Abstract
Perception of the plant steroid hormone brassinolide (BL) by the membrane-associated receptor kinase BRI1 triggers the dephosphorylation and accumulation in the nucleus of the transcriptional modulators BES1 and BZR1. We identified bsu1-1D as a dominant suppressor of bri1 in Arabidopsis. BSU1 encodes a nuclear-localized serine–threonine protein phosphatase with an N-terminal Kelch-repeat domain, and is preferentially expressed in elongating cells. BSU1 is able to modulate the phosphorylation state of BES1, counteracting the action of the glycogen synthase kinase-3 BIN2, and leading to increased steady-state levels of dephosphorylated BES1. BSU1 belongs to a small gene family; loss-of-function analyses unravel the extent of functional overlap among members of the family and confirm the role of these phosphatases in the control of cell elongation by BL. Our data indicate that BES1 is subject to antagonistic phosphorylation and dephosphorylation reactions in the nucleus, which fine-tune the amplitude of the response to BL.
Keywords: Arabidopsis, brassinosteroids, protein phosphatase, Kelch repeat, BES1, BIN2
Brassinosteroids (BRs) are a group of plant-specific polyhydroxylated sterols first identified by their ability to promote stem elongation when applied topically (Grove et al. 1979). Genetic defects in the synthesis of BRs result in dwarfism, abnormal vascular development, reduced male fertility, and altered responses to growth factors and environmental stimuli, attesting to the importance of these hormones for normal plant morphogenesis.
Unlike animal steroid hormones, BRs are perceived at the cell surface by a receptor that includes the BRI1 protein. BRI1 was originally identified in studies of dwarf mutants that resemble BR-deficient mutants but are not rescued by treatment with brassinolide (BL), the end product of the biosynthetic pathway (Clouse et al. 1996; Kauschmann et al. 1996). BRI1 encodes a membrane protein composed of an extracellular leucine-rich repeat domain linked by a single transmembrane pass to an intracellular Ser/Thr kinase (Li and Chory 1997). BL is perceived by the extracellular domain of BRI1, leading to its autophosphorylation (Wang et al. 2001). Saturating screens for BL insensitivity yielded a wealth of loss-of-function alleles for BRI1 in Arabidopsis and other species (Friedrichsen et al. 2000; Nomura et al. 2003), suggesting that signaling components downstream of BRI1 are redundant or essential for viability.
Other BL signaling components have been identified using gain-of-function genetic screens. BAK1, a leucine-rich repeat receptor kinase that interacts with BRI1, was identified by its ability to suppress the phenotype of a weak bri1 mutant when overexpressed (Li et al. 2002; Nam and Li 2002). Semidominant, BL-insensitive mutants carry mutations in the BIN2 gene (also known as UCU1 or DWF12), which encodes glycogen synthase kinase-3 (GSK3; Choe et al. 2002; Li and Nam 2002, Pérez-Pérez et al. 2002). Because the bin2-1 mutation increased the in vitro activity of the enzyme and overexpression of the wild-type protein reproduced the mutant phenotype in a dose-dependent manner (Li and Nam 2002), BIN2 is considered a negative regulator of the pathway.
Semidominant mutations that suppressed weak bri1 mutants or conferred resistance to the biosynthesis inhibitor brassinazole identified two nearly identical genes, BES1 and BZR1. Each mutant carries the same missense mutation, which has been shown to have a stabilizing effect on the proteins (Wang et al. 2002; Yin et al. 2002a; Zhao et al. 2002). In vivo, BES1 and BZR1 proteins appear as a series of phosphorylated and dephosphorylated forms; the latter rapidly and specifically accumulate at the expense of the former after BL treatment (Wang et al. 2002; Yin et al. 2002a). Both proteins interact with and are phosphorylated by BIN2 in vitro (Yin et al. 2002a; Zhao et al. 2002). Moreover, the levels of BES1 are reduced in bin2-1 plants (He et al. 2002; Yin et al. 2002a), whereas overexpression of wild-type BES1 or BZR1 suppresses the bin2 phenotype (He et al. 2002; Zhao et al. 2002); in fact, BES1 and BZR1 mutant proteins overaccumulate regardless of their phosphorylation state (Wang et al. 2002; Yin et al. 2002a). The recurrent finding of the same mutation suggests that uncoupling phosphorylation from the control of protein abundance is highly restricted, and that phosphorylation by BIN2 determines BES1 and BZR1 half-lives.
BES1 and BZR1 are found in the nucleus and modulate the transcription of BL-regulated genes, whose expression is altered in bes1 mutants (Yin et al. 2002a). Control of the half-life of transcriptional modulators by a GSK3 is reminiscent of the β-catenin-dependent Wnt signaling pathway, which plays a major role in embryo patterning and cell proliferation in metazoans (Pires-daSilva and Sommer 2003). As for β-catenin, dephosphorylated BES1 and BZR1, kept in check by BIN2, fine-tune the signal strength.
BIN2 and BES1–BZR1 belong to small gene families, some of whose members appear to have overlapping functions. To identify new components in a context of genetic redundancy, we searched for dominant suppressors of a weak bri1 mutant using activation tagging (Weigel et al. 2000). We present evidence for the role of a small family of Ser/Thr protein phosphatases in BL signaling. Overexpression of one of them, BSU1, suppressed bri1 and bin2 phenotypes, allowing the accumulation of dephosphorylated BES1; conversely, reduced expression of two BSU1 homologs led to a semi-dwarf phenotype. These phosphatases are thus positive effectors of BL signaling, counteracting the action of BIN2. In addition, we found that BSU1 is a nuclear protein, suggesting that dephosphorylation of BES1 takes place in the nucleus. The emerging picture is one of a dynamic balance of processes controlling the levels of dephosphorylated BES1 in the nucleus, and thereby, the magnitude of the response to BL.
Results
bsu1-1D, a dominant suppressor of bri1 identified by activation tagging
Genetic redundancy and embryonic lethality can prevent the analysis of gene function by traditional loss-of-function mutant screens. As an alternative to circumvent this problem, the random insertion of strong viral enhancers in the genome can be exploited to produce dominant phenotypes by overexpression of nearby genes, a technique called activation tagging (Weigel et al. 2000). Based on the evidence for functional overlap, we searched for dominant suppressors of a weak bri1 mutant. We screened ∼7000 T1 lines and identified 14 independent lines (called bsu, for bri1 suppressors) that partially suppressed, in a dominant way, the dwarf phenotype of bri1-5. Here, we describe the detailed characterization of one of them, BSU1.
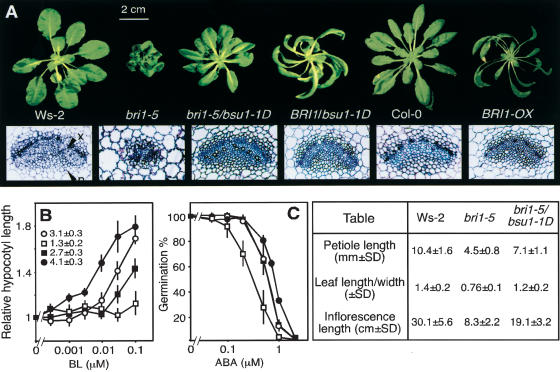
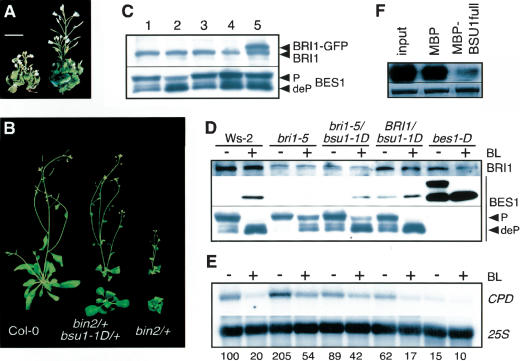
The suppression of bri1's pleiotropic phenotype by bsu1-1D extended to all stages of the life cycle (Fig. 1). bri1-5/bsu1-1D seedlings had longer hypocotyls and reduced cotyledon epinasty than bri1 single mutants; in adults, petiole length and leaf blade expansion were restored, relaxing the proximo–distal shortening typical of BR-related mutants; inflorescence length was also intermediate between bri1-5 and the wild-type, Ws-2(Fig. 1A; Fig. 1 included table). Plants carrying bsu1-1D in a BRI1 background, in addition, displayed outwardly curving leaves and long inflorescences, reminiscent of plants overexpressing BRI1 or the BR-biosynthetic gene DWF4 (Fig. 1A; Choe et al. 2001; Wang et al. 2001).
Figure 1.
bsu1-1D partly suppresses the bri1-5 mutant phenotype. (A, from left to right) Wild-type (Ws-2), bri1-5, bri1-5/bsu1-1D, BRI1/bsu1-1D, Col-0, and Col-0/proBRI1:BRI1–GFP (BRI1-Ox; Wang et al. 2001) grown for 35 d under short-day conditions (9 h light/15 h darkness). (Below) Transverse sections of the petioles of fully expanded rosette leaves from the same plants (all pictures at the same magnification). (x) Xylem; (p) phloem. (Table) Measurements were taken from the fourth fully expanded leaves and fully dried inflorescences of some of these plants. (B,C) Hormone responses of Ws-2(open circles), bri1-5 (open squares), bri1-5/bsu1-1D (filled squares), and BRI1/bsu1-1D (filled circles). Results are the average of three independent experiments, with 20 seedlings per condition and experiment. (B) Hypocotyl elongation in response to BL. Seedlings were grown on plates with different concentrations of BL for 5 d at 21°C under long-day conditions (16 h light/8 h darkness, 30 μmole/m2 · sec white light). The inset displays hypocotyl lengths (in millimeters ±S.D.) without BL. (C) Germination in response to ABA. Seeds were germinated at 21°C under continuous light, and the germination was scored at the fourth day. The curves corresponding to Ws-2and bsu1-1D overlap.
BRs have been implicated in xylem differentiation (Yamamoto et al. 2001) and vascular patterning, which prompted us to check the effects of bsu1-1D on vascular anatomy (Fig. 1A, bottom). Wild-type petioles (Ws-2and Col-0) have a single, oval bundle composed of adaxial xylem and abaxial phloem. bri1-5 revealed a rounded and disorganized bundle, with reduced numbers of phloem and differentiated xylem cells. bsu1-1D significantly restored a wild-type looking anatomy in the bri1-5 background, although xylem cells displayed remarkably thickened walls. Finally, the vasculature of BRI1/bsu1-1D plants resembled that of BRI1-overexpressing plants, with reduced xylem differentiation in a crescent-shaped bundle, probably caused by different rates of elongation on both sides of the blade.
To determine if bsu1-1D had effects on BL signaling, we performed dose-response curves with BL and other hormones known to interact with BL. Seedlings carrying bsu1-1D in the bri1-5 or BRI1 backgrounds were more sensitive to exogenous BL; hypocotyls of bri1-5/bsu1-1D responded to relatively low concentrations of BL, whereas those of BRI1/bsu1-1D swelled and wilted at lower concentrations than their wild-type counterparts. This latter phenotype is typical of seedlings exposed to high concentrations of BL, and is probably induced by ethylene overproduction (Fig. 1B; Mandava 1988). Abscisic acid (ABA) and BL have been reported to act antagonistically in many responses: the expression of some BL-induced genes is repressed by ABA (Friedrichsen et al. 2002), whereas BR-related mutants are overly sensitive to ABA in germination assays (Steber and McCourt 2000). In this regard, bsu1-1D seeds were able to germinate at higher ABA concentrations than their wild-type siblings (Fig. 1C). In summary, several developmental and physiological responses associated to BL were enhanced in bsu1-1D plants.
BSU1 encodes a Ser–Thr phosphatase with a long N-terminal extension
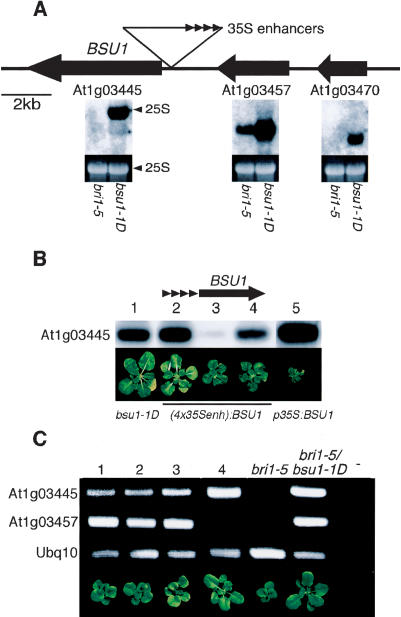
Cosegregation of the suppressor phenotype with the selection marker, Southern blot analysis, and plasmid rescue (data not shown) indicated a single T-DNA insertion in an intergenic region at the top of Chromosome I. Unexpectedly, three genes on both sides of the T-DNA were overexpressed: at the left border, At1g03445, encoding a putative Ser/Thr phosphatase, and at the right border, adjacent to the enhancers, At1g03457 and At1g03470, coding for an RNA-binding protein and a putative protein of unknown function, respectively (Fig. 2A).
Figure 2.
Identification of BSU1. (A) Three genes surrounding the T-DNA insertion were overexpressed in bsu1-1D. Arrows indicate the direction of transcription. (Below) Northern blots probed with the 3′-UTR of each gene in bri1-5 and bri1-5/bsu1-1D, and EtBr-stained 25S rRNA as control. (B) Recapitulation of the bsu1-1D phenotype. (Lane 1) bri1-5/bsu1-1D. (Lanes 2–4) Three independent T2 bri1-5 plants transformed with the BSU1 genomic fragment downstream from 35S enhancers (schematized on top), showing different degrees of expression and phenotypic suppression. (Lane 5) Expression of the genomic coding sequence of BSU1 under the 35S promoter in the bri1-5 background. All plants depicted at the same age. (C) Reducing the expression of BSU1 reverts the bsu1-1D phenotype. RT–PCR analysis of T2 lines of bri1-5/bsu1-1D plants transformed with RNAi constructs against At1g03445 (lanes 1,2,3) and At1g03457 (lane 4). Controls from bri1-5, bri1-5/bsu1-1D, and without template (–) are shown. Ubq10 serves as an internal control.
Neither the genomic nor the cDNA sequences of At1g03457 or At1g03470, driven by the 35S promoter or by their own promoters downstream of the 35S enhancers, were able to restore the suppressor phenotype in the bri1-5 background (data not shown). Overexpression of At1g03445 under the 35S promoter, on the other hand, had deleterious effects: few transformants survived, and those that did showed very small leaves (Fig. 2B), elongated inflorescences, and reduced fertility. However, the expression of the genomic fragment adjacent to the T-DNA left border, downstream of the 35S enhancers, recapitulated the bsu1-1D phenotype, and the degree of suppression correlated with the transcript levels of At1g03445 (Fig. 2B).
As an alternative to define the gene responsible for the phenotype, we attempted to revert the phenotype of bri1-5/bsu1-1D plants by reducing the mRNA levels of each of the three genes one at a time with RNA interference. Neither RNAi against At1g03457 (shown for comparison in Fig. 2C) nor against At1g03470 (data not shown) caused dwarfing; in contrast, reducing the levels of At1g03445 caused reversion to a bri1-5-like phenotype (Fig. 2C). Although we cannot eliminate side effects of the combined overexpression of three genes of unknown function, overexpression of At1g03445 appears to be the major cause of the phenotypic change, and we therefore refer to it as BSU1.
When we started this study, the product of BSU1 was classified as hypothetical, because no expression evidence for this gene was available. Indeed, we couldn't detect any signal in Northern blots of Ws-2or bri1-5 mRNA, even after long exposures. We therefore obtained the full-length cDNA from bsu1-1D plants, which allowed us to correct misannotations in the database and determine that the T-DNA was inserted 313 bases upstream of the probable translation start site. The BSU1 gene, thus defined, consists of 22 exons over 5290 bp, coding for a protein composed of a long N-terminal domain, a connecting middle region, and a C-terminal phosphatase domain (Fig. 3D). Three homologs of BSU1 are present in Arabidopsis, At4g03080, At1g08420, and At2g27210, hereafter dubbed, respectively, BSL1, BSL2, and BSL3 (for BSU1-like). BSU1–BSL1 and BSL2–BSL3 are located in two pairs of chromosomal repeats.
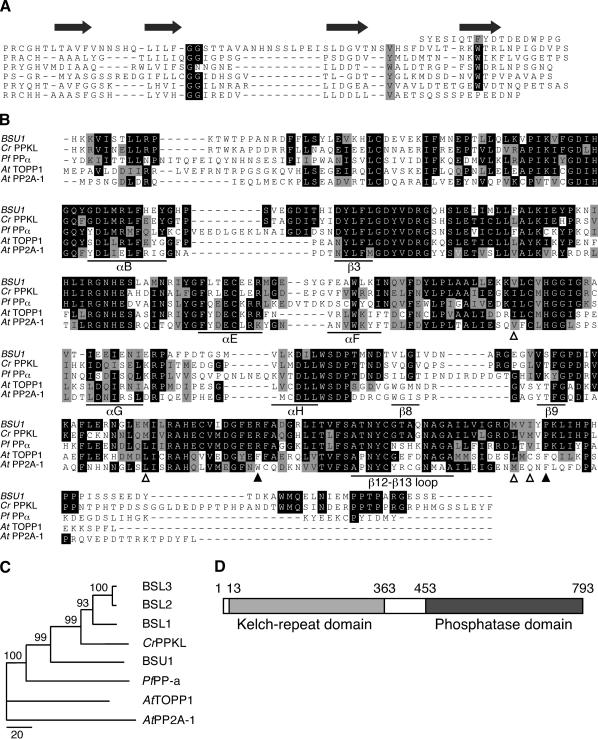
Figure 3.
BSU1 belongs to a small family of Ser/Thr protein phosphatases. (A) Alignment of the Kelch repeats at the N terminus of BSU1 (residues 13–363) highlighting its diagnostic features: a pair of Gly followed, at a variable distance, by a hydrophobic residue and a Trp (Adams et al. 2000). The arrows indicate the predicted location of the β-strands of the repeats. (B) Alignment of the catalytic domains of BSU1 (residues 453–793) and homologs from Chlamydomonas reinhardtii (CrPPKL, AV639890, residues 520–880) and Plasmodium falciparum (PfPPα, U88869, residues 519–875), and Arabidopsis thaliana PP1 (TOPP1, S20882, residues 15–318) and PP2A-1 (Q07098, residues 1–306). The alignment was generated with ClustalX and manually adjusted. The secondary-structure elements flanking the insertions and the β12–β13 loop are labeled as in PP1 (Goldberg et al. 1995). The empty and filled arrowheads point to preserved hydrophobic residues and nonconservative substitutions along the RVxF motif interaction surface, respectively. (C) Phylogenetic relationship of the sequences shown in B plus those of the BSL proteins. Sequences were aligned with ClustalX and manually adjusted, and the N-J tree with 1000 bootstrap repetitions was calculated with MEGA2.1 (Kumar et al. 2001). The bar indicates the number of substitutions per 100 residues, and values at each node, the percentage bootstrap support value. (D) Schematic representation of the domain structure of BSU1, with the corresponding residue numbers.
The N-terminal domain of BSU1 is predicted to adopt a Kelch-repeat β-propeller structure (Adams et al. 2000). The Kelch domain of BSU1 likely consists of six β-sheets with N-terminal closure, that is, the closing blade formed by the first N-terminal and the last three C-terminal β-strands (Fig. 3A). The first Ser/Thr phosphatase with a Kelch-repeat domain at its N terminus, PfPPα, was identified in Plasmodium falciparum (Li and Baker 1998). Homologous sequences were later found in many, probably all, angiosperms, and we also identified a related sequence in the alga Chlamydomonas reinhardtii (Fig. 3B). In a recent overview, it was suggested that these proteins be called PPKL, for protein phosphatases with Kelch-like repeats (Kutuzov and Andreeva 2002).
The C-terminal domains of PPKLs correspond to members of the Ser/Thr protein phosphatase superfamily. Overall sequence conservation suggests that their tertiary structure and catalytic properties should be similar to other related enzymes like PP1 and PP2A (Barford et al. 1998). However, extensions at the C terminus and at certain loops connecting secondary-structure elements (Fig. 3B), as well as changes in conserved residues important for regulation (arrowheads in Fig. 3B), make these phosphatases characteristically divergent among other proteins of their ilk (Fig. 3C; see Kerk et al. 2002).
BSU1 is a functional Ser/Thr phosphatase
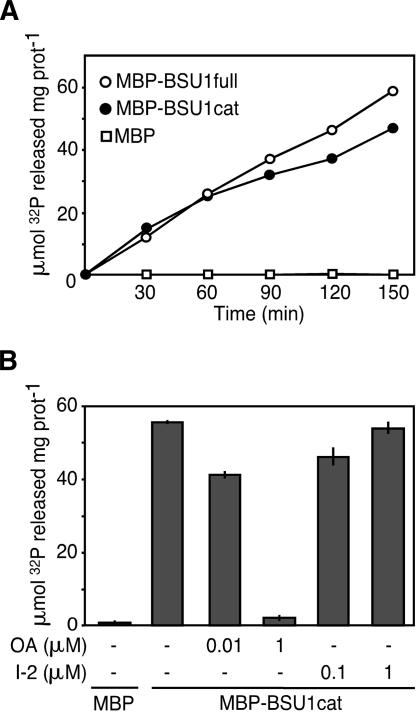
To determine whether BSU1 was a functional protein phosphatase, we expressed the full-length and catalytic domain sequences in Escherichia coli as maltose-binding protein (MBP) fusions, and characterized their activity toward phosphorylated myelin-basic protein (MyBP). Both forms were equally able to dephosphorylate MyBP in vitro (Fig. 4A). The Kelch-repeat domain did not interfere with the enzymatic activity nor did it interact with the catalytic moiety in yeast two-hybrid assays (data not shown), suggesting that both domains behave independently.
Figure 4.
BSU1 is a functional phosphatase. (A) MBP and MBP fusions to the full-length (full) or catalytic moiety (cat) of BSU1, expressed in E. coli, were assayed with 32P-labeled MyBP. Activity values were corrected for equal amounts of the catalytic subunit. Shown is the average of two independent measurements. (B) MBP-BSU1 cat was incubated with the indicated amounts of okadaic acid (OA) or Inhibitor-2(I-2) for 3 h. Data are means ± deviation of the amount of 32P released, quantified in two independent experiments.
Different families of phosphatases can be distinguished by their sensitivity to inhibitors. BSU1's catalytic domain was inhibited only at high concentrations of okadaic acid, a signature of PP1 as compared with PP2A, which is ∼100 times more sensitive to this toxin (Fig. 4B). In fact, the sequence of the β12–β13 loop of BSU1, the target for okadaic acid, is more similar to PP1 than to PP2A (underlined in Fig. 3B). On the other hand, PP1s, as opposed to PP2As, are highly sensitive to Inhibitor-2(I-2), a protein that acts both as a folding mediator and as an inhibitor (Alessi et al. 1993). Contrary to PP1, however, the catalytic portion of BSU1 appeared to be quite insensitive to I-2(Fig. 4B). In sum, BSU1 codes for a functional phosphatase displaying features of both PP1 and PP2A activities.
BSU1 is a nuclear protein expressed in young, elongating tissues
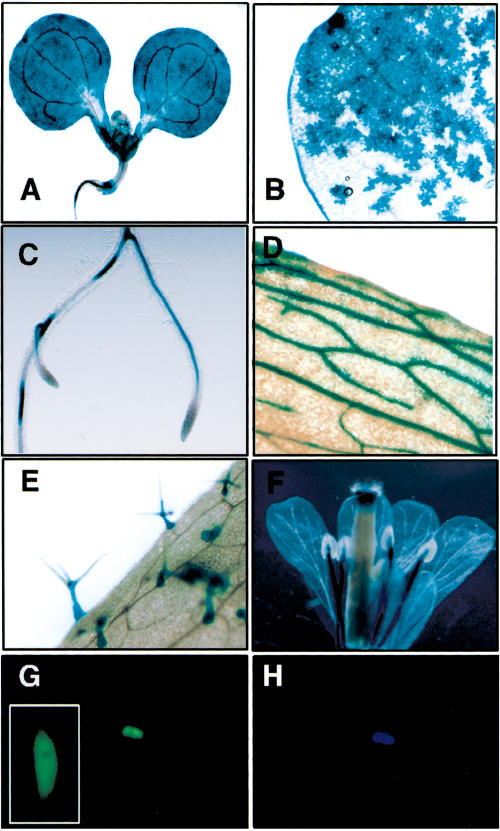
To gain further insight into the role of BSU1 in growth processes, we examined its expression pattern and subcellular localization. The BSU1 promoter region was fused to β-glucuronidase, and the expression pattern was assessed in T2 plants at different developmental stages. In young seedlings, GUS staining was found at the base of the hypocotyl, at the tip and most peripheral cell layers of cotyledons (Fig. 5A,B), and in the vascular cylinder of roots, particularly in the elongation zone and at the point of emergence of lateral roots (Fig. 5C). In mature plants, the BSU1 promoter was still active in the root vasculature, but was almost completely silenced in fully expanded stems and leaves, except in minor veins (Fig. 5D,E) of some lines where the expression of the transgene was highest. Staining reappeared in flowers, mainly in sepal veins, anther filaments, and in the style (Fig. 5F). This expression pattern suggests that BSU1 is expressed in actively growing regions and apparently enriched in vascular tissues.
Figure 5.
BSU1 is expressed in the nuclei of young tissues. (A–F) Seven-day-old Arabidopsis seedlings expressing a proBSU1:GUS reporter construct showed promoter activity in cotyledons, the base of the hypocotyl, and the root vasculature (A); staining was more intense at the tip and the rim of cotyledons (B) and at emerging lateral roots (C). In adults, staining was observed in the leaf vasculature (D) and trichomes (E), and in the vasculature of sepals and stamens and in the style (F). (G) BSU1 fused to GFP localizes to the nuclei (DAPI-stained in H) of dark-grown hypocotyls, but is excluded from the nucleoli (inset).
To investigate the intracellular localization of BSU1, we inserted the GFP sequence N-terminal to the full BSU1 genomic coding region, and we expressed the fusion construct under the control of BSU1's native promoter downstream of the 35S enhancers, as we had done to recapitulate the bsu1-1D phenotype (Fig. 2B). Transgenic Ws-2plants expressing the GFP–BSU1 fusion protein displayed outwardly curving leaves similar to those of BRI1/bsu1-1D, indicating that the fusion protein was functional (data not shown). Although BSU1 lacks recognizable nuclear localization signals, the fusion protein was found in the nuclei of dark-grown hypocotyls (Fig. 5G) and roots (data not shown). BL has been reported to trigger the accumulation of BES1 in the nucleus (Yin et al. 2002a), but we couldn't detect any effects of such treatment on BSU1 localization or abundance (data not shown).
bsu1-1D suppresses bri1 and bin2 by accumulation of dephosphorylated BES1
To determine the function of BSU1 in BL responses, we analyzed the epistasis of bsu1-1D with mutants in other components of the signaling pathway. bsu1-1D partly suppressed the phenotype of a null bri1 mutant (bri1-4; Noguchi et al. 1999), albeit more weakly than in bri1-5. In bri1-4/bsu1-1D plants, leaves and inflorescences were more elongated and fertility restored to some extent, indicating that BSU1 acts downstream or in parallel to BRI1 (Fig. 6A).
Figure 6.
BSU1 modulates the amount of dephosphorylated BES1. (A) Overexpression of BSU1 partly suppressed the bri1-4 mutant phenotype. Mature bri1-4 (left) and bri1-4/bsu1-1D (right) plants are shown. Bar, 1 cm. (B) Overexpression of BSU1 partly suppressed the bin2-1 mutant phenotype. The bin2-1/+ plant derives from a cross between Ws-2and bin2-1 (in Col-0), to control for ecotype variability. (C,D) Phosphorylation state of BES1 in bsu1-1D. Because BRI1 migrates as an ∼120-kD protein and BES1 between ∼30 and 50 kD, the lower parts of the blots were probed for BES1 and the upper parts were probed for BRI1 kinase. (C) Proteins extracted from 2-week-old seedlings of Ws-2 (lane 1), BRI1/bsu1-1D (lane 2), Col-0 (lane 3), 35SE-DWF4 (lane 4; Wang et al. 2001), and BRI1-Ox (lane 5). (D) Proteins extracted as before from seedlings mock-treated (–) or treated for 2h with 1 μM BL (+). Short (middle panel) and long (lower panel) exposures of similar samples from two independent experiments probed for BES1. The bes1-D lanes were eliminated in the longer exposure panel. (E) Northern blot of total mRNA extracted from seedlings treated as in D, and loaded in the same order, probed for CPD. Below, expression percentage referred to untreated Ws-2, relative to 25S rRNA. (F) BES1 protein (1.25 μg) was radiolabeled using BIN2 and [γ-32P]ATP, and then treated with MBP or MBP-BSU1 full (5 μg) for 12h. (Upper panel) Autoradiography. (Lower panel) Coomassie brilliant blue-stained BES1.
Overexpression of wild-type BES1 and BZR1 rescued the dwarfism and infertility of bin2 mutants, supporting their roles as positive effectors of BL signaling (Wang et al. 2002; Zhao et al. 2002). Likewise, overexpression of BSU1 significantly restored overall vegetative size in homozygous (data not shown) and heterozygous bin2-1 mutants (Fig. 6B). In the latter, inflorescence length and seed set were mostly recovered, although leaf rolling, typical of this mutant, was only partly suppressed. Therefore, BSU1 can be formally described as a positive modulator of BL signaling, acting at the level or downstream of BIN2.
We next checked the effects of bsu1-1D on the phosphorylation state of BES1. In the absence of ligand, BES1 and BZR1 are found as phosphorylated species, that after BL or in vitro phosphatase treatments coalesce into a dephosphorylated, apparently homogeneous form of faster electrophoretic mobility (He et al. 2002; Yin et al. 2002a). Consistent with the physiological and morphological evidence, dephosphorylated BES1 was more abundant in all bsu1-1D plants. The overall levels of BES1 increased in plants overexpressing BRI1 and DWF4, but the proportion of the different forms did not depart from that of wild-type plants (Fig. 6C, cf. lanes 4,5 and 1,3); in contrast, the dephosphorylated species were significantly enriched in BRI1/bsu1-1D plants (Fig. 6C, lane 2). This result confirms that BSU1 acts downstream of BRI1, and implies that inputs at various levels of the pathway, leading to different BES1 phosphorylation state equilibria, can produce similar phenotypic outputs. The effects on BES1 phosphorylation were also highlighted in the comparison of bri1-5 with its bsu1-1D siblings (Fig. 6D). BES1 was less abundant in bri1-5 plants, and the protein was mostly found in the phosphorylated form; treatment with BL resulted in a limited accumulation of dephosphorylated BES1 (Fig. 6D, lanes 3,4). In contrast, the state of BES1 in bri1-5/bsu1-1D plants was more similar to that of wild-type plants, and more dephosphorylated BES1 accumulated after BL treatment, although some phosphorylated forms remained (Fig. 6D, lanes 5,6). bes1-D mutants, on the other hand, accumulated high levels of both phosphorylated and dephosphorylated BES1, but all phosphorylated forms disappeared after BL treatment (Fig. 6D, lanes 9,10), further supporting the role of a phosphatase in the response.
The transcription of a BL-regulated gene, CPD, correlated with the amount of dephosphorylated BES1. CPD catalyzes a crucial step in the biosynthesis of BL (Szekeres et al. 1996). CPD mRNA abundance is under strong feedback inhibition by BL: transcript levels fall sharply in wild-type plants after BL treatment, are constitutively reduced in bzr1-D (Wang et al. 2002), but are unresponsive and remain high in bri1 mutants (Mathur et al. 1998). The accumulation of CPD mRNA was reduced in bsu1-1D plants compared with isogenic, non-tagged siblings (Fig. 6E). The effect was more evident in the bri1-5 background, which showed a limited response to BL. As expected from the role of BES1 as a central modulator of the responses to BL, we found that CPD mRNA reached a saturated minimal level in bes1-D plants (Fig. 6E). Taken together, these results indicate that BSU1 acts at the level of BES1, modulating its phosphorylation state.
Members of the BSU family are involved in the control of cell elongation
Because increased BSU1 expression partly rescued the phenotype of bri1-5 (Fig. 1A) and bri1-4 mutants (Fig. 6A), we hypothesized that loss of function of BSU1 would result in dwarfed plants. We therefore identified a T-DNA insertion in the phosphatase domain of BSU1 (SALK_030721; Alonso et al. 2003). Because BSU1 and its closest homolog BSL1 are encoded in duplicated chromosomal segments, we presumed that their functions might overlap, and identified an insertion in the phosphatase domain of this gene as well (SALK_033442). Homozygous plants for the insertion in either gene had no noticeable phenotype, but neither did double-homozygous F2 siblings from a cross between both lines (data not shown).
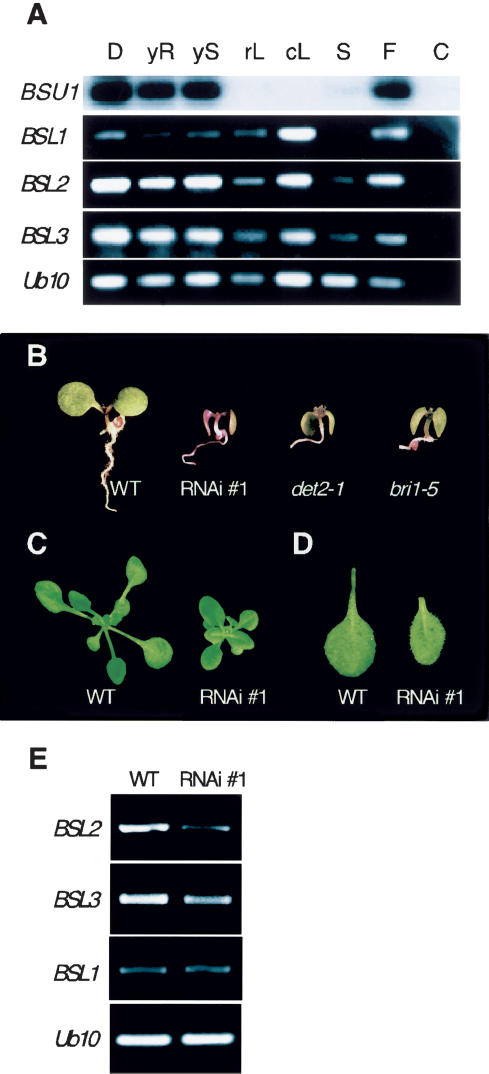
To assess the level of redundancy in the family, we analyzed the expression pattern of the four homologous genes in various organs at different developmental stages (Fig. 7A). We confirmed that BSU1 was very weakly expressed and could only be detected after probing cDNA-derived amplification products. Consistent with the promoter activity pattern (Fig. 5A), BSU1 was expressed in young shoots, roots, and flowers and in light- and dark-grown seedlings, but was absent from mature leaves and stems (Fig. 7A). BSU1 homologs were all expressed at higher levels. Two of them (BSL2 and BSL3) were expressed throughout the plant and, like BSU1, were more abundant in younger parts. In contrast, BSL1 seems to be expressed at lower levels in young tissues relative to older ones (Fig. 7A).
Figure 7.
Functional redundancy in the BSU family. (A) Expression pattern of BSU1 and its homologs evaluated by RT–PCR in Ws-2. (D) Five-day-old seedlings grown in the dark; (yR) roots of 12-day-old seedlings grown in the light on vertical plates; (yS) shoots of 12-day-old seedlings grown in the light on vertical plates; (Rl) mature rosette leaves; (Cl) mature cauline leaves; (S) mature stem; (F) the tip of the inflorescence, including flowers at different developmental stages; (C) negative control with no template added. (B) Phenotype of 10-day-old seedling of Ws-2(WT), RNAi knockdown T2 plants for BSL2 and BSL3, det2-1 and bri1-5, grown under long-day conditions. (C) Rosette phenotype of 20-day-old WT and RNAi plants. (D) Comparison of the first true leaves in WT and RNAi plants. (E) Expression analysis by RT–PCR of BSL1, BSL2, and BSL3 in both WT and RNAi plants. Ubq10 serves as a control.
BSL2 and BSL3 are also encoded in chromosomal repeats, and their coding sequences are 88% identical. Both are closely related to the only BSU1 ortholog that appears to be present in rice (J013001J19), their phosphatase domains being 79% identical. We therefore decided to knock down both genes at the same time, using a fragment in the Kelch domain as a target for RNAi. The sequences of both genes in this region are 89% identical, include stretches of >30 identical contiguous nucleotides, and at the same time depart from the corresponding sequences in BSU1 and BSL1 (59% and 70% identity, respectively). Because Kelch-repeat domains are characterized by low primary sequence similarity, we assumed that this construct would not affect other Kelch-containing genes. T2 and T3 knock-down seedlings displayed dwarfism, as well as short hypocotyls and epinastic cotyledons that resembled the phenotype of bri1-5 or the BR-biosynthesis defective mutant det2-1 (Fig. 7B). Later in development, the phenotypic effect was weaker, but transformants still showed a compact rosette (Fig. 7C) composed of leaves with shortened petioles and blades (Fig. 7D). When grown under short-day conditions, knock-down plants displayed leaves rolling spirally downward (data not shown), similar to those described for the weak alleles of ucu1/bin2 mutants (Pérez-Pérez et al. 2002). We confirmed that the expression of both BSL2 and BSL3 was reduced in the RNAi lines, whereas expression of BSL1 was unaffected (Fig. 7E). This result demonstrates that members of the BSU family are involved in elongation processes throughout the life of Arabidopsis, and that their functions overlap to some extent. This may explain why no loss-of-function phenotypes have been reported so far for any of these genes.
Discussion
Our present understanding of BL signal transduction suggests that accumulation of the dephosphorylated forms of BES1 and BZR1 in the nucleus is the key control point of the pathway. This logic is similar to that of the β-catenin-dependent Wnt pathway in animal cells (Yin et al. 2002a). In the absence of stimulus, β-catenin is phosphorylated by GSK3β and directed to a proteasome-dependent degradation fate (Salic et al. 2000). Perception of Wnt at the plasma membrane causes the disassociation of GSK3β from β-catenin, which then builds up in a dephosphorylated form in the nucleus, where it modulates the transcription of target genes (Staal et al. 2002). Rapid biochemical responses to the Wnt stimulus suggest a role for protein phosphatases. Indeed, indirect evidence points at an as-yet-unidentified phosphatase acting on β-catenin (Sadot et al. 2002). In Arabidopsis, inhibitors of the proteasome activity do not alter the disappearance of phosphorylated BZR1 after BL treatment, indicating that the accumulation of the dephosphorylated form depends not only on de novo synthesis in the absence of active BIN2, but also on a dephosphorylating activity (He et al. 2002).
We present strong evidence for the role of protein phosphatases belonging to the PPKL family in regulating the amounts of dephosphorylated BES1 in the nucleus. Increased BSU1 expression caused the accumulation of dephosphorylated BES1 and led to a partial phenotypic suppression in mutants impaired in BL signaling. BSU1 expression in young tissues and in the vasculature, where BL effects are hypothesized to be strongest, also supports a role for this protein in cell elongation. In a search for comparable expression patterns, we found striking similarities to the promoter activity of GASA1 (Raventos et al. 2000). GASA1 encodes a protein of unknown function, likely targeted to the secretory pathway. GASA1 expression is correlated with cell elongation, and is antagonistically regulated by gibberellins (GAs) and BRs (Bouquin et al. 2001).
The overall design of the pathway argues that BL signaling, as determined by the amount of dephosphorylated BES1, is poised for rapid responses. The regulation of BSU1 should shed some light on this matter, but we are so far clueless concerning the mechanisms that modulate its activity. We did not detect direct in vitro interactions between BSU1 and BES1 or BIN2, nor did we observe changes in the transcript or protein levels or subcellular localization in response to BL. Instead, we favor the idea that modulation of the enzyme's activity by interacting partners will be more relevant. This scenario is not without antecedents: our finding that the in vitro dephosphorylation of BES1 by BSU1 is fairly inefficient (Fig. 6F) may mirror the fact that, in vitro, the interaction between GSK3β and β-catenin is very weak and phosphorylation only proceeds in the presence of the scaffolding protein axin (Ikeda et al. 1998).
The structure, regulatory properties, and subcellular localization of BSU1 also hint at the existence of additional players in the BL signaling pathway. Generally, protein phosphatases incorporate domains or recruit partners that direct the catalytic subunit to specific substrates at defined subcellular locations (Bollen 2001). This is likely true for BSU1 as well, as Kelch domains are usually implicated in protein–protein interactions (Adams et al. 2000). Also, the extended loops in the catalytic domain may introduce new surface determinants in an otherwise highly conserved core (Fig. 3B): the β8–β9 loop faces the active site, whereas the αE–αF loop adjoins a region critical for the interaction between PP1 and Sds22, involved in the nuclear targeting of the phosphatase (Ceulemans et al. 2002). Keeping in mind that the enzymatic properties of recombinant Ser/Thr phosphatases like PP1 are not always comparable to those of the native proteins (Endo et al. 1997), we show that recombinant BSU1 displays a sui generis sensitivity toward inhibitors (Fig. 4B). Many PP1 regulatory subunits, including I-2, bear a loosely conserved sequence known as the RVxF motif that interacts with a hydrophobic groove on the surface of the phosphatase (Egloff et al. 1997). Contacts between two neighboring Phe residues in the phosphatase and the aromatic residue of the RVxF motif are critical for docking (Wu and Tatchell 2001). In BSU1, the hydrophobic residues lining the groove are preserved (empty arrowheads in Fig. 3B), but, significantly, the two Phe residues are replaced by Arg and Pro (filled arrowheads). These changes probably account for the insensitivity of BSU1 to I-2(Fig. 4B) and, by extension, impair the association of other RVxF-containing proteins, keeping BSU1 out of the regulatory circuits of other phosphatases.
The nuclear localization of BSU1 (Fig. 5G) was surprising and could not be predicted from its sequence, meaning that it has noncanonical nuclear localization signals or that other proteins anchor it. This finding raises questions on where BIN2, BES1, and BSU1 meet in the cell. Previous results showed that BES1 accumulates in the nucleus in response to BL (Yin et al. 2002a). By analogy to β-catenin, it was assumed that BES1 is dephosphorylated in the cytosol, thereafter moving into the nucleus to modulate the expression of BL-responsive genes. However, our results, together with the fact that BES1 and BZR1 were recently described as exclusively nuclear in root tips (Zhao et al. 2002), support a nuclear dephosphorylation of BES1 by members of the BSU family. We thus suggest (Fig. 8) that a key step in the regulation of BES1 takes place in the nucleus, probably associated to a multiprotein complex. The subcellular localization of BIN2 has not been reported, but the BIN2 homolog AtSKθ was found predominantly in nuclei of developing cells (Tavares et al. 2002). In this model, further genetic and biochemical approaches will be required to connect the membrane-localized perception of BL to the nuclear events.
Figure 8.
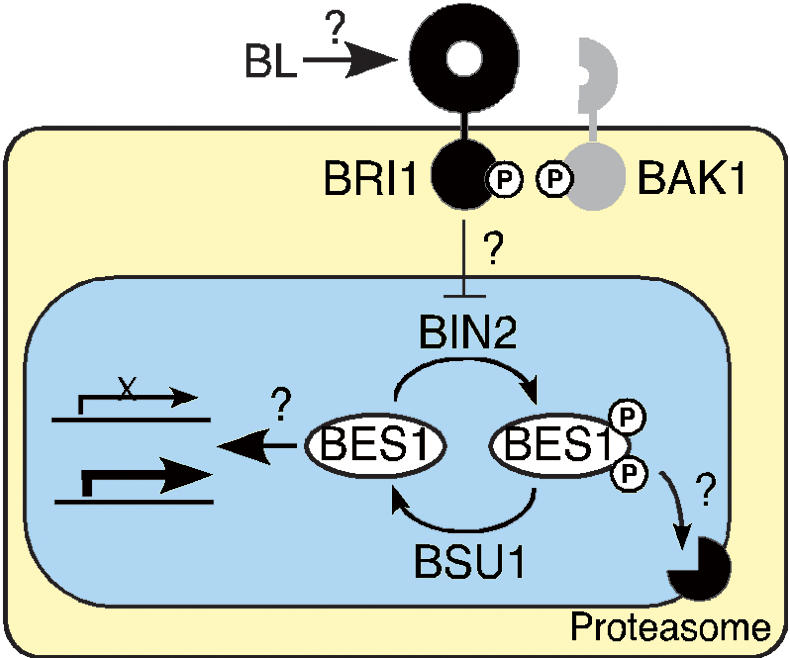
Membrane perception of BL by BRI1 and a coreceptor, BAK1, triggers auto- and transphosphorylation of the receptor kinases (Li et al. 2002). When the signal reaches the nucleus, the activity of BIN2 is depressed, thus allowing PPKL phosphatases to dephosphorylate BES1, which accumulates and regulates the expression of target genes. The subcellular localizations of BIN2 and the proteasome involved in the degradation of phosphorylated BES1 remain undefined, but they are tentatively placed inside the nucleus as are several steps along the pathway.
A major caveat of activation tagging is to show that the effects of overexpression are specific. This was particularly important in our case, as BSU1 doesn't appear to have internal inhibitory domains (Fig. 4A), and consequently, overabundant catalytic domains could escape regulatory partners or titrate out regulators of other phosphatases, triggering unspecific side reactions. Indeed, overexpression by the strong 35S promoter was detrimental (Fig. 2B). However, the phenotype associated with the knock-down in expression of BSU1 homologs suggests a positive role for PPKLs in BL signaling (Fig. 7B). We are in the process of generating multiple knockout lines for the whole family to extend these observations.
Functional overlap appears to be significant in BL signaling downstream from BRI1. BIN2 belongs to a family of 10 genes (Charrier et al. 2002); although reduced expression of BIN2 alleviated the phenotype of a weak bri1 mutant (Li and Nam 2002), the recessive mutation ucu1-3 produced only weak effects in a wild-type background (Pérez-Pérez et al. 2002). Also, the phosphorylation state of four additional BES1/BZR1 homologs is similarly affected by BL treatment (Y. Yin, unpubl.). It has been estimated that >80% of Arabidopsis genes are included in chromosomal duplications (Simillion et al. 2002). As an example, BES1 and BZR1 and the BL-induced transcription factors BEE1 and BEE3 (Friedrichsen et al. 2002) are encoded in chromosomal repeats. In gene families like AG/STK/SHP1/SHP2 (Pinyopich et al. 2003), Athb-8/REV/PHV/PHB (Sessa et al. 1998), or BRI1/BRL2/BRL1/BRL3 (Yin et al. 2002b), at least a couple of their members are paralogs, and genetic evidence has shown that functional overlap of these duplicated genes is relevant: only hypermorphic mutations have been identified for PHB and PHV (McConnell et al. 2001), and combined loss of function was needed to reveal phenotypic effects of SHP1 and SHP2 (Liljiegren et al. 2000) and BRL1 and BRL3 (A. Caño-Delgado, unpubl.). The BSU family is composed of four genes codified in two pairs of duplicated regions. BSU1 and BSL1, at the top of Chromosomes I and IV, diverge in sequence, expression pattern (Figs. 3C, 7A; data not shown), and probably also in function, as suggested by the lack of phenotypic effects in double-knockout mutants. BSL2 and BSL3, on the other hand, are 88% identical, appear to have broadly similar expression domains (Fig. 7A), and appear, to some extent, to act redundantly (Fig. 7B). As such, this family may therefore be a suitable model to gauge the mechanisms involved in the maintenance of duplicated genes against mutational drift: BSU1–BSL1 may be an example of functional diversification, whereas BSL2–BSL3 may reflect a process of subfunctionalization (Lynch and Force 2000). In this context, a pending question is the relative contribution of BSU1 to BL signaling. Its remarkably weak expression indicates that BSL2 and BSL3 probably account for most of the BES1 dephosphorylating activity.
This work represents the first functional characterization of a PPKL. Members of this family can be detected in angiosperms, in green algae, and in all the species related to Plasmodium falciparum analyzed. This phylogenetic distribution prompts interesting evolutionary inferences. P. falciparum belongs to the phylum Apicomplexa, which groups intracellular parasites carrying a nonphotosynthetic but essential plast, the apicoplast, derived from a secondary endosymbiosis, probably with a red alga (Maréchal and Cesbron-Delauw 2001). Whether Apicomplexa incorporated the PfPPα gene (Li and Baker 1998) from red algae or through lateral gene transfer from Viridiplantae, as described for other species that underwent higher-order endosymbiosis (Archibald et al. 2003), these proteins must have appeared in unicellular photosynthetic organisms, and were later coopted for cell-to-cell signaling upon the transition to multicellularity in the plant lineage.
Our data reinforce the idea that the response to BL is determined by the amount of dephosphorylated BES1, fine-tuned by a complex balance of synthesis, degradation, and antagonistic phosphorylation and dephosphorylation, organized in a web of redundant players. Dissecting the regulatory properties of this network, as well as the intracellular dynamics of its components, should be the next steps in the study of BL signal transduction.
Materials and methods
Plant handling and transformation and general molecular biology manipulations followed standard procedures (Ausubel et al. 1999; Weigel and Glazebrook 2001) or manufacturers' instructions. Mutants in the Ws-2background were kindly provided by Frans Tax (University of Arizona).
Screen for bsu
Arabidopsis thaliana bri1-5 (Noguchi et al. 1999) plants were transformed with Agrobacterium GV3101 carrying pSKI015 (Weigel et al. 2000) by flower spraying. Transformants were germinated on soil and selected by watering with a solution of 0.006% glufosinate-ammonium (Finale Basta, Bayer Cropscience). Phenotypes were scored at the seedling and adult stages. Plants showing some suppression of the bri1-5 phenotype were back-crossed to the parental line, and the segregation of the selection marker and the phenotype was followed in the progeny.
Sequences flanking the T-DNA insertion were identified by TAIL-PCR (Lin et al. 1995) and by plasmid rescue from genomic DNA digested with EcoRI, HindIII, and BamHI, as described (Weigel et al. 2000). The 5′- and 3′-ends of BSU1 mRNA were obtained by RACE-PCR (First-choice RLM-RACE; Ambion), and the full-length cDNA by RT–PCR from total RNA isolated from bsu1-1D plants, amplified with VentR DNA-polymerase (New England Biolabs).
Recapitulation of the bsu1-1D phenotype
A genomic fragment ranging from the sequence adjacent to the T-DNA insertion up to 242 bp of the 3′-UTR region was amplified from BAC F21B7 using VentR DNA polymerase (New England Biolabs) with primers 5′-CAGGTACCGCACTTTT TAAAACGCGTCGTATTTC-3′ and 5′-CAGGTACCGAACA CAACAATCAGAATGATAATG-3′, introducing flanking KpnI sites. The amplification product was sequenced, cloned into pMN20 (Weigel et al. 2000), and transformed into bri1-5 plants by flower dipping. Transgenic plants were selected on 0.5× MS (Murashige and Skoog) plates with 50 μg/mL kanamycin. For the expression under the control of the 35S promoter, a genomic fragment comprising only the coding region, including the 5′-UTR, was similarly amplified with primers 5′-CTGGATCCGAGGAATGGCTCCTGATCAA-3′ and 5′-CTGTCGACTTAACTTGTGCTCCTCAT-3′ and cloned into the BamHI–SalI sites of pCHF3.
RNAi
All the RNAi constructs were made in pHannibal (Wesley et al. 2001). The inverted repeats, flanked by the 35S promoter and the rbcS terminator, were inserted in the NotI site of the binary vector pART27 for plant transformation. To knock down BSU1 in bsu1-1D (Fig. 2C), we chose the linker region connecting the Kelch repeat and the phosphatase domains because it is very specific for At1g034445. A 282-bp fragment was amplified from bsu1-1D cDNA with primers 5′-CTGGATCCTCGAGAAGAT AATCCTGAT-3′ and 5′-CCGAATTCATCGATGCAAGTCTT GAGT-3′. To knock down the BSU1 homologs, we chose a region of 306 bp in the Kelch domain of BSL2. The fragment was amplified from Ws-2cDNA with primers 5′-CTGGATCCTCGAGAAGTGGACTAGGCT-3′ and 5′-CCGAATTCATCGATGGCAGTATCCAAGG-3′.
Transcript analysis
Typically 10–15 μg of total RNA, extracted with TRIzol Reagent (Invitrogen), was run in formaldehyde denaturing gels, transferred to Hybond-N+ membranes (Amersham), and hybridized with [α-32P]dCTP randomly radiolabeled probes. Probes hybridizing to the 3′-UTR regions of At1g03445, At1g03457, At1g03470, and CPD were amplified from Col-0 genomic DNA. Results were visualized by autoradiography or using a PhosphorImager (Molecular Dynamics).
GUS and GFP fusions
For plant transformation, 2.12 kb of the genomic sequence upstream of the BSU1 start codon was amplified from the BAC F21B7 using PfuTurbo DNA polymerase (Stratagene) and primers 5′-CGGGATCCCTTCAATATCCTATGGCATTTCC3′ and 5′-CATGCCATGGCACAATATTTTGTGGTGGAG-3′. The BamHI–NcoI fragment was fused to the GUSPlus gene in pCAMBIA1305.1. The GFP sequence was fused by chimeric PCR at the N terminus of BSU1 in the context of the genomic fragment used for recapitulation, after the second codon. An additional Gly codon was introduced 3′ of GFP, linking it to the rest of the BSU1 coding sequence. The fused fragment was inserted into pMN20 for plant transformation.
Petiole sections
Plants were grown for 35 d in short days (9 h light/15 h darkness). Petioles from the fourth rosette leaf were cut and fixed in 1% glutaraldehyde in PBS buffer and embedded in Technovit 7100 resin. Then 4-μm transverse sections were cut with glass knives using a Leica autocut microtome, stained with Toluidine blue, and photographed in a Zeiss microscope under bright field.
Protein analysis
For protein extraction, 10-day-old seedlings grown under strong light (16 h light/8 h darkness, 90 μmole/m2 · sec) were used. For BL treatments, whole seedlings were soaked in 0.5× MS liquid medium with 1 μM BL (CIDtech Research Inc.) dissolved in 80% ethanol or with an equal volume of solvent, frozen, and ground. Proteins were extracted by boiling the homogenate with two volumes/fresh weight of 2× Laemmli buffer, run in Novex Pre-Cast 4%–20% Tris-Gly gels, and transferred to nitrocellulose. Anti-BES1 serum was obtained by immunizing rats with an MBP–BES1 fusion protein expressed in E. coli, encompassing the full-length BES1. The serum was affinity-purified by adsorption to a similarly obtained GST–BES1 fusion protein immobilized onto CNBr-activated Sepharose 4B (Amersham); the bound antibodies were eluted with glycine buffer (pH 2.5), neutralized with Tris-base, and equilibrated in PBS. Anti-BRI1 kinase serum was obtained from rabbits (Wang et al. 2001). The signals in Western blots were detected with horseradish peroxidase-linked secondary antibodies by chemiluminiscence (Supersignal West Pico Chemiluminiscence Substrate; Pierce).
Protein expression and activity
The full-length BSU1 and phosphatase sequences were amplified from BSU1 cDNA with primers 5′-GTGAATTCGCTCCTGATCAATCTTATC-3′ and 5′-GAGAATTCCATAAGAAGG TCATTTCGA-3′ for the respective 5′-ends, and primer 5′-CGAGTCGACCCTTTATTCACTTGACTC-3′ for the 3′-end. The fragments were cloned into the EcoRI/SalI sites of pMAL-C (New England Biolabs). Cultures of transformed E. coli BL21-CodonPlus-RIPL cells (Stratagene) were grown at 18°C in YEP medium supplemented with 0.2% glucose and 1 mM MnCl2 until they reached an OD600 of 0.6, induced with 40 mM IPTG, and grown for an additional 10 h at the same temperature. The fusion proteins were purified and their phosphatase activity assayed according to the manufacturer's specifications (PSP Assay System; New England Biolabs). Inhibition studies were performed using similar procedures, adding okadaic acid (Sigma) or Inhibitor-2(New England Biolabs) to the reaction.
Acknowledgments
We thank Pablo Cerdán and Javier Palatnik for insightful suggestions; and Danielle Friedrichsen, Jennifer Nemhauser, and Takeshi Nakano for their help during the first stages of this work. This work was supported by Human Frontier Science Program Organization long-term fellowships to S.M.-G., G.V., and A.C.-D., and grants from the USDA and HFSP to J.C. J.C. is an investigator of the Howard Hughes Medical Institute. The GenBank accession number for the mRNA sequence of BSU1 is AY372269.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1174204.
References
- Adams J., Kelso, R., and Cooley, L. 2000. The Kelch repeat superfamily of proteins: Propellers of cell function. Trends Cell Biol. 10: 17–24. [DOI] [PubMed] [Google Scholar]
- Alessi R.R., Street, A.J., Cohen, P., and Cohen, P.T.W. 1993. Inhibitor-2functions like a chaperone to fold three expressed isoforms of mammalian protein phosphatase-1 into a conformation with specificity and regulatory properties of the native enzyme. Eur. J. Biochem. 213: 1055–1066. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Archibald J.M., Rogers, M.B., Toop, M., Ishida, K., and Keeling, P.J. 2003. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl. Acad. Sci. 100: 7678–7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Stuhl, K. 1999. Current protocols in molecular biology. John Wiley & Sons Inc., New York.
- Barford D., Das, A.K., and Egloff, M.-P. 1998. The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Ann. Rev. Biophys. Biomol. Struct. 27: 133–164. [DOI] [PubMed] [Google Scholar]
- Bollen M. 2001. Combinatorial control of protein phosphatase-1. Trends Biochem. Sci. 26: 426–431. [DOI] [PubMed] [Google Scholar]
- Bouquin T., Meier, C., Foster, R., Nielsen, M.E., and Mundy, J. 2001. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 127: 450–458. [PMC free article] [PubMed] [Google Scholar]
- Ceulemans H., Vulsteke, V., De Maeyer, M., Tatchell, K., Stalmans, W., and Bollen, M. 2002. Binding of the concave surface of the Sds22 superhelix to the α4/α5/α6–triangle of Protein Phosphatase-1. J. Biol. Chem. 49: 47331–47337. [DOI] [PubMed] [Google Scholar]
- Charrier B., Champion, A., Henry, Y., and Kreis, M. 2002. Expression profiling of the whole Arabidopsis Shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 130: 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Fujioka, S., Noguchi, T., Takatsuto, S., Yoshida, S., and Feldmann, K. 2001. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26: 573–582. [DOI] [PubMed] [Google Scholar]
- Choe S., Schmitz, R., Fujioka, S., Takatsuto, S., Lee, M.-O., Yoshida, S., Feldmann, K., and Tax, F. 2002. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3β-like kinase. Plant Physiol. 130: 1506–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D., Langford, M., and McMorris, T.C. 1996. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.-P., Johnson, D., Moorhead, G., Cohen, P., Cohen, P., and Barford, D. 1997. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16: 1876–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S., Connor, J.H., Forney, B., Zhang, L., Ingebritsen, T.S., Lee, E.Y.C., and Shenolikar, S. 1997. Conversion of protein phosphatase-1 catalytic subunit to a Mn2+ dependent enzyme impairs its regulation by inhibitor-1. Biochemistry 36: 6986–6992. [DOI] [PubMed] [Google Scholar]
- Friedrichsen D.M., Joazeiro, C.A., Li, J., Hunter, T., and Chory, J. 2000. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D.M., Nemhauser, J., Muramitsu, T., Maloof, J.N., Alonso, J., Ecker, J.R., Furuya, M., and Chory, J. 2002. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J., Huang, H.B., Kwon, Y.G., Greengard, P., Naim, A.C., and Kuriyan, J. 1995. Three-dimensional structure of the catalytic subunit of protein Ser/Thr phosphatase-1. Nature 376: 745–753. [DOI] [PubMed] [Google Scholar]
- Grove M.D., Spencer, F.G., Rohwededer, W.K., Mandava, N.B., Worley, J.F., Warthen Jr., J.D., Steffens, G.L., Flippen-Anderson, J.L., and Cook Jr., J.R. 1979. Brassinolide, a plant growth promoting steroid isolated from Brassica napus pollen. Nature 281: 216–217. [Google Scholar]
- He J.-X., Gendron, J., Yang, Y., Li, J., and Wang, Z.-Y. 2002. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. 99: 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Kishida, S., Yamamoto, H., Murai, H., Koyam, S., and Kikuchi, A. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 17: 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauschmann A., Jessop, A., Koncz, C., Szekeres, M., Willmitzer, L., and Altmann, T. 1996. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 9: 701–713. [Google Scholar]
- Kerk D., Bulgrien, J., Smith, D., Barsam, B., Veretnik, S., and Gribskov, M. 2002. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 129: 908–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinf. Appl. Notes 12: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Kutuzov M.A. and Andreeva, A.V. 2002. Protein Ser/Thr phosphatases with Kelch-like repeat domains. Cell. Signal. 14: 745–750. [DOI] [PubMed] [Google Scholar]
- Li J.L. and Baker, D.A. 1998. A putative serine/threonine phosphatase from Plasmodium falciparum contains a large N-terminal extension and five unique inserts in the catalytic domain. Mol. Biochem. Parasitol. 95: 287–295. [DOI] [PubMed] [Google Scholar]
- Li J. and Chory, J. 1997. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Li J. and Nam, K.H. 2002. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li J., Wen, J., Lease, K., Doke, J., Tax, F., and Walker, J.C. 2002. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Liljiegren S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. 2000. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770. [DOI] [PubMed] [Google Scholar]
- Lin Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463. [DOI] [PubMed] [Google Scholar]
- Lynch M. and Force, A. 2000. The possibility of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava N.B. 1988. Plant growth-promoting brassinosteroids. Ann. Rev. Plant Physiol. Plant Mol. Biol. 39: 23–52. [Google Scholar]
- Maréchal E. and Cesbron-Delauw, M.F. 2001. The apicoplast: A new member of the plastid family. T. Plant Sci. 6: 200–205. [DOI] [PubMed] [Google Scholar]
- Mathur J., Molnar, G., Fujioka, S., Takatsuto, S., Sakurai, A., Yokota, T., Adam, G., Voigt, B., Nagy, F., Maas, C., et al. 1998. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 14: 593–602. [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713. [DOI] [PubMed] [Google Scholar]
- Nam K.H. and Li, J. 2002. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., Feldmann, K., and Tax, F.E. 1999. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Phys. 121: 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Bishop, G.J., Kaneta, T., Reid, J.B., Chory, J., and Yokota, T. 2003. The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J. 36: 291–300. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Ponce, M., and Micol, J.L. 2002. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 242: 161–173. [DOI] [PubMed] [Google Scholar]
- Pinyopich A., Ditta, G.S., Savidge, B., Liljiegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. 2003. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88. [DOI] [PubMed] [Google Scholar]
- Pires-daSilva A. and Sommer, R.J. 2003. The evolution of signalling pathways in animal development. Nat. Rev. Gen. 4: 39–49. [DOI] [PubMed] [Google Scholar]
- Raventos D., Meier, C., Mattsson, O., Jensen, A.B., and Mundy, J. 2000. Fusion genetic analysis of gibberellin signaling mutants. Plant J. 22: 427–438. [DOI] [PubMed] [Google Scholar]
- Sadot E., Conacci-Sorell, M., Zhurinsky, J., Shnizer, D., Lando, Z., Zharhary, D., Kam, Z., Ben-Ze'ev, A., and Geiger, B. 2002. Regulation of S33/S37 phosphorylated β-catenin in normal and transformed cells. J. Cell Sci. 115: 2771–2780. [DOI] [PubMed] [Google Scholar]
- Salic A., Lee, E., Mayer, L., and Kirschner, M.W. 2000. Control of β-catenin stability: Reconstitution of the cytoplasmic steps of the Wnt pathway in Xenopus egg extracts. Mol. Cell 5: 523–532. [DOI] [PubMed] [Google Scholar]
- Sessa G., Steindler, C., Morelli, G., and Ruberti, I. 1998. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 38: 609–622. [DOI] [PubMed] [Google Scholar]
- Simillion C., Vanderpoele, K., Van Montagu, M.C.E., Zabeau, M., and Van der Peer, Y. 2002. The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. 99: 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal F.J.T., van Noort, M., Strouss, G.J., and Clevers, H.C. 2002. Wnt signals are transmitted through N-terminally dephosphorylated β-catenin. EMBO Rep. 31: 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber C. and McCourt, P. 2000. A role of brassinosteroids in germination of Arabidopsis. Plant Physiol. 125: 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G., Nagy, F., Schell, J., and Koncz, C. 1996. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and deetiolation in Arabidopsis. Cell 85: 171–182. [DOI] [PubMed] [Google Scholar]
- Tavares R., Vidal, J., van Lammeren, A., and Kreis, M. 2002. AtSKθ, a plant homologue of SGG/GSK-3 marks developing tissues in Arabidopsis thaliana. Plant Mol. Biol. 50: 261–271. [DOI] [PubMed] [Google Scholar]
- Wang Z.-Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. 2001. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383. [DOI] [PubMed] [Google Scholar]
- Wang Z.-Y., Nakano, T., Gendron, J., He, J.-X., Chen, M., Vafeados, D., Yang, Y., Fujioka, S., Yoshida, S., Asami, T., et al. 2002. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Weigel D. and Glazebrook, J. 2001. Arabidopsis: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Weigel D., Ahn, J.H., Blazquez, M.A., Borevitz, J.O., Christensen, S.K., Fankhauser, C., Ferrandiz, C., Kardailsky, I., Malancharuvil, E.J., Neff, M.M., et al. 2000. Activation tagging in Arabidopsis. Plant Phys. 122: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., Helliwell, C.A., Smith, N.A., Wang, M.B., Rouse, D.T., Lin, Q., Gooding, P.S., Singh, S.P., Abbott, D., Stoutjesdijk, P.A., et al. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590. [DOI] [PubMed] [Google Scholar]
- Wu X. and Tatchell, K. 2001. Mutations in yeast protein phosphatase type 1 that affect targeting subunit binding. Biochemistry 40: 7410–7420. [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Fujioka, S., Demura, T., Takatsuto, S., Yoshida, S., and Fukuda, H. 2001. Brassinosteroids levels increase drastically prior to morphogenesis of tracheary elements. Plant Phys. 125: 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang, Z.-Y., Mora-García, S., Li, J., Yoshida, S., Asami, T., and Chory, J. 2002a. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wu, D., and Chory, J. 2002b. Plant receptor kinase: systemin receptor identified. Proc. Natl. Acad. Sci. 99: 9090–9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Peng, P., Schmitz, R., Decker, A., Tax, F., and Li, J. 2002. Two putative BIN2 substrates are nuclear encoded components of brassinosteroid signaling. Plant Physiol. 130: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]