Abstract
Pioneering work in model organisms reveals that the reproductive system is involved not only in propagation of the species but also regulates organismal metabolism and longevity. In C. elegans, prevention of germline stem cell proliferation results in a 60% extension of lifespan, termed gonadal longevity. Gonadal longevity relies on the transcriptional activities of steroid nuclear receptor DAF-12, the FOXO transcription factor homolog DAF-16, the FOXA transcription factor homolog PHA-4, and the HNF-4-like nuclear receptor NHR-80. These transcription factors work in an integrated transcriptional network to regulate fatty acid lipolysis, autophagy, stress resistance and other processes, which altogether enhance homeostasis and extend life. Because the reproductive system also regulates longevity in other species, studies in C. elegans may shed light on ancient mechanisms governing reproduction and survival.
Keywords: gonad, hormone, steroid, fat metabolism, aging
1.0 Biology of Aging
Aging is a fundamental biological process that is all too familiar. It manifests as the gradual loss of physical and mental capabilities with time, accompanied by a decline in function in nearly all organs and physiologic systems of the body, eventually leading to demise. Aging is the major risk factor for the major killer diseases in humans, including cardiovascular disease, cancer, diabetes and neurodegenerative diseases. As the elderly become an ever greater proportion of society, it is critical to address how to increase health into old age. Thus elucidating the underlying causes of aging, and longevity mechanisms that oppose it, should help understand a fascinating question in biology with enormous societal impact.
Over the last few decades, groundbreaking work in model genetic organisms have shown that single gene mutations can enhance both organismal health and lifespan. Such studies have brought to light evolutionarily conserved signaling pathways, which mobilize natural defense and stress resistance mechanisms to combat aging (Kenyon, 2010). For example, a modest reduction of insulin/IGF signaling (IIS) leads to longevity in a manner dependent on the FOXO forkhead transcription factor. Components of IIS have been shown to regulate lifespan in yeast, worms, flies, and mice, demonstrating remarkable conservation. Even in humans, multiple independent genetic association studies implicate FOXO homolog with exceptional longevity (Kenyon, 2010). Importantly, such pathways also ameliorate animal models of tumorigenesis, heart disease, proteotoxicity and neurodegenerative disease (Kenyon, 2010). Thus health and life enhancing mechanisms are triggered by signaling pathways, and point in particular to endocrine systems in coordinating lifespan regulation.
From a wealth of studies in C. elegans, Drosophila, yeast, and mice, we now know that scores of genes and multiple conserved pathways can extend lifespan. These include regulatory pathways that reduce mitochondrial function, decrease protein synthesis, mediate nutrient sensing and energy homeostasis, and interpret signals from the reproductive system, to name a few (Kenyon, 2010). A unifying idea is that signaling mechanisms coordinate metabolism towards growth and reproduction or towards maintenance and survival. While often conceptualized as a tradeoff, where energy and resources used for one process come at the cost of another, the emerging view is that these rather represent different signaling states of the organism that can be uncoupled from explicit costs or tradeoffs. To illustrate these ideas, we focus below specifically on the role of the reproductive system in regulating lifespan in C. elegans.
2.0 Gonadal longevity: control of lifespan through the reproductive system
Specific cells and tissues in the body not only serve as endocrine sources to regulate metabolism, behavior, development and reproduction, but also the lifespan of the organism. In particular, the gonad, comprised of the germline and the somatic gonad play a critical role. Classic experiments by the Kenyon laboratory showed that when C. elegans germline precursor cells are removed from the gonad by laser microsurgery, germlineless animals live 60% longer than mock ablated controls (Hsin and Kenyon, 1999). Longevity triggered by germline elimination arises not as a simple consequence of a resource tradeoff with fertility, since additional removal of the somatic gonadal precursors, which comprise the support tissues surrounding the germ cells, abrogates this life extension. In this case, animals completely lacking the gonad are also infertile yet nevertheless not long lived. These observations instead suggest that the somatic gonad generates life lengthening signals, while the germline antagonizes this, but the nature of these signals remain poorly understood.
Within the germline, the origin of life shortening signals is likely germline stem cells (Arantes-Oliveira et al., 2002). In this context, mutations that reduce oocyte or sperm formation have little effect on longevity. This is unexpected especially because vast resources (yolk, fat, mitochondria, maternal message and protein) are invested in oocytes. By contrast, mutations that reduce the number of germline stem cells such as glp-1 loss of function induce longevity, while mutations that cause germline overproliferation, such as gld-1, shorten lifespan (Arantes-Oliveira et al., 2002). Glp-1 encodes a homolog of Notch that is expressed in germline stem cells, whose normal role is to promote mitotic proliferation and delay the transition to meiosis. GLP-1 responds to LAG-2/delta ligands produced in somatic gonadal distal tip cells, which constitutes the niche encasing the germline stem cell population. glp-1 mutants are temperature sensitive for germline proliferation defects. Even late upshifts in the adult to the restrictive temperature impact metabolism and longevity, albeit not to the same extent as earlier upshifts (Arantes-Oliveira et al., 2002; Wang et al., 2008). Importantly, work from Drosophila suggests that gonadal longevity is evolutionarily conserved, since adult flies lacking germline stem cells live longer than controls (Flatt et al., 2008). In mice, transplantation of young ovaries into older females increases the lifespan (Cargill et al., 2003), revealing that the mammalian gonad produces unknown life enhancing factors.
In the wild, of course, animals that lack the germline do not exist, so what is the meaning of germline loss and gonadal longevity? Elegant studies on germline stem cell proliferation by Hubbard and colleagues reveal that a number of important signaling pathways regulate this process (Korta et al., 2012; Michaelson et al., 2010). In particular, their work implicates nutrient sensing pathways including IIS, steroidal signaling, TOR and S6kinase signaling, as well as dietary restriction and amino acid availability, as important modulators of germline proliferation, cell cycle, differentiation and apoptosis. As described below many of these pathways are implicated in longevity induced by germline absence, which provokes signaling in the somatic tissues. However, it is unknown whether such pathways also work specifically within the germline to influence organismal life span. A simple hypothesis is that when the germline encounters unfavorable conditions, such as nutrient deprivation, DNA damage, or infection, germline stem cell proliferation is prevented. Quiescent stem cells trigger signaling towards catabolism and survival modes throughout the body. This ensures coordinated preservation of germline and soma. Upon return to favorable conditions, cells throughout the body can then resume growth and reproductive modes. In support of this idea, C. elegans can enter into an adult reproductive diapause in response to starvation just prior to maturation, during which the germline is whittled down to a handful of germline stem cells (Angelo and Van Gilst, 2009). Remarkably, such animals can survive without food for weeks—much longer than in replete conditions--yet when returned to nutrient rich conditions, will reconstitute the germline and other tissues and produce viable progeny. Although a plausible hypothesis, it remains to be seen whether adult reproductive diapause and gonadal longevity are equivalent.
Conceivably, another aspect of gonadal longevity may reflect the coordination of somatic life span and reproductive life span. As in mammals, germline cells within the C. elegans gonad have a finite reproductive span, the time period of fertility. Typically, C. elegans hermaphrodites produce progeny up to day 4-5 of adulthood, and thereafter live another 15 days. A decline in fertility arises due to defects in germline stem cells, including granularization and cellularization, as well as a decrease in oocyte quality, chromosomal stability and segregation (Luo et al., 2010). Additionally late oocytes inappropriately undergo endoreplication, possibly reflecting a loss of checkpoint control. A number of signaling pathways enhance reproductive longevity, and extend the length of the reproductive period during adulthood. They include those that affect both somatic life span and reproductive span such as reduced IIS and dietary restriction (Hughes et al., 2007; Luo et al., 2009). Others specifically promote extended reproductive span, including reduced serotonergic and TGF-beta/Sma signaling (Luo et al., 2009; Sze et al., 2000), with the latter doubling the reproductive span to 8-9 days or more. Pathways specific for reproductive span may reflect the special mechanisms required to maintain oocyte quality and chromosomal stability in meiotic cells. Genes identified as upregulated in oocytes with enhanced reproductive span have a striking correspondence with mammalian factors associated with youthful oocyte profiles, which include functions involved in cell cycle control, DNA maintenance, and pluripotency (Luo et al., 2010). Surprisingly, TGF-beta and IIS regulate reproductive span cell non-autonomously, from somatic tissues including hypodermis (TGF-beta), gut and muscle (IIS), revealing complex tissue interplay (Luo et al., 2010). It will be interesting to identify the hormonal signals that coordinate these processes.
Recently a number of chromatin modifying factors have been implicated in transgenerational inheritance of longevity, demonstrating that epigenetic mechanisms of germline transmission affect the life span of progeny. Depletion of components of the H3K4 trimethylation complex, including ASH-2, WDR-5, as well as the histone methyl transferase SET-2, extend C. elegans life span 20-30% (Greer et al., 2010). Brunet and colleagues asked whether such life span extension could be transmitted transgenerationally, by crossing in wild-type copies of chromatin modifying factor genes and examining life span in subsequent generations (Greer et al., 2011). Remarkably, genetically wild-type animals that were F3 descendents of wdr-5 mutants remained just as long lived as Po wdr-5 mutants, living 20% longer lived than controls. Longevity was transmitted 3-4 generations, but not longer, revealing a striking reset of phenotype. Transgenerational longevity is dependent on the H3K4m3 demethylase RBR-2, which catalyzes the production of H3K4m2, somehow implicating such chromatin marks in transgenerational inheritance (Greer et al., 2011). Longevity also depends on the presence of the germline: fem-3 mutants, which harbor oocytes but lack sperm, suppress the longevity effect, as do pgl-1 mutants, which have defects in germline development. Thus, transgenerational longevity, which depends on the presence of gametes, differs from gonadal longevity arising from loss of germline stem cells, since the latter is independent of ooctye and sperm. It will be fascinating to understand the molecular mechanisms underlying the transgenerational effect.
3.0 Steroid hormone control of gonadal longevity
For the remainder of the review we focus our attention on the dissection of gonadal longevity. In the absence of the germline stem cells, what are the molecular mechanisms in the soma that regulate life span? The observation that the gonad regulates the longevity of the entire organism implies that hormonal mechanisms are at work. Indeed, genetic experiments in C. elegans reveal that endocrine signaling, including components of steroidal signaling, IIS and fatty acid metabolism function in gonadal longevity pathways. Presumably their respective receptors work within mainly somatic tissues to regulate longevity throughout the body. Below we highlight the interplay of these signal transduction mechanisms.
The longevity of animals lacking the germline depends on a steroid hormone signaling pathway typified by C. elegans nuclear hormone receptor DAF-12 (Hsin and Kenyon, 1999). Nuclear hormone receptors are transcription factors that bind to fat soluble hormones such as steroids and fatty acids to directly regulate gene transcription. DAF-12’s closest vertebrate relatives include Liver-X, Farnesoid-X (FXR), and Vitamin-D receptors (Antebi et al., 2000). Like its vertebrate cousins, DAF-12 responds to ligands that are bile acid-like steroids, in this case called the dafachronic acids (DA) (Motola et al., 2006), which regulate key aspects of C. elegans life history. During larval development DAF-12/FXR serves as a stage selector, mediating the choice between reproductive development or arrest at the dauer diapause (Antebi et al., 1998; Riddle et al., 1981), a long-lived alternate third larval stage specialized for survival (Fielenbach and Antebi, 2008). It also regulates developmental timing in the heterochronic circuit, promoting transitions from second to third larval stage in various tissues by turning on let-7 microRNA homologs, mir-84 and mir-241 (Antebi et al., 1998; Bethke et al., 2009; Hammell et al., 2009). Finally, DAF-12 extends adult lifespan when germline stem cells are removed (Hsin and Kenyon, 1999). Thus, DAF-12 stands at the nexus of pathways linking developmental progression and longevity.
Work from my laboratory has shown that in addition to DAF-12, multiple components of steroidal signaling including the dafachronic acids, and activities involved in DA biosynthesis, such as the DAF-36/Rieske-like oxygenase and DAF-9/CYP27A1, all contribute to the gonadal longevity pathway (Gerisch et al., 2007; Gerisch et al., 2001; Rottiers et al., 2006). Notably, loss of function mutations in daf-12, daf-9 and daf-36 abrogate the longevity of germlineless glp-1 mutants or animals whose germline precursors have been removed by laser microsurgery. A recently discovered hormone biosynthetic gene, dhs-16, which functions biochemically as a 3-hydroxysteroid dehydrogenase, has been shown to behave similarly in these circuits (Wollam et al., 2012). Genetic studies suggest that longevity requires the transcriptional activity of DAF-12 bound to dafachronic acids. Whereas DA supplementation restores longevity to daf-9;glp-1 and daf-36;glp-1back to that of glp-1, it has no effect on shortlived daf-12;glp-1 double mutants (Gerisch et al., 2007).
4.0 Life extending signals from the somatic gonad include the DAs
As mentioned above, removal of the somatic gonadal precursors abrogates the longevity of germlineless animals, revealing the somatic gonad as a source of life extending signals. What is the molecular nature of these life extending signals? New evidence suggests they include the DAs themselves: DA supplementation restores longevity of animals lacking germline and somatic gonad, in a fashion dependent on the presence of daf-12 (Yamawaki et al., 2010). Moreover, DA supplementation does not further extend the lifespan of germline deficient animals, or that of gonad intact wild-type animals, revealing that DA is required but not sufficient for lifespan extension. If the somatic gonad is involved in DA production, then tissues within this organ would be expected to express endogenous hormone biosynthetic genes. Indeed, daf-9/CYP27A1, an essential hormone biosynthetic gene, is found in the spermatheca of the somatic gonad, though its levels of expression are unaffected by germline removal (Gerisch et al., 2001; Jia et al., 2002). In fact the source of the hormone is not as important as the level, since overexpression of daf-9/CYP27A1 from other steroidogenic tissues such as the epidermis, the nervous system, or specialized neuroendocrine cells in the head called the XXX suffice to restore longevity to gonadless worms (Yamawaki et al., 2010). Other tissues must also be involved since distinct steps of DA biosynthesis take place outside of the somatic gonad. For example, daf-36/Rieske-like oxygenase, which catalyzes the first step in the conversion of cholesterol to 7-dehydrocholesterol, is expressed mainly in the intestine (Rottiers et al., 2006; Wollam et al., 2011; Yoshiyama-Yanagawa et al., 2011), revealing DA biosynthesis is distributed in different tissues. DAF-12 is found in the nucleus of most tissues throughout the body, but the cellular focus of its activity for gonadal longevity has not been determined.
If somatic gonad signaling and DA production are congruent, another prediction is that DAF-12 target gene expression should show dependence on the gonad. Consistent with this, germline absence stimulates DAF-12 transcriptional activity, as measured by the upregulation of the DAF-12 dependent target cdr-6, as well as other genes (McCormick et al., 2012; Yamawaki et al., 2010). Conversely, further removal of the somatic gonad diminishes DAF-12 dependent gene expression, but can be rescued by exogenous DA. Taken together these data suggest that when the gonad is intact, signals from the germline impinge on the somatic gonad to inhibit DA/DAF-12 signaling. When the germline is absent, DA signaling is derepressed.
5.0 DAF-16/FOXO, a central regulator of gonadal longevity
A second critical integrator of gonadal signaling, DAF-16/FOXO is a forkhead transcription factor that serves as a central regulator of longevity (Kenyon, 2010). DAF-16/FOXO is best known as a transducer of IIS. Molecular genetic studies have led to a model whereby stimulation of the insulin/IGF receptor results in activation of a PI3K/PDK/AKT kinase cascade, which phosphorylates DAF-16/FOXO, resulting in its nuclear exclusion in most tissues and inhibition of its transcriptional activity (Kenyon, 2010). A reduction of IIS results in FOXO accumulation in the nucleus, where it turns on genes involved in stress resistance, quality control, immunity, and longevity. In particular, mild loss-of-function mutations in the daf-2, insulin/IGF receptor ortholog, provoke a remarkable extension of lifespan by 2-3 fold, in a manner wholly dependent on DAF-16/FOXO (Kenyon et al., 1993).
Gonadal longevity also strictly requires daf-16/FOXO as null mutants completely abolish the longevity of germlineless animals (Hsin and Kenyon, 1999). Knockdown of activity only during adulthood is sufficient to suppress life extension (Arantes-Oliveira et al., 2002). The evidence suggests that DAF-16/FOXO responds to germline ablation differently from reduced IIS. First, the longevity of reduced IIS is additive with that of gonadal longevity, since daf-2;glp-1 double mutants live 4-5 fold longer than wild-type (Hsin and Kenyon, 1999). Second, gonadal longevity shows strict dependence on a number of other genes, including tcer-1 and kri-1 (Berman and Kenyon, 2006; Ghazi et al., 2009), described below, which have little effect on IIS longevity. Third, gonadal signaling affects DAF-16 activity in a stage and tissue specific manner distinct from IIS. In particular, germline ablation causes DAF-16/FOXO to localize to the nucleus of intestinal cells during the first day of adulthood (Berman and Kenyon, 2006; Lin et al., 2001), whereas IIS reduction causes nuclear localization in most cell types (Henderson and Johnson, 2001; Lee et al., 2001; Lin et al., 2001). Fourth, only a subset of FOXO target genes are induced upon germline ablation including sod-3, dod-8, gpd-2, etc. (Ghazi et al., 2009). The intestine is a key metabolic and endocrine organ that stores fat, produces yolk, and relays hormonal signals to the rest of the organism. Expression of constitutively nuclear localized mutant forms of DAF-16 (referred to as DAF-16AM) within the intestine fully rescues the longevity of daf-16;glp-1 double mutants, showing that nuclear localization within this tissue is necessary for lifespan extension (Berman and Kenyon, 2006) (Libina et al., 2003).
6.0 Cell autonomous and non-autonomous signals affect intestinal DAF-16/FOXO activity
The experiments described above raise several key questions: How is DAF-16/FOXO localization and activity controlled by the gonadal longevity pathway? What is the relationship between steroidal signaling and DAF-16/FOXO? Both cell non-autonomous and autonomous mechanisms appear to play a role. An elegant study by Kenyon and colleagues found that components of the DA/DAF-12 signaling pathway visibly facilitate DAF-16/FOXO nuclear accumulation and activity (Berman and Kenyon, 2006). RNAi knockdown or mutation of daf-9/CYP27A1, as well as daf-12/FXR substantially reduce the nuclear localization of daf-16∷gfp in germlineless animals. Other hormone biosynthetic genes, such as daf-36/Rieske-like oxygenase and dhs-16/hydroxysteroid dehydrogenase also show this phenotype, and DA supplementation restores DAF-16/FOXO nuclear localization and longevity in germlineless animals lacking these hormone biosynthetic genes (Gerisch et al., 2007; Wollam et al., 2012).
Interestingly, constitutively nuclear localized daf-16AM/FOXO bypasses the requirement for daf-9/CYP27A1, but not for daf-12/FXR. Thus, DAF-12 has at least two activities. First, it partly induces DAF-16/FOXO nuclear localization. Second, it influences DAF-16/FOXO transcriptional activity and/or has critical downstream transcriptional targets of its own, independent of daf-9 and DA (Berman and Kenyon, 2006), although DA independence should be interpreted with caution, since alleles of daf-9 examined were non-null. FOXO proteins can form complexes with the mammalian nuclear receptor PPAR gamma, and DAF-12/FXR and DAF-16/FOXO physically interact by GST-pulldown experiments in vitro, but whether this occurs in vivo is yet unknown (Dowell et al., 2003).
Though steroidal signaling is important for DAF-16/FOXO nuclear localization and activity, DAF-12/FXR and DAF-16/FOXO do not work in a simple linear pathway. Evidence indicates that they exhibit distinct, only partially shared transcriptional outputs. For example sod-3 and lipl-4 are daf-16/FOXO dependent target genes, mostly independent of daf-12 (Wang et al., 2008; Yamawaki et al., 2010). Conversely, cdr-6 appears daf-12 dependent but daf-16 independent. Global transcriptional profiles of glp-1 mutants reveal 230 genes that are daf-16 dependent, and 130 that are daf-12 dependent. Nine of these genes are shared between the two, although more sensitive measures reveal a greater overlap for the targets of these transcription factors (McCormick et al., 2012).
Recently, the microRNA mir-71 has emerged as an important regulator of survival and longevity (Boulias and Horvitz, 2012; de Lencastre et al., 2010; Zhang et al., 2011). In aging wild-type animals, mir-71 expression increases during adulthood. Consistent with a role in aging, deletion of mir-71 hastens age-related decline in body movement and pharyngeal pumping and results in a 40% decrease of wild-type lifespan (Boulias and Horvitz, 2012; de Lencastre et al., 2010). While mir-71 loss fully abolishes longevity of glp-1 or germline precursor ablation, it proportionally shortens the lifespan of daf-2/InsR and cco-1/cytochrome C oxidase RNAi treated animals as much as wild-type, arguing that it may work more specifically within the gonadal pathway (Boulias and Horvitz, 2012). mir-71 overexpression modestly extends lifespan in the wild-type background, and further enhances glp-1 lifespan. This longevity is daf-16, but not daf-12 dependent, and requires daf-16 activity within the intestine. While mir-71 deletion has little effect on total DAF-16 protein levels, it reduces DAF-16 nuclear accumulation and target gene expression of lipl-4 and sod-3 (Boulias and Horvitz, 2012). Surprisingly, an analysis of the tissue requirements of mir-71 demonstrates that neural expression rescues gonadal longevity. Thus mir-71 works through an indirect cell non-autonomous mechanism to influence daf-16, and reveals that a complex tissue interplay between gonad, nervous system and intestine governs gonadal longevity. In the future it will be interesting to understand the regulatory targets of this microRNA and how it signals to other tissues.
How then is DAF-16/FOXO localization and activity regulated exclusively in the intestine? An important clue came from the discovery of KRI-1, a homolog of the human KRIT/CCM1 protein, a disease locus implicated in cerebral cavernous malformations (Berman and Kenyon, 2006). KRIT is an ankyrin repeat containing protein whose expression is limited to the C. elegans intestine and pharynx, and which localizes predominately to the apical junctions of cells as well as the nucleus. Similar to daf-9/CYP27A1, loss of kri-1 function abolishes gonadal longevity as well as DAF-16/FOXO nuclear localization, yet constitutively nuclear localized DAF-16AM bypasses the requirement for kri-1 (Berman and Kenyon, 2006). Moreover, kri-1 loss does little to diminish daf-2/InsR longevity, and in fact in some backgrounds can actually further extend lifespan. These findings suggest that kri-1 works specifically in the gonadal longevity pathway to regulate the tissue-appropriate localization of DAF-16/FOXO within the nuclei of intestinal cells.
Given that daf-16/FOXO behaves differently in response to reduced IIS and germline removal, a plausible hypothesis is that it also works in specialized transcriptional complexes to regulate gonadal longevity. One such component may be TCER-1, whose mammalian homolog TCERG-1 associates with the C-terminal region RNA polymerase II and co-purifies with the splicing complex (Ghazi et al., 2009; Goldstrohm et al., 2001; Sune and Garcia-Blanco, 1999). TCERG1 is thought to enhance elongation efficiency and couple transcription to splicing and RNA processing. Somewhat surprisingly, tcer-1(+) makes animals specifically competent for gonadal longevity, as tcer-1 RNAi or mutation significantly reduces the longevity of glp-1 germlineless animals, but has little effect on wild-type, daf-2/InsR mutants or other models of longevity including a genetic model dietary restriction (eat-2) or reduced mitochondrial respiration (isp-1) (Ghazi et al., 2009). Despite widespread expression, tcer-1 becomes upregulated specifically in the nuclei of intestine and neurons in germlineless animals. This upregulation depends on kri-1 but not on daf-12 or daf-16; in kri-1 mutants tcer-1∷gfp upregulation is not seen. Thus, kri-1 confers tissue specific regulation to both TCER-1 and DAF-16. Whereas tcer-1 loss of function abrogates gonadal longevity, overexpression is sufficient to modestly extend the lifespan of gonad intact animals. This longevity is dependent on kri-1 and daf-16. Although tcer-1 does not affect DAF-16/FOXO localization, it does impact the expression of some daf-16 target genes (e.g. dod-8) but not others (e.g. sod-3), suggesting its main responsibility lies in facilitating specific DAF-16 transcriptional complexes (Ghazi et al., 2009). A simple model is that upregulation of tcer-1 ensures productive transcriptional elongation and processing of a subset of daf-16 target genes. It is unknown, however, if DAF-16 and TCER actually physically interact. It is also unclear whether tcer-1 affects other transcription factors in the pathway.
Several other gene products collaborate with tcer-1 to regulate gonadal longevity. phi-62 encodes a highly conserved RNA binding protein of unknown function which was found as upregulated in glp-1 microarrays relative to wild-type (McCormick et al., 2012). RNAi knockdown abrogates glp-1 longevity, while having only a minor effect on wild-type. phi-62 and tcer-1 affect the same subset of daf-16 target genes in a similar way, and phi-62 is required for the longevity of tcer-1 overexpressors. ftt-2 encodes a 14-3-3 scaffold protein homolog whose expression was also found to be regulated in microarray experiments (McCormick et al., 2012). It is predicted to physically interact with PHI-62 based on yeast two hybrid screens from Drosophila. Like phi-62, ftt-2 knockdown reduces the longevity of glp-1 animals and tcer-1 overexpressors. ftt-2 has been previously shown to work in a complex with DAF-16/FOXO, and regulates its nuclear localization (Berdichevsky et al., 2006; Li et al., 2007; Wang et al., 2006). Conceivably, all these proteins collaborate in transcriptional complexes to regulate gonadal longevity. In the future it will be important to unravel the molecular mechanisms underlying these various components.
7.0 Autophagy and fat metabolism: proximal mechanisms of longevity?
Much of the discussion above has focused on endocrine mechanisms that coordinate the response to gonadal signals. But what might be the proximal processes and mechanisms that confer longevity? Recent work reveals that fat metabolism and autophagy play a critical role. One of the salient features of germlineless glp-1 mutants is that they accumulate substantial amounts of fat in intestine and epidermis, as measured by oil red O staining and triglyceride levels. On the other hand, they have lower levels of Nile red staining, which labels a lysosomal like organelle involved in fat metabolism (O’Rourke et al., 2009; Wang et al., 2008). To see whether metabolism might contribute to the longevity phenotype, Wang and Ruvkun screened through an RNAi library containing various metabolic genes, and found that knockdown of the fatty acid lipase, lipl-4, increased the Nile red staining of glp-1 animals (Wang et al., 2008). Moreover, lipl-4 RNAi disrupts glp-1 longevity and lipl-4 overexpression robustly extends lifespan in gonad intact wild-type animals. Fatty acid lipases are known to liberate fatty acids from triglycerides and fatty acyl cholesterol esters in response to energy demand. Correlatively, glp-1 mutants exhibit high levels of lipase activity partly dependent on lipl-4 (Hansen et al., 2008). These studies suggest that despite the fat accumulation phenotypes of glp-1, the liberation of free fatty acids functions somehow to extend lifespan; possibly this is linked to the Nile red staining acidic compartment. How free fatty acids extend life span remains enigmatic. Conceivably, they work as signaling molecules themselves relaying messages to other tissues. Alternately, they may stimulate the turnover of fat stores, or beta-oxidation of fat, which could exert beneficial effects. Clearly further studies are merited to understand this important question.
How might lipl-4 expression link to upstream endocrine signaling pathways? Apparently lipl-4 is upregulated in germlineless animals by daf-16/FOXO activity (Wang et al., 2008). lipl-4 may not be specific to the gonadal pathway, however, since it is also partially required for daf-2/InsR longevity. Although daf-12 does not substantially affect lipl-4 expression, daf-12 regulates other fat metabolism genes, such as lips-17, a triacylglycerol lipase, and fard-1, a fatty acid CoA-reductase, which are also required for gonadal longevity (McCormick et al., 2012).
Aguilaniu and colleagues discovered that the nuclear hormone receptor, nhr-80, regulates another aspect of fat metabolism and longevity (Goudeau et al., 2011). nhr-80 is one of many C. elegans homologs of HNF4-like nuclear hormone receptors, whose mammalian counterpart regulates liver development, fat metabolism, and glucose homeostasis (Brock et al., 2006). Loss of nhr-80 function reportedly abrogates lifespan extension of germlineless animals, but has little effect on wild-type animals, as well as on the extended longevity due to reduced IIS, reduced mitochondrial function (cyc-1 RNAi), and dietary restriction. Accordingly, nhr-80 mRNA is induced upon germline ablation in a manner largely independent on daf-12 and daf-16. The protein resides in the nucleus of intestinal and neuronal cells, and increases in expression specifically within the intestine upon germline ablation; however, it is unknown if this is kri-1 regulated. Loss of nhr-80 has no effect on daf-16/FOXO nuclear localization (Goudeau et al., 2011). Moreover, nhr-80 RNAi further shortens the life span of daf-16;glp-1. Taken together these results suggest that nhr-80 represents a novel and independent input into the pathway.
NHR-80 is one of handful of transcription factors that regulate the delta-9 fatty acid CoA-desaturase genes, fat-5, fat-6, and fat-7, and functions in homeostatic circuits to maintain fatty acid composition (Brock et al., 2006). These desaturases convert the saturated fats, palmitic and stearic acid, to their respective monounsaturated fats, palmitoleic and oleic acid. Accordingly, nhr-80 mutants exhibit increased levels of C18:0 stearic acid and decreased levels of C18:1 oleic acid (Brock et al., 2006). In animals that lack a germline, nhr-80(+) functions mainly to turn on fat-6/SteroylCoA-Desaturase 1 (Goudeau et al., 2011). Correlatively, germlineless animals contain higher levels of oleic acid, in a nhr-80 dependent fashion. These desaturases contribute to gonadal longevity, since fat-6;fat-7 double mutants abolish lifespan extension of glp-1, but have little effect on wild-type. Remarkably, lifespan extension is restored in triple mutants if such animals are provided with exogenous oleic acid, showing that this monounsaturated fat is important (Goudeau et al., 2011). At the moment it is unclear how oleic acid wields its effects. Oleic acid is not sufficient to extend wild-type lifespan, nor is it able to overcome the life shortening effects of nhr-80 deficiency in the germlineless background. Oleic acid and NHR-80 are thought to work in concert to extend life. Conceivably, oleic acid or its metabolites could serve as a signaling molecule that binds to NHR-80 to activate gene expression. Alternately, oleic acid could enhance triglyceride storage, stimulate lipolysis, or beta-oxidation to exert beneficial effects.
nhr-80 overexpression also has life extending properties. Although nhr-80 overexpression does not extend the lifespan of gonad intact wild-type animals, it further extends the lifespan of germlineless glp-1 mutants (Goudeau et al., 2011). It is unclear if this represents a novel type of longevity layered upon glp-1 or represents a maximization of gonadal longevity. This longevity is independent of daf-16/FOXO or daf-9/CYP27A1, but dependent on daf-12/FXR. Thus, nhr-80 and daf-12 may work in overlapping pathways. Perhaps daf-12 shares responsibilities in regulating downstream genes important for fatty acid metabolism, or in other tissues in the body important for longevity. Regardless it seems quite likely that the various transcription factors work in a complex network, which mutually enforce the longevity state, yet have somewhat independent outputs.
Other transcription factors that regulate the delta-9 desaturases include SBP-1, NHR-49 and MDT-15. SBP-1 is a homolog of the SREBP1 transcription factor that governs lipogenesis (Yang et al., 2006). NHR-49 is a functional homolog of the nuclear receptor PPAR alpha, which regulates beta-oxidation, as well as fatty acid desaturation, binding and transport (Van Gilst et al., 2005). NHR-49 apparently works in a complex with NHR-80 (Pathare et al., 2012), so is likely to play a role in the gonadal longevity pathway. Similarly, MDT-15 is a component of the mediator complex that putatively works as a co-activator of NHR-49 and SBP-1 (Taubert et al., 2006) (Yang et al., 2006). Whereas nhr-49 and sbp-1 mutants have not been tested in the gonadal pathway, mdt-15 mutations suppress glp-1 longevity (McCormick et al., 2012), with the caveat that these mutants also dramatically shorten wild-type lifespan (Van Gilst et al., 2005).
Autophagy is a process whereby cells internally phagocytose intracellular vesicular compartments, organelles, and protein aggregates, and deliver them to the lysosome for degradation. This process rids the cell of toxic lipophilic and proteinaceous waste, and is essential to healthy turnover of vesicular proteins and mitochondria, but is also used to extract energy from raw materials under nutrient limitation. Autophagy is required for longevity in a number of models including reduced IIS and dietary restriction (Hansen et al., 2008; Toth et al., 2008). As seen with other long lived models, Hansen and colleagues observed that germlineless animals accumulate autophagic particles in epidermal cells and intestine, as measured by electron microscopy and lgg-1∷gfp, a marker of such foci (Lapierre et al., 2011). Additionally, germlineless animals induce expression of autophagy genes and foci in a manner dependent on pha-4, but independent of daf-16/FOXO. PHA-4 is a FOXA forkhead transcription factor homolog that mediates longevity induced by dietary restriction (Panowski et al., 2007). Adult knockdown of pha-4 as well as various autophagy genes abolishes glp-1 longevity, yet has only minor effects on wild-type (Lapierre et al., 2011). Thus, both DAF-16/FOXO and PHA-4/FOXA homologs are required for gonadal longevity but for seemingly different processes, i.e. lipolysis and autophagy. Autophagy is required but not sufficient for longevity since daf-16;glp-1 double mutants still induce autophagy, yet are not long lived. This implies other processes must be rate limiting.
One of the major regulators of autophagy and longevity is TOR. TOR is a PI3 related kinase associated with dietary restriction mediated longevity (McCormick et al., 2011). In response to nutrient limitation, TOR downregulation stimulates autophagy and suppresses protein synthesis. As might be predicted, germline absence also triggers downregulation of TOR (Lapierre et al., 2011). Consistent with a role in the gonadal pathway, TOR RNAi extends the lifespan of wild-type animals but does not further extend the lifespan of glp-1 mutants. Finally, TOR downregulation induces pha-4 expression in germlineless animals. Thus, gonadal longevity requires the concerted downregulation of TOR, upregulation of PHA-4 and autophagic processes.
Recently, autophagy has been implicated in lipid remodeling whereby it stimulates lipolysis in lysosomal-like acidic compartments (Singh et al., 2009), thereby liberating free fatty acids for fuel. Prompted by these findings Hansen and colleagues explored the role of autophagy and lipolysis in gonadal longevity (Lapierre et al., 2011). They first observed that TOR downregulation results in induction of lipl-4 expression; this induction is daf-16 dependent. Interestingly, autophagic vesicle formation also depends on the presence of lipl-4, as RNAi to this gene reduced lgg-1∷gfp puncta. Conversely, lipl-4 overexpression stimulates lgg-1∷gfp puncta, as well as autophagic gene expression (Lapierre et al., 2011). The longevity of lipl-4 overexpression itself depends on the presence of autophagy genes and pha-4. These studies provide an important link between lipolysis and autophagy, and reveal that autophagy is critical to the life extending properties of fatty acid liberation. Thus, an alternative explanation for how fat metabolism genes might influence longevity may lie in lipophagy, the interplay between autophagy and lipolysis (Lapierre et al., 2011).
8.0 Working model and perspectives
Prevention of germline stem cell proliferation initiates a dramatic cascade of events, which include endocrine coordination of metabolic states favoring extended survival, fatty acid lipolysis and autophagy. Putting together these observations, we suggest the following integrated working model (Figure 1). In favorable conditions, when the germline is actively proliferating, it suppresses, through unknown mechanisms, DA/DAF-12 and DAF-16 longevity promoting activities in the soma. Under adverse conditions, perhaps in response to nutrient stress or other types of stress, germline proliferation slows or comes to a halt. Germline quiescence results in derepression of DA/DAF-12 signaling, possibly in the intestine and/or throughout the body, and the prompting of DAF-16 activity primarily in the intestine. Control of DAF-16 nuclear accumulation is stimulated by steroidal signaling as well as neural inputs from mir-71, and within the intestine itself by KRI-1. DAF-16 nuclear activity is promoted by postulated complexes comprised of FTT-2, PHI-62, TCER-1, and perhaps DAF-12. In parallel, NHR-80 transcriptional complexes are activated by independent inputs from the gonad, perhaps in a manner dependent on oleic acid (OA) or free fatty acids (FFA). DAF-16 and NHR-80 upregulate aspects of fatty acid desaturation and oleic acid production, while DAF-16 and DAF-12 turn on various fatty acid lipases and other important targets. Germline loss also triggers TOR downregulation which in turn stimulates PHA-4 and autophagy, as well as DAF-16 and LIPL-4 expression. Interlinked autophagy and lipolysis somehow promote a healthful state either through effects on metabolism, turnover, or as yet unknown downstream pathways that signal throughout the body. Conceivably, these various pathways are specialized in sensing or reacting to specific nutrient signals (fats, sterols, amino acids, carbohydrates) or other stress signals.
Figure 1.
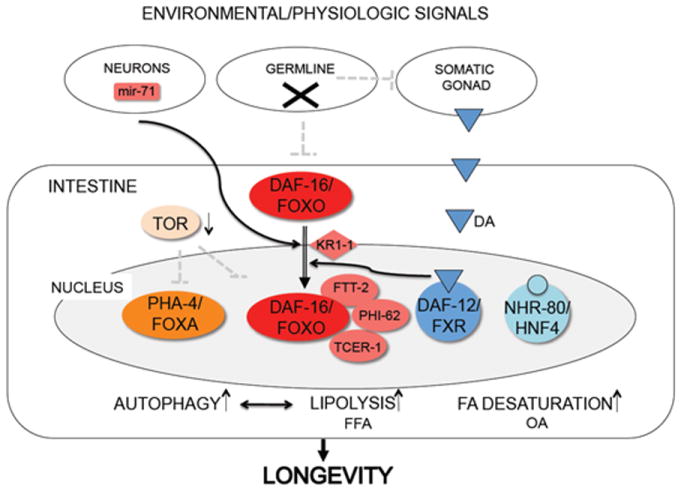
Model for gonadal longevity (see Section 8.0 for text)
Many aspects of this model are speculative, and clearly, many questions remain. Foremost among them, what cues does the germline respond to? What signals emanate from germline stem cells to cause life shortening? What are the primary sensors of such signals and what are the immediate pathways activated by germline removal? What is the order of events and nature of the regulatory hierarchy governing gonadal longevity? How do the various pathways interact? What are the tissue requirements of the various factors, and do they work in a cell autonomous or cell non-autonomous manner? How does fat metabolism extend lifespan? What are the proximal processes mediating lifespan extension? Is there feedback of metabolism onto regulatory events? What natural ecological process does germline removal mimic? How well conserved are the pathways and processes in evolution? It will be particularly important to understand this in context of mammalian biology. Does a similar phenomenon still exist in female mammals that lack a proliferating pool of adult germline stem cells, and set aside their gametes early in development? Or does menopause correspond to such a state? Even if there is not a one to one correspondence to mammals, the pathways deployed may still reveal important longevity mechanisms used in other contexts, as well as illuminate fundamental relationships between the reproductive system and longevity.
Highlights.
This work highlights recent discoveries on genes and processes involved in regulation of C. elegans life span by the reproductive system.
It focuses on how hormonal signaling pathways and transcription factors coordinate metabolism, lipolysis, and autophagy to regulate the life span.
Acknowledgments
I would like to thank members of the Antebi lab for critical reading of the manuscript and my sources of funding for their support (Max Planck Gesellschaft, CECAD, DFG/SFB635, BMBF/SYBACOL, NIH/NIA, Ellison Medical Foundation).
Abbreviations
- DA
Dafachronic acid
- NHR
Nuclear Hormone Receptor
- IIS
Insulin/IGF signaling
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angelo G, Van Gilst MR. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science. 2009;326:954–958. doi: 10.1126/science.1178343. [DOI] [PubMed] [Google Scholar]
- Antebi A, Culotti JG, Hedgecock EM. daf-12 regulates developmental age and the dauer alternative in C. elegans. Development. 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K, Horvitz HR. The C. elegans MicroRNA mir-71 Acts in Neurons to Promote Germline-Mediated Longevity through Regulation of DAF-16/FOXO. Cell Metab. 2012;15:439–450. doi: 10.1016/j.cmet.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TJ, Browse J, Watts JL. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006;2:e108. doi: 10.1371/journal.pgen.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill SL, Carey JR, Muller HG, Anderson G. Age of ovary determines remaining life expectancy in old ovariectomized mice. Aging cell. 2003;2:185–190. doi: 10.1046/j.1474-9728.2003.00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol. 2010;20:2159–2168. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell P, Otto TC, Adi S, Lane MD. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem. 2003;278:45485–45491. doi: 10.1074/jbc.M309069200. [DOI] [PubMed] [Google Scholar]
- Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Min KJ, D’Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, Jones DL, Tatar M. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci U S A. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, Mangelsdorf DJ, Antebi A. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci U S A. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- Ghazi A, Henis-Korenblit S, Kenyon C. A transcription elongation factor that links signals from the reproductive system to lifespan extension in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000639. doi: 10.1371/journal.pgen.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Albrecht TR, Sune C, Bedford MT, Garcia-Blanco MA. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol Cell Biol. 2001;21:7617–7628. doi: 10.1128/MCB.21.22.7617-7628.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudeau J, Bellemin S, Toselli-Mollereau E, Shamalnasab M, Chen Y, Aguilaniu H. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol. 2011;9:e1000599. doi: 10.1371/journal.pbio.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18668–18673. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode C. elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hughes SE, Evason K, Xiong C, Kornfeld K. Genetic and pharmacological factors that influence reproductive aging in nematodes. PLoS Genet. 2007;3:e25. doi: 10.1371/journal.pgen.0030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Albert PS, Riddle DL. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development. 2002;129:221–231. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Korta DZ, Tuck S, Hubbard EJ. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development. 2012;139:859–870. doi: 10.1242/dev.074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- Li J, Tewari M, Vidal M, Lee SS. The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Developmental biology. 2007;301:82–91. doi: 10.1016/j.ydbio.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the C. elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Luo S, Kleemann GA, Ashraf JM, Shaw WM, Murphy CT. TGF-beta and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell. 2010;143:299–312. doi: 10.1016/j.cell.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Shaw WM, Ashraf J, Murphy CT. TGF-beta Sma/Mab signaling mutations uncouple reproductive aging from somatic aging. PLoS Genet. 2009;5:e1000789. doi: 10.1371/journal.pgen.1000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick M, Chen K, Ramaswamy P, Kenyon C. New genes that extend Caenorhabditis elegans’ lifespan in response to reproductive signals. Aging cell. 2012;11:192–202. doi: 10.1111/j.1474-9726.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MA, Tsai SY, Kennedy BK. TOR and ageing: a complex pathway for a complex process. Philos Trans R Soc Lond B Biol Sci. 2011;366:17–27. doi: 10.1098/rstb.2010.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Korta DZ, Capua Y, Hubbard EJ. Insulin signaling promotes germline proliferation in C. elegans. Development. 2010;137:671–680. doi: 10.1242/dev.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma K, Sunino K, Xu E, Auchus R, Antebi A, Mangelsdorf M. Identification of DAF-12 ligands that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- O’Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Pathare PP, Lin A, Bornfeldt KE, Taubert S, Van Gilst MR. Coordinate regulation of lipid metabolism by novel nuclear receptor partnerships. PLoS Genet. 2012;8:e1002645. doi: 10.1371/journal.pgen.1002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle DL, Swanson MM, Albert PS. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Rottiers V, Motola DL, Gerisch B, Cummins CL, Nishiwaki K, Mangelsdorf DJ, Antebi A. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev Cell. 2006;10:473–482. doi: 10.1016/j.devcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sune C, Garcia-Blanco MA. Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner. Mol Cell Biol. 1999;19:4719–4728. doi: 10.1128/mcb.19.7.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Victor M, Loer C, Shi Y, Ruvkun G. Food and metabolic signalling defects in a C. elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, O’Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, Tissenbaum HA. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Wollam J, Magner DB, Magomedova L, Rass E, Shen Y, Rottiers V, Habermann B, Cummins CL, Antebi A. A novel 3-hydroxysteroid dehydrogenase that regulates reproductive development and longevity. PLoS Biol. 2012;10:e1001305. doi: 10.1371/journal.pbio.1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollam J, Magomedova L, Magner DB, Shen Y, Rottiers V, Motola DL, Mangelsdorf DJ, Cummins CL, Antebi A. The Rieske oxygenase DAF-36 functions as a cholesterol 7-desaturase in steroidogenic pathways governing longevity. Aging cell. 2011;10:879–884. doi: 10.1111/j.1474-9726.2011.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki TM, Berman JR, Suchanek-Kavipurapu M, McCormick M, Gaglia MM, Lee SJ, Kenyon C. The somatic reproductive tissues of C. elegans promote longevity through steroid hormone signaling. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- Yoshiyama-Yanagawa T, Enya S, Shimada-Niwa Y, Yaguchi S, Haramoto Y, Matsuya T, Shiomi K, Sasakura Y, Takahashi S, Asashima M, et al. The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol metabolizing enzyme. J Biol Chem. 2011 doi: 10.1074/jbc.M111.244384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zabinsky R, Teng Y, Cui M, Han M. microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc Natl Acad Sci U S A. 2011;108:17997–18002. doi: 10.1073/pnas.1105982108. [DOI] [PMC free article] [PubMed] [Google Scholar]


