Abstract
Vascular smooth muscle cell (VSMC) phenotypic modulation and proliferation are critical cellular events in the development of a variety of proliferative vascular diseases. However, the molecular mechanisms involved in cellular events are still unclear. MicroRNAs (miRNAs) represent a novel class of small, non-coding RNAs that negatively regulate gene expression via degradation or translational inhibition of their target mRNAs. In a previous study, we identified that miR-145 is the most abundant miRNA in normal arteries and VSMCs. However, the roles of miR-145 in VSMC biology and vascular disease are unknown. In our recent Circulation Research article, we found that the expression of miR-145 is significantly downregulated in dedifferentiated VSMCs and in balloon-injured arteries. Moreover, both in vitro and in vivo studies demonstrated that miR-145 is a critical modulator of VSMC phenotype and proliferation. This review article summarizes the current research progress regarding the roles of miR-145 in VSMC biology and discusses the potential therapeutic opportunities surrounding this miRNA in vascular disease.
Keywords: microRNA, microRNA-145, smooth muscle cells, vascular disease, proliferation, differentiation
Introduction
Vascular smooth muscle cells (VSMCs) are not terminally differentiated and are able to modulate their phenotype in response to changing local environmental cues. It is well known that the phenotypic modulation of VSMCs from a differentiated phenotype to a dedifferentiated state, accompanied by accelerated VSMC proliferation, plays a critical role in the pathogenesis of a variety of proliferative vascular diseases such as atherosclerosis, hypertension, restenosis after angioplasty or bypass, diabetic vascular complications, and transplantation arteriopathy.1-3 However, the molecular mechanisms involved in VSMC phenotypic modulation and proliferation are still unclear.
MicroRNAs (miRNAs) represent a novel class of endogenous, small, non-coding RNAs that negatively regulate gene expression via degradation or translational inhibition of their target mRNAs.4 More importantly, one miRNA is able to regulate the expression of multiple genes because it can bind to its mRNA targets as either an imperfect or perfect complement. Thus, one miRNA is as functionally important as a transcription factor.5 As a group, miRNAs may directly regulate at least 30% of the genes in a cell.6 It is therefore not surprising that miRNAs are involved in the regulation of all major cellular functions.
microRNA-145 (miR-145) is a 22-nt, highly conserved miRNA. Recently, miR-145 has been highlighted, because it is among the most downregulated miR-NAs in diverse cancers including bladder cancer,7,8 breast cancer,9,10 colon cancer,11,12 colorectal cancer,13-16 gastric cancers,17 hepatocellular carcinoma,18,19 lung cancer,20,21 nasopharyngeal carcinoma,22 oral cancer,23 ovarian cancer,24,25 pituitary tumors26 and prostate cancer.27 miR-145 has a strong inhibitory effect on cancer cell proliferation and is a novel tumor suppressor.20,21 However, the roles of miR-145 in VSMC biology and vascular disease have not been explored until recently. In our two recent Circulation Research articles, we have identified that miR-145 is the most abundant miRNA in normal arteries and in differentiated VSMCs.28,29 Interestingly, the expression of this VSMC-enriched miRNA is significantly down-regulated in dedifferentiated VSMCs, balloon-injured arteries, and atherosclerotic arteries. Moreover, we have demonstrated both in vitro and in vivo that miR-145 is critical modulator of VSMC phenotype and proliferation. The biological roles of miR-145 in VSMCs have been further verified by a recent research article in Nature.30 This review article summarizes the current research progress regarding the roles of miR-145 in VSMC biology and discusses the potential therapeutic opportunities surrounding this miRNA in vascular disease.
miR-145 is the Most Abundant miRNA in Normal Arteries and is Mainly Localized in VSMCs
Tissue-specific expression is one important characteristic of miRNA expression. One miRNA may be highly expressed in one tissue but have no or low expression in other tissues. To study the biological functions of miR-145 in vascular disease, we first determined the relative miR-145 expression in rat carotid arteries through miRNA microarray analysis. Among the arrayed miRNAs, miR-145 was the most abundant miRNA in normal rat carotid arteries.28 As we know, VSMCs, endothelial cells (ECs), and fibroblasts are the three major cell types in normal vascular walls. To further determine the cellular distribution of miR-145 expression in the vascular wall, we isolated VSMCs, ECs, and fibroblasts from rat arteries and measured miR-145 levels in these cells by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). As shown in Figure 1, miR-145 was highly expressed in VSMCs. However, it was almost undetectable in ECs. In addition, miR-145 was also expressed in fibroblasts, but the expression level was lower than that of VSMCs. The cellular distribution of miR-145 was further confirmed by in situ hybridization of rat carotid arteries. Thus, miR-145 is mainly expressed in the VSMCs of the normal artery.
Figure 1.
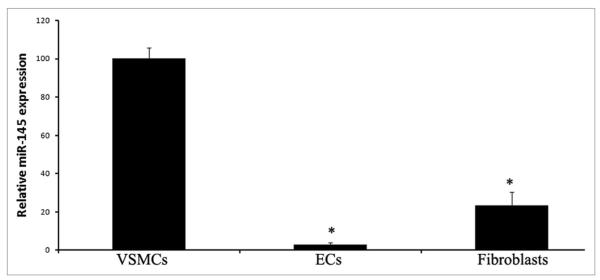
Relative expression of miR-145 in rat vascular VSMCs, ECs and fibroblasts. Note: n = 6; *p < 0.05 compared with that in VSMCs.
miR-145 is the Most Abundant miRNA in Primary Cultured VSMCs and its Expression is Quickly Downregulated in Dedifferentiated VSMCs
In our recent study, we identified that miR-145 is the most abundant miRNA in primary cultured rat VSMCs.29 The expression of this miRNA was significantly downregulated in sub-cultured dedifferentiated VSMCs and VSMCs in a platelet-derived growth factor (PDGF)-induced dedifferentiated state.29 Cordes et al. found that, during the development of arteries, the expression of miR-145 is associated with the state of VSMC differentiation.30 miR-145 expression is notably absent in the aorta and pulmonary arteries in later cardiogenesis, during which VSMCs and arteries are developing. In contrast, high transcript levels of miR-145 in VSMCs of the arteries are demonstrated postnatally, after VSMCs and arteries completed development.30
miR-145 is a Critical Switch for VSMC Phenotypic Modulation
The positive correlation of miR-145 expression and VSMC differentiation encouraged us to determine the biological role of miR-145 in VSMC phenotypic modulation. In a PDGF-induced phenotypic modulation model, we tested whether or not miR-145 had an effect on the VSMC phenotype. We found that overexpression of miR-145 increased expression of VSMC differentiation marker genes such as SM α-actin, calponin, and SM-MHC. In contrast, levels of these marker genes were decreased in VSMCs treated with a miR-145 inhibitor, 2′OMe-miR-145.29 Similar effects of miR-145 modulation were also found in quiescent, non-stimulated VSMCs: we found that modulating the miR-145 level itself is sufficient to elicit phenotypic changes. The regulatory effect of miR-145 on VSMC phenotype was further verified in a Nature article, in which a strong regulatory effect of miR-145 on VSMC differentiation was identified.30 In an in vivo VSMC phenotypic modulation model, induced by balloon-injury of rat carotid arteries, we confirmed via adenovirus-mediated miR-145 gene transfer that miR-145 is a critical regulator of VSMC phenotypic modulation in arterial walls.29
miR-145 is Involved in VSMC Proliferation
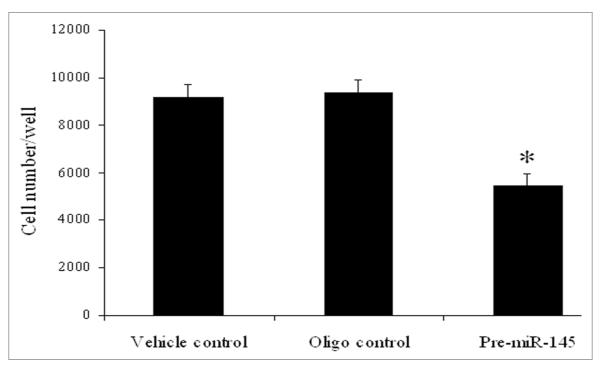
VSMC phenotypic modulation is accompanied by changes in VSMC proliferation. The differentiated VSMCs, also referred to as the “contractile” phenotype, often have a lower proliferative rate. In contrast, a higher proliferative rate is often found in dedifferentiated VSMCs, also referred to as the “synthetic” phenotype. To further determine the biological roles of miR-145 in VSMCs, the effect of upregulation of miR-145 via pre-miR-145 (100 nM) on cultured VSMC proliferation was determined. As shown in Figure 2, VSMC proliferation induced by PDGF was significantly inhibited by overexpression of miR-145. The inhibitory effect of miR-145 on VSMCs was further verified by the Nature article.30
Figure 2.
The effect of pre-miR-145 (100 nM) on cell proliferation in cultured VSMCs at 24 h after treatment with PDGF (20 ng/ml). Note: n = 6; *p < 0.05 compared with oligo control.
Potential Target Genes Involved in miR-145-Mediated Cellular Effects
miRNAs achieve their biological functions via their multiple target genes. Currently, we are just beginning to understand which target genes are involved in miR-145-mediated cellular effects. It should be noted that target genes of miRNAs are cell-type specific. In cancer cells, c-Myc, rhotekin (RTKN), insulin receptor substrate-1, and beta-actin are target genes of miR-145 that are related to its inhibitory effect on cancer cell growth.31-33 In human embryonic stem cells, OCT4, SOX2 and kruppel-like factor 4 (KLF4) were found to be the miR-145 target genes.34 In addition, superoxide dismutase-2 was found to be a target gene of miR-145 in brain cells.35
In VSMCs, we have identified that kruppel-like factor 5 (KLF5) is a target gene of miR-145 that is responsible for its regulatory effect on VSMC phenotypic modulation and VSMC proliferation.29 Although KLF4 is also a potential target gene of miR-145 in VSMCs, the time course change of its expression in cultured VSMCs after PDGF stimulation and in balloon-injured rat carotid arteries does not match perfectly with the expression change of miR-145.29 Thus, the role of KLF4 is miR-145-mediated biological effects on VSMCs should be further studied. In the recent Nature article, Cordes et al. found that KLF4 and Calmodulin kinase IIdelta (CamkIId) are two target genes of miR-145 in VSMCs.30 In addition, they also found that myocardin also may be a direct target gene of miR-145 in VSMCs. However, in contrast to the inhibitory effect of a miRNA on its target genes, overexpression of miR-145 increased the expression of myocardin.30 In our Circulation Research article, we also found that the expression of myocardin was increased in miR-145-overexpressed VSMCs. However, our explanation is that, at least in part, the upregulated myocardin is induced by indirect effect of miR-145 through its target gene KLF5.29
Downregulation of miR-145 in Proliferative Vascular Diseases
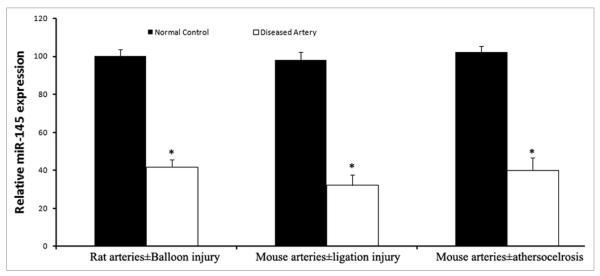
To uncover the potential roles of miRNAs in proliferative vascular diseases, miR-145 expression in rat balloon-injured arteries, mouse ligation-injured carotid arteries, and atherosclerotic aortas of ApE knock-out mice was determined. As shown in Figure 4, miR-145 is significantly down-regulated in these arteries with intimal hyperplasia. The results in the diseased mouse arteries are consistent with the recent study by Cordes et al.30 Notably, in their study, transcripts of miR-145 were downregulated to nearly undetectable levels in atherosclerotic arteries, an even more pronounced change compared to the result we obtained (Fig. 1). One possible reason for the discrepancy may be related to the tissue selection. As we described above, the expression of miR-145 is mainly localized in VSMCs. If the selected atherosclerotic tissue had fewer VSMCs, the expression level of miR-145 could be lower.
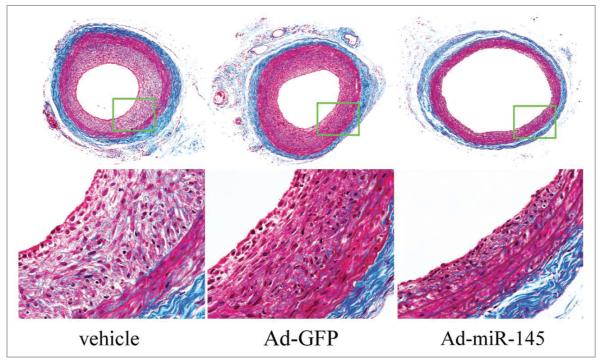
Figure 4.
Representative Masson’s trichrome stained photomicrographs of rat carotid arteries treated with vehicle, control adenovirus (Ad-GFP) or adenovirus expressing miR-145 (Ad-miR145).
Potential Therapeutic Opportunity of the miR-145 in Vascular Disease
VSMC phenotypic modulation and proliferation are critical cellular events in the pathogenesis of a variety of proliferative vascular diseases with neointimal lesion formation. As the recent studies have demonstrated that miR-145 is an important regulator for VSMC phenotype and proliferation, we predict that miR-145 may represent a novel therapeutic target for vascular disease. Indeed, as demonstrated in our Circulation Research article, restoration of the downregulated miR-145 is sufficient to inhibit neointimal lesion formation in rat carotid arteries after angioplasty (Fig. 4). However, we should be aware that we are at very early stage in studying the roles of miR-145 in VSMCs and vascular disease. We still have a long way to go before miR-145-related therapies can be applied to clinical vascular disease. The detailed target genes, the potential side effects, and a therapeutic strategy to upregulate the downregulated miR-145 in the vessels under the disease condition should first be identified.
Figure 3.
The expression of miR-145 in rat balloon-injured carotid arteries, mouse ligated carotid arteries, atherosclerotic aortas, compared with that in their normal controls. Note: n = 5; *p < 0.05 compared with oligo control.
Acknowledgements
This work was supported by a National Institutes of Health Grant HL080133 and a grant from American Heart Association 09GRNT2250567.
Abbreviations
- miRNAs
microRNAs
- miR-145
microRNA-145
- VSMCs
vascular smooth muscle cells
- PDGF
platelet-derived growth factor
- ECs
endothelial cells
- RTKN
rhotekin
- KLF4
kruppel-like factor 4
- KLF5
kruppel-like factor 5
- CamkIId
calmodulin kinase IIdelta
References
- 1.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 2.Rzucidlo EM, Martin KA, Powell RJ. Regulation of vascular smooth muscle cell differentiation. J Vasc Surg. 2007;45:25–32. doi: 10.1016/j.jvs.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Cai X. Regulation of smooth muscle cells in development and vascular disease: current therapeutic strategies. Expert Rev Cardiovasc Ther. 2006;4:789–800. doi: 10.1586/14779072.4.6.789. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–52. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 8.Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–60. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 10.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell sub-populations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 11.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 12.Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–24. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 13.Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29–32. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 15.Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, et al. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26:27–34. doi: 10.3233/DMA-2009-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, et al. Over- and under- expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–75. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 17.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 18.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently downregulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 19.Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–32. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Sempere LF, Galimberti F, Freemantle SJ, Black C, Dragnev KH, et al. Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer Res. 2009;15:1177–83. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45:2197–206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 22.Chen HC, Chen GH, Chen YH, Liao WL, Liu CY, Chang KP, et al. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer. 2009;100:1002–11. doi: 10.1038/sj.bjc.6604948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu T, Wang XY, Gong RG, Li A, Yang S, Cao YT, et al. The expression profile of microRNAs in a model of 7,12-dimethyl-benz[a]anthrance-induced oral carcinogenesis in Syrian hamster. J Exp Clin Cancer Res. 2009;28:64–70. doi: 10.1186/1756-9966-28-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 25.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–5. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 26.Amaral FC, Torres N, Saggioro F, Neder L, Machado HR, Silva WA, Jr, et al. MicroRNAs differentially expressed in ACTH-secreting pituitary tumors. J Clin Endocrinol Metab. 2009;94:320–3. doi: 10.1210/jc.2008-1451. [DOI] [PubMed] [Google Scholar]
- 27.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–93. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 28.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–88. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 29.Cheng Y, Liu X, Yang J, Lin Y, Xu D, Lu Q, et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–66. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207–12. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng L, et al. miR-145 inhibits breast cancer cell growth through RTKN. Int J Oncol. 2009;34:1461–6. [PubMed] [Google Scholar]
- 33.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 34.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2 and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 35.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29:675–87. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]