional complexity of AP-1 remains largely elusive.
In the present study, we employed a systems biology network approach and identified AP-1 as the core complex modulating the fundamental processes of inflammation, including inflammatory signaling initiation, the signaling cascade, and inflammatory mediator releases, in response to C. pneumoniae infection. The model predicted a role for TLR3 in initiating the inflammatory response, which was confirmed by infecting cells expressing different TLRs. Our results provide a unifying mechanism by which C. pneumoniae regulates inflammation.
Results
AP-1 members are the core components of a TF network Initiated by C. pneumoniae infection
To systematically decode a TF network involved in inflammation initiated by C. pneumoniae infection, we applied a protein-based network approach to analyze expression profiling altered by C. pneumoniae infection. We first assembled a protein interaction network by integrating the known protein-interaction databases as we previously published (Wang et al., 2010). We then mapped the network with the transcriptome data that was significantly altered by C. pneumoniae infection in HCAEC, a primary cell line used as a model for studying arthrosclerosis and C. pneumoniae infection (Molestina et al., 1998, Kaul et al., 2001, Hogdahl et al., 2008). HCAEC were infected with live C. pneumoniae at five time points, representing the developmental cycle of the organism. The time points corresponded to bacterial attachment (5 min post infection), entry (25 min), initial transformation into metabolically active reticulate bodies and replication (2 hrs), metabolism and replication (24 hrs), and transformation into infectious particles for release and infection of adjacent cells (60 hrs). The transcription alterations during infection with living bacteria were compared with those from UV-treated C. pneumoniae and mock-infected HCAEC.
The TFs with significantly altered expression during C. pneumoniae infection were mapped to their corresponding proteins in the above network to integrate the gene expression profiling and network database as we previously described (Wang et al., 2010). The overlaid network became a dynamic TF network activated by C. pneumoniae infection (Figure 1A and Figure 2).
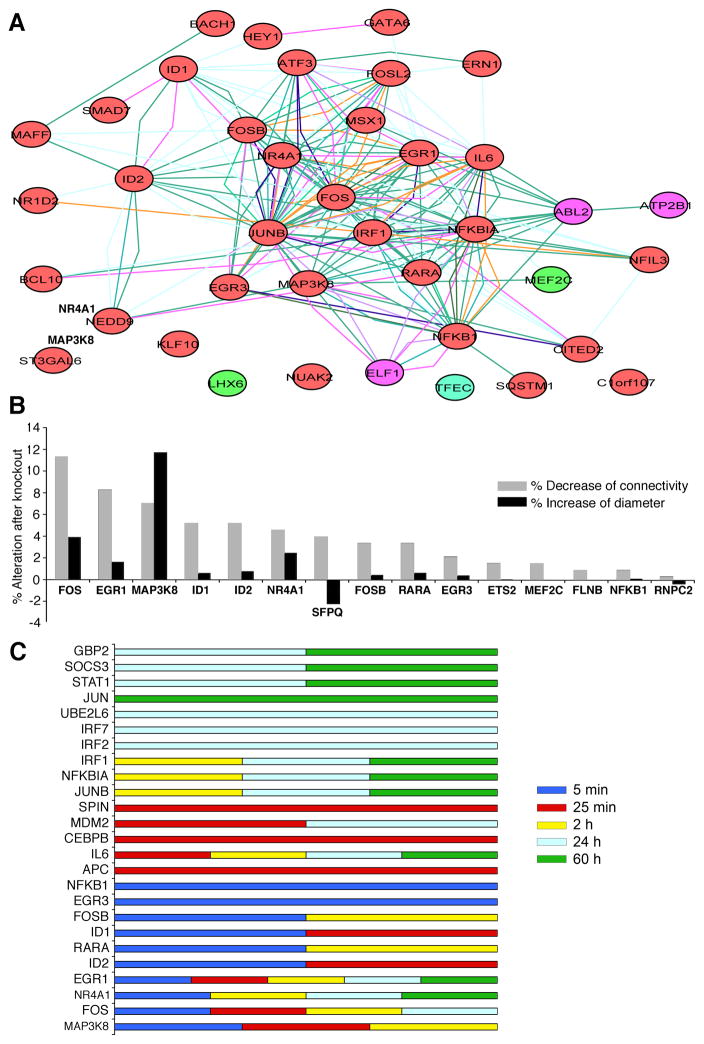
Fig. 1. AP-1 components dominate the Transcription Factor (TF) network initiated by C. pneumoniae infection of HCAEC.
A. AP-1 components, FOS, FOSB and JUNB dominate the TF network initiated by C. pneumoniae infection. The TF network was derived from a genome-wide protein interaction network overlapped with transcriptome data from this study (see Material and Methods). Nodes and edges donate proteins and interactions, respectively. The color of the node represents the gene expression level: Red for up-regulation, green for down-regulation and violet for equivocal at 5 mim..
B. The contribution of TFs to network connectivity and to the diameter of the network activated at 5 min post infection. TFs were systematically knocked out in silico, and the alterations in network connectivity (measured as average number of neighbors) and network diameter (measured as average shortest path length) were calculated (see Materials and Methods). The top TFs that contributed most to the network are shown. FOS and MAP3K8 contributed the most to both connectivity and diameter, respectively, suggesting that AP-1 dominates the TF network for C. pneumoniae.
C. Summary of the top 10 TFs from our transcriptome data at each time point that contributed the most to the diameter of the network activated at 5 min, 25 min, 2H, 24H, and 60H. From top to bottom, GBP2, SOCS3, STAT1, JUN, UBE2L6, IRF7, and IRF2 predominated in late infection (24 hrs and 60 hrs), while IRF1, NFKIA, and JUNB were featured at 2 hrs, 24 hrs and 60 hrs. SPIN, MDM2, CEBPB, and APC predominated at 25 min. EGR1, NR4A1, IL6, and FOS were primarily activated from 5 min to 24 hrs or to 60 hrs.
Fig. 2. In silico knockout of hubs and bottlenecks in the TF network.
A. Hubs and bottleneck and their neighbors for the wild-type network activated at 5 min. The green arrow highlights a hub (FOS). The pink arrow highlights a bottleneck (SFPQ).
B. The network was locally disconnected after the combination of knocking out the first 5 genes, FOS, EGR1, MAP3K8, ID1, and ID2, compared to the wild-type network shown in (A).
To identify the key TFs in the infection model, we searched for the hubs and bottlenecks (Yu et al., 2007) in the TF network as we previously described (Wang et al., 2010). We systematically in silico knocked out each TF and examined their contribution to network connectivity—the distance between a node and every other node in the transmission range—calculated as the average number of neighbors (that is, average distance). The TFs that contributed most to the connectivity of the network at 5min were FOS (v-fos FBJ murine osteosarcoma viral oncogene homolog), EGR1 (early growth response), and MAP3K8 (mitogen-activated protein kinase kinase kinase 8) (Figure 1B, grey bars). In addition, we further calculated the contribution of a single TF to the network diameter—the average of the shortest path length, which essentially characterized the network’s interconnectivity (Wang et al., 2010). The longer the diameter, the less interconnectivity there is in the network. Knocking out a hub (highly connected protein) would increase the diameter because of the loss of short paths in a network, whereas knocking out a bottleneck [node with many short paths going through them, analogous to key bridges that link sub-networks to a whole map network] (Yu et al., 2007), would decrease the diameter because the network would be broken down and the long path that normally links sub-networks would be lost. Knocking out FOS dramatically increased the network diameter (Figure 1B, black bar), indicating that it is a hub in this network. However, knocking out SFPQ (splicing factor proline/glutamine rich - polypyrimidine tract binding protein associated) remarkably decreased the network diameter (Figure 1B, black bar), and SFPQ also notably contributed to network connectivity (Figure 1B, grey bar), suggesting that SFPQ is a bottleneck in this network. Deleting hub FOS (Figure 2, green arrow) caused a huge loss in network connectivity. This suggested that the FOS hub and SFPQ bottleneck (Figure 2, pink arrow) are critical coordinators in the network activated by C. pneumoniae at 5 min.
Similarly, the top 10 nodes that contributed to the network diameter for the rest of the time points are summarized in Figure 1C. The top two hubs at 25min were identified as FOS and AP-C (Activated Protein C), FOS and JUNB at 2 hrs, FOS and STAT1 at 24 hrs, and JUN and STAT1 at 60 hrs (Figure 1C). Clearly, from the frequency of each TF in the entire profiling, FOS and JUN predominated in these dynamic networks, indicating that the AP-1 complex is likely the core component of the inflammatory network initiated by C. pneumoniae. The microarray data has been deposited in GEO. The accession number is GSE27008 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=vduzheioawcigtm&acc=GSE27008).
AP-1 regulatory network
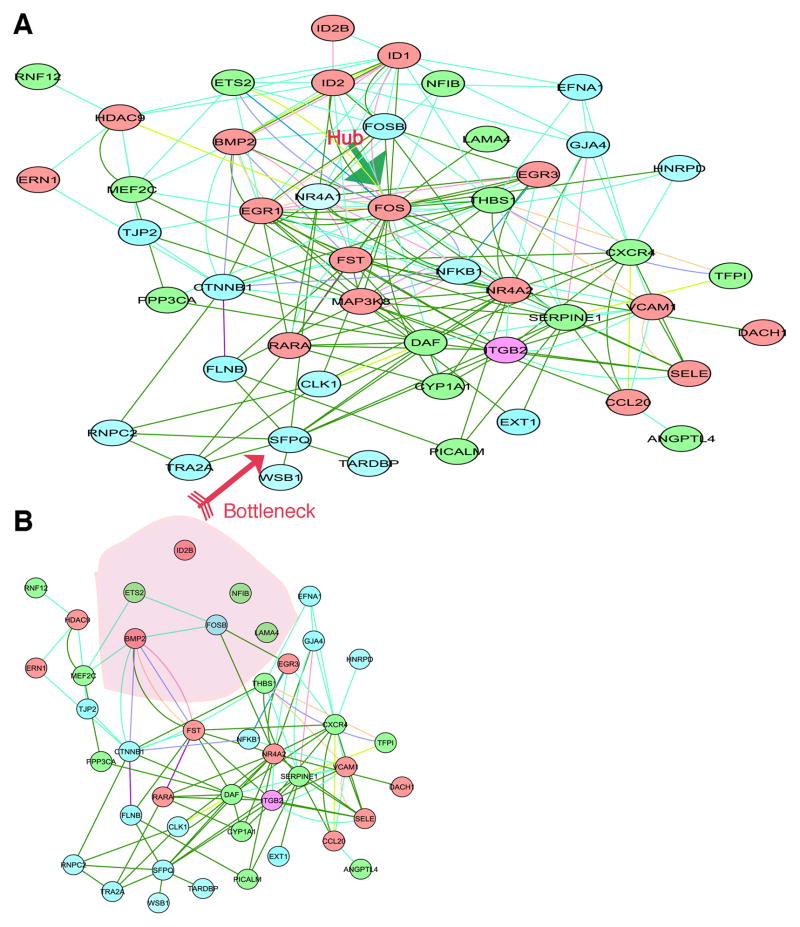
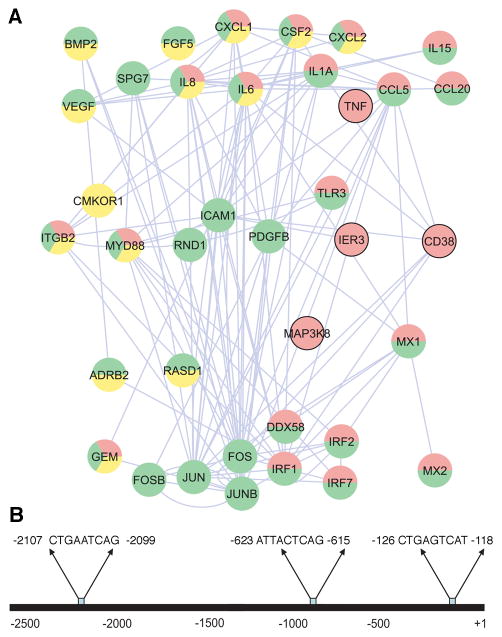
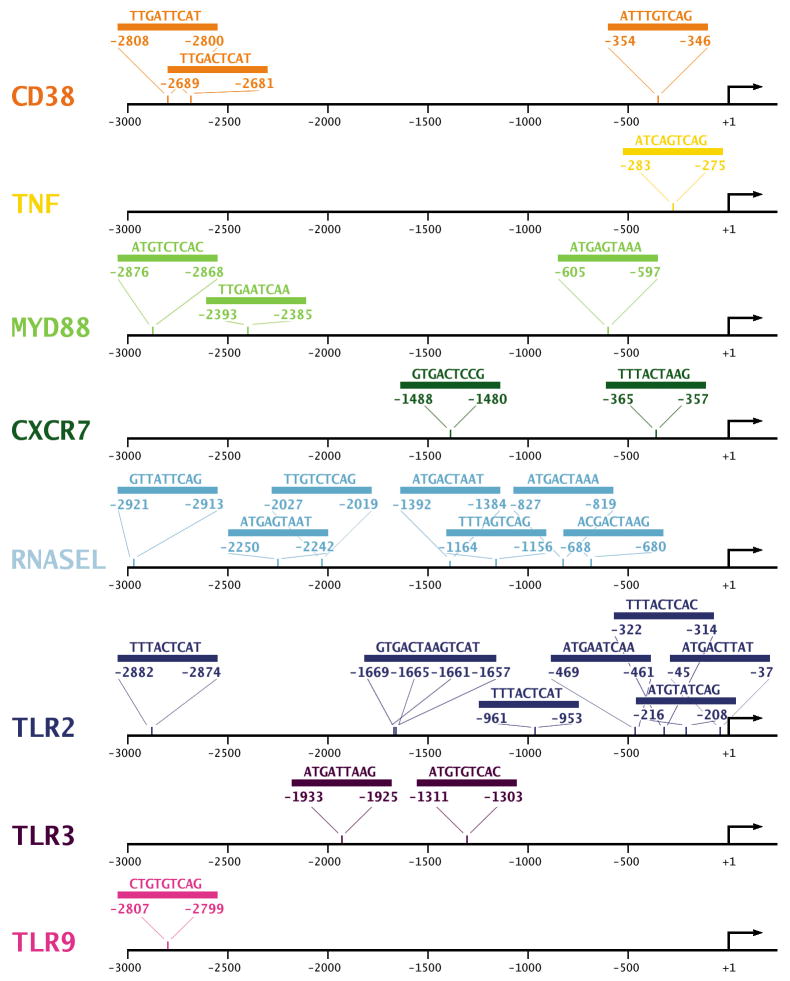
After deducing that the AP-1 complex is likely the core component of the inflammatory network stimulated by C. pneumoniae, we searched for additional members of the AP-1 regulatory network. We mapped the known protein–protein interactions with inflammatory genes that are associated with the AP-1 complex and that were significantly altered after C. pneumoniae infection of HCAEC (Figure 3A). Special attention was paid to genes that have putative AP-1 binding motifs predicted from the motif-binding matrix (http://xerad.systemsbiology.net/MotifMogulServer/) with constraint criteria as shown in Figure 4. Most of these inflammatory genes exhibit more than two putative AP-1 binding sites located within 3000 base pairs upstream of the transcriptional start sites. Examples are shown in Figure 3B and Figure 4. The mapped network containing these AP-1 targets became an AP-1 regulatory network that was further classified into functional protein complexes based on gene functions and cellular locations in the gene ontology database (Figure 3A) (www.geneontology.org). This network, then, contained a number of primary complexes central to inflammation, including receptor (yellow), binding and adhesion (green), and immune response (red) complexes (Figure 3A). These protein interactions have largely been known to be dynamically activated by the AP-1 complex. For example, the features of inflammation induced by C. pneumoniae infections in in vitro studies and murine models, such as inflammatory mediators IL6, IL8, CD38, and TNFα (Selzman et al., 2003, Belland et al., 2004, Campbell et al., 2004) and key signaling kinases like MAP3K8, were among the factors included in these complexes.
Fig. 3. AP-1 target gene predictions using the protein-protein interaction network and promoter sequence analysis.
A. Predicting AP-1-associated genes using the protein interaction network. The genes were determined from the network assembled from known transcription factor protein–protein interactions and transcriptome data as described in Materials and Methods. This network was organized by the gene ontology database, in which gene functions and localizations were included. AP-1 (nucleus) is predicted to interact with a number of major functional modules, including those for receptor complexes (yellow), immune response (red), and binding and adhesion (green).
B. Computational analysis of AP-1 binding sites for inflammatory mediators. These AP-1 binding sites were identified by scanning the 5′ upstream promoter regions of each gene with MotifMogul. For illustration, only the one for IL8 is shown; the rest are shown in Figure 4.
Fig. 4.
AP-1 binding sites for inflammatory mediators predicted by MotifMogul. MotifMogul was used to predict AP-1 binding sites for inflammatory mediators as described in detail in Materials and Methods.
AP-1 complex mediates expressions of inflammatory mediators
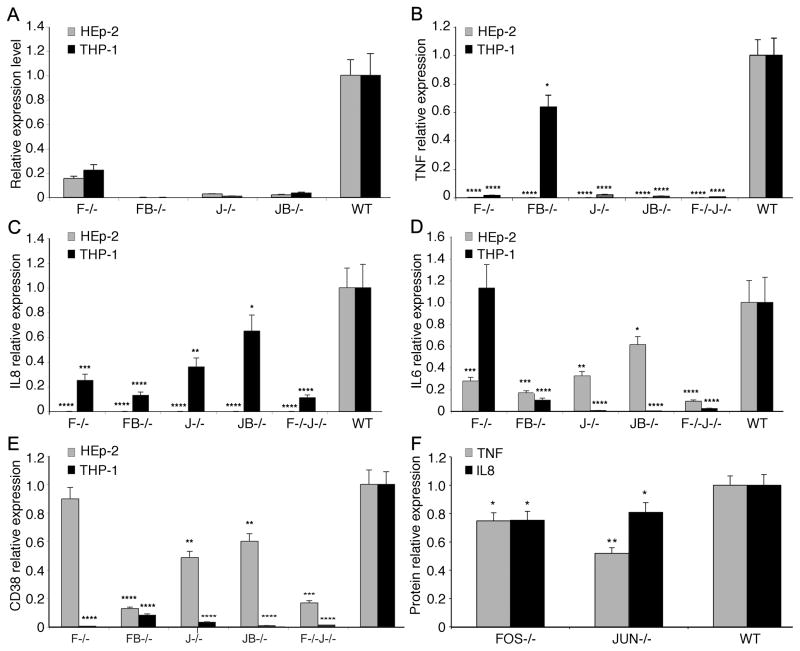
To confirm the role of AP-1 in inflammation as predicted above, we experimentally examined the effect of AP-1 on the expression of inflammatory factors IL6, IL8, TNFα, and CD38 during C. pneumoniae infection of HEp-2 and human peripheral blood monocytic leukemia (THP-1; ATCC TIB-202) cells. siRNA designed against AP-1 members FOS, FOSB, JUN, and JUNB (Table S1) were used to knock down expression of each in addition to the combination knockdown of FOS and JUN. The knockdown efficiency was determined by measuring gene expression alteration of IL6, IL8, TNFα, and CD38 by qRT-PCR (Table S2).
siRNA treatment successfully knocked down at least 75% of the gene expression for AP-1 components FOS, FOSB, JUN, and JUNB (Figure 5A). Knockdown of AP-1 significantly reduced expression of almost all examined inflammatory factors (Figure 5B–5E) in both HEp-2 and THP-1 cells except for IL6 in THP-1 cells with FOS− (Figure 5D). Notably, the combined knockdown of AP-1 components produced, in general, a greater reduction of inflammatory gene expression than any single AP-1 member knockdown (Figure 5B–5E). For example, IL6 expression was not significantly altered under single FOS knockdown in THP-1 cells, but was dramatically repressed under the combination knockdown of FOS−JUN−. These findings suggested that AP-1 dimers are crucial TFs for inflammatory mediator release during C. pneumoniae infection.
Fig. 5. AP-1 regulates gene expression of inflammatory mediators in C. pneumoniae infected HEp-2 and THP-1 cells.
A. AP-1 components FOS, FOSB, JUN and JUNB were knocked down by siRNA. The knockdown efficiency was determined by measuring gene expression alteration by qRT-PCR after siRNA knockdown of each AP-1 member (FOS, F−/−; FOSB, FB−/−; JUN, J−/−; JUNB, J−/−; FOS and JUN, F−/−J−/−) in C. pneumoniae infected HEp-2 and THP-1 cells. At least 75% of the gene expression of AP-1 members was knocked down when compared with the scrambled siRNA control.
B–E. Knockdown of AP-1 members FOS and JUN during C. pneumoniae infection significantly decreased key pro-inflammatory mediators TNF, IL8, IL6 and CD38, but not IL6, measured by qRT-PCR in both HEp-2 and THP-1 cells compared to wild type (WT).
F. Knockdown of AP-1 components JUN and FOS downregulate inflammatory factors TNF and IL8 compared to WT measured during infection. *p<0.045, **p<0.01, ***p<0.001, and ****p<0.0005.
The expression levels of selected inflammatory genes were also verified at the protein level using a Luminex multiplex protein quantitative assay (Figure 5F). In general, results measured at the protein and mRNA level were consistent. Both FOS− and JUN− resulted in significantly lower protein levels of IL8 and TNFα compared with the control. Together, our data suggest that AP-1 is a key TF complex regulating inflammation initiated by C. pneumoniae infection.
AP-1 complex mediates inflammatory signaling via TLR3 and MAP3K8
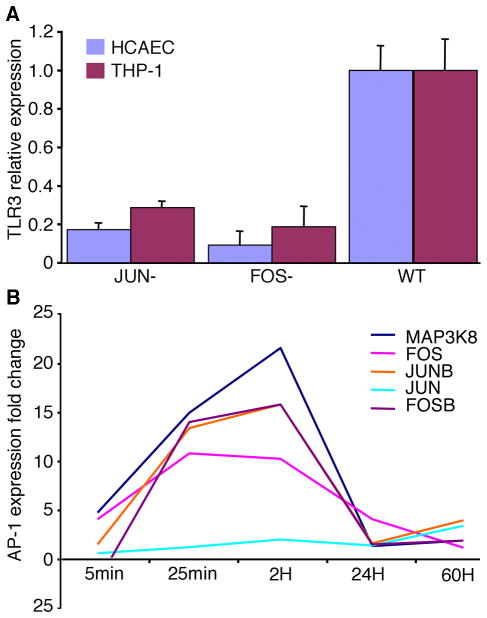
We next explored the mechanisms for AP-1 regulation of inflammation by examining the key inflammatory pathway regulated by AP-1 during C. pneumoniae infection. Inflammatory signaling is initiated by pattern recognition receptors (PRR), which then precipitate a signaling cascade via protein kinases. To identify crucial genes primarily involved in inflammation regulated by AP-1, we systematically screened genes with the highest correlation with AP-1 based on both the AP-1 target network (Figure 3) and the mathematical model analysis of co-expression profiling. Gene expression alterations of all components of the AP-1 regulatory network (Figure 3A) were measured by qRT-PCR for both over-expression of AP-1 (wild-type) and knockdown of AP-1 (FOS−, JUN− and combination FOS− JUN−) via siRNA during C. pneumoniae infection of HCAECs. A standard stepwise regression model was used to select genes with the largest partial correlation with AP-1. The results showed that AP-1 strongly associates with TLR3 and MAP3K8. Based on gene ontology (Figure 3A), TLR3 appeared to be targeted by AP-1, while MAP3K8 appeared to be the key kinase for signaling transduction. We found that JUN and FOS knockdown significantly inhibited TLR3 expression (Figure 6A) but not TLR4. TLR2 expression was similarly inhibited. By plotting the dynamic gene expression pattern of AP-1 components and MAP3K8 for 5min, 25min, 2H, 24H and 60H, we found similar expression patterns between AP-1 complex TFs and MAP3K8 in HCAECs (Figure 6B), which is consistent with the general regulatory pathway induced by pathogens as previously reported by Nijhara et al. (Nijhara et al., 2001). Together, these data demonstrate that the AP-1 complex mediates inflammatory signaling via TLR3 and MAP3K8.
Fig. 6. AP-1 Complex mediates inflammatory signaling via TLR3 and MAP3K8.
A. JUN and FOS knockdown by siRNA dramatically alters gene expression of TLR3 in C. pneumoniae infected HCAEC and THP-1 cells as measured by qRT-PCR.
B. Expression of AP-1 components correlates with MAPK signaling pathway during C. pneumoniae infection of HCAEC measured by both microarray and qRT-PCR.
TLR3 regulates activation of NF-κB and expression of the chemokine IL8
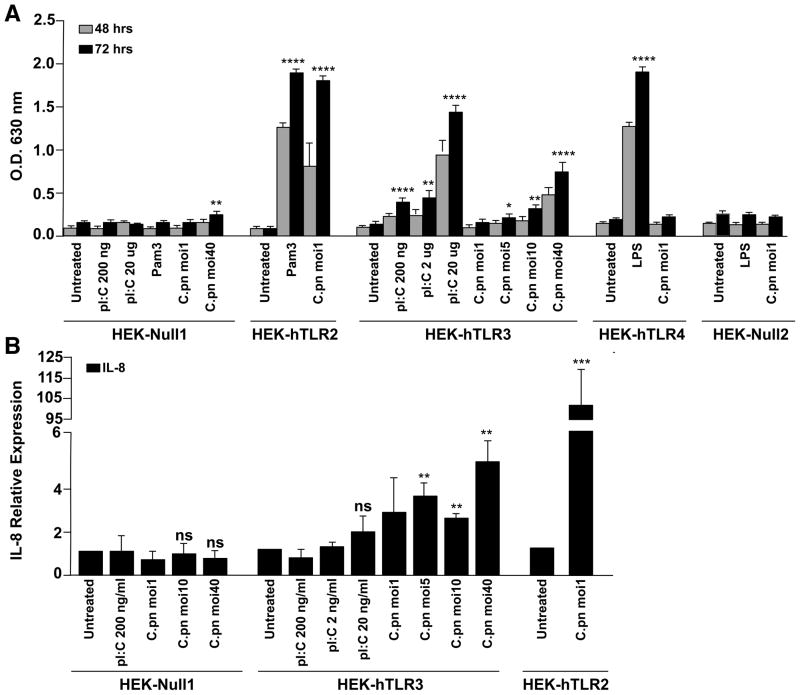
TLR2 was previously shown to stimulate the innate immune response during infection by C. muridarum and C. pneumoniae, while TLR4 is thought to have no effect (Prebeck et al., 2001, Darville et al., 2003, Naiki et al., 2005, Cao et al., 2007). A role for TLR3 has been reported for infection of an oviduct cell line by the murine pathogen, C. muridarum (Derbigny et al., 2005, Derbigny et al., 2007, Derbigny et al., 2010) but never in animal studies. TLR3 effects on infection by the human pathogens, C. trachomatis or C. pneumoniae, have never been studied. Given our unexpected finding that the AP-1 complex regulates signaling via TLR3 during C. pneumoniae infection, we screened the TLRs likely to be involved in the response to C. pneumoniae using HEK293 cells that are stably transfected with control vector or human TLR2, TLR3 or TLR4. The cells, including control cells, stably co-express an NF-κB-inducible SEAP (secreted embryonic alkaline phosphatase) reporter gene whose enzymatic activity can be conveniently and quantitatively monitored through a colorimetric assay.
As expected, HEK cells expressing NF-κB-induced reporter gene but not TLR (HEK-Null1) did not show NF-κB activation in response to stimulation with ligands for TLR2 (Pam3), TLR4 (LPS) or TLR3 (Pam3), or infection with C. pneumoniae (Figure 7A). However, C. pneumoniae infection or Pam3-stimulation induced strong NF-κB activation in HEK-TLR2 cells, in agreement with previous reports. C. pneumoniae infection had no effect in HEK-TLR4 cells, although the positive control, LPS, stimulated NF-κB activation in these cells. C. pneumoniae infection had no effect at low MOIs in TLR3-expressing cells, but activated NF-κB at significant levels at an MOI of 10 or 40. Even at the highest MOI, there was no NF-κB in the HEK-Null1 cells.
Fig. 7. TLR3 mediates NF-κB activation and IL8 expression.
A. Control HEK cells (HEK-Null) or HEK cells expressing TLR2, TLR3 or TLR4 were infected with C. pneumoniae (MOI of 1, 5, 10 or 40) or stimulated with the TLR ligands, 5 mg/mL Pam3CSK4 (TLR2 ligand), 500 ng/mL LPS (TLR4 ligand) or 200 ng/mL, 2 mg/mL or 20 mg/mL poly(I:C) (TLR3 ligand) for 48 hrs or 72 hrs. NF-κB activation was assessed by measuring SEAP activity colorimetrically at 630 nm. (
B. HEK cells expressing TLR3 or TLR2 were infected with C. pneumoniae at the indicated MOI or stimulated with poly(I:C), and IL8 expression was measured by qPCR. *p < 0.045, **p < 0.01, ***p < 0.001, and ****p < 0.0005.
The same cells were also used to measure IL8 gene expression during C. pneumoniae infection (Figure 7B). IL8 expression in HEK-TLR3 cells was observed even at low MOIs of infection. Much higher levels of expression were found in HEK-TLR2 cells, also consistent with previous studies, although TLR3-dependent IL8 production during chlamydial infection has not been reported until now. C. pneumoniae infection, even at MOI = 40, had no effect on IL8 expression in HEK cells in the absence of TLR.
Discussion
Inflammation is fundamentally modulated by TFs. However, the challenge has been to identify the critical TFs that regulate inflammation, because a complex network is usually activated during pathogenic infections. The NF-κB complex, a well-characterized TF complex, has been considered key in regulating inflammatory mediators (for example, IL6 and IL8) during chlamydial infections (Gencay et al., 2003, Buchholz et al., 2006). However, recent studies argue against this (Le Negrate et al., 2008). Significantly, chlamydial infection has been reported to interfere with NF-κB signaling (Lad et al., 2007, Le Negrate et al., 2008, Betts et al., 2009, Wolf et al., 2009).
In the present study, we employed a systems biology approach and found that AP-1 contributes to the major structure of the TF network initiated by C. pneumoniae infection (Figure 1) and regulates many inflammatory mediators (Figure 3). RNAi knockdown of single components of AP-1 or the AP-1 dimer combination significantly reduced the expression of inflammatory mediators, such as IL6, IL8, CD38 and TNFα, at both the mRNA and protein level (Figure 5), although the latter levels were not as low as might be expected given the corresponding gene knockdown. The inflammatory mediator IL8 has been reported to be regulated by NF-κB, but IL8 release also occurs through the MAPK pathway independently of the NF-κB complex during C. trachomatis infection (Lad et al., 2007). Here, we revealed that IL8 is primarily regulated by AP-1. Our findings suggest that AP-1 is a key TF complex that regulates the inflammatory network activated by C. pneumoniae infection.
AP-1 is a ubiquitous dimeric protein complex composed of different Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, Fra-1, Fra-2, and FosB) subfamilies (Curran et al., 1988) and commonly activatedduring microbial infection with bacteria and viruses (Seo et al., 2004, Xie et al., 2005). AP-1 complexes normally functionas positive factors in regulating inflammation and the cell cycle. Yet, different combinations of AP-1 members express differential biological effects. While JUN, FOS and FOSB are often positively associated with inflammation, cell growth, cellular transformation, tumor formation and tumor progression, JUNB performs a negative regulatory role in mediating cell proliferation (Shaulian et al., 2002). AP-1 mediates the release of inflammatory mediators such as IL8 (Yeo et al., 2004) and regulates angiogenesis caused by pathogen infection as in the case of herpesvirus infection (Ye et al., 2007). AP-1 also interacts with other TFs to cope with pathogen development (Ravichandran et al., 2006) and modulates expression of inflammatory mediators during infection with pathogens such as Group B Streptococcus (Vallejo et al., 2000). This suggests that the biological functions of AP-1 are complex and that much of its functional complexity remains to be elucidated.
The function of AP-1 is known to vary depending on cell type (Chinenov et al., 2001, Eferl et al., 2003). We used three different cell types to examine the AP-1 function during C. pneumoniae infection and, surprisingly, found that inflammatory mediators, IL6, IL8, CD38, and IFN, were primarily regulated by most of the AP-1 members examined here. In fact, other inflammatory mediators predicted in Figure 3A were also regulated by at least one AP-1 member (either FOS or JUN) for a given cell line. This suggests that AP-1 likely regulates inflammation stimulated by C. pneumoniae infection. Our results nicely parallel the recent observations showing that AP-1 serves as a crucial modulator in regulating inflammation initiated by other pathogens such as Group B streptococcus, Herpes simplex virus 1 and Helicobacter pylori (Zachos et al., 1999, Vallejo et al., 2000, Wu et al., 2006).
The underlying molecular mechanism(s) whereby AP-1 mediates inflammation during infections remains largely unknown. Here, our data revealed that AP-1 is activated primarily from TLR3 and MAP3K8 signaling pathways during C. pneumoniae infection. TLR3 is a host PRR that recognizes dsRNA derived from many different viral pathogens (Takeuchi et al., 2009). The interaction of dsRNA with TLR3 and its ectodomain results in dimer formation and recruitment of TIR-domain-containing adapter-inducing interferon-β (TRIF) that initiates signaling pathways and ultimately downstream TF activation, including AP-1 (Botos et al., 2009). While dsRNA is commonly produced by viral replication and is not known to be associated with chlamydiae, we speculate that it is possible that the organism generates a unique molecular ligand that is recognized by TLR3. In a murine oviduct cell line that was infected with C. muridarum, TLR3 was expressed in infected cells and served as a trigger for IFNβ production (Derbigny et al., 2005), which was subsequently found to be dependent on interferon regulatory factor 3 (IRF3) and TRIF (Derbigny et al., 2007). Recently, the IFNβ response was shown to be dependent on TLR3 in the same cell line (Derbigny et al., 2010). In another study. C. pneumoniae induced foam cell formation was influenced by TLR2 and TLR4 but not TLR3 (Chen et al., 2008). Our data revealed that TLR3 as well as interferon regulatory factors were activated during C. pneumoniae infection (Figure 3) and regulated by JUN and FOS (Figure 6). AP-1 may regulate inflammation via regulating TLR3, which appears to be one key receptor initiating inflammatory signaling triggered by C. pneumoniae infection, although signaling initiated by TLR3 can cascade via various pathways (Vercammen et al., 2008).
In addition, our data showed that MAP3K8 is a key enzyme regulating AP-1 signaling during C. pneumoniae infection (Figures 2 and 4), which is consistent with previous reports showing that AP-1 is regulated by the MAPK pathway (Chinenov et al., 2001, Eferl et al., 2003). Previous studies demonstrated that release of inflammatory mediators such as IL8 are dependent on the MAP pathway independent of NF-κB (Cho et al., 2001, Balasubramanian et al., 2003). We found that IL8 release was dependent on AP-1. Thus, our findings suggest a model wherein C. pneumoniae initiates inflammatory signaling via TLR3 and the MAPK pathway, which in turn activates AP-1, triggering inflammatory mediator and chemokine release that are linked to chlamydial disease (Darville et al., 2003, Buchholz et al., 2006, Sessa et al., 2009).
The functions of different AP-1 components could vary considerably, as inducers or inhibitors depending on specific conditions. There are many pathways that can transmit the signaling, once initiated (Figure 3). Our data showed that the FOS and JUN complex serves as a central mediator in regulating inflammation initiated by C. pneumoniae, and this complex appears to perform its functions primarily via a potential feedback loop that may interact with TLR3 and MAP3K8. This does not exclude other TF complexes or other signaling pathway components that may be involved in the inflammatory process. In fact, many of the intermediate steps and components in the complex network (Figure 3) remain to be experimentally discovered. However, we consider our results to be less biased than other biological and experimental conditions because they are drawn from a systems biology based approach using established and primary cell lines, including experimental validation. Thus, our results are supporting a role for AP-1 in regulating inflammation induced by C. pneumoniae. Our findings lay the foundation for further research to expand our understanding of the AP-1 complex and other TF networks in C. pneumoniae-stimulated inflammation using appropriate animal models and in vitro model systems. In addition, our approach will be important for identifying immunotherapeutic markers as targets for interventions to prevent the inflammatory diseases caused by C. pneumoniae such as atherosclerosis (Campbell et al., 2004) and asthma (Hahn et al., 2012). Indeed, while there are many publications regarding the role of this pathogen in atherosclerosis, the present work may also be important in understanding the pathogenesis of asthma, given the growing body of evidence supporting an association between C. pneumoniae and asthma (Metz et al., 2010, Olivares-Zavaleta et al., 2011, Senn et al., 2011, Hahn et al., 2012, Patel et al., 2012).
Experimental Procedures
Network assembly
We combined existing network databases and systems network approaches using our previously described methods (Wang et al., 2010). Briefly, our network database included proteins and interactions from BIND (http://bond.unleashedinformatics.com/Action), DIP (http://dip.doe-mbi.ucla.edu/), HPRD (http://hprd.org/), PreBIND (http://www.blueprint.org/products/prebind/index.html), curated inflammatory disease databases (Calvano et al., 2005, Reiss et al., 2005a, Reiss et al., 2005b, von Mering et al., 2005), biocarta (http://www.biocarta.com/pathfiles/h_inflamPathway.asp, http://www.biocarta.com/pathfiles/h_LairPathway.asp), KEGG (http://www.genome.jp/kegg/pathway.html), EMBL human database (http://www.embl-heidelberg.de/), and two cytokine databases (http://cytokine.medic.kumamoto-u.ac.jp/ and http://people.bu.edu/gilmore/nf-kb/). This network was then used as a basis for building a C. pneumoniae network from whole human genome microarray data of C. pneumoniae infection of HCAEC over five experimental time points representing the developmental cycle of the organism (see below).
Chlamydia pneumoniae propagation
C. pneumoniae strain A03 (a gift from Dr. James Summersgill) was previously isolated from the coronary artery of a patient with atherosclerosis (Ramirez, 1996). This clinical isolate was propagated in HEp-2 cells (ATCC CCL-23; Manassas, VA) following standard laboratory protocols as we have described previously (Ramirez, 1996, Molestina et al., 1998, Mukhopadhyay et al., 2004, Wang et al., 2010). Isolates were purified using 30% RenoCal centrifugation gradients as described (Li et al., 2005, Wang et al., 2010) to eliminate ~0.1% of contaminating human cellular material.
HCAEC culture and infection
The cell culture was performed using a previously reported method (Wang et al., 2010). Briefly, Endothelial Cell Basal Medium-2 with hEGF, Hydrocortisone, GA-1000, FBS, VEGF, hFGF-B, R3-IGF-1, and Ascorbic Acid (Clonetics™ EGM®-2-MV BulletKit, East Rutherford, NJ) were used to grow HCAEC (Clonetics™) according to the manufacturer’s instructions. The cells were grown in T25 flasks in 37°C with 5% CO2 to a confluence of ~80% and ~55%, respectively, for subsequent group 1 (0 min, 5 min, 25 min, and 2 hrs) and group 2 (24 hrs and 60 hrs) infections with C. pneumoniae. A C. pneumoniae multiplicity of infection (MOI) of 100 and 5 was used for group 1 and group 2 infections, respectively. The higher confluence and MOI for group 1 was used to ensure sufficient infection for a short period of C. pneumoniae growth for RNA extraction. The lower confluence and MOI for group 2 allowed the cells to grow and for C. pneumoniae to infect these cells for the 24 hrs and 60 hrs time points in such a way that neither the cells nor C. pneumoniae would overgrow the cell culture system, which could result in apoptosis and loss of infection. For UV-inactivation, C. pneumoniae was treated with a UV germicidal light for 3 hrs (30 w, 15 cm). HCAEC were infected as above with UV-treated C. pneumoniae at the same MOI as for viable C. pneumoniae. Mock-infected HCAEC were used as controls for each time point.
RNA extraction, microarray hybridization and array detection
Infected and uninfected HCAEC at 0 min, 5 min, 25 min, 2 hrs, 24 hrs and 60 hrs post infection were trypsinized and collected by centrifugation. RNA was purified using the RNeasy RNA purification kit (QIAGEN Inc., Valencia, CA), including an on column treatment with DNase to eliminate all traces of DNA, according to the manufacturer’s instructions and as we have previously described (Gomes et al., 2005). Affymetrix Human Genome U133 Plus 2.0 Arrays, which contain over 47,000 transcripts that completely cover the whole human genome, were employed in this study. GeneChip® One-Cycle Target Labeling and Control Reagents (Affymetrix, Santa Clara, CA) were used to process RNA and for hybridization to the microarrays following the manufacturer’s protocols. Briefly, cRNA was generated from total RNA. The resultant biotinylated cRNA was fragmented and hybridized to the microarrays. The arrays were washed, stained, and scanned with the Affymetrix scanner using the manufacturer’s recommended protocols as per the Stanford University Gene and Protein Expression Core Facility (Palo Alto, CA).
Network analysis
Network analysis was performed as we described in detail previously (Wang et al., 2010). Briefly, genes with significant alterations in gene expression compared to controls in the microarray [t test, p < 0.05, as per the Bioconductor package] (Gentleman et al., 2004) were used to overlay the network described above. These overlaid networks became the inflammatory networks activated (up- and downregulated) by C. pneumoniae. The activated networks were decomposed into functional modules based on topological interconnection intensity and gene functions (http://www.geneontology.org/) (Bader et al., 2003, Maere et al., 2005, Singhal et al., 2007a, Singhal et al., 2007b). Genes were classified according to the gene ontology database (http://www.geneontology.org/) (Garcia et al., 2007).
Standard stepwise regression, which adds and removes variables, was used to select the variables (genes, in our case). The basic method of stepwise regression is to calculate an F-statistic for each variable in the model (Hennekens et al., 1987). If the F-statistic for any variable is less than F, the variable with the smallest F is removed from the model. If no variable can be removed, the procedure attempts to add a variable. An F-statistic is calculated for each variable not yet in the model. The variable with the largest F-statistic is then added, provided its F-statistic is larger than F to enter. Adding this variable is equivalent to choosing the variable with the largest partial correlation or the variable that most effectively reduces the error sum of squares. The regression equation is then calculated, results are displayed, and the procedure continues to a new step. If no variable meets the criteria for addition to the network, the stepwise procedure is finished.
AP-1 binding sites were predicted using the motif Mogul (http://xerad.systemsbiology.net/MotifMogulServer/), with human stringent matrices, stringent MotifLocator Scans at 0.01%, p < 0.05, and others as per the default setting in the programs.
Small interfering RNA knockdown
Small interfering RNA (siRNA) knockdown experiments were performed as we previously described (Wang et al., 2010). Briefly, based on the results of the microarray analyses, siRNAs were selected to target AP-1 members, FOS, FOSB, JUN and JUNB (Table S1). To avoid cell line bias, RNAi experiments were also performed in HEp-2 and THP-1 cell lines in addition to HCAEC, which are primary cells. Since results were similar between THP-1 and HCAEC experiments, some experiments were performed with only HEp-2 and THP-1 cell lines owing to the difficulty in working with primary cells.
Cells were cultured to 60%, and siRNAs including scrambled siRNA as a control, were transfected at a 75 nM final concentration, using 0.45% Oligofectamine (Invitrogen, Carlsbad, CA) in a 48-well format. The plates were placed in a tissue culture incubator at 37°C and 5% C02. After 48 hrs of siRNA-mediated gene knockdown, the medium was removed and the cells were infected with C. pneumoniae at an MOI of 5. After an additional 24 hrs of incubation as above, each experiment was analyzed for quantitative gene expression and protein concentrations as described below. All experiments were performed in triplicate.
Quantitative RT-PCR
Quantitative RT-PCR was performed as we have previously described (Gomes et al., 2005, Wang et al., 2010). Total RNA was extracted using an RNeasy RNA isolation kit (QIAGEN). DNA was digested with RQ DNase (Promega, Madison, WI) at 37°C for 30 min, and cDNA was generated using a Reverse Transcription kit (ABI, Carlsbad, CA). Quantitative real time reverse transcriptase PCR (qRT-PCR) was run using the Power SYBR Green PCR Kit (ABI) and reagents as we described (Gomes et al., 2005, Wang et al., 2010). The primers for each analyte are shown in Table S2. Each experiment contained negative controls including no template controls, mock-infected HCAEC, HEp-2 or THP-1 cell cDNA, and RNA samples without RT. PCR parameters consisted of 1 cycle of 95°C for 15 min, followed by 40 cycles of PCR at 95°C for 15s, 55°C for 30s, and 72°C for 30s. The relative amount of target gene mRNA and 16S bacterial rRNA was normalized to beta-actin mRNA. These experiments were performed in triplicate.
Cytokine protein quantitation
Cytokine concentrations for IL2, IL4, IL6, IL8, IL10, IL12, GM-CSF, IFNγ, TNFα, and IL1β were tested using Bio-Plex 10-plex kits (Bio-Rad, Hercules, CA) as per the manufacturer’s protocols. Briefly, 50 ul of cell culture supernatant for each sample were collected and assayed in duplicate. A range (1.95–32,000 pg/mL) of standards for each cytokine were re-suspended in diluent and used to plot standard curves. Data obtained from Bio-Plex Manager software program (Bio-Rad) for standardization and standard curves were converted to Excel™ (Microsoft Corporation, Seattle, WA) to determine the final concentration for each protein.
NF-κB activity and chemokine expression in HEK-TLR cells
HEK-Blue SEAP reporter cell lines, Null1, Null2, hTLR2, hTLR3 or hTLR4 (Invivogen, San Diego, CA), were grown in T75 flasks with DMEM high glucose media supplemented with 10% FCS, 50 U/mL penicillin, 50 mg/mL streptomycin and selected with the appropriate antibiotic according to the manufacturer’s instructions. Twelve hours prior to stimulation, 1×105 HEK-Blue cells/well (24-well plate) were plated. At ~50% confluency, cells were left untreated or infected with C. pneumoniae at an MOI of 1, 5, 10 or 40; or treated with 0.2, 2.0, or 20 mg/mL of poly I:C (pI:C), 5 mg/mL Pam3SCK4, or 0.5 mg/mL ultra pure E. coli lipopolysaccharide (LPS) as indicated (Invivogen). Supernatants were collected at 48 and 72 hpi for the NF-κB activity assay. Whole cells were collected for RNA isolation (below) at 72 hpi.
Collected supernatants from each sample were mixed at a ratio of 9:1 with QUANTI-Blue solution (Invivogen) in a flat-bottom 96-well plate. The samples were incubated at 37°C for 2 hrs and then the secretion level of Secreted Embryonic Alkaline Phosphatase (SEAP) was measured at 630 nm using a spectrophotometer.
For measurement of cytokine gene expression by real-time PCR, mRNA was isolated from cells after the indicated treatments using the Qiagen RNeasy kit (QIAGEN) following the manufacturer’s instructions. The synthesis of the complementary DNA (cDNA) template was conducted according to the manufacturer’s instruction (TaqMan, Roche, Pleasanton, CA). qPCR analysis using Mx3000P (Stratagene, La Jolla, CA) was conducted in triplicates in a 20 mL final volume with Brilliant III Ultra-Fast SYBR Green qPCR master mix (Stratagene). Real-time PCR included initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 5 s, 60°C for 20 s, and one cycle of 95°C for 1 min, 55°C for 30 s, 95°C for 30 s. The average for each treatment was normalized to the activity of a house-keeping gene, GAPDH. The relative expression of IL-8 for each cell type was normalized to the untreated samples. The statistical analysis was carried out using the unpaired t test where * p < 0.045, ** p < 0.01, *** p < 0.001, and **** p < 0.0005. Data were collected from three independent experiments.
Supplementary Material
Acknowledgments
We would like to thank Iong Iong Ip, Jiaxin Li and Tigist Mehari for excellent technical assistance. This research was supported in part by the Pacific Vascular Foundation (to SCJ and DD) and by the National Institutes of Allergy and Infectious Diseases sponsored Pathogen Functional Genomics Resource Center at JCVI (to DD).
Footnotes
Conflict of Interest
The authors have no conflict of interest to declare.
Additional Supporting Information may be found in the online version of this article:
References
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian A, Ganju RK, Groopman JE. Hepatitis C virus and HIV envelope proteins collaboratively mediate interleukin-8 secretion through activation of p38 MAP kinase and SHP2 in hepatocytes. J Biol Chem. 2003;278:35755–35766. doi: 10.1074/jbc.M302889200. [DOI] [PubMed] [Google Scholar]
- Belland RJ, Ouellette SP, Gieffers J, Byrne GI. Chlamydia pneumoniae and atherosclerosis. Cell Microbiol. 2004;6:117–127. doi: 10.1046/j.1462-5822.2003.00352.x. [DOI] [PubMed] [Google Scholar]
- Betts HJ, Wolf K, Fields KA. Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr Opin Microbiol. 2009;12:81–87. doi: 10.1016/j.mib.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Botos I, Liu L, Wang Y, Segal DM, Davies DR. The toll-like receptor 3:dsRNA signaling complex. Biochim Biophys Acta. 2009;1789:667–674. doi: 10.1016/j.bbagrm.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz KR, Stephens RS. Activation of the host cell proinflammatory interleukin-8 response by Chlamydia trachomatis. Cell Microbiol. 2006;8:1768–1779. doi: 10.1111/j.1462-5822.2006.00747.x. [DOI] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Campbell LA, Kuo CC. Chlamydia pneumoniae--an infectious risk factor for atherosclerosis? Nat Rev Microbiol. 2004;2:23–32. doi: 10.1038/nrmicro796. [DOI] [PubMed] [Google Scholar]
- Cao F, Castrillo A, Tontonoz P, Re F, Byrne GI. Chlamydia pneumoniae-induced macrophage foam cell formation is mediated by Toll-like receptor 2. Infect Immun. 2007;75:753–759. doi: 10.1128/IAI.01386-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sorrentino R, Shimada K, Bulut Y, Doherty TM, Crother TR, Arditi M. Chlamydia pneumoniae-induced foam cell formation requires MyD88-dependent and -independent signaling and is reciprocally modulated by liver X receptor activation. J Immunol. 2008;181:7186–7193. doi: 10.4049/jimmunol.181.10.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Cho NH, Seong SY, Choi MS, Kim IS. Expression of chemokine genes in human dermal microvascular endothelial cell lines infected with Orientia tsutsugamushi. Infect Immun. 2001;69:1265–1272. doi: 10.1128/IAI.69.3.1265-1272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran T, Franza BR., Jr Fos and Jun: the AP-1 connection. Cell. 1988;55:395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- Darville T, O’Neill JM, Andrews CW, Jr, Nagarajan UM, Stahl L, Ojcius DM. Toll-like receptor-2, but not Toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J Immunol. 2003;171:6187–6197. doi: 10.4049/jimmunol.171.11.6187. [DOI] [PubMed] [Google Scholar]
- Dechend R, Maass M, Gieffers J, Dietz R, Scheidereit C, Leutz A, Gulba DC. Chlamydia pneumoniae infection of vascular smooth muscle and endothelial cells activates NF-kappaB and induces tissue factor and PAI-1 expression: a potential link to accelerated arteriosclerosis. Circulation. 1999;100:1369–1373. doi: 10.1161/01.cir.100.13.1369. [DOI] [PubMed] [Google Scholar]
- Derbigny WA, Hong SC, Kerr MS, Temkit M, Johnson RM. Chlamydia muridarum infection elicits a beta interferon response in murine oviduct epithelial cells dependent on interferon regulatory factor 3 and TRIF. Infect Immun. 2007;75:1280–1290. doi: 10.1128/IAI.01525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbigny WA, Johnson RM, Toomey KS, Ofner S, Jayarapu K. The Chlamydia muridarum-induced IFN-beta response is TLR3-dependent in murine oviduct epithelial cells. J Immunol. 2010;185:6689–6697. doi: 10.4049/jimmunol.1001548. [DOI] [PubMed] [Google Scholar]
- Derbigny WA, Kerr MS, Johnson RM. Pattern recognition molecules activated by Chlamydia muridarum infection of cloned murine oviduct epithelial cell lines. J Immunol. 2005;175:6065–6075. doi: 10.4049/jimmunol.175.9.6065. [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Garcia M, Jemal A, Ward EM, Center MM, Hao Y, Siegel RL, Thun MJ. Global Cancer Facts & Figures 2007. Atlanta, GA: American Cancer Society; 2007. [Google Scholar]
- Gencay MM, Tamm M, Glanville A, Perruchoud AP, Roth M. Chlamydia pneumoniae activates epithelial cell proliferation via NF-kappaB and the glucocorticoid receptor. Infect Immun. 2003;71:5814–5822. doi: 10.1128/IAI.71.10.5814-5822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes JP, Hsia RC, Mead S, Borrego MJ, Dean D. Immunoreactivity and differential developmental expression of known and putative Chlamydia trachomatis membrane proteins for biologically variant serovars representing distinct disease groups. Microbes Infect. 2005;7:410–420. doi: 10.1016/j.micinf.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Hahn DL, Schure A, Patel K, Childs T, Drizik E, Webley W. Chlamydia pneumoniae-specific IgE is prevalent in asthma and is associated with disease severity. PLoS One. 2012;7:e35945. doi: 10.1371/journal.pone.0035945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennekens CH, Buring JE. Medicine (Baltimore) Boston: Little, Brown and Co; 1987. Epidemiology in Medicine; pp. 1–383. [Google Scholar]
- Hogdahl M, Soderlund G, Kihlstrom E. Expression of chemokines and adhesion molecules in human coronary artery endothelial cells infected with Chlamydia (Chlamydophila) pneumoniae. APMIS. 2008;116:1082–1088. doi: 10.1111/j.1600-0463.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- Huang B, Dong Y, Cheng Y, Xie Q, Lin G, Wu Y, et al. Chlamydia pneumoniae infection and hyperlipidaemia-induced expression of P50 and c-Fos in the heart of C57BL6J mice. Acta Cardiol. 2008;63:175–179. doi: 10.2143/AC.63.2.2029525. [DOI] [PubMed] [Google Scholar]
- Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Kaul R, Wenman WM. Chlamydia pneumoniae facilitates monocyte adhesion to endothelial and smooth muscle cells. Microb Pathog. 2001;30:149–155. doi: 10.1006/mpat.2000.0420. [DOI] [PubMed] [Google Scholar]
- Krull M, Kramp J, Petrov T, Klucken AC, Hocke AC, Walter C, et al. Differences in cell activation by Chlamydophila pneumoniae and Chlamydia trachomatis infection in human endothelial cells. Infect Immun. 2004;72:6615–6621. doi: 10.1128/IAI.72.11.6615-6621.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad SP, Li J, da Silva Correia J, Pan Q, Gadwal S, Ulevitch RJ, Li E. Cleavage of p65/RelA of the NF-kappaB pathway by Chlamydia. Proc Natl Acad Sci U S A. 2007;104:2933–2938. doi: 10.1073/pnas.0608393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Negrate G, Krieg A, Faustin B, Loeffler M, Godzik A, Krajewski S, Reed JC. ChlaDub1 of Chlamydia trachomatis suppresses NF-κB activation and inhibits IκBα ubiquitination and degradation. Cell Microbiol. 2008;10:1879–1892. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- Li D, Vaglenov A, Kim T, Wang C, Gao D, Kaltenboeck B. High-yield culture and purification of Chlamydiaceae bacteria. J Microbiol Methods. 2005;61:17–24. doi: 10.1016/j.mimet.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Metz G, Kraft M. Effects of atypical infections with Mycoplasma and Chlamydia on asthma. Immunol Allergy Clin North Am. 2010;30:575–585. doi: 10.1016/j.iac.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Selzman CH, Shames BD, Barton HA, Johnson SM, Harken AH. Chlamydia pneumoniae activates nuclear factor kappaB and activator protein 1 in human vascular smooth muscle and induces cellular proliferation. J Surg Res. 2000;90:76–81. doi: 10.1006/jsre.2000.5847. [DOI] [PubMed] [Google Scholar]
- Misaghi S, Balsara ZR, Catic A, Spooner E, Ploegh HL, Starnbach MN. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol. 2006;61:142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- Molestina RE, Dean D, Miller RD, Ramirez JA, Summersgill JT. Characterization of a strain of Chlamydia pneumoniae isolated from a coronary atheroma by analysis of the omp1 gene and biological activity in human endothelial cells. Infect Immun. 1998;66:1370–1376. doi: 10.1128/iai.66.4.1370-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Clark AP, Sullivan ED, Miller RD, Summersgill JT. Detailed protocol for purification of Chlamydia pneumoniae elementary bodies. J Clin Microbiol. 2004;42:3288–3290. doi: 10.1128/JCM.42.7.3288-3290.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki Y, Michelsen KS, Schroder NW, Alsabeh R, Slepenkin A, Zhang W, et al. MyD88 is pivotal for the early inflammatory response and subsequent bacterial clearance and survival in a mouse model of Chlamydia pneumoniae pneumonia. J Biol Chem. 2005;280:29242–29249. doi: 10.1074/jbc.M503225200. [DOI] [PubMed] [Google Scholar]
- Nijhara R, Jana SS, Goswami SK, Rana A, Majumdar SS, Kumar V, Sarkar DP. Sustained activation of mitogen-activated protein kinases and activator protein 1 by the hepatitis B virus X protein in mouse hepatocytes in vivo. J Virol. 2001;75:10348–10358. doi: 10.1128/JVI.75.21.10348-10358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Zavaleta N, Carmody A, Messer R, Whitmire WM, Caldwell HD. Chlamydia pneumoniae inhibits activated human T lymphocyte proliferation by the induction of apoptotic and pyroptotic pathways. J Immunol. 2011;186:7120–7126. doi: 10.4049/jimmunol.1100393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KK, Anderson E, Salva PS, Webley WC. The prevalence and identity of Chlamydia-specific IgE in children with asthma and other chronic respiratory symptoms. Respir Res. 2012;13:32. doi: 10.1186/1465-9921-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- Prebeck S, Kirschning C, Durr S, da Costa C, Donath B, Brand K, et al. Predominant role of toll-like receptor 2 versus 4 in Chlamydia pneumoniae-induced activation of dendritic cells. J Immunol. 2001;167:3316–3323. doi: 10.4049/jimmunol.167.6.3316. [DOI] [PubMed] [Google Scholar]
- Ramirez JA. Isolation of Chlamydia pneumoniae from the coronary artery of a patient with coronary atherosclerosis. The Chlamydia pneumoniae/Atherosclerosis Study Group. Ann Intern Med. 1996;125:979–982. doi: 10.7326/0003-4819-125-12-199612150-00008. [DOI] [PubMed] [Google Scholar]
- Ravichandran V, Sabath BF, Jensen PN, Houff SA, Major EO. Interactions between c-Jun, nuclear factor 1, and JC virus promoter sequences: implications for viral tropism. J Virol. 2006;80:10506–10513. doi: 10.1128/JVI.01355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss DJ, Avila-Campillo I, Thorsson V, Schwikowski B, Galitski T. Tools enabling the elucidation of molecular pathways active in human disease: application to Hepatitis C virus infection. BMC Bioinformatics. 2005a;6:154. doi: 10.1186/1471-2105-6-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005b;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzman CH, Netea MG, Zimmerman MA, Weinberg A, Reznikov LL, Grover FL, Dinarello CA. Atherogenic effects of Chlamydia pneumoniae: refuting the innocent bystander hypothesis. J Thorac Cardiovasc Surg. 2003;126:688–693. doi: 10.1016/s0022-5223(03)00738-4. [DOI] [PubMed] [Google Scholar]
- Senn L, Jaton K, Fitting JW, Greub G. Does respiratory infection due to Chlamydia pneumoniae still exist? Clin Infect Dis. 2011;53:847–848. doi: 10.1093/cid/cir515. [DOI] [PubMed] [Google Scholar]
- Seo JH, Lim JW, Kim H, Kim KH. Helicobacter pylori in a Korean isolate activates mitogen-activated protein kinases, AP-1, and NF-kappaB and induces chemokine expression in gastric epithelial AGS cells. Lab Invest. 2004;84:49–62. doi: 10.1038/sj.labinvest.3700010. [DOI] [PubMed] [Google Scholar]
- Sessa R, Di Pietro M, Schiavoni G, Macone A, Maras B, Fontana M, et al. Chlamydia pneumoniae induces T cell apoptosis through glutathione redox imbalance and secretion of TNF-alpha. Int J Immunopath Pharmacol. 2009;22:659–668. doi: 10.1177/039463200902200311. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Singhal M, Domico K. CABIN: collective analysis of biological interaction networks. Comput Biol Chem. 2007a;31:222–225. doi: 10.1016/j.compbiolchem.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Singhal M, Resat H. A domain-based approach to predict protein-protein interactions. BMC Bioinformatics. 2007b;8:199. doi: 10.1186/1471-2105-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo JG, Knuefermann P, Mann DL, Sivasubramanian N. Group B Streptococcus induces TNF-alpha gene expression and activation of the transcription factors NF-kappa B and activator protein-1 in human cord blood monocytes. J Immunol. 2000;165:419–425. doi: 10.4049/jimmunol.165.1.419. [DOI] [PubMed] [Google Scholar]
- Vercammen E, Staal J, Beyaert R. Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin Microbiol Rev. 2008;21:13–25. doi: 10.1128/CMR.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Johnston SC, Chou J, Dean D. A systemic network for Chlamydia pneumoniae entry into human cells. J Bacteriol. 2010;192:2809–2815. doi: 10.1128/JB.01462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Plano GV, Fields KA. A protein secreted by the respiratory pathogen Chlamydia pneumoniae impairs IL-17 signaling via interaction with human Act1. Cell Microbiol. 2009;11:769–779. doi: 10.1111/j.1462-5822.2009.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Lu H, Sun Y, Graham DY, Cheung HS, Yamaoka Y. Balance between polyoma enhancing activator 3 and activator protein 1 regulates Helicobacter pylori-stimulated matrix metalloproteinase 1 expression. Cancer Res. 2006;66:5111–5120. doi: 10.1158/0008-5472.CAN-06-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Pan H, Yoo S, Gao SJ. Kaposi’s sarcoma-associated herpesvirus induction of AP-1 and interleukin 6 during primary infection mediated by multiple mitogen-activated protein kinase pathways. J Virol. 2005;79:15027–15037. doi: 10.1128/JVI.79.24.15027-15037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, Blackbourn DJ, Mengel M, Xie JP, Qian LW, Greene W, et al. Kaposi’s sarcoma-associated herpesvirus promotes angiogenesis by inducing angiopoietin-2 expression via AP-1 and Ets1. J Virol. 2007;81:3980–3991. doi: 10.1128/JVI.02089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M, Park HK, Lee KM, Lee KJ, Kim JH, Cho SW, Hahm KB. Blockage of HSP 90 modulates Helicobacter pylori-induced IL-8 productions through the inactivation of transcriptional factors of AP-1 and NF-kappaB. Biochem Biophys Res Commun. 2004;320:816–824. doi: 10.1016/j.bbrc.2004.05.214. [DOI] [PubMed] [Google Scholar]
- Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLOS Comp Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos G, Clements B, Conner J. Herpes simplex virus type 1 infection stimulates p38/c-Jun N-terminal mitogen-activated protein kinase pathways and activates transcription factor AP-1. J Biol Chem. 1999;274:5097–5103. doi: 10.1074/jbc.274.8.5097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.