Abstract
Tea tree oil (TTO) is a steam distillate of Melaleuca alternifolia that demonstrates broad-spectrum antibacterial activity. This study was designed to document how TTO challenge influences the Staphylococcus aureus transcriptome. Overall, bioinformatic analyses (S. aureus microarray meta-database) revealed that both ethanol and TTO induce related transcriptional alterations. TTO challenge led to the down-regulation of genes involved with energy-intensive transcription and translation, and altered the regulation of genes involved with heat shock (e.g. clpC, clpL, ctsR, dnaK, groES, groEL, grpE and hrcA) and cell wall metabolism (e.g. cwrA, isaA, sle1, vraSR and vraX). Inactivation of the heat shock gene dnaK or vraSR which encodes a two-component regulatory system that responds to peptidoglycan biosynthesis inhibition led to an increase in TTO susceptibility which demonstrates a protective role for these genes in the S. aureus TTO response. Agene (mmpL) encoding a putative resistance, nodulation and cell division efflux pump was also highly induced by TTO. The principal antimicrobial TTO terpene, terpinen-4-ol, altered ten genes in a transcriptional direction analogous to TTO. Collectively, this study provides additional insight into the response of a bacterial pathogen to the antimicrobial terpene mixture TTO.
Keywords: tea tree oil, Staphylococcus aureus, transcriptomics, heat shock, vra
INTRODUCTION
Tea tree oil (TTO) extracted from Melaleuca alternifolia has been shown to exhibit broad-spectrum antimicrobial activity in vitro (Carson et al., 1995) and is comprised of hundreds of hydrocarbon components (Southwell and Lowe, 1999), including the major antimicrobial terpene, terpinen-4-ol (Carson and Riley, 1995). Preliminary trials suggest that TTO formulations may be effective in the treatment of acne and fungal infections, and bacterial pathogen decolonization protocols (Carson et al., 2006). The overall bactericidal activity of TTO is attributed to its ability to denature proteins and alter membrane and cell wall structure and function (Carson et al., 2002; Carson et al., 2006; Carson and Riley, 1995; Cox et al., 1998; Cox et al., 2000; Gustafson et al., 1998; Sikkema et al., 1995).
Infections caused by the bacterial pathogen Staphylococcus aureus are attributed to a mortality rate comparable to the deaths caused by HIV/AIDS, tuberculosis and viral hepatitis combined in the United States (Boucher and Corey, 2008). The relatively high incidence of infections caused by multiply antibiotic-resistant methicillin-resistant S. aureus (MRSA) strains remains a major concern within the medical community (Bubacz, 2007). For instance, there were an estimated 14 million healthcare visits for suspected skin and soft tissue infections caused by S. aureus in 2005 (Hersh et al., 2008), and MRSA can cause a high percentage of these infections (Moran et al., 2006). S. aureus and MRSA are susceptible to TTO and have been targeted in TTO decolonization clinical trials (Dryden et al., 2004).
In an effort to better understand the anti-staphylococcal activity of TTO, we have performed a transcriptional profiling experiment with S. aureus exposed to a growth inhibitory concentration of TTO. To our knowledge, this is the first report of a bacterial pathogen’s genome-wide TTO transcriptional response.
MATERIALS AND METHODS
Chemicals, TTO and bacterial strains utilized
All chemicals were purchased from Sigma-Aldrich Co. (St Louis, MO) except TTO, which was obtained as a gift from Paul Bolster (P. Guinane Pty. Ltd. New South Wales, Australia; M. alternifolia oil sample A354) (Table 1). Mueller–Hinton broth (MHB) and bacteriological grade agar were purchased from Becton Dickinson and Company (Sparks, MD). S. aureus strains SH1000, SH1000dnaK::kan, N315, N315vraSR::cat, COL, and clinical MRSA strains LP9 and MM66 have been previously described (Delgado et al., 2007; Horsburgh et al., 2002; Gustafson et al., 1992; Kuroda et al., 2003; Singh et al., 2007).
Table 1.
TTO chemical composition
| Component | A354 (%) | ISO4730 (% range) |
|---|---|---|
| α-pinene | 2.4 | 1–6 |
| sabinene | 0.4 | trace–3.5 |
| α-terpinene | 9.8 | 5–13 |
| limonene | 0.8 | 0.5–1.5 |
| p-cymene | 2.2 | 0.5–8 |
| 1,8-cineole | 1.7 | trace–15 |
| γ-terpinene | 20.6 | 10–28 |
| terpinolene | 3.4 | 1.5–5 |
| terpinen-4-ol | 41.5 | 30–48 |
| α-terpineol | 2.9 | 1.5–8 |
| aromadendrene | 1.5 | trace–3 |
| ledene | 0.9 | trace–3 |
| δ-cadinene | 1.1 | trace–3 |
| globulol | 0.3 | trace–1 |
| viridiflorol | 0.4 | trace–1 |
TTO and ethanol susceptibility testing, TTO kill curve and whole cell autolysis
Overnight, S. aureus cultures were incubated for 24 h (37 °C, 200 rpm) and used to initiate growth in all cultures required for the following experiments. TTO minimum inhibitory concentration (MIC) measurements were performed by adding 1ml of diluted overnight culture (final OD580 = 0.01) into 1ml of MHB containing 0.01% (v/v) Tween 80 and 0.05% to 0.65% TTO (v/v), and after 24 h incubation at 37 °C, MICs were determined. Ethanol MICs were performed as described above except; MHB containing 3% to 15% filter sterilized (0.22 μm) (Nalge Nunc International, Rochester, NY) ethanol (v/v) without Tween 80 addition was utilized. Minimum bactericidal concentrations (MBC) were determined by streaking 100 μl aliquots of the MIC tube, and all MIC tubes containing higher drug concentrations, onto MH agar and scoring for the lowest drug concentration with no observable growth following 24 h incubation (37 °C). The TTO kill curve was carried out utilizing early exponential SH1000 cultures (OD580 = 0.4) and the same culture challenged with 0.25% TTO (v/v). Colony forming units per ml (CFUs/ml) were then determined in untreated and TTO-challenged cultures 15, 30, 60 and 120 min after TTO exposure. Triton X-100-induced whole cell autolysis was performed as previously described (Gustafson et al., 1992). Briefly, individual 25 ml mid-exponential phase SH1000 cultures (OD580 = 0.4) were subjected to no challenge or 0.25% TTO challenge for 15 min. The cells were then harvested by centrifugation (8200 × g, 4 °C, 10 min), washed once with 5ml of ice-cold water and repelleted. The cell pellets were then suspended in 25 ml of Tris–HCl (0.05 M, pH 7.2) or the same buffer containing 0.05% Triton X-100. The cell suspensions were then incubated with gentle agitation at 37 °C, and OD580 was read every h for 8 h.
Microarray, quantitative real-time polymerase chain reaction and S. aureus microarray meta-database
Total RNA was isolated as previously described (Riordan et al., 2007), and DNA microarrays (TIGR version 6) were used for transcriptional profiling of laboratory strain SH1000 (MHB cultures OD580= 0.4 at 37 °C, 200 rpm) following 0.25% TTO v/v upshock for 15 min using protocols provided by Pathogen Functional Genomics Resource Center (http://pfgrc.jcvi.org/index.php/microarray/array_description/staphylococcus_aureus/version6.html). RNA isolated from untreated and TTO-challenged SH1000 cultures were then converted to fluorescently labeled cDNA and hybridized to S. aureus microarrays version 6 as previously described (Riordan et al., 2007; Delgado et al., 2008). Duplicate microarrays of SH1000 were hybridized and analyzed, and ORFs altered 1.5-fold and above were determined to be significant (p<0.05) (Riordan et al., 2007). The S. aureus TTO transcriptome data in this publication is deposited in NCBI’s Gene Expression Omnibus accessible through GEO series accession number GSE31554 (http://www.ncbi.nlm.nih.gov/geo/). Based on our laboratory preference, when possible, all genes were given S. aureus strain COL locus ID numbers. The S. aureus microarray meta-database (SAMMD) analysis was then used to compare the SH1000 TTO stimulon to 93 publicly available experimental S. aureus transcriptional responses (Nagarajan and Elasri, 2007). Initially, the ORF IDs of genes differentially regulated following TTO challenge were converted to strain COL locus IDs, and non-redundant IDs were then used to search against SAMMD. Select genes altered in the TTO microarray data were then validated by quantitative real-time polymerase chain reaction (qRT-PCR) as previously described (Riordan et al., 2007) using primers found in Table 2 in SH1000 cultures challenged with TTO as described above. In addition, qRT-PCR experiments were also performed with MHB cultures (OD580 = 0.4, 37 °C, 200 rpm) challenged with the major TTO antimicrobial component terpinen-4-ol (0.25% v/v final concentration) alone (Carson and Riley, 1995; Cox et al., 2001). Critical cycle threshold values were normalized using 16 S rRNA as an internal reference, and changes in gene expression were reported using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Table 2.
Primers used for qRT-PCR experiments
| Gene | Function | Locus ID | Primer sequence (forward and reverse) |
|---|---|---|---|
| betB | betaine aldehyde dehydrogenase | SACOL2628 | 5′-AATTGCTGTTGGTGGTAAACG-3′ |
| 5′-TAACGACAGGTCCGAAAACC-3′ | |||
| dnaK | DnaK protein | SACOL1637 | 5′-CCGGTGACAACAAACTTGG-3′ |
| 5′-TCAGCAGCATCTTTCAAACG-3′ | |||
| mmpL | MmpL efflux pump, putative | SACOL2566 | 5′-GGAATGACATCTACAGAAGTAGGC-3′ |
| 5′-AACTGCTAGTCCAATCATTACGG-3′ | |||
| purA | adenylosuccinate synthase | SACOL0018 | 5′-GAGGTTGGTCGTGAATACGG-3′ |
| 5′-TGGGTACTCAGTAATTTCTTTACCG-3′ | |||
| purM | phosphoribosylaminoimidazole synthetase | SACOL1080 | 5′-AATATGGGTATTGGCTATACGG-3′ |
| 5′-CACAATATGACCAATTTGATAGGC-3′ | |||
| rpmI | ribosomal protein L35 | SACOL1726 | 5′-TGCCAAAAATGAAAACTCACC-3′ |
| 5′-GAGATGTGAAAGCTCTTGAACG-3′ | |||
| tenA | transcriptional regulator, TenA family | SACOL2086 | 5′-TAGGAGCTGACGCATTACGC-3′ |
| 5′-CCCATTGTTCTAGTGTCATAGCC-3′ | |||
| vraR | DNA-binding response regulator VraR | SACOL1942 | 5′-AAAGAAGCAATTGCCAAAGC-3′ |
| 5′-TGAGTCGTCGCTTCTACACC-3′ | |||
| vraS | histidine kinase sensor | SACOL1943 | 5′-AGTGCCGATGAAAGTTGTGC-3′ |
| 5′-TTTTGTACCGTTTGAATGACG-3′ | |||
| vraX | VraX protein | SACOL0625 | 5′-TCGACAGTATCACCATGAAGG-3′ |
| 5′-TTTCAGTATCACTAAATGAATCGTCAC-3′ |
RESULTS AND DISCUSSION
SH1000 viable cell counts were determined following the addition of TTO at various time points. At 15 min, SH1000 cell population numbers had not decreased following TTO challenge (control 2.2 × 106 vs TTO challenged 2.0 × 106 CFUs/ml). Therefore, the cell population used in the microarray experiment described here does not represent a population that is experiencing events leading to immediate cell death. At all future time points investigated, the TTO-challenged cultures demonstrated a significant fall in surviving CFUs/ml only after 30min (30min – 1.3 × 106, 60 min – 5.2 × 105 and 120 min – 6.9 × 104 CFUs/ml).
Following 15 min of 0.25% TTO challenge, 312 genes were up-regulated, and 324 genes were down-regulated ≥1.5-fold (Table 3). The directional alteration in expression of ten of these genes (betB, dnaK, mmpL, purA, purM, rpmI, tenA, vraR, vraS and vraX) was confirmed by qRT-PCR (Table 3). qRT-PCR performed with cultures challenged with 0.25% v/v terpinen-4-ol also resulted in a similar direction of altered gene expression of these ten genes as TTO challenge (Table 3).
Table 3.
Representative S. aureus genes altered by TTO challenge
| Gene | Function | Locus ID | Fold change in gene expression
|
||
|---|---|---|---|---|---|
| Microarray | qRT- PCR (TTO) | qRT- PCR (Terpinen-4-ol) | |||
| Up-regulated genes | |||||
| vraX | VraX protein | SACOL0625 | 39.0 | 3.0 | 2.3 |
| mmpL | MmpL-like RND efflux pump | SACOL2566 | 14.7 | 2.8 | 1.6 |
| hypothetical protein (72 aa) | SACOL1033 | 13.1 | |||
| grpE | GrpE protein | SACOL1638 | 11.8 | ||
| hypothetical protein (39 aa) | SAR1729a | 10.7 | |||
| putative short chain oxidoreductase | SACOL2594 | 10.7 | |||
| hypothetical protein (138 aa) | SACOL2621 | 10.6 | |||
| hrcA | heat-inducible transcriptional repressor | SACOL1639 | 10.5 | ||
| cwrA | CwrA, conserved hypothetical protein | SACOL2571 | 10.4 | ||
| hypothetical protein (188 aa) | SACOL0568 | 9.4 | |||
| putative ATP:guanido phosphotransferase | SACOL0569 | 9.0 | |||
| csb7 | alkyl hydroperoxidase | SACOL2484 | 8.7 | ||
| clpC | ATP-dependent Clp protease, ATP-binding subunit ClpC | SACOL0570 | 8.3 | ||
| hypothetical protein (71 aa) | SAS1587 | 8.2 | |||
| acetoin reductase | SACOL0111 | 8.2 | |||
| ctsR | putative DNA-binding protein | SACOL0567 | 8.1 | ||
| hypothetical protein (146 aa) | SACOL0768 | 7.6 | |||
| epiE | epidermin immunity protein F | SACOL1872 | 7.4 | ||
| alpha/beta fold family hydrolase | SACOL2597 | 7.0 | |||
| clpL | ATP-dependent Clp proteinase chain | SACOL2563 | 6.9 | ||
| groEL | GroEL protein | SACOL2016 | 4.9 | ||
| groES | GroES protein | SACOL2017 | 4.1 | ||
| vraR | DNA-binding response regulator | SACOL1942 | 3.9 | 2.2 | 2.0 |
| dnaK | DnaK protein | SACOL1637 | 3.5 | 1.7 | 1.8 |
| dnaJ | DnaJ protein | SACOL1636 | 3.0 | ||
| vraS | histidine kinase sensor | SACOL1943 | 1.7 | 1.8 | 1.9 |
| Down-regulated genes | |||||
| purA | adenylosuccinate synthase | SACOL0018 | −13.1 | −2.2 | −2.1 |
| purM | phosphoribosylaminoimidazole synthetase | SACOL1080 | −11.5 | −1.7 | −1.6 |
| sle1 | N-acetylmuramyl-L-alanine amidase | SACOL0507 | −10.8 | ||
| isaA | lytic transglycosylase | SACOL2584 | −10.8 | ||
| purL | phosphoribosylformylglycinamidine synthetase | SACOL1078 | −10.5 | ||
| dltA | D-alanine-D-alanyl carrier protein ligase | SACOL0935 | −10.1 | ||
| purF | amidophosphoribosyltransferase | SACOL1079 | −9.8 | ||
| purK | phosphoribosylaminoimidazole carboxylase CO2-fixation | SACOL1074 | −8.9 | ||
| rplD | 50S ribosomal protein L4 | SACOL2238 | −8.6 | ||
| purE | phosphoribosylaminoimidazole carboxylase, catalytic subunit | SACOL1073 | −8.3 | ||
| purN | phosphoribosylaminoimidazole carboxylase, catalytic subunit | SACOL1072 | −8.0 | ||
| tenA | transcriptional regulator, TenA family | SACOL2086 | −7.9 | −2.9 | −1.8 |
| purH | phosphoribosylaminoimidazolecarboxamide formyltransferase | SACOL1082 | −7.8 | ||
| betA | choline dehydrogenase | SACOL2627 | −7.6 | ||
| ndk | nucleoside diphosphate kinase | SACOL1509 | −7.6 | ||
| purS | phosphoribosylformylglycinamidine synthase | SACOL1076 | −7.2 | ||
| purF | phosphoribosylpyrophosphate amidotransferase | SACOL1079 | −7.2 | ||
| similar to xanthine/uracil permease family protein | SACOL2242 | −7.1 | |||
| betB | betaine aldehyde dehydrogenase | SACOL2628 | −7.0 | −1.5 | −1.6 |
| hypothetical protein (191 aa) | SACOL1086 | −7.0 | |||
| rpmI | ribosomal protein L35 | SACOL1726 | −5.9 | −2.0 | −1.8 |
| thiD | phosphomethylpyrimidine kinase | SACOL2085 | −5.1 | ||
| purQ | phosphoribosylformylglycinamidine synthase I | SACOL1077 | −4.9 | ||
| purC | phosphoribosylaminoimidazole-succinocarboxamide synthase | SACOL1075 | −4.6 | ||
| thiE | putative thiamine-phosphate pyrophosphorylase | SACOL2083 | −3.8 | ||
| thiM | hydroxyethylthiazole kinase | SACOL2084 | −3.2 | ||
| adk | adenylate kinase | SACOL2218 | −2.5 | ||
Overall SAMMD analysis revealed the highest overlap between the TTO stimulon and the response of two unrelated MRSA strains (MM66 and LP9) (Delgado et al., 2008) to a growth-inhibiting ethanol concentration (10% v/v) (GEO series accession number GSE17391 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17391). The SH1000 TTO transcriptional response shared 425 and 469 altered genes with the LP9 and MM66 ethanol transcriptional responses, respectively. It is of interest to note that S. aureus strains demonstrating relatively high TTO (% v/v) MICs (SH1000 = 0.28 ± 0.01, LP9 = 0.2 ± 0) also demonstrated relatively high ethanol (%v/v)MICs (SH1000 = 9.0 ± 1.0, LP9 = 9.1 ± 1.0). Conversely, S. aureus strains that exhibited relatively low TTO MICs (COL= 0.15 ± 0, MM66 = 0.15 ± 0.03) also displayed relatively low ethanol MICs (COL= 6.9 ± 1.2, MM66 = 7.2 ± 1.0).
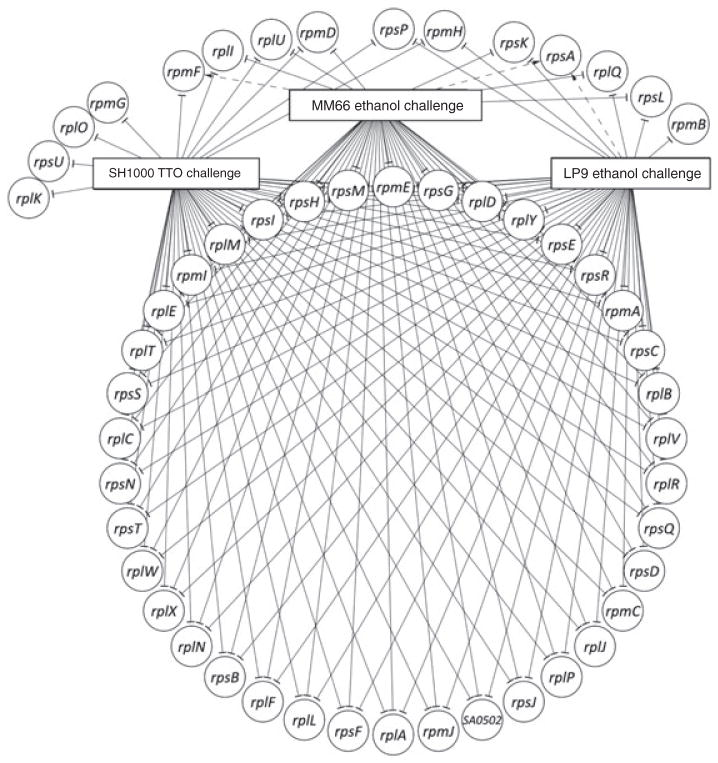
A total of 19 genes involved with transcription were down-regulated (−2.3- to −24.2-fold) following TTO-challenge including rpoA, rpoB, rpoC and rpoE which encode the DNA-directed RNA polymerase alpha-, beta- and delta-subunits, as well as a sigma factor. In addition, of the 149 translational genes found within S. aureus, 73 are down-regulated (−1.5- to −18.6-fold) in SH1000 following TTO challenge (Fig. 1). These down-regulated genes included 27 genes (≥ −1.6-fold) of the large 29 gene ribosomal operon (Wang et al., 2004). Many translation genes were similarly downregulated by ethanol exposure (Fig. 1).
Figure 1.
Translational genes similarly affected by TTO and ethanol challenge generated with SAMMD data and Cytoscape (Shannon et al., 2003). Solid lines ending in a perpendicular line denote gene down-regulation, dashed lines ending in an arrow denote gene up-regulation.
The classic heat shock genes encode proteases and molecular chaperones which collectively contribute to the continued maintenance and remodeling of denatured proteins following exposure to stress, such as ethanol or heat treatment (Chastanet et al., 2003; Gottesman et al., 1997; Qoronfleh et al., 1990). The processing of denatured proteins is energetically costly and powered by ATP hydrolysis (Kenniston et al., 2003; Lin and Rye, 2006). Some of the highest TTO up-regulated genes in SH1000 included the heat shock genes dnaK, groEL, groES, dnaJ and grpE (Table 3), which can be controlled by the synergistically acting HrcA and CtsR DNA-binding proteins (Chastanet et al., 2003). Both hrcA and ctsR were also up-regulated following TTO challenge (Table 3). DnaK is important to the general stress response, assists in denatured protein refolding and may be involved in membrane assembly, maintenance and structure (de Crouy-Chanel et al., 1999). It is also important in the response of S. aureus to TTO, since an insertionally inactivated dnaK mutant of SH1000 (SH1000dnaK::kan) displayed a lower TTO (% v/v) MIC (0.25 ± 0) and MBC (0.30 ± 0), compared to parent strain SH1000 (MIC = 0.28 ± 0.01, MBC= 0.32 ± 0.01, p<0.05). CtsR regulates the expression of the heat shock responsive clp ATP-binding protease genes (Derre et al., 1999), and clpB, clpC, clpL and clpP were all up-regulated following TTO challenge (Table 3). Two additional heat-responsive genes within the ctsR operon (SACOL0568 and SACOL0569) (Wang et al., 2004) were also up-regulated following TTO challenge (Table 3).
Many of the genes that demonstrated the greatest down-regulation following TTO challenge include those required for purine biosynthesis (purA, purC, purD, purE, purF, purH, purK, purL, purM, purN, purQ and purS) (Table 3). Nucleotide metabolism in bacteria is affected by the activity of adenylate kinase Adk and di-phosphate kinase Ndk (Willemoes and Kilstrup, 2005), and both adk and ndk were also down-regulated following TTO challenge (Table 3). SACOL2242, which encodes a xanthine/uracil permease family protein, was also down-regulated by TTO challenge (Table 3). Following TTO exposure, one of the thiamine (vitamin B1) biosynthetic operons (tenA-thiM-thiD-thiE) (Muller et al., 2009) was also down-regulated in SH1000. Bacterial purine biosynthetic pathways have been linked to thiamine biosynthesis (Petersen et al., 1996), so the TTO-induced down-regulation of both purine and thiamine biosynthetic genes might be linked.
The gene that demonstrated the greatest increase in expression following TTO challenge was vraX, which encodes a 55 aa protein (McAleese et al., 2006; Scherl et al., 2006). The function of VraX is presently unknown; however, it has been proposed that the first 19 N-terminal aa sequence of this protein is a signal peptide, suggesting that VraX is exported (Scherl et al., 2006). SAMMD analysis revealed that vraX expression is also up-regulated by multiple cell wall and/or membrane active compounds (bacitracin, d-cycloserine, oxacillin, tunicamycin, flavomycin, fosfomycin, teicoplanin, vancomycin, daptomycin, lysostaphin, epicatechin gallate, ranalexin and antimicrobial peptides) (Bernal et al., 2010; Dengler et al., 2011; Overton et al., 2011; Pietiainen et al., 2009; Utaida et al., 2003). Our results and those previously reported support the notion that vraX up-regulation follows all forms of cell membrane and/or cell wall metabolism insult. Genes encoding the two-component system VraSR (vancomycin-resistance associated sensor/regulator) are both up-regulated following vancomycin exposure (Kuroda et al., 2003) and in S. aureus strains exhibiting decreased susceptibility to vancomycin (Kuroda et al., 2000). Our data indicated that vraSR is also up-regulated following TTO insult (1.7- and 3.9-fold) (Table 3). Since vraR is required for the expression of vraX (Dengler et al., 2011), it is possible that the up-regulation of this gene contributes to increased vraX expression following TTO challenge. The expression of additional genes whose regulation is altered by vraSR (e.g. ctpA, drp35, fmtA, opuD, pbp2, prsA and sgtB) (McAleese et al., 2006; Utaida et al., 2003) was also altered in expression following TTO challenge (Table 3). In support of a hypothesis that vraSR plays a protective role in the the response of S. aureus to TTO, we determined that N315vraSR::cat demonstrated a reduced TTO (% v/v) MIC (0.15 ± 0) and MBC (0.20 ± 0), compared to parent strain N315 (MIC = 0.23 ± 0.01, MBC= 0.26 ± 0.01, p<0.05). Two genes encoding known peptidoglycan hydrolases or autolysins (sle1 and isaA) (Kajimura et al., 2005; Stapleton et al., 2007) were also down-regulated, and cwrA, a gene that is induced by cell wall active antimicrobials (Balibar et al., 2010), was up-regulated by TTO challenge (Table 3).
Since cell wall metabolism genes were affected by TTO challenge, we performed whole cell autolytic assays with TTO-treated and untreated SH1000 cell populations. While we detected a slight reduction in unstimulated whole cell autolysis in SH1000 challenged with TTO compared to untreated SH1000, Triton X-100 stimulated autolysis occurred at a similar rate for TTO-treated and untreated cells (data not shown).
One of the most highly TTO-induced genes (mmpL) (Table 3) encodes a protein that is a member of the resistance, nodulation and cell division (RND) family of proteins. mmpL produces a product that demonstrates the greatest identity (31%) across its entire length with the Mycobacterium tuberculosis RND family protein MmpL7 that is required for virulence (Perez et al., 2006). It is of interest to note that the Pseudomonas aeruginosa RND-protein MexB is part of the MexAB-OprM efflux pump that is required for the full expression of TTO tolerance by this organism (Papadopoulos et al., 2008).
CONCLUSIONS
Our SAMMD findings indicate that both the commonly employed antiseptic ethanol and TTO induce analogous transcriptional responses which may be related to the ability of these substances to cause similar damage to both membrane and protein structures (Carson et al., 2006; Gustafson et al., 1998; McDonnell and Russell, 1999). This finding strengthens the idea that these two substances have similar mechanisms of anti-staphylococcal activity.
Another major finding is that TTO challenge leads to the down-regulation of a large cadre of the genes involved with both transcription and translation. The down-regulation of these genes may occur since these processes become futile in a TTO-challenged cell population. Since the synthesis of ribosomal components and translation consumes the majority of energy in growing cells (Dethlefsen and Schmidt, 2007; Russell and Cook, 1995), we propose that the down-regulation of these genes during TTO challenge allows the cell to conserve and/or relocate energy resources to processes designed to protect the cell from TTO cidal activity, such as the heat shock response. A protective role for the heat shock response against TTO challenge is supported by the finding that the inactivation of dnaK in SH1000 leads to an increase in TTO susceptibility.
The TTO up-regulation of vraX and vraSR, and the altered regulation of other genes involved with cell wall metabolism following TTO challenge indicates that cell wall metabolism is affected by TTO and terpinen-4-ol as was previously suggested for TTO at least (Carson et al., 2002; Gustafson et al., 1998). This finding led us to investigate the importance of vraSR in the response of strain N315 to TTO. vraSR inactivation in N315 led to an increase in TTO susceptibility, demonstrating that this two-component regulatory system is required for the TTO protective response. Interestingly, whole cell autolysis experiments suggest that autolysin production and/or activity is not irreversibly altered following TTO challenge.
It is possible that the up-regulation of mmpL represents an effort by S. aureus to reduce TTO and antimicrobial terpene accumulation. Research is presently underway to determine if mmpL inactivation leads to a reduction in susceptibility to TTO and its antimicrobial terpenes.
Furthermore, qRT-PCR experimentation revealed that ten genes altered by TTO challenge also responded in a similar transcriptional direction to the major TTO antimicrobial terpinen-4-ol. These genes included the TTO and terpenin-4-ol up-regulated dnaK, mmpL, vraR, vraS and vraX, and the down-regulated betB, purA, purM, rpml and tenA. These equivalent transcriptional responses indicate that terpinen-4-ol alone probably contributes to the TTO-induced transcriptome alterations observed.
Acknowledgments
All authors wish to acknowledge the former and ongoing support from the National Institutes of Health: SC1GM083882-01 (JEG); S06 GM61222-05 (JC, NMSU-MBRS-RISE PROGRAM); R25 GM07667-30 (NMSU-MARC PROGRAM) and 1R15AI084006 (BJW). This project was also supported by grants from the National Center for Research Resources (5P20RR016480-12) and the National Institute of General Medical Sciences (8 P20 GM103451-12) from the National Institutes of Health. Special thanks to Professor Thomas V. Riley, Dr. Christine Carson (University of Western Australia) and Paul Bolster (P. Guinane Pty. Ltd. New South Wales, Australia) for their help in the acquisition of TTO utilized in this study.
Footnotes
Conflict of Interest
All authors declare no financial/commercial conflicts of interest.
References
- Balibar CJ, Shen X, McGuire D, Yu D, McKenney D, Tao J. cwrA, a gene that specifically responds to cell wall damage in Staphylococcus aureus. Microbiology. 2010;156 (Pt 5):1372–1383. doi: 10.1099/mic.0.036129-0. [DOI] [PubMed] [Google Scholar]
- Bernal P, Lemaire S, Pinho MG, Mobashery S, Hinds J, Taylor PW. Insertion of epicatechin gallate into the cytoplasmic membrane of methicillin-resistant Staphylococcus aureus disrupts penicillin-binding protein (PBP) 2a-mediated beta-lactam resistance by delocalizing PBP2. J Biol Chem. 2010;285:24055–24065. doi: 10.1074/jbc.M110.114793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(S5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- Bubacz MR. Community-acquired methicillin-resistant Staphylococcus aureus: an ever-emerging epidemic. AAOHN J. 2007;55:193–194. doi: 10.1177/216507990705500504. [DOI] [PubMed] [Google Scholar]
- Carson CF, Cookson BD, Farrelly HD, Riley TV. Susceptibility of methicillin-resistant Staphylococcus aureus to the essential oil of Melaleuca alternifolia. J Antimicrob Chemother. 1995;35:421–424. doi: 10.1093/jac/35.3.421. [DOI] [PubMed] [Google Scholar]
- Carson CF, Hammer KA, Riley TV. Melaleuca alternifolia (Tea Tree) oil: a review of antimicrobial and other medicinal properties. Clin Microbiol Rev. 2006;19:50–62. doi: 10.1128/CMR.19.1.50-62.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CF, Mee BJ, Riley TV. Mechanismof action of Melaleuca alternifolia (tea tree) oil on Staphylococcus aureus determined by time-kill, lysis, leakage, and salt tolerance assays and electronmicroscopy. Antimicrob Agents Chemother. 2002;46:1914–1920. doi: 10.1128/AAC.46.6.1914-1920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CF, Riley TV. Antimicrobial activity of the major components of the essential oil of Melaleuca alternifolia. J Appl Bacteriol. 1995;78:264–269. doi: 10.1111/j.1365-2672.1995.tb05025.x. [DOI] [PubMed] [Google Scholar]
- Chastanet A, Fert J, Msadek T. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol Microbiol. 2003;47:1061–1073. doi: 10.1046/j.1365-2958.2003.03355.x. [DOI] [PubMed] [Google Scholar]
- Cox SD, Gustafson JE, Mann CM, et al. Tea tree oil causes K+leakage and inhibits respiration in Escherichia coli. Lett Appl Microbiol. 1998;26:355–358. doi: 10.1046/j.1472-765x.1998.00348.x. [DOI] [PubMed] [Google Scholar]
- Cox SD, Mann CM, Markham JL. Interactions between components of the essential oil of Melaleuca alternifolia. J Appl Microbiol. 2001;91:492–497. doi: 10.1046/j.1365-2672.2001.01406.x. [DOI] [PubMed] [Google Scholar]
- Cox SD, Mann CM, Markham JL, et al. The mode of antimicrobial action of the essential oil of Melaleuca alternifolia (tea tree oil) J Appl Microbiol. 2000;88:170–175. doi: 10.1046/j.1365-2672.2000.00943.x. [DOI] [PubMed] [Google Scholar]
- de Crouy-Chanel A, Kohiyama M, Richarme G. Interaction of DnaK with native proteins and membrane proteins correlates with their accessible hydrophobicity. Gene. 1999;230:163–170. doi: 10.1016/s0378-1119(99)00083-9. [DOI] [PubMed] [Google Scholar]
- Delgado A, Riordan JT, Lamichhane-Khadka R, et al. Hetero-vancomycin-intermediate methicillin-resistant Staphylococcus aureus isolate from a medical center in Las Cruces, New Mexico. J Clin Microbiol. 2007;45:1325–1329. doi: 10.1128/JCM.02437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado A, Zaman S, Muthaiyan A, et al. The fusidic acid stimulon of Staphylococcus aureus. J Antimicrob Chemother. 2008;62:1207–1214. doi: 10.1093/jac/dkn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler V, Meier PS, Heusser R, Berger-Bachi B, McCallum N. Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics. BMC Microbiol. 2011;11:16. doi: 10.1186/1471-2180-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derre I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol Microbiol. 1999;31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Schmidt TM. Performance of the translational apparatus varies with the ecological strategies of bacteria. J Bacteriol. 2007;189:3237–3245. doi: 10.1128/JB.01686-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden MS, Dailly S, Crouch M. A randomized, controlled trial of tea tree topical preparations versus a standard topical regimen for the clearance of MRSA colonization. J Hosp Infect. 2004;56:283–286. doi: 10.1016/j.jhin.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Wickner S, Maurizi MR. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- Gustafson JE, Berger-Bachi B, Strassle A, Wilkinson BJ. Autolysis of methicillin-resistant and -susceptible Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:566–572. doi: 10.1128/aac.36.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson JE, Liew YC, Chew S, et al. Effects of tea tree oil on Escherichia coli. Lett Appl Microbiol. 1998;26:194–198. doi: 10.1046/j.1472-765x.1998.00317.x. [DOI] [PubMed] [Google Scholar]
- Hersh AL, Chambers HF, Maselli JH, Gonzales R. National trends in ambulatory visits and antibiotic prescribing for skin and soft-tissue infections. Arch Intern Med. 2008;168:1585–1591. doi: 10.1001/archinte.168.14.1585. [DOI] [PubMed] [Google Scholar]
- Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. SigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325–4. J Bacteriol. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura J, Fujiwara T, Yamada S, et al. Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol Microbiol. 2005;58:1087–1101. doi: 10.1111/j.1365-2958.2005.04881.x. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol. 2003;49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Kuwahara-Arai K, Hiramatsu K. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun. 2000;269:485–490. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- Lin Z, Rye HS. GroEL-mediated protein folding: making the impossible, possible. Crit Rev Biochem Mol Biol. 2006;41:211–239. doi: 10.1080/10409230600760382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McAleese F, Wu SW, Sieradzki K, et al. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J Bacteriol. 2006;188:1120–1133. doi: 10.1128/JB.188.3.1120-1133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 1999;12:147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran GJKA, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- Muller IB, Bergmann B, Groves MR, et al. The vitamin B1 metabolism of Staphylococcus aureus is controlled at enzymatic and transcriptional levels. PLoS One. 2009;4:e7656. doi: 10.1371/journal.pone.0007656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan V, Elasri MO. SAMMD: Staphylococcus aureus microarray meta-database. BMC Genomics. 2007;8:351. doi: 10.1186/1471-2164-8-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton IM, Graham S, Gould KA, et al. Global network analysis of drug tolerance, mode of action and virulence in methicillin-resistant S. aureus. BMC Syst Biol. 2011;5:68. doi: 10.1186/1752-0509-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos CJ, Carson CF, Chang BJ, Riley TV. Role of the MexAB-OprM efflux pump of Pseudomonas aeruginosa in tolerance to tea tree (Melaleuca alternifolia) oil and its monoterpene components terpinen-4-ol, 1,8-cineole, and alpha-terpineol. Appl Environ Microbiol. 2008;74:1932–1935. doi: 10.1128/AEM.02334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J, Garcia R, Bach H, et al. Mycobacterium tuberculosis transporter MmpL7 is a potential substrate for kinase PknD. Biochem Biophys Res Commun. 2006;348:6–12. doi: 10.1016/j.bbrc.2006.06.164. [DOI] [PubMed] [Google Scholar]
- Petersen L, Enos-Berlage J, Downs DM. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics. 1996;143(1):37–44. doi: 10.1093/genetics/143.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietiainen M, Francois P, Hyyrylainen HL, et al. Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics. 2009;10:429. doi: 10.1186/1471-2164-10-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qoronfleh MW, Streips UN, Wilkinson BJ. Basic features of the staphylococcal heat shock response. Antonie Van Leeuwenhoek. 1990;58:79–86. doi: 10.1007/BF00422721. [DOI] [PubMed] [Google Scholar]
- Riordan JT, Muthaiyan A, Van Voorhies W, et al. Response of Staphylococcus aureus to salicylate challenge. J Bacteriol. 2007;189:220–227. doi: 10.1128/JB.01149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JB, Cook GM. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A, Francois P, Charbonnier Y, et al. Exploring glyco-peptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics. 2006;7:296. doi: 10.1186/1471-2164-7-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikkema J, de Bont JA, Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology. 2007;153(Pt 9):3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- Southwell I, Lowe R, editors. Tea Tree: The genus Melaleuca (v 9) Hardwood Academic Publishers; Amsterdam: 1999. [Google Scholar]
- Stapleton MR, Horsburgh MJ, Hayhurst EJ, et al. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J Bacteriol. 2007;189:7316–7325. doi: 10.1128/JB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utaida S, Dunman PM, Macapagal D, et al. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology. 2003;149(Pt 10):2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- Wang L, Trawick JD, Yamamoto R, Zamudio C. Genome-wide operon prediction in Staphylococcus aureus. Nucleic Acids Res. 2004;32:3689–3702. doi: 10.1093/nar/gkh694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemoes M, Kilstrup M. Nucleoside triphosphate synthesis catalysed by adenylate kinase is ADP dependent. Arch Biochem Biophys. 2005;444:195–199. doi: 10.1016/j.abb.2005.10.003. [DOI] [PubMed] [Google Scholar]