Abstract
In cycling cells, transcription of ribosomal RNA genes by RNA polymerase I (Pol I) is tightly coordinated with cell growth. Here, we show that the mammalian target of rapamycin (mTOR) regulates Pol I transcription by modulating the activity of TIF-IA, a regulatory factor that senses nutrient and growth-factor availability. Inhibition of mTOR signaling by rapamycin inactivates TIF-IA and impairs transcription-initiation complex formation. Moreover, rapamycin treatment leads to translocation of TIF-IA into the cytoplasm. Rapamycin-mediated inactivation of TIF-IA is caused by hypophosphorylation of Ser 44 (S44) and hyperphosphorylation of Ser 199 (S199). Phosphorylation at these sites affects TIF-IA activity in opposite ways, for example, phosphorylation of S44 activates and S199 inactivates TIF-IA. The results identify a new target for mTOR-signaling pathways and elucidate the molecular mechanism underlying mTOR-dependent regulation of rRNA synthesis.
Keywords: RNA polymerase I, TIF-IA, mTOR, transcription, phosphorylation, PP2A
In multicellular organisms, a cell is exposed to multiple, potentially contradictory signals. Therefore, the cell must have mechanisms for converting these signals into a defined response. In one mode of integration, activation of a key metabolic process requires input from several distinct biochemical pathways. Transcription by RNA polymerase I (Pol I) exhibits such behavior. Conditions that harm cell growth and proliferation, such as growth-factor deprivation, amino acid withdrawal, cell confluence, or inhibition of protein synthesis down-regulate Pol I transcription (for review, see Grummt 2003). Stimulation of cell proliferation by mitogenic signals, on the other hand, results in an increase in ribosomal RNA synthesis (Zhao et al. 2003). This growth-dependent regulation of rRNA synthesis is mediated by TIF-IA, an essential initiation factor that is associated with the initiation-competent form of Pol I (Buttgereit et al. 1985; Schnapp et al. 1990). Extracts prepared from quiescent cells lack TIF-IA activity and, therefore, are transcriptionally inactive. Transcriptional activity of extracts from growth-arrested cells can be restored by recombinant TIF-IA (Bodem et al. 2000; Cavanaugh et al. 2002). The mechanism of growth-dependent regulation of Pol I transcription by TIF-IA is poorly understood and, in particular, the signaling events that lie upstream of TIF-IA remain obscure.
It is well established that starvation or lack of nutrients inhibits signaling events that require the mammalian target of rapamycin, mTOR (also known as FRAP, RAFT, and RAPT). Proteins in the mTOR family all have a C-terminal kinase domain that phosphorylates serine and threonine residues. mTOR has been demonstrated to control diverse readouts, all of which are related to cell growth, including transcription, translation, PKC signaling, protein degradation, membrane traffic, or actin organization (for review, see Gingras et al. 2001; Proud 2002). The multitude and diversity of growth-related readouts controlled by mTOR indicate that this conserved kinase may not be simply part of a single, linear growth-controlling pathway, but can be regarded as a central controller of cell growth. Mammalian TOR phosphorylates at least two regulators of protein synthesis, S6K1 (p70 ribosomal S6 kinase) and 4E-BP1, an inhibitor of translation initiation (Brunn et al. 1997; Burnett et al. 1998; Isotani et al. 1999). Amino acids positively regulate mTOR signaling and activate S6Ks. Conversely, amino acid deprivation impairs the activity and the phosphorylation state of both downstream targets of mTOR (Hara et al. 1998; Lynch 2001). These effects can be mimicked by adding rapamycin, a potent inhibitor of TOR, which suggests that the effects of amino acids are mediated via the mTOR pathway.
In both yeast and mammals, rapamycin has been shown to impair ribosome biosynthesis by inhibiting transcription of rRNA genes by Pol I, ribosomal protein genes by Pol II, and 5S and tRNA synthesis by Pol III (Mahajan 1994; Zaragoza et al. 1998; Powers and Walter 1999; Claypool et al. 2003; Hannan et al. 2003). Loss of TOR results in phenotypes characteristic of amino acid deprivation, including reduced nucleolar size and cell-cycle arrest at the G1/S boundary (Zhang et al. 2000). Cell-cycle arrest may be mediated either via inhibition of protein synthesis or via effects on the enzymatic activity of G1-specific Cdks. Rapamycin accelerates the degradation of cyclin D1, resulting in decreased Cdk4 kinase activity, decreased phosphorylation of the retinoblastoma protein pRb, and increased association of the Cdk inhibitor p27Kip1 with Cdk2/cyclin E (Hashemolhosseini et al. 1998; Kawamata et al. 1998).
To explore the mechanisms by which growth-controlling pathways feed into the regulation of ribosomal RNA gene transcription, we have investigated the role of mTOR/S6K1-signaling pathways on Pol I transcription. The results demonstrate that mTOR-dependent regulatory pathways target the growth- and nutrient-responsive factor TIF-IA. Inhibition of mTOR signaling by rapamycin inactivates TIF-IA by decreasing phosphorylation at Ser 44 (S44) and enhancing phosphorylation at Ser 199 (S199). Phosphorylation of these serines affects TIF-IA activity in opposite ways, that is, phosphorylation of S44 activates and phosphorylation of S199 inactivates TIF-IA. Our results reveal a functional interplay between mTOR-dependent protein kinases and phosphatases that positively and negatively regulate RNA Pol I transcription, and provide a mechanistic explanation of how mTOR coordinates rRNA and protein synthesis.
Results
Rapamycin treatment inactivates TIF-IA
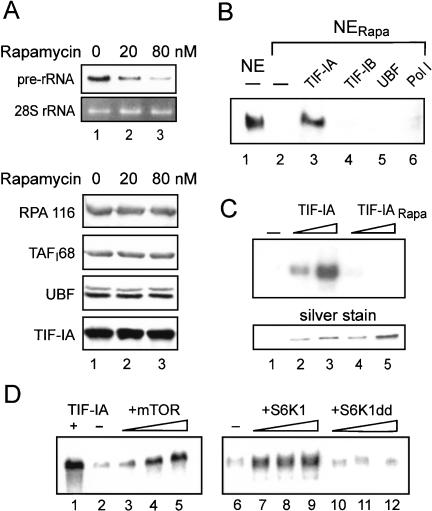
Previous studies in yeast and mammals have established that rapamycin treatment, which inhibits TOR signaling, decreases rRNA gene transcription. In support of this, treatment of Hela cells with rapamycin inhibits Pol I transcription in a concentration-dependent manner, resulting in an almost complete shut-off of cellular 45S pre-rRNA synthesis after 60 min (Fig. 1A). The level of cellular Pol I, UBF, TIF-IA, and TIF-IB was not affected by rapamycin treatment, indicating that mTOR signaling regulates the activity but not the amount of Pol I or basal transcription factors.
Figure 1.
Rapamycin treatment impairs Pol I transcription by inactivating TIF-IA. (A) Northern blot. 45S pre-rRNA levels were monitored in 15 μg of total RNA from untreated Hela cells or cells that were treated with 20 or 80 nM rapamycin for 60 min. Western blots demonstrating that the amount of Pol I, TIF-IB/SL1, TIF-IA, and UBF remained unaffected by rapamycin treatment are shown below. (B) Complementation of transcriptional activity in nuclear extracts from rapamycin-treated cells. Transcription reactions contained nuclear extracts (30 μg) from untreated (NE) or rapamycin-treated (NERapa) FM3A cells. (Lanes 3–6) Fractions containing partially purified Pol I transcription factors (Schnapp and Grummt 1996) were added to NERapa-containing reactions. (C) TIF-IA from rapamycin-treated cells is transcriptionally inactive. Twenty nanograms and 40 ng of TIF-IA immunopurified from untreated (lanes 2,3) and rapamycin-treated NIH3T3 cells (lanes 4,5) were added to transcription reactions containing nuclear extracts (30 μg protein) from density-arrested cells. A silver-stained SDS–polyacrylamide gel showing the amount of TIF-IA added to the transcription assays is shown below. (D) Exogenous mTOR and S6K restore transcriptional activity in nuclear extracts from rapamycin-treated cells. Reactions were supplemented with 50 ng of TIF-IA (lane 1), 10, 20, and 30 ng of HA-mTOR (lanes 3–5), 100, 150, and 200 ng of GST–S6K1 (lanes 7–9) or the same amounts of GST–S6K1dd (lanes 10–12).
To explore the mechanisms by which mTOR signaling controls pre-rRNA synthesis, we compared Pol I transcription in nuclear extracts from untreated or rapamycin-treated FM3A cells. Both extracts exhibited marked differences in transcriptional activity. Extracts from untreated, exponentially growing cells efficiently transcribed rDNA, whereas extracts from rapamycin-treated cells were virtually inactive (Fig. 1B, lanes 1,2). To examine which component(s) of the Pol I transcription machinery was inactivated by rapamycin treatment, the basal factors TIF-IA (Fig. 1B, lane 3), TIF-IB/SL1 (Fig. 1B, lane 4), UBF (Fig. 1B, lane 5), or Pol I (Fig. 1B, lane 6) were assayed for their capability to rescue transcriptional activity of extracts from rapamycin-treated cells. This complementation assay revealed that TIF-IA, but none of the other factors tested, was capable of relieving transcriptional repression in extracts from rapamycin-treated cells. Thus, inhibition of mTOR-dependent pathways inactivates TIF-IA and down-regulates Pol I transcription.
If this conclusion was correct, then TIF-IA from rapamycin-treated cells should be transcriptionally inactive. To test this hypothesis, TIF-IA was immunopurified from rapamycin-treated and untreated cells, and the activity of bead-bound TIF-IA was assayed in vitro. Immunopurified TIF-IA from exponentially growing cells enhanced Pol I transcription in extracts from density-arrested cells in a dose-dependent manner (Fig. 1C, lanes 2,3). A comparable amount of TIF-IA from rapamycin-treated cells, however, was transcriptionally inactive (Fig. 1C, lanes 4,5). This finding underscores the key role of TIF-IA in growth- and nutrient-dependent regulation of Pol I transcription and demonstrates that mTOR-dependent signaling pathways regulate TIF-IA activity.
To elucidate the molecular mechanisms underlying mTOR-dependent regulation of TIF-IA activity, we studied whether mTOR itself or downstream kinases target TIF-IA. Given that inhibition of mTOR signaling inactivates TIF-IA, exogenous mTOR should be capable of restoring transcriptional activity of extracts from rapamycin-treated cells. Immunopurified mTOR activated Pol I transcription in a dose-dependent manner (Fig. 1D, lanes 3–5). The downstream kinase S6K1 also relieved rapamycin-mediated transcriptional repression (Fig. 1D, lanes 7–9), whereas S6K1dd, a kinase-deficient S6K1 mutant (Dennis et al. 1998), did not rescue transcription (Fig. 1D, lanes 10–12). The fact that both mTOR and S6K1, but not mutant S6K1, restore Pol I transcription in extracts from rapamycin-treated cells suggests that both mTOR and S6K1, or S6K1 alone, is required for maintaining TIF-IA activity.
Rapamycin treatment alters the phosphorylation pattern of TIF-IA
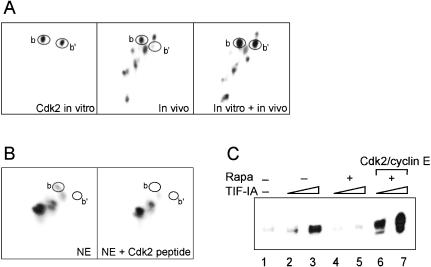
An important prediction of TIF-IA activity being regulated by mTOR-signaling pathways is that the phosphorylation pattern of TIF-IA is altered in rapamycin-treated cells. To test this, we compared the tryptic phosphopeptide pattern of TIF-IA derived from exponentially growing and rapamycin-treated cells. Cells were transfected with a vector encoding Flag-tagged TIF-IA, labeled with [32P]orthophosphate, and immunopurified TIF-IA was subjected to two-dimensional tryptic phosphopeptide mapping (Fig. 2A). We have shown previously that TIF-IA from growing cells is phosphorylated at multiple sites and exhibits a characteristic phosphorylation pattern (Zhao et al. 2003). After rapamycin treatment, phosphorylation of two peptides was markedly altered. Phosphorylation of one peptide, spot a (Fig. 2), was enhanced, whereas that of spot b (and b′, a partial tryptic digestion product) was suppressed (Fig. 2A, middle). Thus, inhibition of mTOR signaling results in hyperphosphorylation of peptide a and hypophosphorylation of peptide b.
Figure 2.
Rapamycin treatment alters the phosphorylation pattern of TIF-IA. (A) Two-dimensional tryptic phosphopeptide maps of TIF-IA labeled in vivo. HEK293T cells overexpressing Flag-tagged TIF-IA or TIF-IAS44A, respectively, were labeled for 2 h with [32P]orthophosphate in the absence or presence of 20 nM rapamycin. TIF-IA was immunopurified and subjected to two-dimensional tryptic phosphopeptide mapping. (B) Nuclear extracts from rapamycin-treated cells fail to phosphorylate TIF-IA at S44. Immobilized TIF-IA was incubated with [γ-32P]ATP and nuclear extract from untreated, rapamycin-treated, or amino acid-starved cells as indicated. The tryptic phosphopeptide map of in vitro phosphorylated TIF-IA was compared with the phosphorylation pattern of TIF-IA labeled in vivo.
After having established that mTOR either directly or indirectly activates TIF-IA, it was important to determine which phosphoamino acids are targeted in vivo and to identify the kinase(s)/phosphatase(s) that modify TIF-IA in a rapamycin-sensitive manner. Previous studies have revealed that phosphorylation of TIF-IA at two serine residues, S633 and S649, by MAPK-dependent signaling cascades is important for cell proliferation (Zhao et al. 2003). Moreover, analysis of tryptic peptides by mass spectroscopy revealed three serine residues, S44, S170, and S172 that were phosphorylated in TIF-IA overexpressed in HEK293T cells (Schlosser et al. 2002). S170 and S172 are contained in genuine target sites for casein kinase II (CKII), and in vitro phosphorylation of TIF-IA by CKII-labeled spots h, i, and j (J. Zhao and I. Grummt, unpubl.), that is, peptides that are not affected by rapamycin treatment. To examine whether S44 is the phosphoacceptor site in peptide a or b, we mutated S44 to alanine (TIF-IAS44A) and compared the phosphorylation pattern of wild-type and mutant TIF-IA (Fig. 2A, right). This analysis revealed that phosphopeptides b and b′ were lacking in TIF-IAS44A, demonstrating that phosphopeptide b contains S44.
To recapitulate rapamycin-sensitive phosphorylation of S44 in vitro, we utilized nuclear extracts from untreated or rapamycin-treated cells to phosphorylate immunopurified TIF-IA. As shown in Figure 2B, extracts from exponentially growing cells labeled spot b (S44), as well as spots f, g, and d, for example, the phosphopeptides that contain S633 and S649 and are phosphorylated after mitogenic stimulation by MAPK kinases (Zhao et al. 2003). The phosphorylation pattern with extract from rapamycin-treated cells closely resembled that of untreated cells, except that spot b, the rapamycin-sensitive phospopeptide was lacking. Notably, the same phosphorylation pattern was obtained with extract from amino acid-starved cells, a finding that is consistent with TIF-IA being regulated by nutrient availability that feeds into TOR signaling pathways.
Phosphorylation at S44 and S199 affects TIF-IA activity in opposite ways
The fact that phosphorylation of peptide a was increased and peptide b was decreased after rapamycin treatment, suggests that phosphorylation at these two sites affect TIF-IA activity in opposite ways. Phosphorylation of peptide a should inhibit TIF-IA activity, whereas phosphorylation of peptide b, for example, S44, should augment TIF-IA activity. Consistent with this prediction, the S44 point mutant (TIF-IAS44A) was transcriptionally inactive in the cell-free transcription system (Fig. 3A, lanes 4,5). To monitor TIF-IA activity in vivo, NIH3T3 cells were cotransfected with a Pol I reporter plasmid and increasing amounts of an expression vector encoding wild-type or mutant TIF-IA. Although wild-type TIF-IA enhanced transcription of the Pol I reporter in a dose-dependent manner (Fig. 3A, lanes 7–9), no activation was observed with TIF-IAS44A (Fig. 3A, lanes 10–12). This demonstrates that phosphorylation at S44 is required for TIF-IA activity.
Figure 3.
Phosphorylation of S44 activates, whereas phosphorylation at S199 inactivates TIF-IA. (A) Transcriptional activity of TIF-IAS44A in vitro and in vivo. (Lanes 1–5) To assay transcriptional activity in vitro, TIF-IA and TIF-IAS44A were immunopurified from HEK293T cells, and assayed for their ability to rescue Pol I transcription in a nuclear extract from density-arrested cells. (Lanes 6–12) To monitor TIF-IA activity in vivo, NIH3T3 cells were cotransfected with 2.5 μg of Pol I reporter plasmid (pMr1930-BH) and 0.25, 0.5, or 1 μg of pcDNA3.1-TIF-IA or TIF-IAS44A, and transcripts from the reporter plasmid were analyzed on Northern blots. (B) Transcriptional activity of S199 point mutants in vitro and in vivo. (Lanes 1–7) Flag-tagged wild-type TIF-IA and the indicated mutants were immunopurified from HEK293T cells, and 30 and 60 ng were assayed for their capability to restore transcriptional activity of nuclear extract from density-arrested FM3A cells. The amount and purity of TIF-IA were estimated on silver-stained SDS–polyacrylamide gels (data not shown). (Lanes 8–12) NIH3T3 cells were cotransfected with the Pol I reporter plasmid pMr1930-BHand 0.5 or 1 μg of the expression vector encoding TIF-IA or TIF-IAS199A, respectively, and transcripts from the reporter plasmid were monitored on Northern blots.
In addition to hypophosphorylation of S44, rapamycin-mediated inactivation of TIF-IA correlated with enhanced phosphorylation of peptide a. To identify the phosphoacceptor site in peptide a, we generated a set of point mutants, in which evolutionarily conserved serine residues were replaced by alanine or aspartic acid, and tested the mutants for their capability to rescue Pol I transcription in extracts from density-arrested cells. The rationale of this approach was to identify S ® A mutants that are transcriptionally active while the corresponding S ® D are transcriptionally inactive. This assay, together with two-dimensional phosphopeptide maps of the mutants (data not shown), revealed that Ser 199 (S199) is phosphorylated in tryptic peptide a. Mutation of S199 to alanine (TIF-IAS199A) did not impair TIF-IA activity in vitro (Fig. 3B, lanes 4,5) and in vivo (Fig. 3B, lanes 11,12). In contrast, replacement of S199 by aspartic acid (TIF-IAS199D) inactivated TIF-IA (Fig. 3B, lanes 6,7). Taken together, these results reveal that phosphorylation at S44 and S199 affect TIF-IA activity in opposite ways, phosphorylation at S44 activating and phosphorylation at S199 inactivating TIF-IA.
Cdk2/cyclin E-mediated phosphorylation at Ser 44 activates TIF-IA
The observation that rapamycin treatment decreases phosphorylation of S44 suggests that inhibition of mTOR either inactivates rapamycin-sensitive kinase(s) that phosphorylate S44 or, alternatively, activates phosphatase(s) that dephosphorylate S44. S44 resides in a sequence (FNSPPRKTV) that is a potential target site of Cdks (Kitagawa et al. 1996). We have shown previously that the gradual increase of rDNA transcription during G1 progression correlates with the activation of G1-specific Cdk/cyclin complexes (Voit et al. 1999). We therefore reasoned that phosphorylation by G1-phase Cdks may activate TIF-IA. To examine whether Cdks phosphorylate TIF-IA at S44, we phosphorylated TIF-IA by immunopurified Cdk2/cyclin E in vitro and compared the tryptic peptides with the two-dimensional map of TIF-IA labeled in vivo. Two peptides were radiolabeled by Cdk2/cyclin E in vitro (Fig. 4A, left), which comigrated with spots b and b′, the rapamycin-sensitive peptides that harbor phospho-S44 (Fig. 4A, right). Moreover, if phosphorylated by nuclear extract in the presence of a synthetic peptide harboring the Cdk2/cyclin E target sequence, spots b and b′ were not labeled in vitro (Fig. 4B). This result demonstrates that Cdk2/cyclin E (and other G1-phase Cdks) phosphorylate TIF-IA at S44.
Figure 4.
Cdk2/cyclin E-mediated phosphorylation at Ser 44 activates TIF-IA. (A) Cdk2/cyclin E phosphorylates S44 (phosphopeptide b). GST–TIF-IA was phosphorylated with immunopurified Cdk2/cyclin E in vitro and subjected to tryptic phosphopeptide mapping. To prove that the labeled phosphopeptides that represent spots b and b′, TIF-IA labeled in vitro was mixed with TIF-IA purified from metabolically labeled HEK293T cells, and tryptic phosphopeptides were analyzed by two-dimensional electrophoresis and chromatography. (B) A synthetic peptide harboring the target sequence for Cdk2/cyclin E competes for phosphorylation at S44. TIF-IA was incubated with [γ-32P]ATP and nuclear extract from growing cells in the absence or presence (10 μM) of a synthetic Cdk2/cyclin E target peptide (YGRKKRRQRRRGPVKRRLDL, Calbiochem). (C) Phosphorylation by Cdk2/cyclin E is required for TIF-IA activity. Thirty nanograms and 90 ng of Flag-tagged TIF-IA from untreated (lanes 2,3) or rapamycin-treated (lanes 4–7) HeLa cells were assayed for their capability to restore transcription of nuclear extracts from rapamycin-treated cells. (Lanes 6,7) TIF-IA was phosphorylated with immunopurified Cdk2/cyclin E in the presence of ATPγS (100 μM) before adding to the transcription reactions.
As rapamycin-treatment caused hypophosphorylation of S44, we next studied whether phosphorylation at S44 by Cdk2/cyclin E may activate TIF-IA. For this, we phosphorylated TIF-IA from rapamycin-treated cells with Cdk2/cyclin E in vitro and assayed TIF-IA activity in nuclear extracts from rapamycin-treated cells. Consistent with the results above, TIF-IA from rapamycin-treated cells was transcriptionally inactive and did not activate rDNA transcription (Fig. 4C, lanes 1,4,5). However, after phosphorylation by Cdk2/cyclin E, TIF-IA activity was fully restored and even exceeded that of TIF-IA isolated from exponentially growing cells (Fig. 4C, cf. lanes 2,3 and 6,7). This result demonstrates that Cdk-dependent phosphorylation of S44 is necessary for TIF-IA activity.
Protein phosphatase 2A regulates TIF-IA activity in an mTORp-dependent manner
Two mechanisms could be responsible for the decreased phosphorylation of S44 after rapamycin treatment, for example, inhibition of the respective kinase(s) or activation of counteracting phosphatase(s). Given that inhibition of Pol I transcription is observed as early as 60 min after rapamycin administration, it seems unlikely that inhibition of Cdk(s) would account for the rapid hypophosphorylation of S44 and, hence, inactivation of TIF-IA. We therefore reasoned that S44 may be specifically dephosphorylated by a phosphatase that is activated by rapamycin treatment. A likely candidate is phosphatase 2A (PP2A), which is known to be down-regulated by mTOR (Peterson et al. 1999). To test whether PP2A antagonizes Cdk-mediated phosphorylation of TIF-IA, we incubated immobilized TIF-IA with limiting amounts of either purified PP2A or calf intestine alkaline phosphatase (CIAP), and compared the activity of untreated and phosphatase-treated TIF-IA in the cell-free transcription system (Fig. 5A). Significantly, dephosphorylation by PP2A strongly reduced TIF-IA activity (Fig. 5A, lanes 4,5), whereas treatment with CIAP did not affect the activity of TIF-IA (Fig. 5A, lanes 6,7). The specific inactivation of TIF-IA by PP2A supports the view that phosphorylation of S44 by Cdks and dephosphorylation by PP2A plays an important role in nutrient-dependent regulation of TIF-IA activity.
Figure 5.
PP2A counteracts S44 phosphorylation by Cdk2/cyclin E. (A) Dephosphorylation by PP2A inactivates TIF-IA. Immobilized Flag-tagged TIF-IA was incubated with 0.003 units of either PP2A (Upstate Biochemicals) or calf intestine alkaline phosphatase (CIAP, Roche Molecular Biochemicals) for 30 min at 30°C in the supplied phosphatase buffer. Thirty nanograms and 90 ng of untreated (lanes 2,3), PP2A-treated (lanes 4,5), and CIAP-treated (lanes 6,7) TIF-IA were assayed for their capability to activate transcription in nuclear extracts from rapamycin-treated cells. (B) Phosphatase inhibitors alleviate rapamycin-mediated inactivation of TIF-IA. TIF-IA was purified from cells that were treated for 1 h with either okadaic acid (1 μM), rapamycin (20 nM), or calyculin A (50 nM), in the absence or presence of rapamycin as indicated. Thirty nanograms (even numbers) and 90 ng (odd numbers except 1) of TIF-IA were assayed for their capability to activate transcription of nuclear extracts from rapamycin-treated cells. (C) Inhibition of PP2A/PP1 in vivo alleviates rapamycin-dependent hypophosphorylation of S44. Hela cells overexpressing Flag-tagged TIF-IA were labeled for 2 h with [32P]orthophosphate in the absence (left) or presence (middle) of rapamycin. (Right) Metabolic labeling was performed in the presence of both rapamycin (20 nM) and calyculin (50 nM). Radiolabeled TIF-IA was immunopurified and subjected to two-dimensional tryptic phosphopeptide mapping.
If PP2A dephosphorylates and thus inactivates TIF-IA in vivo, inhibition of PP2A should protect TIF-IA from inactivation by rapamycin. Treatment of HEK293T cells overexpressing Flag-TIF-IA with okadaic acid or calyculin A, inhibitors of both PP2A and PP1 with a strong preference for PP2A (Yan and Mumby 1999) prevented rapamycin-mediated inactivation of TIF-IA (Fig. 5B).
To directly demonstrate that inhibition of PP2A alleviates rapamycin-mediated dephosphorylation of TIF-IA, we compared the tryptic phosphopeptide pattern of TIF-IA that was in vivo labeled in the presence of rapamycin or rapamycin plus calyculin A (Fig. 5C). Significantly, rapamycin-induced hypophosphorylation of S44 (spot b) was partially reverted upon inhibition of PP2A by calyculin A. Although treatment with phosphatase inhibitors in vivo may have a broad range of effects on signaling pathways, the finding that inhibitors of PP2A override rapamycin-mediated hypophosphorylation of S44, which correlates with inactivation of TIF-IA, supports the view that mTOR signaling activates PP2A, which, in turn, dephosphorylates S44 and inactivates TIF-IA.
Inhibition of mTOR signaling impairs the interaction between TIF-IA and Pol I
TIF-IA has been shown to interact with both Pol I and the TBP-containing promoter recognition factor TIF-IB/SL1 (Miller et al. 2001; Cavanaugh et al. 2002; Yuan et al. 2002). Thus, by linking both protein complexes, TIF-IA may play a key role in recruiting Pol I to the rDNA promoter. To investigate whether the interaction between TIF-IA and Pol I was affected by inhibition of mTOR signaling, TIF-IA was immunoprecipitated from mock-treated and rapamycin-treated HeLa cells expressing Flag-tagged TIF-IA. Coprecipitated Pol I was visualized on immunoblots using antibodies against the second largest subunit of Pol I (anti-RPA116) and against PAF67, a Pol I-associated factor that decorates the initiation-competent subpopulation of Pol I (Seither et al. 2001). In mock-treated cells, both RPA116 and PAF67 coprecipitated with TIF-IA (Fig. 6A, lane 3). In rapamycin-treated cells, however, the interaction of TIF-IA with both RPA116 and PAF67 was markedly decreased (Fig. 6A, lane 4). This demonstrates that inhibition of mTOR signaling impairs the interaction between TIF-IA and Pol I.
Figure 6.
Phosphorylation of S199 impairs the interaction of TIF-IA with Pol I. (A) Rapamycin treatment inhibits the interaction between TIF-IA and Pol I. Lysates from mock- or rapamycin-treated HeLa cells expressing Flag-tagged TIF-IA were incubated with anti-Flag antibodies, and immunoprecipitated TIF-IA and Pol I were visualized on Western blots using anti-Flag, anti-RPA116, or anti-PAF67 antibodies. (B) A mutant that mimics phosphorylation of TIF-IA at S199 fails to interact with Pol I and TIF-IB. Flag-tagged TIF-IA or the indicated point mutants were overexpressed in HEK293T cells and immunoprecipitated with anti-Flag antibodies. Coprecipitated Pol I and TIF-IB/SL1 were visualized on Western blots with anti-RPA116 and anti-TAFI110 antibodies.
Given the importance of mTOR-mediated alterations in the phosphorylation pattern on TIF-IA activity, one could predict that either phosphorylation of S199, dephosphorylation of S44, or both, regulate the interaction of TIF-IA with the Pol I transcription apparatus. We therefore examined whether the S ® A or S ® D point mutant(s) were capable of interacting with Pol I and/or TIF-IB/SL1. For this, TIF-IA was precipitated from cells overexpressing either Flag-tagged wild-type TIF-IA or the respective S199 and S44 mutant(s), and coprecipitated Pol I and TIF-IB/SL1 were visualized on Western blots. As shown in Figure 6B, wild-type TIF-IA, TIF-IAS44A, and TIF-IAS199A interacted with both Pol I and TIF-IB/SL1. In contrast, no association of either Pol I or TIF-IB/SL1 could be detected with TIF-IAS199D. This result implies that the interaction of TIF-IA with the two main players of the Pol I transcription machinery, that is, Pol I and the promoter selectivity factor TIF-IB/SL1, is regulated by the phosphorylation state of S199.
The cellular localization of TIF-IA is controlled by mTOR
TOR-dependent signaling pathways have been shown to control the subcellular localization of several nutrient-regulated transcription factors (Beck and Hall 1999). To examine whether inactivation of TIF-IA by rapamycin was accompanied by changes in the cellular localization of TIF-IA, we visualized Flag-tagged TIF-IA in untreated and rapamycin-treated HeLa cells by immunofluorescence. In untreated cells, overexpressed TIF-IA was uniformly distributed throughout the nucleus (Fig. 7A, top). After 2 h of rapamycin treatment, however, staining of the the nucleus was reduced, whereas fluorescence of the cytoplasm was increased (Fig. 7A, bottom). Apparently, inhibition of mTOR signaling promoted translocation of TIF-IA into the cytoplasm.
Figure 7.
mTOR controls the nucle(ol)ar localization of TIF-IA. (A) Rapamycin treatment mediates translocation of TIF-IA into the cytoplasm. HeLa cells expressing Flag-tagged TIF-IA were incubated with 20 nM rapamycin for 2 h and immunostained with Cy3-conjugated anti-Flag antibodies. Fluorescence and merged images with phase contrast from the same view are shown. (B) TIF-IA accumulates in the cytoplasm after rapamycin treatment. The levels of TIF-IA and Pol I (RPA116) were analyzed by Western blotting in whole-cell extracts (lanes 1,2), nuclear extract (lanes 3,4), and the respective S-100 fraction (lanes 5,6) from untreated or rapamycin-treated FM3A cells. (C) Rapamycin-treatment inactivates both nuclear and cytoplasmic TIF-IA. Transcription reactions contained 50 μg of nuclear extract from growing (lanes 1–3) or rapamycin-treated (lanes 4–6) cells. The assays were supplemented with 50 μg of the respective S-100 fraction (lanes 2,5) or 3 ng of cellular TIF-IA (lanes 3,6).
To demonstrate changes in the nuclear-cytoplasmic distribution of TIF-IA after rapamycin treatment by a different experimental approach, we determined the amount of TIF-IA in nuclear and cytoplasmatic extracts from untreated and rapamycin-treated cells. Consistent with the results in Figure 1A, the amount of TIF-IA was the same in lysates from untreated and rapamycin-treated cells (Fig. 7B, lanes 1,2). However, in nuclear extracts from rapamycin-treated cells, the amount of TIF-IA was approximately twofold lower than from untreated cells (Fig. 7B, lanes 3,4). Reciprocally, TIF-IA levels were increased in the cytoplasmic fraction from rapamycin-treated cells compared with control cells (Fig. 7B, lanes 5,6). Thus, inhibition of mTOR signaling down-regulates Pol I transcription by three interrelated mechanisms that involve hypophosphorylation of S44, hyperphosphorylation of S199, and shuttling of TIF-IA from the nucle(ol)us into the cytoplasm.
The finding that nuclear extracts from rapamycin-treated cells contain less TIF-IA than extracts from un-treated cells, raised the question of whether the impaired transcriptional activity of nuclear extracts from rapamycin-treated cells was due to depletion rather than inactivation of TIF-IA. Although most of our experiments exclude this possibility, we examined transcriptional activity of nuclear extracts from untreated and rapamycin-treated cells after supplementating with the corresponding cytoplasmic fraction (S-100 extract). Notably, S-100 extracts from rapamycin-treated cells that contain more TIF-IA than control extracts did not activate transcription (Fig. 7C, lane 5). This result is consistent with inactivation of TIF-IA by rapamycin and suggests that translocation into the cytoplasm occurs after hypophosphorylation of S44 and hyperphosphorylation of S199.
Discussion
Studies in yeast and mammals have demonstrated that TOR signaling regulates cell growth and couples nutrient availability to the regulation of ribosome synthesis (Schmelzle and Hall 2000; Kim et al. 2003). A variety of extracellular stimuli along with intracellular metabolites, such as glucose, ATP, and amino acid levels, feed into mTOR, which, in turn, relays them to the down-stream effectors S6K1 and 4E-BP (Jacinto and Hall 2003). The complex signaling cascades that sense nutrient availability are integrated by TOR, and a functional TOR pathway is required for the coordinated regulation of transcription by all three classes of RNA polymerase, for example, the synthesis of pre-rRNA by Pol I, ribosomal protein genes by Pol II, and tRNA and 5S RNA by Pol III (Mahajan 1994; Zaragoza et al. 1998; Hardwick et al. 1999; Powers and Walter 1999).
In this study, we have investigated the contribution of mTOR-dependent pathways in conveying external growth signals to the Pol I transcription machinery. Inhibition of mTOR by rapamycin is known to decrease Pol I transcription both in vivo and in vitro. In exploring the mechanism underlying mTOR regulation of rDNA transcription, we found that rapamycin-mediated inhibition of Pol I transcription is due to inactivation of the basal transcription factor TIF-IA (or Rrn3p in yeast), the factor that adapts Pol I transcription to cell growth (for review, see Grummt 2003). Pol I transcription is down-regulated in nuclear extracts from rapamycin-treated cells and transcriptional activity can be restored by affinity-purified recombinant TIF-IA. Neither Pol I, TIF-IB/SL1, or UBF were capable of relieving transcription inhibition, indicating that the activity of TIF-IA, but none of the other components of the Pol I transcription machinery, is regulated by mTOR-sensitive signaling pathways. Transcriptional activity was also rescued by exogenous mTOR or S6K1, but not by a kinase-deficient S6K1 mutant. This indicates that either S6K1 itself or S6K1-dependent enzymes regulate TIF-IA activity.
Previous studies have established that TIF-IA is phosphorylated at multiple sites and down-regulation of Pol I transcription in confluent, serum-starved, or cycloheximide-treated cells TIF-IA is accompanied by alterations in the phosphorylation pattern and activity of TIF-IA. Consistent with TIF-IA responding to a variety of external signals, the phosphorylation pattern of TIF-IA depends on the physiological state of the cells. In a previous study, we have shown that activation of rDNA transcription by growth factors correlates with phosphorylation of TIF-IA at two specific serine residues (S633 and S649) by ERK and RSK kinases. Phosphorylation at these serine residues activates TIF-IA and promotes cellular pre-rRNA synthesis (Zhao et al. 2003). Our finding that inhibitors of ERK- and mTOR-dependent signaling inactivate TIF-IA and abrogate Pol I transcription underscores the molecular cross-talk between mTOR and ERK signal-transduction pathways (Wang et al. 2001), and demonstrates that TIF-IA is a common final target for growth factor- and nutrient-dependent activation of ribosome biogenesis.
Given the essential role of TIF-IA in bridging Pol I to promoter-bound TIF-IB/SL1, one would expect that signaling pathways modulate the interaction of TIF-IA with Pol I and/or TIF-IB/SL1. Interactions between TIF-IA with Pol I were found to be affected by diverse regulatory pathways that link the cell's biosynthetic activities to environmental conditions. Noteworthy, only the fraction of Pol I that is associated with TIF-IA (or Rrn3p in yeast) is capable of assembling into productive initiation complexes (Tower and Sollner-Webb 1987; Schnapp et al. 1990; Miller et al. 2001). The amount of TIF-IA/Rrn3p associated with Pol I, but not the overall level of TIF-IA/Rrn3p, was decreased in growth-arrested cells, indicating that TIF-IA/Rrn3p either fails to associate with Pol I or dissociates from Pol I (Milkereit and Tschochner 1998; Cavanaugh et al. 2002; Yuan et al. 2002). It has not been known, however, whether the interaction of TIF-IA with promoter-bound TIF-IB/SL1 is also regulated by external signals. Our finding that rapamycin treatment abolishes the association of TIF-IA with both Pol I and TIF-IB/SL1 reveals that mTOR signaling affects transcription-initiation complex formation. Thus, by facilitating recruitment of Pol I to the rDNA promoter, mTOR signaling may be a key event in the complex pathways that the cell uses to adapt Pol I transcription to nutrient availability.
The molecular basis of TOR-mediated regulation of TIF-IA was pinpointed to the reversible phosphorylation of two opposing serine residues, S44 and S199. Phosphorylation of S44 is required for TIF-IA function, whereas phosphorylation of S199 impedes TIF-IA activity. S44 is evolutionarily conserved and mass spectrometry has demonstrated phosphorylation of this site in vivo (Schlosser et al. 2002). Substitution of S44 by alanine inactivates TIF-IA without affecting its capability to interact with either Pol I or TIF-IB/SL1. This indicates that phosphorylation of S44 is indispensable for TIF-IA function by an unknown mechanism. In contrast to S44, phosphorylation of S199 exerts a negative effect on TIF-IA activity. Whereas replacement of S199 by alanine did not affect TIF-IA activity, the respective mutant containing an aspartic acid at residue 199 was transcriptionally inactive and failed to associate with both Pol I and TIF-IB/SL1. This suggests that rapamycin inhibits Pol I transcription by disturbing the balanced interplay of positive and negative phosphorylations that are required for interactions with the Pol I transcription machinery.
An important question is whether mTOR itself, or a downstream kinase phosphorylates TIF-IA. S6K1 is known to act downstream of mTOR (Pullen and Thomas 1997), raising the possibility that S6K1 directly phosphorylates TIF-IA. However, TIF-IA contains no consensus recognition motif for S6K1, and neither immunopurified mTOR nor S6K1 were capable of phosphorylating TIF-IA in vitro (C. Mayer and I. Grummt, unpubl.). Ser 44 resides within a recognition motif for Cdks (SPxR). This raises the possibility that Cdk-mediated phosphorylation of S44 links nutrient-sensing pathways to cellcycle control. Pol I transcription oscillates during the cell cycle, being highest in S2 and G2 phase, shutting down at mitosis and recovering during G1-phase progression (Klein and Grummt 1999). It is not yet known whether or not TIF-IA is inactivated during mitosis and is reactivated by phosphorylation by G1-specific Cdks. Our finding that immunopurified Cdk2/cyclin E phosphorylates S44 and phosphorylation at this site is required for TIF-IA function, supports the view that phosphorylation by Cdks is essential for regulation of TIF-IA activity and, hence, cell cycle-dependent fluctuations of Pol I transcription. On the other hand, although rapamycin inhibits Cdk activation and compromises cell-cycle progression (Albers et al. 1993; Chou and Blenis 1996; Kawamata et al. 1998; Hidayat et al. 2003), inhibition of S44 phosphorylation by G1-phase Cdks is unlikely to be responsible for the rapid switch-off of pre-rRNA synthesis, because TIF-IA inactivation was observed as early as 1 h after rapamycin administration. Inactivation of TIF-IA by rapamycin has not been observed in the presence of protein phosphatase inhibitors, indicating that activation of phosphatase(s) rather than inhibition of Cdks induced rapamycin-dependent hypophosphorylation of S44. In support of this, PP2A is very rapidly activated by rapamycin (Zaragoza et al. 1998; Jiang and Broach 1999; Peterson et al. 1999; Hartley and Cooper 2002), and the regulated activity of PP2A has been proposed as a master switch to regulate the activity of downstream effectors of mTOR, such as S6K1 and 4E-BP. Like TIF-IA, S6K1 and 4E-BP are phosphorylated at multiple sites and are dephosphorylated within 30 min after rapamycin treatment (Jacinto and Hall 2003).
Together, our results demonstrate that mTOR signaling regulates TIF-IA activity by positive- and negative-acting phosphorylations. Whereas phosphorylation of S44 is indispensable for TIF-IA activity, phosphorylation at S199 inhibits TIF-IA activity. This indicates that mTOR-responsive kinase(s) and phosphatase(s) modulate the activity of this central growth-dependent transcription factor and implies that balance of antagonizing phosphorylations may play a key role in the regulation of Pol I transcription. Interestingly, mTOR signaling not only controls the activity of TIF-IA, but also its intracellular localization. Once inactivated by rapamycin treatment, a significant part of TIF-IA is released from the nucleus and accumulates in the cytoplasm. mTOR-sensitive sequestration of TIF-IA in the cytoplasm is reminiscent of studies in yeast that have shown that the TOR-signaling pathway broadly controls nutrient metabolism by sequestering several transcription factors in the cytoplasm (Di Como and Arndt 1996; Beck and Hall 1999; Jiang and Broach 1999).
While this paper was in revision, two studies have been published that address the molecular mechanisms underlying TOR-dependent regulation of rRNA synthesis in yeast and mammalian cells. Consistent with our data, rapamycin treatment of yeast decreased the amount of the Rrn3p–Pol I complex, demonstrating that the TOR-dependent phosphorylation regulates the recruitment of Pol I to the rDNA promoter (Claypool et al. 2003). This finding underscores the evolutionary conservation of a fundamental mechanism that regulates appropriate rRNA synthesis in response to nutrient availability. Another study, on the other hand, showed that in NIH3T3 cells the activity of UBF, and not TIF-IA, is regulated by mTOR signaling pathways via a mechanism that requires S6K1 activation (Hannan et al. 2003). Rapamycin treatment led to rapid dephosphorylation of UBF, which reduced its ability to associate with TIF-IB/SL1. In our hands, however, rapamycin-mediated repression of rDNA transcription was exclusively rescued by TIF-IA, and no additive or synergistic effect of UBF was observed. A detailed analysis of the different experimental setups will be necessary to explain this obvious discrepancy to our results. Taken together, our results are consistent with the following model: Under favorable conditions, mTOR promotes rRNA synthesis by activating Cdk-mediated phosphorylation of TIF-IA at S44, while preventing phosphorylation at S199. These modifications are required for tethering TIF-IA to Pol I and retaining TIF-IA in the nucleus and nucleolus. Upon unfavorable conditions, mTOR is inactive, PP2A is activated, and the unleashed PP2A phosphatase dephosphorylates TIF-IA. In addition, S199 is hyperphosphorylated by a yet to be identified kinase, leading to sequestration of TIF-IA in the cytoplasm. Thus, mTOR signaling occupies a central position in the global regulation of rRNA synthesis, and TIF-IA, which is conserved from yeast to man (Moorefield et al. 2000), appears to be a key signaling molecule utilized by many, if not all, eukaryotes to control growth in response to nutrient availability.
Materials and methods
Plasmids
cDNAs encoding wild-type (DDBJ/EMBL/GenBank accession no. AJ272050) and mutant TIF-IA were tagged at the 5′ end with sequences encoding the Flag epitope peptide and cloned into pcDNA3.1 (Invitrogen) and baculovirus-based vectors. To generate mutant forms of TIF-IA, Ser 44 or Ser 199 were replaced by alanine or aspartic acid, respectively, by overlap extension PCR. pMr600, the template used for in vitro transcription, contains a 622-bp PvuII fragment harboring murine rDNA sequences from –324 to +292. The reporter plasmid pMr1930-BHcontains a 5′-terminal mouse rDNA fragment (from –1930 to +292) fused to a 3′-terminal rDNA fragment including two “Sal box” terminator elements (Budde and Grummt 1999). Vectors encoding HA-tagged mTOR (Dennis et al. 2001) and GST/myc-tagged S6K1 and S6K1dd (K100Q) have been described (Dennis et al. 1998).
Cell culture, transfections, and RNA analysis
NIH3T3 and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% FCS and 2 mM glutamine. To inhibit mTOR signaling, cells were treated with 20 nM rapamycin for 1–2 h. Cellular pre-rRNA was monitored on Northern blots by hybridization to antisense RNA complementary to the first 155 nucleotides of unprocessed 45S pre-RNA (Voit et al. 1999). For reporter assays, 5 × 105 NIH3T3 cells were cotransfected with 2.5 μg of reporter plasmid pMr1930-BHand different amounts of pcDNA3.1-Flag-TIF-IA. RNA was isolated after 48 h, and transcripts from the rDNA reporter plasmid were monitored by hybridization to a riboprobe that is complementary to pUC sequences inserted between the rDNA promoter and terminator sequences of pMr1930-BH.
In vitro transcription assays
Nuclear extracts were prepared from exponentially growing, rapamycin-treated (20 nM for 1 h), amino acid-starved, or density-arrested FM3A cells. For density arrest, cells were cultured for 12–16 h at a density of 2 × 106 cells/mL; 25 μL assays contained 50 ng of template DNA (pMr600/EcoRI), 30 μg nuclear extract proteins, 12 mM HEPES-KOH, (pH 8.0), 0.1 mM EDTA, 5 mM MgCl2, 80 mM KCl, 10 mM creatine phosphate, 12% (v/v) glycerol, 0.66 mM each of ATP, GTP, and CTP, 0.01 mM UTP, and 0.5 μCi [α32P]UTP (5000 Ci/mmole). After incubation for 60 min at 30°C, RNA was extracted and analyzed on non-denaturing 5% polyacrylamide gels.
Metabolic labeling and tryptic phosphopeptide analysis of TIF-IA
For metabolic labeling and tryptic phosphopeptide analysis of TIF-IA, 3–5 × 106 HEK293T cells overexpressing Flag-tagged TIF-IA were cultured in DMEM/10% FCS, transferred to phosphate-free DMEM with 10% dialyzed FCS for 45 min before labeling with 0.5 mCi/mL [32P]orthophosphate for 2–4 h. Cells were lysed in RIPA buffer (20 mM Tris-HCl at pH 7.4, 200 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% Triton X-100, 0.1% SDS, 10 mM β-glycerophosphate, 10 mM KH2PO4, 1 mM Na3VO4, 0.5 mM PMSF, 1 μg/mL aprotinin, 5 μg/mL leupeptin, and 1 μg/mL pepstatin A). TIF-IA was immunoprecipitated, separated by SDS-PAGE, transferred to nitrocellulose, and visualized by autoradiography. TIF-IA was digested overnight at 37°C with trypsin (Promega, sequencing grade) in 50 mM ammonium bicarbonate. After lyophilization, peptides were resolved on cellulose thin-layer plates by electrophoresis for 25 min at 1000 V in 1% (w/v) ammonium carbonate (pH8.9), followed by ascending chromatography in a buffer containing 62.5% (v/v) isobutyric acid, 1.9% (v/v) n-butanol, 4.8% (v/v) pyridine, and 2.9% (v/v) acetic acid.
To phosphorylate TIF-IA in vitro, 200 ng of immobilized Flag-tagged TIF-IA was incubated with 100 μg nuclear extract in 30 μL of kinase buffer (30 mM Tris-HCl at pH 8.0, 10 mM MgCl2, 20% glycerol) containing 100 μM ATP and 5 μCi of [γ-32P]ATP (5000 Ci/mmole) for 30 min at 30°C.
Purification of TIF-IA, mTOR, S6K1, and Cdk2/cyclinE
Cellular TIF-IA was purified from nuclear extracts of FM3A cells by chromatography on DEAE-Sepharose and Heparin-Ul-trogel (Schnapp and Grummt 1996), followed by fractionation on polylysine–agarose (PL-650 fraction). Recombinant TIF-IA was immunopurified either from baculovirus-infected Sf9 cells or from HEK293T cells overexpressing Flag-tagged TIF-IA. The 4 × 106 cells were lysed in 0.8 mL of IP buffer (20 mM Tris-HCl at pH7.4, 200 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% Triton X-100) in the presence of protease inhibitors. After centrifugation at 10,000 × g for 30 min, the supernatants were incubated for 4 h at 4°C with anti-Flag (M2) agarose (Sigma). The immunoprecipitates were successively washed with buffer AM (20 mM Tris-HCl at pH 8.0, 5 mM MgCl2, 0.1 mM EDTA, 20% glycerol) containing 800 and 100 mM KCl, respectively. The amount and purity of TIF-IA was estimated on silver-stained SDS-polyacrylamide gels.
mTOR and S6K1 were purified from HEK293T cells overexpressing HA-tagged mTOR or GST/myc-tagged S6K1, respectively. The 5 × 106 cells were lysed in 0.8 mL of IP buffer, centrifuged at 10,000 × g for 20 min, and the supernatants were incubated for 4 h at 4°C with either 15 μg of anti-HA antibodies bound to 20 μL of protein G-agarose or 20 μL of GST–glutathione Sepharose. Beads were washed three times with AM-400 and AM-100 and twice in 1.5× kinase buffer (37 mM Tris-HCl at pH7.8, 10 mM MgCl2, 1.5 mM DTT, 1.5 mM β-glycerophosphate, 0.1 mM Na3VO4) before adding to transcription or kinase assays. Immunopurification of Cdk2/cyclin E from Sf9 cells and in vitro kinase assays have been described (Voit and Grummt 2001). Bead-bound cdk2/cyclin E was washed with IP buffer containing 300 mM NaCl and 0.5% Triton X-100, and finally with IP buffer containing 10% glycerol. To phosphorylate TIF-IA, 0.5 μg of GST/TIF-IA was incubated in 15 μL of kinase buffer containing 20 μCi [γ-32P]ATP (5,000 Ci/mmole) and 10 fmoles Cdk2/cyclin E.
Coimmunoprecipitation and immunoblotting
To analyze the interaction of TIF-IA with Pol I and TIF-IB/SL1, cells overexpressing Flag-tagged TIF-IA were lysed in IP buffer, cleared by centrifugation at 10,000 × g for 30 min, and the supernatants were incubated for 4 h at 4°C with anti-Flag (M2) agarose (Sigma). The immunoprecipitates were washed with IP buffer (for associated TIF-IB/SL1) or with IP buffer containing 600 mM NaCl (for Pol I). TIF-IB/SL1 was visualized on immunoblots with α-TAFI110 antibodies and Pol I with antibodies against the second largest subunit of Pol I (RPA116) or PAF67 (Seither et al. 2001).
Immunofluorescence
Cells grown on coverslips were fixed with 1% paraformaldehyde for 20 min. After permeabilization with methanol for 1 min, cells were washed twice with cold TBS containing 0.1% BSA. Flag-tagged TIF-IA was detected with Cy3-conjugated anti-Flag monoclonal antibodies (M2, Sigma). Immunofluorescence was monitored by microscopy and images were digitally recorded.
Acknowledgments
We thank Bettina Dörr for preparation and fractionation of cell extract and G. Thomas and Y.-G. Gangloff for the S6K1 expression vectors. We thank all members of the lab for sharing reagents and advice. This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.285504.
References
- Albers M.W., Williams, R.T., Brown, E.J., Tanaka, A., Hall, F.L., and Schreiber, S.L. 1993. FKBP-rapamycin inhibits a cyclin-dependent kinase activity and a cyclin D1-Cdk association in early G1 of an osteosarcoma cell line. J. Biol. Chem. 268: 22825–22829. [PubMed] [Google Scholar]
- Beck T. and Hall, M.N. 1999. The Tor signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. [DOI] [PubMed] [Google Scholar]
- Bodem J., Dobreva, G., Hoffmann-Rohrer, U., Iben, S., Zentgraf, H., Delius, H., Vingron, M., and Grummt, I. 2000. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1: 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn G.J., Hudson, C.C., Sekulic, A., Williams, J.M., Hosoi, H., Houghton, P.J., Lawrence Jr., J.C., and Abraham, R.T. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277: 99–101. [DOI] [PubMed] [Google Scholar]
- Budde A. and Grummt, I. 1999. p53 represses ribosomal gene transcription. Oncogene 18: 1119–1124. [DOI] [PubMed] [Google Scholar]
- Burnett P.E., Barrow, R.K., Cohen, N.A., Snyder, S.H., and Sabatini, D.M. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. 95: 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit D., Pflugfelder, G., and Grummt, I. 1985. Growth-dependent regulation of rRNA synthesis is mediated by a transcription initiation factor (TIF-IA). Nucleic Acids Res. 13: 8165–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh A.H., Hirschler-Laszkiewicz, I., Hu, Q., Dundr, M., Smink, T., Misteli, T., and Rothblum, L.I. 2002. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 277: 27423–27432. [DOI] [PubMed] [Google Scholar]
- Chou M.M and Blenis, J. 1996. The 70 kDa S6 kinase complexes with and is activated by the Rho family G proteins Cdc42 and Rac1. Cell 85: 573–583. [DOI] [PubMed] [Google Scholar]
- Claypool J.A., French, S.L., Johzuka, K., Eliason, K., Vu, L., Dodd, J.A., Beyer, A.L., and Nomura, M. 2003. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Cell. Biol. 15: 946–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P.B., Pullen, N., Pearson, R.B., Kozma, S.C., and Thomas, G. 1998. Phosphorylation sites in the autoinhibitory domain participate in p70s6k activation loop phosphorylation. J. Biol. Chem. 273: 14845–14852. [DOI] [PubMed] [Google Scholar]
- Dennis P.B., Jaeschke, A., Saitoh, M., Fowler, B., Kozma, S.C., and Thomas, G. 2001. Mammalian TOR: A homeostatic ATP sensor. Science 294: 1102–1105. [DOI] [PubMed] [Google Scholar]
- Di Como C.J. and Arndt, K.T. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes & Dev. 10: 1904–1916. [DOI] [PubMed] [Google Scholar]
- Gingras A.C., Raught, B., and Sonenberg, N. 2001. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 15: 807–826. [DOI] [PubMed] [Google Scholar]
- Grummt I. 2003. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes & Dev. 17: 1691–1702. [DOI] [PubMed] [Google Scholar]
- Hannan K.M., Brandenburger, Y., Jenkins, A., Sharkey, K., Cavanaugh, A., Rothblum, L., Moss, T., Poortinga, G., McArthur, G.A., Pearson, R.B., et al. 2003. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxyterminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 23: 8862–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yonezawa, K., Weng, Q.P., Kozlowski, M.T., Belham, C., and Avruch, J. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273: 14484–14494. [DOI] [PubMed] [Google Scholar]
- Hardwick J.S., Kuruvilla, F.S., Tong, J.K., Shamji, A.F., and Schreiber, S.L. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. 96: 14866–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley D. and Cooper, G. 2002. Role of mTOR in the degradation of IRS-1: Regulation of PP2A activity. J. Cell. Biochem. 85: 304–314. [DOI] [PubMed] [Google Scholar]
- Hashemolhosseini S., Nagamine, Y., Morley, S.J., Desrivieres, S., Mercep, L., and Ferrari, S. 1998. Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J. Biol. Chem. 273: 14424–14429. [DOI] [PubMed] [Google Scholar]
- Hidayat S., Yoshino, K., Tokunaga, C., Hara, K., Matsuo, M., and Yonezawa, K. 2003. Inhibition of amino acid-mTOR signaling by a leucine derivative induces G1 arrest in Jurkat cells. Biochem. Biophys. Res. Commun. 301: 417–423. [DOI] [PubMed] [Google Scholar]
- Isotani S., Hara, K., Tokunaga, C., Inoue, H., Avruch, J., and Yonezawa, K. 1999. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase α in vitro. J. Biol. Chem. 274: 34493–34498. [DOI] [PubMed] [Google Scholar]
- Jacinto E. and Hall, M.N. 2003. TOR signaling in bugs, brain and brawn. Nat. Rev. 4: 117–126. [DOI] [PubMed] [Google Scholar]
- Jiang Y. and Broach, J.R. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18: 2782–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata S., Sakaida, H., Hori, T., Maeda, M., and Uchiyama, T. 1998. The upregulation of p27Kip1 by rapamycin results in G1 arrest in exponentially growing T-cell lines. Blood 91: 561–569. [PubMed] [Google Scholar]
- Kim D.H., Sarbassov dos, D., Ali, S.M., Latek, R.R., Guntur, K.V., Erdjument-Bromage, H., Tempst, P., and Sabatini, D.M. 2003. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 11: 895–904. [DOI] [PubMed] [Google Scholar]
- Kitagawa M., Higashi, H., Jung, H.K, Suzuki-Takahashi, I., Ikeda, M., Tamai, K., Kato, J., Segawa, K., Yoshida, E., Nishimura, S., et al. 1996. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15: 7060–7069. [PMC free article] [PubMed] [Google Scholar]
- Klein J. and Grummt, I. 1999. Cell cycle-dependent regulation of RNA polymerase I transcription: The nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci.. 96: 6096–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C.J. 2001. Role of leucine in the regulation of mTOR by amino acids: Revelations from structure-activity studies. J. Nutrit. 131: 861–865. [DOI] [PubMed] [Google Scholar]
- Mahajan P.B. 1994. Modulation of transcription of rRNA genes by rapamycin. Int. J. Immunopharmacol. 16: 711–721. [DOI] [PubMed] [Google Scholar]
- Milkereit P. and Tschochner, H. 1998. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 17: 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Panov, K.I., Friedrich, J.K., Trinkle-Mulcahy, L., Lamond, A.I., and Zomerdijk, J.C. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 20: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorefield B., Greene, E.A., and Reeder, R.H. 2000. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad. Sci. 97: 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R.T., Desai, B.N., Hardwick, J.S., and Schreiber, S.L. 1999. Protein phosphatase 2A interacts with 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycin associated protein. Proc. Natl. Acad. Sci. 96: 4438–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. and Walter, P. 1999. Regulation of the ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 104: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud C.G. 2002. Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. 269: 5338–5349. [DOI] [PubMed] [Google Scholar]
- Pullen N. and Thomas, G. 1997. The modular phosphorylation and activation of p70S6k. FEBS Lett. 410: 78–82. [DOI] [PubMed] [Google Scholar]
- Schmelzle T. and Hall, M.N. 2000. TOR, a central controller of cell growth. Cell 103: 253–262. [DOI] [PubMed] [Google Scholar]
- Schnapp A. and Grummt, I. 1996. Purification, assay, and properties of RNA polymerase I and class I-specific transcription factors in mouse. Methods Enzymol. 273: 233–248. [DOI] [PubMed] [Google Scholar]
- Schnapp A., Pfleiderer, C., Rosenbauer, H., and Grummt, I. 1990. A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J. 9: 2857–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser A., Bodem, J., Bossemeyer, D., Grummt, I., and Lehmann, W.D. 2002. Identification of protein phosphorylation sites by combination of elastase digestion, immobilized metal affinity chromatography, and quadrupole-time of flight tandem mass spectrometry. Proteomics 2: 911–918. [DOI] [PubMed] [Google Scholar]
- Seither P., Iben, S., Thiry, M., and Grummt, I. 2001. PAF67, a novel protein that is associated with the initiation-competent form of RNA polymerase I. Biol. Chem. 382: 1163–1170. [DOI] [PubMed] [Google Scholar]
- Tower J. and Sollner-Webb, B. 1987. Transcription of mouse rDNA is regulated by an activated subform of RNA polymerase I. Cell 50: 873–883. [DOI] [PubMed] [Google Scholar]
- Voit R. and Grummt, I. 2001. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc. Natl. Acad. Sci. 98: 13631–13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R., Hoffmann, M., and Grummt, I. 1999. Phosphorylation by G1-specific cdk–cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 18: 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Gout, I., and Proud, C.G. 2001. Cross-talk between the ERK and p70 S6 kinase (S6K) signaling pathways. MEK-dependent activation of S6K2 in cardiomyocytes. J. Biol. Chem. 276: 32670–32677. [DOI] [PubMed] [Google Scholar]
- Yan Y. and Mumby, M.C. 1999. Distinct roles for PP1 and PP2A in phosphorylation of the retinoblastoma protein. PP2A regulates the activities of G(1) cyclin-dependent kinases. J. Biol. Chem. 274: 31917–31924. [DOI] [PubMed] [Google Scholar]
- Yuan X., Zhao, J., Zentgraf, H., Hofmann-Rohrer, U., and Grummt, I. 2002. Multiple interactions between RNA polymerase I, TIF-IA, and TAFI subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep. 3: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza D., Ghavidel, A., Heitman, J., and Schultz, M.C. 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18: 4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Stallock, J.P., Ng, J.C., Reinhard, C., and Neufeld, T.P. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes & Dev. 14: 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan, Y., Frödin, M., and Grummt, I. 2003. ERK-dependent phosphorylation of the transcription factor TIF-IA is required for RNA polymerase I transcription. Mol. Cell 11: 405–413. [DOI] [PubMed] [Google Scholar]