Abstract
As an ever-growing number of genome sequences appear, it is becoming increasingly clear that factors other than genome sequence impart complexity to the proteome. Of the various sources of proteomic variability, post-translational modifications most greatly serve to expand the variety of proteins found in the cell. Likewise, modulating the rates at which different proteins are degraded also results in a constantly changing cellular protein profile. While both strategies for generating proteomic diversity are adopted by organisms across evolution, the responsible pathways and enzymes in Archaea are often less well described than are their eukaryotic and bacterial counterparts. Studies on halophilic archaea, in particular Haloferax volcanii, originally isolated from the Dead Sea, are helping to fill the void. In this review, recent developments concerning post-translational modifications and protein degradation in the haloarchaea are discussed.
Keywords: Archaea, haloarchaea, Haloferax volcanii, post-translational modification, protein degradation, proteome
Introduction
The ability to address microorganisms and other life forms at the level of the genome has revolutionized biological research. At the same time, it is becoming increasingly clear that a major proprortion of the diversity that exists within a cell is generated at the level of the proteome. Numerous factors are responsible for the proteome assuming additional levels of complexity not predicted at the genome level. These include alternative RNA splicing, differential expression of a given protein in response to environmental cues or as a function of developmental stage, and the plethora of possible protein-protein interactions, as well as post-translational modifications (PTMs) and regulated protein degradation.
Proteins can be modified post-translationally by the permanent or temporary covalent attachment of one or more of several classes of molecules, including sugars, lipids or other chemical groups. Flexibility in the spatial distribution of such linked moieties on a target protein and in the timing of their addition, together with the ability to introduce changes in the molecular composition of the bound modifying groups, offer further sources of protein variability. Intra-molecular disulphide linkages, formed via the covalent bonding of Cys residues pairs, influence the three-dimensional conformation of a protein. Disulphide bonds formed between different protein subunits can yield multimeric complexes (Fass, 2011). Proteolytic processing likewise can allow for control of the folding and function of a target protein. For instance, the post-translational removal of targeting sequences and inteins respectively permits the cell to control the site where a protein ultimately resides, as well as the timing and manner in which a protein can act (Paulus, 2000; Gogarten et al., 2002; Jarvis and Robinson, 2004; Hegde and Bernstein, 2006). Specifically, any of these PTMs, either alone or in combination, can affect protein function, interaction of the modified protein with binding partners, protein localization or the rate at which a protein is degraded, among other traits.
At the same time, regulated protein degradation is critical for maintaining protein quality and controlling cell functions. Proteases, which can discern and specifically degrade proteins compromised by denaturation, misincorporation of amino acids and other damaging events, are important for regulating cellular homeostasis. Likewise, regulated protein degradation processes, in which the protease destroys key proteins that may be properly folded but must be removed at specific times or moved to locations to enable molecular mechanisms to occur, are also important to cell function. Central to regulated protein degradation are proteases that are coupled to ATP hydrolysis (Gottesman, 2003). These energy-dependent proteases have a self-compartmentalized structure, in which the proteolytic active sites are housed within a protein nanoparticle that is chambered, gated and linked to an ATPase. The ATPase component of such proteases is required for the unfolding of protein substrate, opening of the gate and facilitating the degradation process (Lupas et al., 1997; Maupin-Furlow, 2012).
It is now clear that post-translational modifications and regulated protein degradation transpire in all three domains of life, namely Eukarya, Bacteria and Archaea. Still, current understanding of the archaeal versions of these processes lags behind that of their eukaryal and bacterial counterparts. In the following, we review what is known of post-translational modification and regulated protein degradation in the halophilic archaea, largely focusing on Haloferax volcanii. With the availability of a complete genome sequence (Hartmann et al., 2010), simple growth requirements and advanced genetic, molecular biology, proteomic and biochemical tools and techniques (DasSarma and Fleishmann, 1995; Allers et al., 2004; Soppa 2006; Kirkland et al., 2008b, Dyall-Smith, 2009; Allers et al., 2010), Hfx. volcanii represents a strain of choice for molecular studies in Archaea and has provided considerable insight into the archaeal versions of these protein processing events.
N-glycosylation
N-glycosylation, the covalent linkage of glycan moieties to select Asn residues of a target protein, was among the first haloarchaeal PTMs to be described. The Halobacterium salinarum surface (S)-layer glycoprotein was the first non-eukaryotic protein shown to be N-glycosylated (Mescher and Strominger, 1976). Two different Asn-linked oligosaccharides modify the S-layer glycoprotein, namely a repeating sulfated pentasaccharide linked via N-glycosylamine to Asn-2 and a sulfated glycan linked by a glucose residue to ten other Asn residues (Mescher and Strominger, 1978; Lechner et al. 1985a; Lechner and Wieland, 1989; Wieland et al. 1980; Wieland et al. 1983). The Hbt. salinarum flagellin (since renamed archaellin (Jarrell and Albers, 2012)) was shown to bear the same glycan-linked sulfated polysaccharide (Wieland et al. 1985). Although applying genetics to identify components of the Hbt. salinarum N-glycosylation pathway was not possible at the time, biochemical approaches served to reveal various aspect of the N-glycosylation pathway of this haloarchaeon. As such, it was shown that dolichol pyrophosphate (DolPP) serves as the lipid carrier of the glucose-linked sulfated glycan decorating 10 sites of N-glycosylation, whereas dolichol phosphate (DolP) bears the repeating sulfated pentasaccharide GlcNAc-linked to S-layer glycoprotein Asn-2 (Wieland et al. 1980; Lechner and Wieland, 1989). A link between these phosphodolichol-charged glycans to N-glycosylation was supported by the observation that in Hbt. salinarum, the glycan moiety of the DolPP-bound sulfated polysaccharide is also detected on the S-layer glycoprotein and archaellins in this species (Lechner et al. 1985a; Wieland et al. 1985). Moreover, the sulfated polysaccharide is methylated in the DolPP-linked form but not when protein-bound (Lechner et al. 1985b). The significance of this observation remains unclear. Finally, studies showing the ability of Hbt. salinarum cells to modify cell-impermeable, sequon-bearing hexapeptides with sulfated oligosaccharides localized the N-glycosylation event to the external cell surface (Lechner et al. 1985a).
Despite these advances, the process of N-glycosylation in the haloarchaea is currently best understood in Hfx. volcanii. The S-layer glycoprotein, comprising the sole component of the S-layer, contains seven putative N-glycosylation sites, namely the motif Asn-X-Ser/Thr, where X is any residue but Pro. Early studies reported modification of the S-layer glycoprotein Asn-13 and Asn-498 positions by a linear string of glucose residues, whereas Asn-274 and/or Asn-279 were supposedly decorated by a glycan containing glucose, galactose and idose (Sumper et al., 1990; Mengele and Sumper, 1992). More recently, evidence for N-glycosylation of the Hfx. volcanii archaellins, FlgA1 and FlgA2, was presented (Tripepi et al., 2010; Tripepi et al., 2012). Likewise, currently unidentified Hfx. volcanii glycoproteins of 150, 105, 98, 58, 56, 54 and 52 kDa have been detected (Zhu et al., 1995; Eichler 2000), although some of these species may correspond to the same polypeptide. Morever, it remains to be determined whether these proteins indeed experience N-glycosylation rather than O-glycosylation. The same is true of LccA, a glycosylated laccase secreted by Hfx. volcanii (Uthandi et al., 2010). The Hfx. volcanii S-layer glycoprotein, containing both N- and O-linked glycans, thus remains the best-characterized glycoprotein in this species (Sumper et al., 1990).
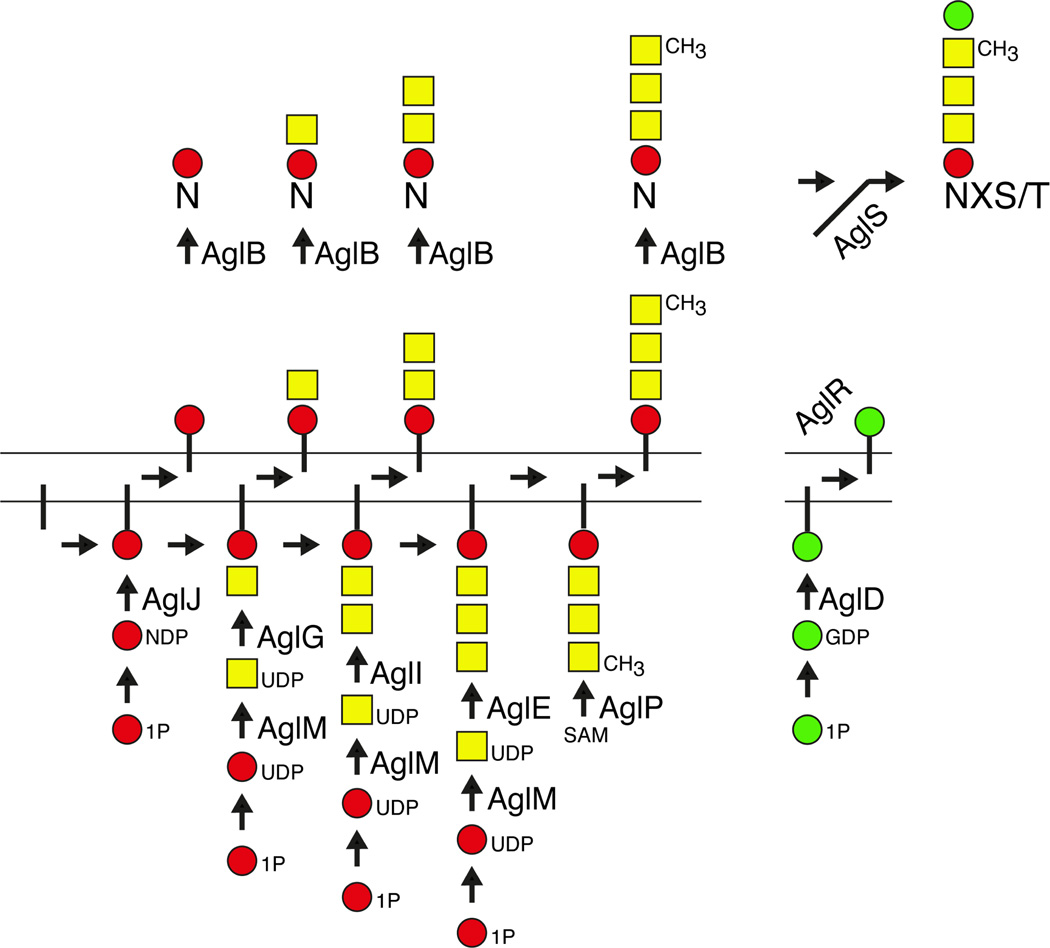
In the last few years, substantial progress in deciphering the pathway of S-layer glycoprotein N-glycosylation has been made, with the identification of a series of agl (archaeal glycosylation) genes encoding proteins involved in the assembly and attachment of a pentasaccharide to select Asn residues of the S-layer glycoprotein. Acting at the cytoplasmic face of the plasma membrane, AglJ, AglG, AglI and AglE sequentially add the first four pentasaccharide residues (i.e. a hexose, two hexuronic acids and the methyl ester of a hexuronic acid) onto a common DolP carrier, while AglD adds the final pentasaccharide residue, mannose, to a distinct DolP (Abu-Qarn et al., 2007; Abu-Qarn et al., 2008b; Yurist-Doutsch et al., 2008; Guan et al. 2010; Kaminski et al., 2010; Magidovich et al., 2010; Yurist-Doutsch et al., 2010). The use of dolichol phosphate by Archaea as the lipid carrier upon which the N-linked glycan is assembled also holds true for eukaryal N-glycosylation (Burda and Aebi, 1999; Hartley and Imperiali, 2012). In contrast, bacterial N-linked glycans are first assembled on a different isoprenoid, undecaprenol phosphate (Szymanski and Wren, 2005; Weerapana and Imperiali, 2006). N-glycosylation roles have also been assigned to AglF, a glucose-1-phosphate uridyltransferase (Yurist-Doutsch et al., 2010), AglM, a UDP-glucose dehydrogenase (Yurist-Doutsch et al., 2010) and AglP, a methyltransferase (Magidovich et al., 2010). Indeed, AglF and AglM were shown to act in a sequential and coordinated manner in vitro, transforming glucose-1-phophosphate into UDP-glucuronic acid (Yurist-Doutsch et al., 2010). In a reaction requiring the archaeal oligosaccharide transferase, AglB (Abu-Qarn and Eichler, 2006; Chaban et al., 2006; Igura et al., 2008), the lipid-linked tetrasaccharide and its precursors are delivered to select Asn residues of the S-layer glycoprotein. The final mannose residue is subsequently transferred from its DolP carrier to the protein-bound tetrasaccharide (Guan et al., 2010) in a reaction requiring AglR, a protein that either serves as the DolP-mannose flippase or contributes to such activity (Kaminski et al., 2012) and AglS, a dolichol phosphate-mannose mannosyltransferase (Cohen-Rosenzweig et al., 2012). Current understanding of Hfx. volcanii N-glycosylation is depicted in Fig. 1.
Fig 1. N-glycosylation in Hfx. Volcanii.
The Hfx. volcanii S-layer glycoprotein, a reporter of N-glycosylation in this species, is modified at Asn-13 and Asn-83 by a pentasaccharide comprising a hexose, two hexuronic acids, the methyl ester of a hexuronic acid and a mannose. The first four subunits of the pentasaccharide are sequentially assembled onto a DolP carrier via the activities of the glycosyltransferases, AglJ, AglG, AglI and AglE. At the same time, AglD adds the final pentasaccharide residue, mannose, onto a distinct DolP. Both charged DolP carriers are reoriented to face the cell exterior, with AglR thought to serve as the DolP-mannose flippase or to contribute to such activity. AglB acts to transfer the DolP-bound tetrasaccharide (and its precursors) to select Asn residues of target proteins, such as the S-layer glycoprotein. The final mannose subunit is then transferred to the protein-bound tetrasaccharide. AglF, AglM and AglP play various sugar-processing roles in the pathway. In the figure, DolP is presented as a vertical line, while hexoses are presented as red circles, hexuronic acids are presented as yellow squares and mannose is presented as a green circle.
Insight gained from N-glycosylation in Hfx. volcanii has served to elucidate aspects of the parallel process in other halophilic archaea. It was initially shown that Hfx. volcanii strains lacking either aglD or aglJ could be functionally complemented by introduction of rrnAC1873 and rrnAC0149, the respective homologues of these genes from Haloarcula marismortui (Calo et al., 2010b; Calo et al., 2011). Like Hfx. volcanii, the haloarchaeon Har. marismortui also originates from the Dead Sea (Oren et al., 1990). Indeed, subsequent efforts revealed the S-layer glycoprotein of both species are decorated with N-linked pentasaccharides comprising a hexose, two hexuronic acids, a methyl ester of hexuronic acid and a mannose (Calo et al., 2011). Still, differences in the N-glycosylation pathways of these two haloarchaea exist. While in Hfx. volcanii the N-linked pentasaccharide is derived from a tetrasaccharide sequentially assembled on a single DolP and a final mannose residue derived from a distinct DolP carrier (Guan et al., 2010), a similar pentasaccharide N-linked to the Har. marismortui S-layer glycoprotein is first fully assembled on a single DolP and only then transferred to the protein target (Calo et al., 2011). The finding that Hfx. volcanii N-glycosylation pathway components can be replaced with homologues from other haloarchaea to yield N-glycan variants has provided a proof-of-concept for developing Hfx. volcanii in a glyco-engineering platform designed to produce tailored glycoproteins (Calo et al., 2011). Such efforts will also exploit the proven ability of Hfx. volcanii to N-glycosylate introduced non-native proteins (Kandiba et al., 2012).
The Hfx. volcanii N-glycosylation pathway, involving multiple glycan-charged DolP carriers, recalls the parallel eukaryal process. In higher Eukarya, the first seven subunits of the 14-meric oligosaccharide assembled in the endoplasmic reticulum are sequentially added to a common phosphodolichol carrier, whereas the second set of seven sugar subunits are derived from single mannose- or glucose-charged DolP (Burda and Aebi, 1999; Helenius and Aebi, 2004; Hartley and Imperiali, 2012). The Hfx. volcanii N-glycosylation pathway further resembles its eukaryal counterpart when one considers that even in cells lacking the oligosaccharyltransferase, AglB, where pentasaccharide-modified DolP would be expected to accumulate, only tetrasaccharide-modified DolP could be detected (Calo et al., 2011). This observation points to the final mannose of the N-linked pentasaccharide as being added to the tetrasaccharide already attached to the S-layer glycoprotein. This same general strategy is employed in Eukarya, where in the Golgi, additional sugar subunits are attached to oligosaccharides already N-linked to the target polypeptide. On the other hand, Har. marismortui N-glycosylation is similar to the parallel bacterial process in which a heptasaccharide is assembled by the sequential addition of seven soluble nucleotide-activated sugars onto a common undecaprenol phosphate carrier (Szymanski and Wren, 2005; Weerapana and Imperiali, 2006; Abu-Qarn et al. 2008a). Yet, although delivered to the lipid-linked rather than the protein-bound tetrasaccharide, the terminal mannose subunit of the pentasaccharide N-linked to the Har. marismortui S-layer glycoprotein is derived from a distinct DolP carrier, as in Hfx. volcanii (Calo et al., 2011).
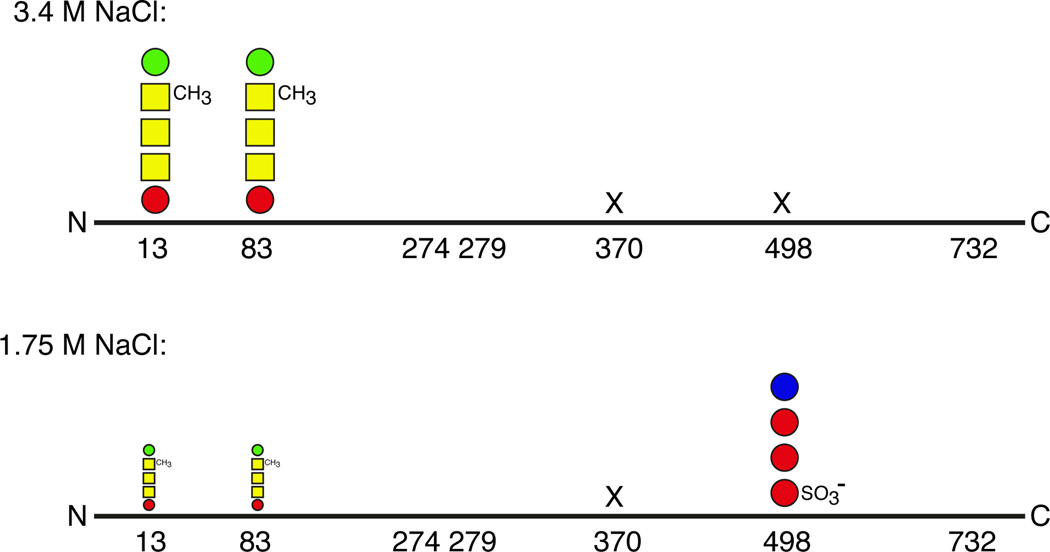
Although the absence or even the perturbation of N-glycosylation compromises the ability of Hfx. volcanii to grow in high salt (Abu-Qarn et al., 2007) and modifies S-layer stability and architecture (Abu-Qarn et al., 2007), as well as S-layer resistance to added protease (Yurist-Doutsch et al., 2008; Kaminski et al., 2010; Yurist-Doutsch et al., 2010), cells lacking AglB, and hence unable to perform N-glycosylation, are viable (Abu-Qarn et al., 2007). As such, it would seem that this post-translational modification is not essential for Hfx. volcanii survival, yet nonetheless is advantageous to Hfx. volcanii in certain scenarios. Thus, one can hypothesize that Hfx. volcanii modifies aspects of N-glycosylation in response to changing growth conditions. This concept has gained support from recent studies comparing N-glycosylation of the S-layer glycoprotein in cells grown in 3.4 or 1.75 M NaCl-containing medium (Guan et al., 2012). At the higher salinity, S-layer glycoprotein Asn-13 and Asn-83 were shown to be modified by the pentasaccharide described above, while DolP was shown to be modified by the tetrasaccharide comprising the first four pentasaccharide residues, again as discussed above. However, cells grown at low salinity contain DolP modified by a distinct tetrasaccharide comprising a sulfated hexose, two hexoses and a rhamnose not seen linked to DolP in cells grown at high salinity. This is likely the same DolP-bound tetrasaccharide observed by Kuntz et al. (1997) in Hfx. volcanii cells grown in 1.25 M NaCl-containing medium. The same tetrasaccharide modified S-layer glycoprotein Asn-498 in cells grown in low salt, whereas no glycan decorated this residue in cells grown in the high salt medium. At the same time, Asn-13 and Asn-83 were modified by substantially less pentasaccharide at the low salt conditions (Fig. 2). Hence, in response to environmental salinity, Hfx. volcanii not only modulates the N-linked glycans decorating the S-layer glycoprotein but also residues subjected to this post-translational modification.
Fig 2. The Hfx. volcanii S-layer glycoprotein undergoes differential N-glycosylation as a function of environmental salinity.
Mass spectrometry was used to reveal that when Hfx. volcanii cells are grown in 3.4 M NaCl-containing medium, Asn-13 and Asn-83 are modified by the pentasaccharide portrayed in Fig 1. In the conditions, Asn-370 and Asn-498 are not modified. When however, the cells are grown at lower salt concentrations (i.e. in medium containing 1.75 M NaCl), S-layer glycoprotein Asn-498 is modified by a ‘low salt’ tetrasaccharide comprising a sulfated hexose, two hexoses and a rhamnose. At the same time, Asn-13 and Asn-83 are still modified by the pentasaccharide described above, albeit much less so. Asn-370 is still not modified. The N-glycosylation status of Asn-274, Asn-279 and Asn-732 was not considered. In the figure, hexoses are presented as red circles, hexuronic acids are presented as yellow squares, mannose is presented as a green circle and rhamnose is presented as a blue circle. Positions where no glycosylation is seen are indicated by ‘x’.
Finally, it should be noted that studies on the methanogens, Methanococcus voltae and Methanococcus maripaludis, and the thermophiles, Sulfolobus acidocaldarius, Pyrococcus furiosus and Archaeoglobus fulgidus have also provided insight into archaeal N-glycosylation (Chaban et al., 2006; Igura et al., 2008; VanDyke et al., 2009; Meyer and Albers, 2011; Jones et al., 2012; Matsumoto et al., 2012).
Phosphorylation
Phosphorylation is widely appreciated as a covalent form of post-translational modification that occurs at His, Asp, Ser, Thr or Tyr residues. Phosphorylation is rapid, reversible and generates conformational changes in protein structure that mediate an array of biological responses from signal transduction to metabolism (Johnson and Barford, 1993). While early studies suggested that Archaea (and Bacteria) use mainly two-component systems of His/Asp phosphorylation, it is now appreciated that Archaea (and Bacteria) also perform Ser/Thr/Tyr phosphorylation (once thought to be restricted to eukaryotes) for creating highly sophisticated regulatory networks (Macek et al., 2008). A number of reviews and genomic surveys are available that highlight the phosphorylation of archaeal proteins and the enzymes (protein kinases/phosphatases) likely to mediate and/or regulate this PTM (Leonard et al., 1998; Kennelly and Potts, 1999; Kennelly, 2003; Eichler and Adams, 2005; Tyagi et al., 2010). In addition, a Phosphorylation Site Database is available online that provides a guide to some of the Ser/Thr/Tyr-phosphorylated proteins in Archaea (Wurgler-Murphy et al., 2004). Thus, the discussion below highlights recent studies on protein phosphorylation in halophilic archaea and how this type of PTM may control cellular function.
i) Phospho-site mapping
Historically, halophilic archaea have provided a useful model system to advance our understanding of how proteins can be phosphorylated across domains of life. In fact, the demonstration that proteins of Hbt. salinarum were reversibly phosphorylated by a light-regulated retinal-dependent mechanism was the first report that proteins in Archaea could be phosphorylated (Spudich and Stoeckenius, 1980). Later use of molecular biology tools revealed a two-component His/Asp phosphorelay system (analogous to the CheA/CheY system of Bacteria) that was responsible for modulating the response of Hbt. salinarum to chemotactic and phototactic stimuli (Rudolph et al., 1995; Rudolph and Oesterhelt, 1995) (Fig. 3). Early study of Hfx. volcanii revealed a Mn2+-stimulated protein phosphatase activity against synthetic protein subtrates phosphorylated at Ser/Thr residues (Oxenrider and Kennelly, 1993). More recent studies using high-throughput methods have facilitated the mapping of phosphorylation sites on proteins of halophilic archaea (9 phospho-sites in Hfx. volcanii and 81 phospho-sites in Hbt. salinarum), thus increasing the number of phospho-sites (from 3 total) previously detected in Archaea (Kirkland et al., 2008a; Aivaliotis et al., 2009). In these high-throughput approaches, wild-type and mutant strains with enhanced levels of phosphoproteins (i.e., Hfx. volcanii ΔpanA (proteasome-activating nucleotidase A) and Hbt. Salinarum ΔserB (OE4405R phosphoserine phosphatase)) were used as input material. Phospho-peptides were enriched from samples by immobilized metal affinity chromatography (IMAC) and metal oxide affinity chromatography (MOAC) (in parallel and sequentially), followed by tandem mass spectrometry (MS/MS). While a phospho-site consensus motif was not apparent, the majority of sites mapped to Ser/Thr/Tyr residues.
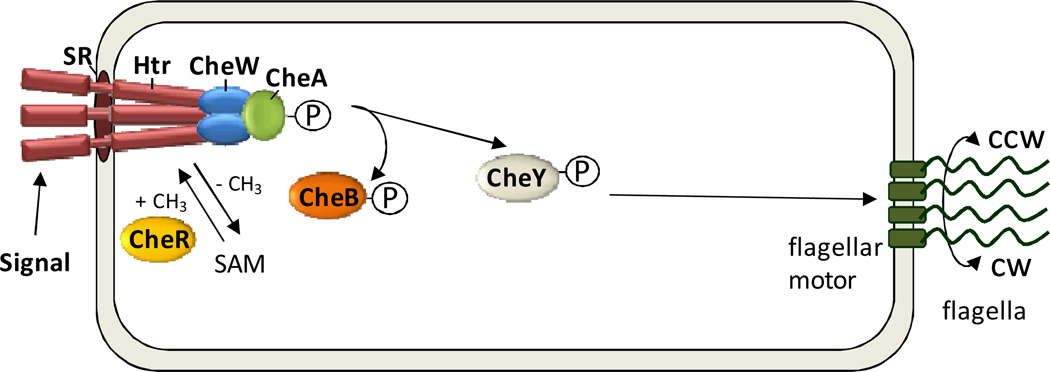
Fig 3. Protein modification in Hbt. salinarum taxis.
Halobacterial transducers (Htrs) are soluble or membrane-bound complexes that associate with signal receptors (SRs). Htrs signal to a two-component regulatory system composed of an autophosphorylating histidine kinase CheA, which mediates phosphotransfer to CheY, the response regulator of the system. CheY targets the flagellar motor and regulates the switch for flagellar rotation (clockwise (CW) vs. counterclockwise (CCW)). Adaptation is promoted by the methylation status of conserved Glu and Gln residues of Htr, where CheB deamidates Htr Gln residues prior to O-methylesterification. Htr is methylated by CheR (+CH3) and demethylated by CheB (−CH3). CheA-mediated phosphorylation regulates the demethylation activity of CheB.
ii) Protein kinases/phosphatases
Halophilic archaea are predicted to encode histidine protein kinases and protein phosphatases of the ‘two-component’ Asp/His phosphorelay system (Koretke et al., 2000; Kim and Forst, 2001) (e.g., Hfx. volcanii is predicted to encode at least 30 histidine protein kinases). The best studied example of an archaeal Asp/His phospho-relay system is that of Hbt. salinarum, in which a CheA histidine protein kinase undergoes ATP-dependent autophosphorylation of a His residue and transfers the phosphoryl group to an Asp residue on CheY, a response regulator thought to be dephosphorylated by the protein phosphatase, CheC (and not CheZ) (Rudolph et al., 1995; Rudolph and Oesterhelt, 1995; Muff and Ordal, 2007; Streif et al., 2010). Ultimately, the phosphorylation status of CheY impacts the ability of this protein to switch the flagellar motor and regulate cellular movement toward favorable light and nutrients (Nutsch et al., 2005). Interestingly, variants of ‘two-component’ histidine protein kinases can act as Ser/Thr/Tyr protein kinases in eukaryotes (Harris et al., 1995) and Bacteria (Min et al., 1993; Yang et al., 1996; Shi et al., 1999; Wu et al., 1999). Whether or not this alternative type of phosphorylation also occurs in Archaea is yet unknown.
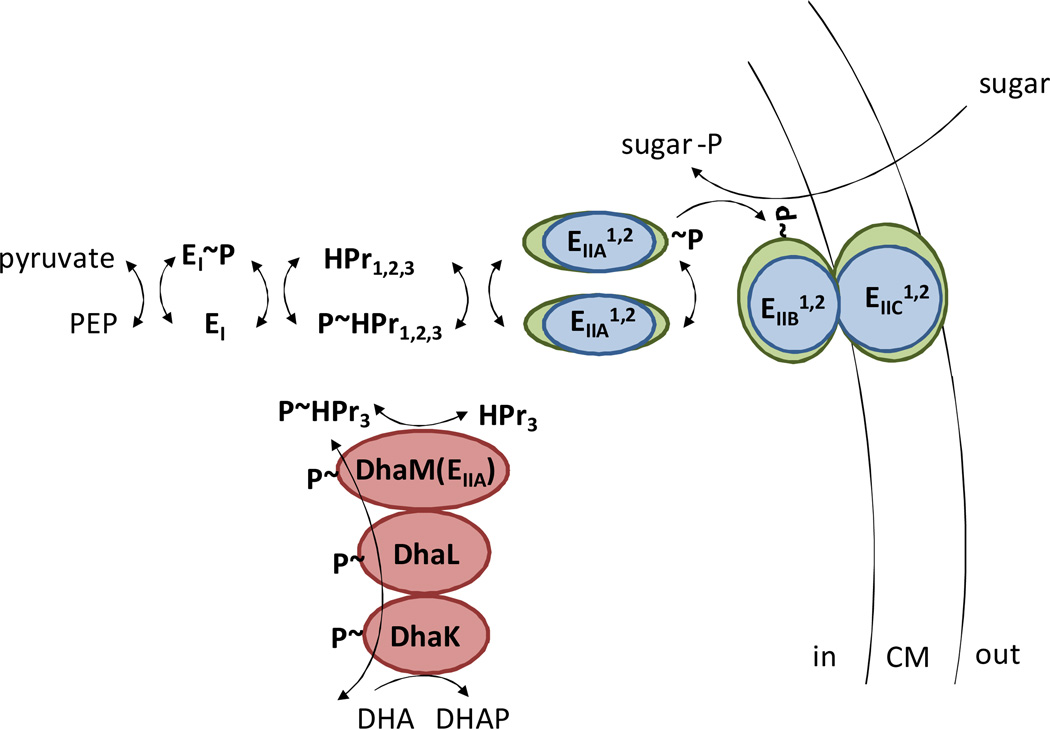
Many halophilic archaea (including Hfx. volcanii) as well as the crenarchaeon Thermofilum pendens harbor homologs of the phosphoenolpyruvate phosphotransferase system (PTS) (Hartman et al., 2010) (Fig. 4). In analogy to what is known for Bacteria (Barabote and Saier, Jr., 2005), the Hfx. volcanii PTS is predicted to mediate transfer of the phosphoryl group on PEP to imported sugars (e.g., fructose, galacticol) or endogenous dihydroxyacetone via dihydroxyacetone kinase (Hartman et al., 2010). Hfx. volcanii PTS homologs include a single enzyme I (PtsI; His~P) and multiple copies of histidine protein (HPr; His~P), enzyme IIA (EIIA; His~P), enzyme IIB (EIIB; Cys~P) and enzyme IIC (EIIC; His~P), with the amino acid residue predicted to be phosphorylated during group translocation provided in parenthesis. Recent work demonstrates that the PTS gene cluster HVO_1495 to HVO_1499, encoding PtsI, EIIB, HPr, EIIA, and EIIC homologs, was highly upregulated as a cotranscript during growth on fructose (Pickl et al., 2012). Deletion of HVO_1499, encoding a homolog of the fructose-specific membrane component EIIC of this cluster, resulted in loss of growth on fructose compared to glucose (Pickl et al., 2012). Thus, the PTS system has a functional involvement in the metabolism of fructose in Hfx. volcanii.
Fig 4. Phosphotransferase system of Hfx. Volcanii.
A schematic diagram of the Hfx. volcanii phosphotransferase (PTS) system predicted to be responsible for responsible for the simultaneous transport and phosphorylation of sugar substrates (e.g., fructose and galacticol) and for the generation of dihydroyacetone phosphate (DHAP) from dihydroxyacetone (DHA) by DHA kinase. A series of enzyme intermediates, including EI, HPr, EIIA, EIIB, EIIC and DHA kinase (DhaM, L, K), are predicted to be phosphorylated.
Atypical RIO-type Ser/Thr protein kinase homologs of the type 1, type 2 and Bud32 (piD261) families are common among the Archaea (including the haloarchaea) (Leonard et al., 1998; Shi et al., 1998; Ponting et al., 1999; LaRonde-LeBlanc and Wlodawer, 2005a,b; Tyagi et al., 2010). Protein kinases of the RIO-type 1 are distinguished by an STGKEA consensus sequence in their N-terminal domain and a second region of homology (IDXXQ, where X represents any amino acid residue) in their C-terminal domain. Kinases of the RIO-type 2 often have an N-terminal helix-turn-helix motif followed by GXGKES and C-terminal IDFPQ sequences. Members of the Bud32 family of Ser/Thr protein kinases are associated with the KEOPS (ECK) complex, composed of three additional subunits (Kae1, Pcc1 and Cgi121) and shown to be required for formation of the tRNA modification threonylcarbamoyladenosine (t6A) in yeast (Srinivasan et al., 2011).
A RIO-type 1 homolog (Rio1p, HVO_0135) of Hfx. volcanii has been characterized at the biochemical level. Rio1p purifies as a monomer and can transfer the γ-phosphoryl group of ATP to α1, a protein that forms the outer rings of 20S proteasomes in Hfx. volcanii (Humbard et al., 2010b). Rio1p-mediated phosphotransfer is not observed and/or is diminished for α1 variants T158A, S58A and T147A. Thus, Rio1p can phosphorylate a substrate protein (α1) at Ser/Thr residues based on in vitro assay.
Homologs of the four subunits of the eukaryotic KEOPS complex are conserved in Archaea, with Hfx. volcanii encoding homologs of Pcc1 (HVO_0652) and Cgi121 (HVO_0013) and a fusion of the Bud32 Ser/Thr protein kinase to Kae1 (HVO_1895). The gene encoding the Bud32-Kae1 homolog and its subdomains appear essential in Hfx. volcanii (Naor et al., 2012), suggesting Bud32-mediated phosphorylation of Ser/Thr residues is important for cell function. Homologs of KEOPS (Bud32, Kae1, Cgi121 and Pcc1) from related Euryarchaeota (Methanocaldococcus jannaschii and Pyrococcus furiosus) have been used for reconstitution of the complex, structural analysis at the atomic level, heterologous complementation and in vitro phosphorylation assays (Hecker et al., 2008; Mao et al., 2008). From this work, Bud32 is suggested to phosphorylate a Thr residue of an insert within the catalytic cleft of Kae1 and, thus, regulate KEOPS function. Whether or not the Bud32 phosphorylates other proteins in Archaea besides Kae1 is not clear. Both yeast Bud32 and its human ortholog PRPK (p53-related protein kinase) can phosphorylate p53 (Abe et al., 2001; Facchin et al., 2003). Likewise, Sulfolobus solfataricus SsoPK5 (a Bud32 homolog) catalyzes the phosphorylation of various proteins in vitro (Haile and Kennelly, 2011).
In addition to the atypical RIO-type Ser/Thr/Tyr protein kinases, homologs of the Bacillus subtilis PrkA (UniProt P39134) are common among halophilic archaea (e.g., Hfx. volcanii HVO_2849 and HVO_2848). Members of the PrkA family possess a Walker A-motif of nucleotide-binding proteins and exhibit distant homology to eukaryotic protein kinases (Fischer et al., 1996). In addition, amino acid residues within the active site of cyclic adenosine 3', 5'-monophosphate (cAMP)-dependent protein kinase are also conserved in PrkA (Fischer et al., 1996). B. subtilis PrkA can phosphorylate a 60 kDa protein at a Ser residue (Fischer et al., 1996). However, further analysis is needed to determine the identity of this protein substrate and confirm that B. subtilis PrkA and its haloarchaeal relatives are Ser/Thr protein kinases.
iii) Phosphorylation of proteasomes
Proteasomes are self-compartmentalized proteases, which undergo a substantial number of post-/co-translational modifications, including phosphorylation. The phosphorylation of these complexes is of interest, since proteasomes are important for cell function (e.g., growth of Hfx. volcanii) (Zhou et al., 2008). Proteasomes are composed of a 20S catalytic core particle (of α- and β-type subunits) and regulatory particles, including AAA+ ATPases (homologs of Cdc48/VAT/p97 and Rpt subunits termed proteasome-associated nucleotidases or PANs) that mediate energy-dependent protein degradation (Maupin-Furlow, 2012; Barthelme and Sauer, 2012). In Hfx. volcanii, proteasomes are modified by phosphorylation in addition to Nα-acetylation, methyl-esterification and cleavage of β subunit precursors that expose the N-terminal threonine residue forming the active sites of 20S proteasomes (Table 2) (Wilson et al., 1999; Humbard et al., 2006; Humbard et al., 2010b). Eukaryotic 20S proteasomes and associated AAA+ ATPases (homologs of PAN termed Rpt1-6 and Cdc48/VAT/p97) are also altered by phosphorylation and other forms of covalent modification, such as the attachment of O-linked N-acetylglucosamine, Nε- and Nα-acetylation, N-myristoylation, and cleavage of β subunit precursors (Zhang et al., 2007; Ewens et al., 2010).
Table 2.
PTMs of Hfx. volcanii proteasomes.
| Subunit | Phosphor- ylated |
Methyl-esterified | Nα- acetylated |
Exposed by autocleavage |
|---|---|---|---|---|
| α1 | Thr147 | Asp20, Glu27, Glu62, Glu112, Glu161 |
Met1 | n.d. |
| α2 | Thr13/Ser14 | n.d. | Met1 | n.d. |
| β | Ser129 | n.d. | n.d. | Thr50 |
| PAN-A | Ser340 | n.d. | n.d. | n.d. |
Residue number according to protein sequence in GenBank [GI:300669661 (α1), GI:12229945 (α2), GI:292655712 (β), GI:302425218 (PAN-A)]
Hfx. volcanii 20S proteasomes and associated PANs are phosphorylated at Ser and Thr residues. To facilitate phospho-site mapping, 20S proteasomes and PAN proteins were purified from Hfx. volcanii by tandem affinity chromatography using His6- and StrepII-tags (Humbard et al., 2006; Humbard et al., 2010b). Sites of phosphorylation were identified by MS/MS (with precursor ion scanning) and included β Ser129, α1 Thr147 and α2 Thr13/Ser14 (not distinguished) of 20S proteasomes and Ser340 of PAN-A (Humbard et al., 2006; Humbard et al., 2010b). A phospho-site for PAN-B was not identified (Humbard et al., 2010b). MS/MS analysis of phosphopeptides enriched from Hfx. volcanii proteomes suggests Cdc48/VAT/p97 homologs are also phosphorylated, however, sites of modification were not identified by this high-throughput method (Kirkland et al., 2008a). In eukaryotes, phosphorylation and acetylation regulate the function of Cdc48/VAT/p97 proteins.
Phosphorylation of the α-type subunits of Hfx. volcanii proteasomes has been investigated at various stages of cell growth and assembly states. Humbard et al. (2010b) separated the α-type subunits (of cell lysate and 20S protesomes purified by affinity chromatography with β subunits) by two-dimensional gel electrophoresis (2DE) and detected α1 and α2 by immunoblot using polyclonal antibodies specific to each protein. Phospohorylated isoforms (two specific for α1 and one specific for α2) were determined by shifting the 2DE-protein spot to a more basic pI after removal of the acidic phosphate groups by phosphatase treatment. Of the two phosphorylated isoforms of α1 detected, the most acidic form was found throughout growth, assembled in 20S proteasomes. In contrast, the least phosphorylated isoform of α1 was present as both unassembled and assembled subunits of 20S proteasomes and was detected at reduced levels in later stages of growth (Humbard et al., 2010b). The phosphorylated isoform of α2 was also found associated with 20S proteasomes. Thus, phosphorylation of the α-type proteins is suggested to influence their assembly and/or be important for 20S proteasome function.
Site-directed mutagenesis has been used to determine the biological role of proteasome phosphorylation in Hfx. volcanii. Hfx. volcanii strains expressing α1 proteins with Ala modifications in Ser/Thr residues likely to be phosphorylated (based on MS analysis and an inability to accept a phosphoryl group from Rio1p, in vitro) display dominant negative phenotypes for cell viability and colony color (i.e., white vs. red) (Humbard et al., 2010b). Thus, phosphorylation of the α1 subunit of 20S proteasomes appears to be closely linked to cell growth and pigmentation (i.e., production of carotenoids) (Humbard et al., 2010b). Interestingly, Hfx. volcanii proteasomal mutant strains deficient in the synthesis of PAN-A display a striking increase in the number of cellular proteins that are phosphorylated, suggesting an added link between protein phosphorylation and proteasome function (e.g., phosphorylation may target proteins for destruction by energy-dependent proteases, in analogy to eukaryotes and Bacteria) (Kirkland et al., 2008a).
Protein acetylation
Protein acetylation is the covalent attachment of an acetyl group to a protein. In general, acetylation can impact protein function, stability and interactions with other molecules. Two types of protein acetylation are known to occur in living cells, Nα-acetylation and Nε- (or lysine) acetylation, with high-energy molecules, such as acetyl-CoA, providing the acetyl group for these modifications. Nα-acetylation is an irreversible mechanism in which an acetyl group is covalently attached to the α-amino group of the N-terminal amino acid of a protein. In contrast, Nε-acetylation occurs when the ε-amino group of a lysine residue is reversibly modified by the covalent attachment of an acetyl group. Studies on Hfx. volcanii have furthered our understanding of both processes in Archaea.
i) Nα-acetylation
Our current understanding of the prevalence of Nα-acetylation in Archaea is largely based on MS-based proteomic surveys (Soppa, 2010; Maupin-Furlow et al., 2012). Like Bacteria, Nα-acetylation of ribosomal proteins appears common among Archaea (Kimura et al., 1989; Hatakeyama and Hatakeyama, 1990; Klussmann et al., 1993; Marquez et al., 2011). Interestingly, the number of proteins reported to be Nα-acetylated varies greatly among the different archaeal groups. In haloarchaea (apparently unlike Bacteria), a relatively high proportion (14–29%) of the proteome is modified by Nα-acetylation (i.e., Hbt. salinarum, Nmn. pharaonis and Hfx. volcanii) (Falb et al., 2006; Aivaliotis et al., 2007; Kirkland et al., 2008b). Likewise, Nα-acetylation appears to impact a large percentage of the S. solfataricus proteome, based on the finding of 17 Nα-acetylated N-termini out of 26 total detected by MS (Mackay et al., 2007). Interestingly, to date, only a single protein (the α subunit of the 20S proteasome) is reported to be Nα-acetylated among the methanogens, suggesting that this form of modification may be rare in this group of Archaea (Zhu et al., 2004; Forbes et al., 2004; Enoki et al., 2011).
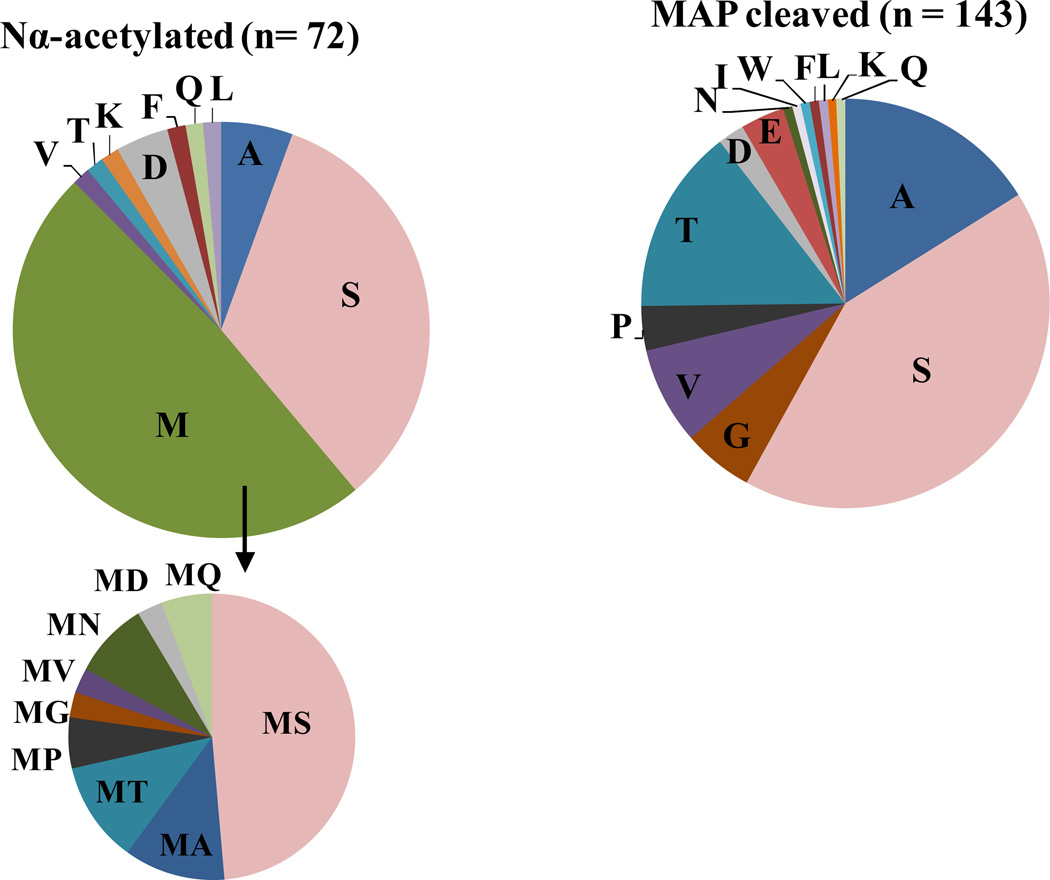
In Hfx. volcanii, like other Archaea, many of the Nα-acetylated proteins appear to be generated by a NatA-type activity (i.e., acetylation of penultimate Ser or Ala residues exposed after removal of N-terminal methionine residues by MAP) (Fig. 5). Nα-acetylated N-terminal methionine residues with penultimate Asp and Asn residues are also detected, consistent with the NatB-like activity of eukaryotes (Kirkland et al., 2008b). Proteins with a Nα-acetylated N-terminal methionine residue followed by a small penultimate residue (Ser, Ala, Thr, Pro and Gly) are also detected, suggesting that Nα-acetylation can restrict their cleavage by MAP. Interestingly, many of the proteins of halophilic archaea (including those of Hfx. volcanii) that are Nα-acetylated are also readily identified by semi-quantitative MS spectral counting in unmodified and/or MAP-cleaved forms (Falb et al., 2006; Aivaliotis et al., 2007; Kirkland et al., 2008b). Liquid chromatography-multiple reaction monitoring (LC-MRM) MS, a technique that provides a more accurate perspective on the abundance of the Nα-acetylated state of a protein, reveals that the α1 proteins that form 20S proteasomes are primarily in an Nα-acetylated Met form, as compared to the MAP cleaved form (103:1 ratio), in Hfx. volcanii (Humbard et al., 2009). Whether or not Nα-acetylation efficiency is also near 100% for other Hfx. volcanii protein substrates and/or can be altered by growth conditions remains to be determined.
Fig 5. Hfx. volcanii N-terminal proteome modified by Nα-acetylation (left) and/or cleavage by methionine aminopeptidase (right).
Pie charts based on the N-terminal proteome of Hfx. volcanii that was detected by MS/MS (Kirkland et al., 2008b). Based on this proteomic analysis, Hfx. volcanii proteins are often cleaved by methionine aminopeptidase (MAP) and/or Nα-acetylated. Penultimate residues exposed by MAP are often small and uncharged (Gly, Ala, Pro, Val, Ser or Thr). Of the N-termini that are acetylated, a relatively equal divide exists between proteins Nα-acetylated at their N-terminal Met versus a residue exposed after MAP cleavage (with Nα-acetylation of exposed Ser and Ala residues common). Among proteins with Nα-acetylated N-terminal Met residues, most (over 80%) have the Met residue followed by a small, uncharged residue (Gly, Ala, Pro, Val, Ser or Thr) typically cleaved by MAP.
While proteins with Nα-acetylated Met-Gln and Met-Asn sequences are not prevalent in Archaea, 20S proteasome α-type subunits with these N-terminal sequences are specifically Nα-acetylated in the Euryarchaeota, Hbt. salinarum, Nmn. pharaonis, Hfx. volcanii and Methanothermobacter thermoautotrophicus (Humbard et al., 2006; Falb et al., 2006; Aivaliotis et al., 2007; Enoki et al., 2011). Site-directed mutagenesis of the N-terminal Met-Gln sequence of the Hfx. volcanii 20S proteasome α1 protein has been used to further investigate this specific Nα-acetylation (Humbard et al., 2009). Variants of α1 were expressed in vivo and analyzed by MS to detect alterations in N-terminal modification (i.e., Nα-acetylation, MAP cleavage). A Q2A substitution rendered the α1 protein susceptible to cleavage by MAP followed by Nα-acetylation of the penultimate Ala by an apparent NatA-type activity similarly to most Nα-acetylated proteins in haloarchaea. However, the N-termini of α1 proteins with the small penultimate amino acid residues Ser and Val were detected predominantly in the uncleaved forms, with Nα-acetylated methionine residues intact. Alteration of penultimate amino acid residues to Asp, Pro and Thr resulted in a mixture of α1 protein in the Nα-acetylated methionine, MAP cleaved and/or unmodified forms. Thus, the enzyme(s) responsible for Nα-acetylation of the α1 N-terminal Met appears to have relaxed sequence specificity with regard to the penultimate residue of the substrate. Furthermore, only the Q2A substitution rendered the N-terminus of α1 fully susceptible to MAP cleavage, suggesting that structural elements of α1 or interacting partners/chaperones may mask primary N-terminal sequences that are otherwise optimal for MAP cleavage (e.g., Met-Ser of the α1 Q2S variant). Interestingly, Nα-acetylation of α1 Met appears important in gating the 20S proteasome, based on the enhanced peptidase activity of 20S proteasomes with the MAP-cleaved α1 Q2A variant and the inability of the gene encoding the α1 Q2A variant to complement the α1 mutation for growth at ‘low’ salt (~1.3 M NaCl).
Insight into archaeal enzyme(s) that mediate Nα-acetylation is provided through comparative genomics, in vitro reconstitution of Nα-acetyltransferase activity and crystallography. Members of the GNAT superfamily that use acetyl-CoAs to acylate their cognate substrates, including the Nα-acetylation of proteins, are widely distributed among Archaea (Vetting et al., 2005). S. solfataricus ssArd1 (SSO0209) is an archaeal GNAT related to Nα-acetyltransferases that has been demonstrated to acetylate the N-terminal residue (Ser) of the DNA-binding protein, Alba (Mackay et al., 2007). Much like the eukaryal Ard1 of NatA, SsArd1 preferentially acetylates N-terminal Ser and Ala residues exposed after methioinine removal (Mackay et al., 2007). SsArd1 also catalyzes appreciable Nα-acetylation of N-terminal Met-Glu and Met-Leu sequences, similar to Nat3 of NatB in eukaryotes (Mackay et al., 2007). While ssArd1 can Nα-acetylate a variety of proteins in vitro, it shows preference for proteins with N-termini that are disordered in crystal structures (Mackay et al., 2007). Thus, it is unclear whether ssArd1functions post- or co-translationally in the cell. The archaeal ssArd1 is thought to represent an ancestral form of some eukaryal Nα-acetyltransferases based on its relaxed sequence specificity. Recent crystallography of SsArd1 now provides structural detail for analysis of this ancestral function (Oke et al., 2010).
ii) Nα-acetylation and protein stability
Based on analogy to eukaryotes, Nα-acetylation is predicted to regulate protein turnover in Hfx. volcanii and other haloarchaea. The long-held argument that Nα-acetylation stabilizes a protein comes from several lines of indirect evidence (Meinnel et al., 2006). For example, proteins modified by Nα-acetylation are often over-represented in protein abundance profiles and comprise a high proportion of the proteome (Falb et al., 2006; Aivaliotis et al., 2007; Martinez et al., 2008; Kirkland et al., 2008b). Futhermore, Nα-acetylation blocks the Nα-amino group of a protein from further modification by destalizing processes such as ‘linear’ ubiquitylation (Meinnel et al., 2005). However, recent evidence suggests Nα-acetylation can mark a protein for destruction by the ubiquitin-proteasome system using a mechanism named the Ac/N-end rule (Hwang et al., 2010). The Ac/N-end rule is based on the finding that an E3 ubiquitin ligase (named Doa10) can recognize proteins with acetylated N-termini and facilitate their ubiquitylation (at internal lysine residues) and degradation by proteasomes in yeast (Hwang et al., 2010). To rationalize this discrepancy between Nα-acetylation in protein stability and degradation,Hwang et al. (2010) provide a model in which nascent proteins can ‘hide’ their acetylated N-termini by rapid folding, interaction with chaperones, and/or assembly into appropriate multi-subunit complexes. Such sequestration of N-termini would render the Nα-acetylation-based degradation signals inaccessible for recognition by the Doa10 E3 ubiquitin ligase, ultimately stabilizing the protein. In contrast, delayed or defective protein folding would expose acetylated N-termini and allow for Doa10-dependent ubiquitylation and proteolysis by proteasomes.
Insight into a potential archaeal Ac/N-end rule pathway are provided by study of the α1 protein of 20S proteasomes in Hfx. volcanii (Humbard et al., 2009; Varshavsky, 2011). Here, the identity of the N-terminal penultimate residue of α1 was found to dramatically alter the concentration of α1 protein in the cell. In particular, the levels of N-terminal α1 variants that were partially non-Nα-acetylated were remarkably higher than the levels of Nα-acetylated (wild type) α1 protein. Furthermore, most of the α1 proteins associated in 20S proteasomes had acetylated N-termini. Thus, proteasomal partners are predicted to obstruct recognition of the acetylated N-terminal domain of α1 and prevent its proteolytic destruction by an Ac/N-end rule pathway.
iii) Nε-acetylation
The reversible and differential Nε-acetylation of proteins can have a major impact on transcription, translation, stress response, detoxification, and carbohydrate and energy metabolism (Hu et al., 2010; Thao and Escalante-Semerena, 2011; Jones and O'Connor, 2011). In eukaryotes, histones are well-known to be modified by Nε-acetylation, in addition to Nα-acetylation, methylation, phosphorylation, ubiquitylation, ADP ribosylation, glycosylation and sumoylation (Shiio and Eisenman, 2003). Numerous bacterial proteins are also differentially Nε-acetylated, with ‘K-acetylomes’ (sub-proteomes composed of proteins with Nε-acetylated lysine residues) thought to rival phospho-proteomes (Aka et al., 2011). In contrast, Nε-acetylation of archaeal proteins is poorly understood.
In Archaea, only a few proteins are known to be Nε-acetylated. A couple of early studies, focused on determining the amino acid sequence of 2Fe-2S ferredoxins from the haloarchaea Hbt. salinarum and Har. marismortui, revealed that a lysine residue near the C-terminus of these proteins is conserved and Nε-acetylated (Hase et al., 1978; Hase et al., 1980). Another archaeal protein that is Nε-acetylated is Alba, a chromatin protein of S. solfataricus. Alba is not only Nα-acetylated on its N-terminal Ser (as discussed above) but is also Nε-acetylated on lysine 16 (Bell et al., 2002). Acetylation of Alba Lys16 is mediated by the protein acetyltransferase, Pat (Marsh et al., 2005). Pat is a homolog of Salomonella Pat (YfiQ) and a member of the family of NDP-forming acetyl-CoA synthetase enzymes with GNAT domains (Starai and Escalante-Semerena, 2004). Alba Lys16 can be deactylated by an NAD+-dependent histone deacetylase class III homolog of S. solfataricus (Sir2) (Bell et al., 2002). Nε-acetylation of Alba reduces its binding affinity for DNA and RNA and strongly prevents its ability to inhibit the DNA helicase activity of the mini-chromosome maintenance (MCM) protein (Bell et al., 2002; Jelinska et al., 2005; Marsh et al., 2006). Thus, Nε-acetylation of Alba is thought to have a global impact on chromatin packaging and gene expression in Crenarchaeota, such as S. solfataricus (Wardleworth et al., 2002; Zhao et al., 2003).
Hfx. volcanii has served as a model for understanding the importance of histone acetyltransferase (HAT) and histone deacetylase (HDAC) gene homologs to archaeal cell function. In particular, gene homologs for three HATs (Pat1, HVO_1756; Pat2, HVO_1821; Elp3, HVO_2888) and two HDACs (Sir2, HVO_2194; HdaI, HVO_0522) were targeted for deletion from the Hfx. volcanii genome (Altman-Price and Mevarech, 2009). Pat1 and Pat2 are related to the S. solfataricus Pat, Elp3 is related to the yeast Elp3 subunit of the elongator complex possessing acetyltransferase activity, Sir2 is a class III HDAC homolog similar to S. solfataricus Sir2, and HdaI is a class II HDAC homolog related to yeast Hda1. Single deletion of sir2, pat1, pat2 or elp3 genes or double deletion of pat1 with either pat2 or elp3 was found to have no detectable impact on the viability of Hfx. volcanii cells. In contrast, hdaI appeared essential, based on the finding that the gene could only be deleted when a wild type copy of hdaI was provided in trans. Attempts to create an elp3 deletion in any of the pat2 null strains were unsuccessful, implying that these two mutations are synthetically lethal and affect a single function or pathway (Altman-Price and Mevarech, 2009). Thus, Elp3- and Pat2-mediated acetylation and HdaI-mediated deacetylation of lysine residues are predicted to be important in Hfx. volcanii.
Hypusine modification
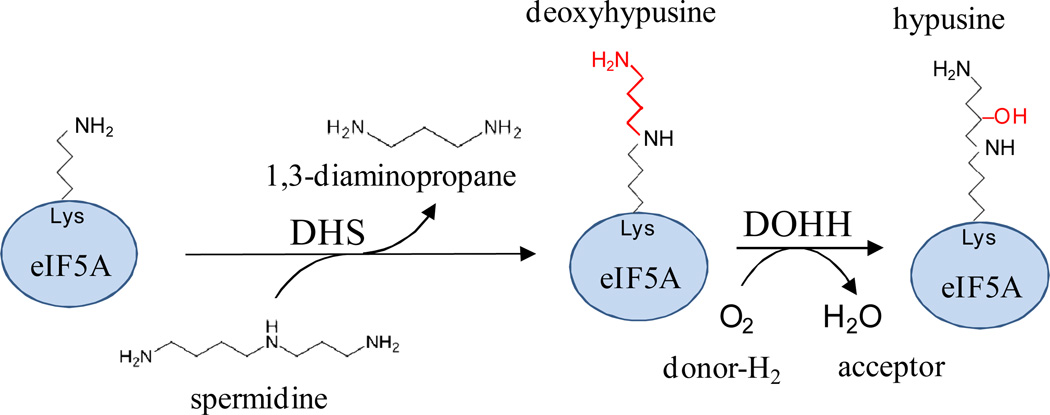
Hypusine [Nε-(4-amino-2-hydroxybutyl)-L-lysine] is formed upon post-translational modification of a conserved lysine residue and is found only in eukaryotic translation ‘initiation’ factor 5A (eIF5A) and the related archaeal aIF5A (Park et al., 1981; Schumann and Klink, 1989; Bartig et al., 1992; Park et al., 2010). Hypusine is essential for activity of eIF5A/aIF5A (including that of Hbt. salinarum), now considered important in translation elongation (Wagner and Klug, 2007; Saini et al., 2009; Park et al., 2010). In hypusine modification of eIF5A, deoxyhypusine synthase (DHS) transfers a 4-aminobutyl moiety from spermidine to the ε-amino group of the conserved lysine residue to form a dexoxyhypusine intermediate, which is hydroxylated to a hypusine residue by deoxyhypusine hydroxylase (DOHH) (Fig. 6). In Archaea, DHS homologs are widespread, suggesting the lysine residue of aIF5A is modified to a deoxyhypusine residue by an enzyme similar to eukaryotic DHS. In contrast, DOHH is an oxygen-dependent enzyme and its homologs are rare in (the often anaerobic) Archaea. Thus, generation of the hypusine-modified form of aIF5A from its deoxyhypusine precursor likely involves a second enzyme distinct from DOHH in Archaea.
Fig 6. Hypusine modification of eIF5A.
Hypusine is a universal modification in archaea and eukaryotes on a single type of protein (eIF5A) by a sequential series of enzyme reactions. Deoxyhypusine synthase (DHS) transfers a 4-aminobutyl moiety from spermidine to the ε-amino group of a specific lysine residue on eIF5A. Deoxyhypusine hydroxylase (DOHH) hydroxylates the modified lysine to form hypusine.
Protein methylation
Protein methylation is a type of post-translational modification that can mediate many important biological processes, spanning from gene regulation and signal transduction to protein stability (Clarke, 1993; Lee et al., 2005; Paik et al., 2007). In this PTM, nucleophilic oxygen, nitrogen and sulfur atoms on the polypeptide chain can be methylated, resulting in the generation of methyl esters, methyl amines, methyl amides and other modifications (with only the hydroxyl groups of Ser, Thr and Tyr not detected in methylated forms). Methylation of the side-chain hydroxyl group of dicarboxylic amino acid residues (Asp, Glu) and the hydroxyl group generated by deamidation of glutamine residues (O-methylesterification) is typically reversible and regulated. Recent evidence also supports the reversible methylation of the amino groups in the side chains of lysine (mono-, di- and tri-methylated forms) and arginine (mono- and di-methylated forms) residues of proteins (Lee et al., 2005). S-adenosylmethionine (SAM) commonly serves as the methyl group donor for the methylation of proteins by methyltransferases that are often specific for their protein substrate, while methylesterases and demethylases can remove the methyl groups. In haloarchaea, O-methylesterification of proteins has been detected and is the focus of the discussion below.
O-methylesterification
The methyl-accepting taxis proteins or transducers are a large group of membrane-associated proteins that are typically methylated at glutamate residues (some of which originate from glutamine residues via deamidation) (Porter et al., 2011). Transducer homologs are relatively widespread among Bacteria and euryarchaeota (including halophiles, methanogens, thermococci and others). In general, transducers act as (or interact with) sensory receptors for a specific stimulus (e.g., light, attractant) and are demethylated in response. The methylation state of the transducer provides a ‘primitive memory’ or adaptation to a two-component regulatory system (CheA/CheY) that switches the flagellar motor and alters cellular movement. Among the archaeal transducers, the best studied are the halobacterial transducers (Htrs) of Hbt. salinarum, which comprise a group of 18 homologs (6 soluble and 12 membrane-spanning) with 1 to 3 conserved methylation sites per protein (Ng et al., 2000; Pfeiffer et al., 2008) (Fig. 3). Early studies of Hbt. salinarum demonstrate methyl-[3H]-labeling of proteins and volatile products of demethylation that are specific to taxis and define the methylated proteins as Htrs (soluble and membrane-spanning), based on the abolishment of methyl-[3H]-labelling after gene deletion and site-directed mutagenesis (Alam et al., 1989; Sundberg et al., 1990; Nordmann et al., 1994; Lindbeck et al., 1995; Brooun et al., 1997; Perazzona and Spudich, 1999; Storch et al., 1999). Recently, MS-based proteomes has been used to map three methylation sites in two of the six soluble Htrs and 19 methylation sites in 10 of the 12 predicted membrane-spanning Htrs (Koch et al., 2008). Sites include singly methylated pairs of Glu and/or Gln, Asp-Glu and Ala-Glu residues (with Glu or deamidated Gln residues methylated). Evidence supports the function of CheB in deamidation and demethylation of the Htrs and CheR in transfer of the methyl group from SAM co-factor to conserved Glu or deamidated Gln residues of Htr (Perazzona and Spudich, 1999; Koch and Oesterhelt, 2005; Koch et al., 2008). Selective demethylation of Htr sites (e.g., one site is demethylated upon stimulation by attractant and another site is demethylated upon simulation with repellent) is proposed to control a diverse array of biological responses, in analogy to the bacterium Bacillus subtilus (Streif et al., 2010).
Like transducers, archaeal 20S proteasomes are modified by O-methylesterification. In particular, five unique acidic residues (Asp20, Glu27, Glu62, Glu112, and Glu161) of the Hfx. volcanii 20S proteasome α1 subunit are methylated (Table 2), as detected by MS/MS with methods devoid of added methanol (Chen et al., 2010; Humbard et al., 2010b). O-linked methylesterification is typically a reversible form of post-translational modification. Furthermore, both methylated and unmethylated forms of α1 are readily detected in the cell (Humbard et al., 2010b). Thus, O-methylesterification of α1 is thought to be regulated and to modulate proteasomal activity (e.g., stability in low salt, high temperature, interaction with AAA+ ATPase partners). While the enzyme responsible for α1 methylation is as yet unknown, a SAM-dependent methyltransferase homolog (HVO_1093) co-transcribed with the α1 gene is a likely candidate (Gil et al., 2007).
N-teminal methionine removal
Methioinine residues can removed from the N-terminus of proteins in all domains of life. Archaea and Eukarya use methionine (Met) for initiation of protein synthesis (Ramesh and RajBhandary, 2001), and this N-terminal methionine can be removed by a methionine aminopeptidase as the protein emerges from the ribosome (Lowther and Matthews, 2000). In contrast to Met, Bacteria and the related organelles of eukaryotes (i.e., mitochondria and chloroplasts) use N-formylmethionine (fMet) for initiation of protein synthesis. fMet is a derivative of Met in which a formyl group has been added to the amino group and is important for bacterial cell function (Guillon et al., 1992). With rare exceptions (Milligan and Koshland, Jr., 1990), the N-formyl group is typically removed from bacterial proteins by a peptide deformylase (Mazel et al., 1994). After deformylation, the resulting N-terminal methionine can be removed (Solbiati et al., 1999).
Methionine aminopeptidases (MAPs; EC 3.4.11.18) are metalloenzymes common to all domains of life that remove N-terminal leading methionine residues from nascent peptides during the early stages of protein synthesis. MAPs generally cleave substrates with penultimate residues that are one of the seven small and uncharged amino acids (i.e., Gly, Ala, Ser, Thr, Pro, Cys and Val). MAPs belong to the MEROPS peptidase family, M24 (clan MG), and are divided into two types (types I and II), with the C-terminal domain of type II distinguished by insertion of an extra ~ 60 amino acid helical sub-domain (Arfin et al., 1995). Bacteria typically only contain the type I MAP (with some exceptions), Archaea contain only type II MAP, while eukaryotes possess both type I and type II MAPs (Bazan et al., 1994; Arfin et al., 1995; Lowther and Matthews, 2002).
Amongst the Archaea, MAPs from the hyperthermophilic P. furiosus (PfMAP) and Thermococcus sp. NA1 have been purified in enzymatically active forms (Tsunasawa et al., 1997; Tahirov et al., 1998; Lee et al., 2006). Of these two enzymes, PfMAP is the most extensively studied. Similar to the bacterial and eukaryal MAPs, PfMAP cleaves N-terminal methionine from substrates whose penultimate amino acid residues are Gly, Ala, Ser, Thr, Pro and Val, based on release of N-terminal methionine from the synthetic peptide, Met-X-Ala-Ala-Ala (where X represents the 19 common amino acids, excluding Cys) (Tsunasawa et al., 1997). PfMAP also provided the first crystal structure of a type II MAP (Tahirov et al., 1998). Those PfMAP residues (Asp82, Asp94, His153, Glu187, Glu280) that coordinate the two catalytic metal ions are conserved in the Escherichia coli type I MAP crystal structure (with the identity of the metal ions used for catalysis still under debate) (Roderick and Matthews, 1993). Likewise, the two positively charged residues (His62 and His161) in the active site cleft of PfMAP that are thought to shuffle protons from the nuclear center to the solvent during catalysis are also conserved in E. coli MAP (Roderick and Matthews, 1993; Tahirov et al., 1998).
While MAPs have not been purified from halophilic archaea, type II MAP homologs are conserved and large-scale proteomic studies provide evidence for MAP activity in this group of microorganisms. In particular, non-redundant N-terminal peptides identified by MS have been reliably mapped to large number of proteins from Hbt. salinarum (606 proteins), Natronomonas pharaonis (328 proteins) and Hfx. volcanii (236 proteins) (Falb et al., 2006; Aivaliotis et al., 2007; Kirkland et al., 2008b). Of these proteins, most (60–70%) were missing an N-terminal methionine and presented N-termini suggesting that MAP cleavage occurred predominantly when the penultimate residue was small and uncharged (Gly, Ala, Pro, Val, Ser or Thr) (for Hfx. volcanii, see Fig. 5). Interestingly, recent comparison of large datasets of non-redundant N-terminal peptides detected by MS from diverse organisms suggests that MAP cleavage efficiency is conserved across all domains of life (Helbig et al., 2010).
Signal peptide cleavage
Proteins destined to reside beyond the confines of the cytoplasm (i.e. secretory and membrane proteins) are often synthesized as preproteins bearing cleavable N-terminal targeting sequences termed signal peptides. In Hfx. volcanii, signal peptides direct preproteins to both the Sec and the Tat translocation systems (Fig. 7) (Pohlschroder et al., 2005; Calo and Eichler, 2011). Although the major protein species of Hfx. volcanii, the S-layer glycoprotein, relies on the Sec translocation system, bioinformatics-based analysis predicts that the Tat translocation system is preferentially used by exported Hfx. volcanii proteins (Dilks et al., 2003; Storf et al., 2010). In each system, once the signal peptide has served its purpose, namely targeting a preprotein to a proteinaceous translocation complex, it is cleaved from the mature protein by the actions of type I signal peptidase (SPase), although, as considered below, the majority of Tat pathway lipoprotein substrates are apparently processed by a type II SPase. While general aspects of archaeal type I SPase biology have been considered in several recent reviews (Ng et al., 2007; Jarrell et al., 2010; Calo and Eichler, 2011), this enyzme has only been biochemically addressed in a limited number of Archaea to date, including Hfx. volcanii (Fine et al., 2006; Fink-Lavi and Eichler, 2008). In Hfx. volcanii, genes encoding two distinct type I SPases, Sec11a (HVO_2603) and Sec11b (HVO_0002) are found (Fine et al., 2006). Of the two, HVO_0002 is an essential gene. Observed differences in the activities of the two purified enzymes towards a signal peptide-bearing reporter protein suggest that Sec11a and Sec11b possess distinct substrate preferences.
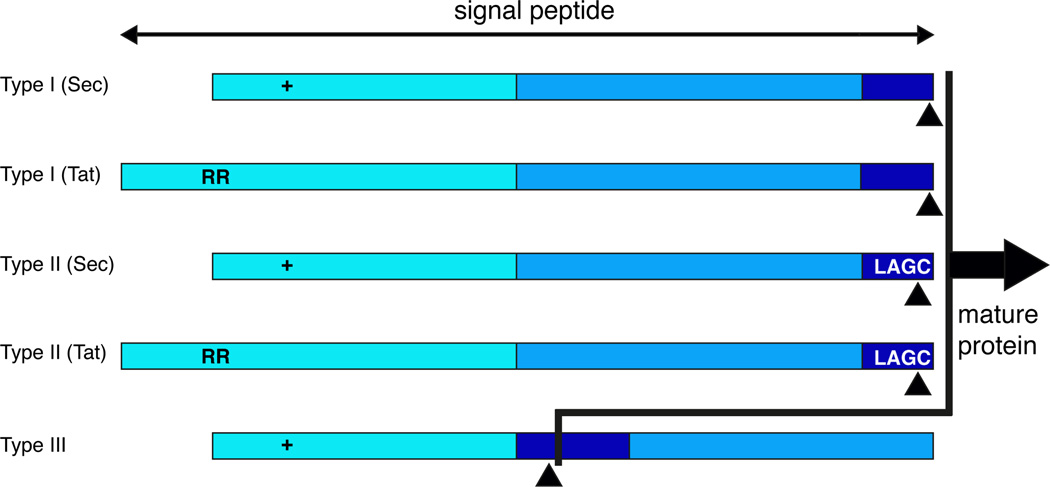
Fig 7. Schematic depiction of haloarchaeal signal peptides and their processing.
Type I and type II signal peptides that target proteins to the Sec or Tat translocation pathways, as well as type III signal peptides, have been reported in haloarchaea. In each signal peptide, the light blue N-terminal region contains positive charges (+) in the case of Sec pathway substrates or the twin arginine residues (RR) characteristic of Tat pathway substrates. Type I signal peptidases cleave the signal peptide after a C-terminal region (dark blue) often ending in alanine(s). Type II signal peptidases cleave the signal peptide upstream of the lipobox cysteine, found in the LAGC consensus sequence. The exposed cysteine that becomes the N-terminal residue of the mature protein may become lipid-modified. Type III signal peptidases act on a C-terminal domain upstream of a hydrophobic stretch (sky blue). The cleavage sites processed by the various signal peptidases are indicated by black triangles. For further details, see Pohlschroder et al. (2005).
At the molecular level, Hfx. volcanii type I SPases present a mosaic of eukaryal, bacterial and archaeal properties (Fine et al., 2006; Calo and Eichler, 2011). Like their eukaryal counterparts, the Hfx. volcanii enzymes have replaced the conserved lysine of the serine-lysine catalytic dyad found in bacterial SPases with a histidine. Site-directed mutagenesis studies revealed that both of these residues are essential for Sec11b activity (Fink-Lavi and Eichler, 2008). Likewise, a third Hfx. volcanii Sec11b residue, Asp-280, was deemed as being essential for catalytic activity of the enzyme. The equivalent residues were also shown to be necessary for Methanococcus voltae SPase activity (Bardy et al., 2005). As the yeast type I SPase also requires these residues for its functionality, it would appear that the archaeal enzyme relies on a catalytic mechanism like that employed by its eukaryal counterpart. On the other hand, yeast SPase also requires the presence of the equivalent of Hfx. volcanii Sec11b Asp-273 (van Valkenburgh et al., 1999), a residue not essential for the activity of the haloarchaeal enzyme (Fink-Lavi and Eichler, 2008). At the same time, the Hfx. volcanii enzymes also share traits with their bacterial counterparts. Tagged versions of Sec11a and Sec11b were purified as single polypeptides, each able to cleave the signal peptide from a reporter protein, suggesting that the Hfx. volcanii enzymes function independently of other proteins, as do bacterial SPases. Eukaryal type I SPases, by contrast, exist as part of a multimeric complex in which additional proteins apart from Sec11 are essential for activity (Evans et al., 1986; YaDeau et al., 1991). Still, the possibility remains that the Hfx. volcanii SPases operate optimally only when in complex with additional components not captured under the conditions employed nor identified in genome searches.
As discussed in more detail below, Hfx. volcanii is thought to contain a large number of lipoproteins. While type II SPases remove characteristic lipoprotein signal sequences, thereby unmasking the site of such lipid-based post-translational modification in target proteins, no such enzyme has been detected in Hfx. volcanii or any other Archaea. It remains unclear whether this reflects the absence of such an archaeal enzyme or whether the archaeal enzyme, possibly designed to act on the ether-based lipids of the archaeal membrane, shares little sequence homology with its bacterial counterpart and, as such, has been overlooked.
Unlike type I SPases that cleave Sec or Tat translocation signal peptides, type III SPases process the unique signal peptides found in archaellins, pili and other cell-surface structures (Albers et al., 2003). Analysis of the Hfx. volcanii genome reveals the presence of several preproteins bearing a type III signal peptide, as well as a single type III signal peptidase (Szabo et al., 2007), a homologue of bacterial PilD, responsible for the processing of bacterial type IV prepilin proteins (Strom et al., 1993). Indeed, Hfx. volcanii cells deleted of pibD (HVO_2993), encoding the pre-pilin peptidase, fail to properly process proteins bearing this class of signal peptide (Tripepi et al., 2010).
Lipid modification
Lipid modification of a protein involves the permanent or temporary covalent attachment of lipid-based groups at various positions within the polypeptide chain and serves a variety of roles, with the most obvious being to enhance the membrane affinity of the modified protein. However, numerous other roles have been assigned to such post-translational modification, including modulation of protein-protein interactions, signal transduction, embryogenesis and pattern formation, protein trafficking through the secretory pathway, evasion of the immune response by infectious parasites and enzyme activation (for review, see Sinensky, 2000; Mann and Beachy, 2004; Pechlivanis and Kuhlmann, 2006; Nadolski and Linder, 2007; Charollais and Van Der Goot, 2009; Greaves, 2009; Aicart-Ramos et al., 2011). In efforts aimed at understanding the modes, pathways and functions of lipid modification in Archaea, studies on Hfx. volcanii have proven to be central.
In addressing the biogenesis of the Hfx. volcanii S-layer glycoprotein, it was shown that the protein is first synthesized as an immature precursor, possessing a lower apparent molecular weight and a less hydrophobic character than the final version of the protein (Eichler, 2001). As the protein can be labeled with [3H] mevalonic acid, an isoprene precursor, the post-translational magnesium-dependent conversion to the mature form apparently involves isoprenylation (Konrad and Eichler, 2002). Moreover, it was shown that such lipid modification of the Hfx. volcanii S-layer glycoprotein only occurs once the protein has traversed the plasma membrane (Eichler, 2001). Similarly, the Hbt. salinarum S-layer glycoprotein also undergoes such maturation and was shown to be modified by a covalently linked diphytanylglyceryl phosphate entity near an O-glycosylated Thr-rich stretch found in the C-terminal region of the protein, upstream of the predicted single transmembrane domain (Kikuchi et al., 1999; Konrad and Eichler, 2002). At the same time, several Hbt. salinarum proteins are modified by a diphytanylglycerol methyl moiety, linked to Cys residues of the protein through a thioetheric bond (Sagami et al., 1994; Sagami et al., 1995).
Since the Hfx. volcanii S-layer glycoprotein includes a predicted membrane-spanning domain (as does its Hbt. salinarum counterpart) (Lechner and Sumper, 1987; Sumper et al., 1990), it is unclear why an additional membrane anchor in the form of a lipid would be required. The recent identification of genes encoding archaeosortases may explain this apparent paradox (Haft et al., 2012). Archaeosortases are predicted by bioinformatics to correspond to the archaeal homologues of bacterial exosortases, enzymes involved in C-terminal anchoring of proteins to membrane-embedded lipids. The archaeosortase, ArtA, is predicted to cleave proteins bearing a C-terminal membrane-spanning domain between this region and the Pro-Gly-Phe motif found immediately upstream. The processed protein is then delivered to the waiting lipid on the external surface of the cell. Accordingly, the Hfx. volcanii S-layer glycoprotein (and its Hbt. salinarum counterpart; Lechner and Sumper, 1987) present a Pro-Gly-Phe motif found immediately upstream of the predicted transmembrane domain (Sumper et al., 1990). Indeed, the coordinated transfer of the Hfx. volcanii S-layer glycoprotein to a lipid moiety following cleavage from the C-terminal membrane-spanning domain could explain several apparent paradoxes, such as the ability of EDTA to solubilize the S-layer glycoprotein, an apparent integral membrane protein, without destroying membrane integrity (Charlebois et al., 1987). In this scenario, EDTA treatment would somehow interfere with the stabilization of newly lipid-modified S-layer glycoprotein in the membrane or in the S-layer, leading to the release of the protein from the cell while leaving the plasma membrane intact.
In addition to such C-terminal post-translational lipid modification, N-terminal lipid modification also occurs, namely in processing of lipoproteins. Based on the presence the so-called ‘lipobox’ sequence motif (composed of the consensus Leu-Ala-Gly-Cys sequence) that includes the cysteine residue that undergoes lipid modification (Hayashi and Wu, 1990) near the start of predicted amino acid sequence, halocyanin, a small blue copper protein of the haloalkaliphile Nmn. pharaonis is thought to undergo such amino-terminal lipid modification (Mattar et al., 1994). Although direct proof for such modification was not provided, analysis of halocyanin by mass spectroscopy was consistent with modification of the N-terminal Cys residue by two C20 phytanyl (glycerol)diether lipids linked via a thioether bond, as well as by acetylation. Indeed, analysis of haloarchaeal genomes predicts numerous lipobox-containing proteins processed by the Tat protein translocation system. In Hfx. volcanii, 72 lipobox-containing proteins are predicted (Gimenez et al., 2007). In the specific cases of the Hfx. volcanii iron-binding protein (Ibp), DsbA-like thioredoxin domain protein (DsbA) and maltose-binding protein (Mbp), replacement of the lipobox cysteine by serine in the precursor forms of these predicted lipoproteins led to defective localization. Mutant Ibp and DsbA were found in the growth medium instead of being largely membrane-associated, as are the wild type proteins; mutant Mpb is apparently unstable and degraded (Gimenez et al., 2007). On the other hand, the predicted lipoproteins, HVO_B0139 and HVO_1242, remained cell-associated in the lipobox cysteine-alanine mutated forms (Storf et al., 2010).
The lipid-based modifications considered above essentially serve to anchor the modified protein to the membrane. The retinal proteins of haloarchaea (Oesterhelt and Stoeckenius, 1971) provide another use for covalently linked lipids. Bacteriorhodopsin, halorhodopsin and the two sensory rhodopsins (and related rhodopsins subsequently reported in bacteria (Beja et al., 2000) and eukaryotes (Bieszke et al., 1999), comprise a family of small, seven trans-membrane domain-containing proteins involved in coupling light energy to the vectorial transport of ions across a membrane. To achieve this feat, these retinal proteins contain covalently linked retinal, an isoprene-derived light-sensitive co-factor. Accordingly, bacteriorhodopsin, the prototype rhodopsin, has served as a useful reporter of the assembly of lipid modification of proteins. For more information on this and related topics, the reader is directed to reviews by Lanyi (2004) and Grote and O’Malley (2011).
Sampylation
SAMPs are small archaeal ubiquitin-like modifier proteins that form isopeptide bonds to lysine residues of archaeal proteins (Fig. 8). While this system was first demonstrated in Hfx. volcanii (Humbard et al., 2010a), it is predicted to exist in all Archaea and appears related to other recently discovered protein conjugation systems, including the TtuBC system of protein conjugation in the hyperthermophilic bacterium, Thermus thermophilus (Humbard et al., 2010a; Shigi, 2012) and the Urm1 system of protein conjugation and sulfur transfer in eukaryotes (Wang et al., 2011). Sampylation is thought to be reversible, based on conservation of JAMM (JAB1/MPN/Mov34 metalloenzyme) domain homologs in all Archaea (Humbard et al., 2010a; Maupin-Furlow, 2012). In eukaryotes, JAMM domain proteins include the Rpn11/Poh1 subunit of 26S proteasomes, required for removal of ubiquitin chains from proteins (Verma et al., 2002; Yao and Cohen, 2002). Likewise, Csn5/Jab1 is a JAMM domain protein subunit of the COP9 signalosome complex that cleaves the ubiquitin-like Nedd8 from proteins (Cope et al., 2002). While archaeal JAMM domain protein function has yet to be demonstrated, the solved crystal structure of an A. fulgidus JAMM domain protein (termed AfJAMM) was used to model the structures of Rpn11/Poh1 and Csn5/Jab1 (Tran et al., 2003; Ambroggio et al., 2004).
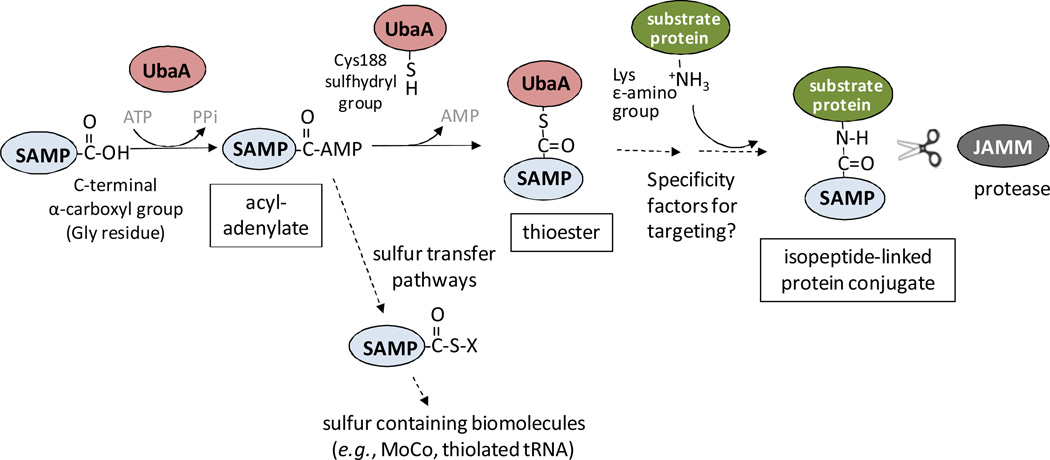
Fig 8. Sampylation in Hfx. Volcanii.
The Hfx. volcanii ubiquitin-like SAMP1/2 and UbaA (a ubiquitin-activating E1 enzyme homolog) function in protein conjugation (sampylation) and sulfur transfer (biosynthesis of MoCo and thiolated tRNA). In these pathways, UbaA is thought to adenylate the C-terminal glycine of the SAMP1/2. In protein conjugation, a thioester intermediate is thought to form between a conserved active site cysteine of UbaA (C188) and the C-terminal carboxyl group of the SAMP. SAMP1/2 are then transferred to lysine residues on protein targets to form an isopeptide bond. Archaeal proteins of the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain superfamily are proposed to cleave isopeptide bonds and, thus, render the pathway reversible.
Much like the ubiquitin and ubiquitin-like protein modifiers of eukaryotes, SAMPs have a β-grasp fold structure and C-terminal diglycine motif that is important to their function in protein conjugation (Ranjan et al., 2010; Humbard et al., 2010a; Jeong et al., 2011). The α-carboxyl of the C-terminal glycine of SAMPs can form an isopeptide bond to the ε-amino group of lysine residues of target proteins by a mechanism that requires UbaA, an ubiquitin-activating E1 enzyme homolog of Archaea (Humbard et al., 2010a; Miranda et al., 2011).
The physiological reason for the formation of these ubiquitin-like protein conjugates in Archaea (and hyperthermophilic bacteria) remains to be determined. However, in analogy to ubiquitin, sampylation is thought to alter the structure, enzymatic activity and types of partners that would associate with the modified protein, including proteasomes, transcription factors and others (Maupin-Furlow, 2012). Interestingly, SAMPs not only form isopeptide bonds with protein targets but also are required for sulfur transfer to biomolecules, including molybdenum cofactor (MoCo) and tRNA (similar to Urm1 of eukaryotes and TtuB of the bacterium T. thermophilus) (Miranda et al., 2011; Shigi, 2012). Sampylation differs from the system of protein conjugation recently predicted for the archaeon Candidatus Caldiarchaeum subterraneum (based on metagenomics) (Nunoura et al., 2011), and some bacteria (based on analogy) (Burroughs et al., 2011). This latter system is rare in Archaea (restricted to Candidatus Caldiarchaeum subterraneum) and appears to incorporate not only ubiquitin-activating E1 homologs but also ubiquitin-conjugating E2 and ubiquitin ligase E3 homologs (Nunoura et al., 2011).
Protein degradation
Proteases are important in maintaining cellular homeostasis, regulating cellular signaling and degrading exogenous proteins for cellular metabolism. In these processes, proteases are needed to catalyze the general turnover of proteins, remove aberrant proteins (e.g., damaged or viral-encoded proteins) and mediate the precisely-timed cleavage or turnover of regulatory proteins important to cell functions ranging from transcription and cell division to metabolism (Gottesman, 2003). Similarly to other Archaea, Hfx. volcanii is predicted to encode a wide variety of endo- and exo-proteases, including energy-dependent and intramembrane-cleaving proteases, typically linked to the early events of protein degradation and regulation (Maupin-Furlow et al., 2005; De Castro et al., 2006). Of the energy-dependent proteases, proteasomes and membrane-associated Lon B-type proteases appear relatively universal in the Archaea (including Hfx. volcanii) (Cha et al., 2010; Maupin-Furlow, 2012).
Among the various regulatory proteases (i.e., excluding signal peptidases) predicted for Hfx. volcanii, only the proteasomes have been studied at the biochemical level (Maupin-Furlow, 2012). 20S proteasomes are purified from Hfx. volcanii in at least three distinct sub-types, composed of four stacked heptameric rings of different subunit composition (i.e., α17β7β7α17, α17β7β7α27 and α27β7β7α27, where subscript represents subunit stoichiometry) (Wilson et al., 1999; Kaczowka and Maupin-Furlow, 2003; Karadzic et al., 2012). The 20S proteasomes are catalytically active in cleaving peptides and small denatured proteins (i.e., oxidized bovine insulin B-chain protein) (Wilson et al., 1999; Kaczowka and Maupin-Furlow, 2003; Karadzic et al., 2012). Hfx. volcanii PANs are purified as PAN-A and PAN-B sub-classes but were not found to be active in stimulating the energy-dependent degradation of proteins by 20S proteasomes (based on in vitro assay) (Reuter et al., 2004; Humbard et al., 2010b). The levels of the α2 subunit of 20S proteasomes and PAN-B are correlated with growth phase (i.e., low levels detected in lag to log phase and high levels detected in stationary phase) suggesting that these two proteins are associated in a biologically-related pathway (Reuter et al., 2004).
Proteasomes appear important in cell division, protein quality control and other functions in Hfx. volcanii. Like eukaryotes, 20S proteasomes of Hfx. volcanii are essential, based on the conditional lethal phenotypes associated with deletion of the β gene or double deletion of the α1 and α2 genes (Zhou et al., 2008). The PANs are not essential (panA panB double mutants are viable) (Zhou et al., 2008), while Cdc48d (one of four Cdc48 homologs in Hfx. volcanii) appears essential, based on the inability to delete this gene from the chromosome (Allers et al., 2010). PAN, Cdc48/p97/VAT, and other members of the AAA ATPase superfamily are thought to form networks of ATPases that associate with and regulate the function of 20S proteasomes in archaeal cells (Maupin-Furlow, 2012; Barthelme and Sauer, 2012; Forouzan et al., 2012). Thus, Cdc48d and 20S proteasomes may regulate functions of cell division. Consistent with a role in protein quality control, proteasomes (PAN-A and α1) are needed for Hfx. volcanii to tolerate stressful conditions, including exposure to L-canavanine (an amino acid analogue that causes protein unfolding) and low salt (Zhou et al., 2008). In addition, α1 is needed to overcome thermal stress (Zhou et al., 2008). Whether sampylation is linked to proteasome-mediated proteolysis, in analogy to the ubiquitin-proteasome system of eukaryotic cells, remains to be determined. Levels of SAMP1-protein conjugates are, however, higher in proteasome mutants, as compared to wild type cells, providing indirect evidence for a connection between these two systems (Humbard et al., 2010a). Still, ubaA (encoding the single ubiquitin-activating E1 homolog) can be deleted without loss of cell viability, suggesting that sampylation is not required for cell division in Hfx. volcanii (Miranda et al., 2011).
Conclusions
Across evolution, proteins can experience any of a long list of PTMs that lead to numerous and varied effects on protein structure, function, localization, oligomerization status and stability. As is the case for much of archaeal biology, the various PTMs performed by Archaea both present traits unique to this life form and others shared by eukaryotes and/or Bacteria. As presented in Table 1, post-translationally-modified proteins abound in Hfx. volcanii, justifying the use of this species for the study of the responsible processes in Archaea. With an ever-growing list of tools available for the genetic, proteomic, biochemical and physiological examination of Hfx. volcanii, it is likely that future studies on this organism, and indeed, on other haloarchaea, will continue to provide novel insight into PTMs in general, and in Archaea, in particular.
Table 1.
List of some PTMs reported in Hfx. volcanii
| PTM | Experimentally-verified target(s) |
|---|---|
| N-glycosylation | S-layer glycoprotein (HVO_2072), FlgA1 (HVO_1210) |
| O-glycosylation | S-layer glycoprotein (HVO_2072) |
| X-glycosylationa | 150, 105, 98, 58, 56, 54 and 52 kDa proteins |
| Lipid modification | S-layer glycoprotein (HVO_2072) |
| Signal peptide cleavage | S-layer glycoprotein (HVO_2072), FlgA1 (HVO_1210), FlgA2 (HVO_1211) |
| Sampylation | SAMP2 (HVO_0202), UbaA, homologs of ribose-1,5-bisphosphate isomerase (HVO_0966), DNA gyrase B subunit (HVO_1572), cysteine hydrolyase (HVO_2328), NADH-quinone oxidoreductase chain c/d (HVO_0980), OsmC (HVO_1289), TBPe (HVO_1727), and tandem rhodanese domain protein (HVO_0025) |
X-glycosylation, form of glycosylation undetermined.
Acknowledgements
J.E. is supported by grants from the Israel Science Foundation (8/11) and the US Army Research Office (W911NF-11-1-520). J. M.-F. is supported by grants from NIH (R01 GM057498) and DOE (DE-FG02-05ER15650).
References
- Abe Y, Matsumoto S, Wei S, et al. Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J Biol Chem. 2001;276:44003–44011. doi: 10.1074/jbc.M105669200. [DOI] [PubMed] [Google Scholar]
- Abu-Qarn M, Eichler J. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol Microbiol. 2006;61:511–525. doi: 10.1111/j.1365-2958.2006.05252.x. [DOI] [PubMed] [Google Scholar]
- Abu-Qarn M, Yurist-Doutsch S, Giordano A, Trauner A, Morris HR, Hitchen P, Medalia O, Dell A, Eichler J. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J Mol Biol. 2007;14:1224–1236. doi: 10.1016/j.jmb.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Abu-Qarn M, Giordano A, Battaglia F, Trauner A, Morris HR, Hitchen P, Dell A, Eichler J. Identification of AglE, a second glycosyltransferase involved in N-glycosylation of the Haloferax volcanii S-layer glycoprotein. J Bacteriol. 2008;190:3140–3146. doi: 10.1128/JB.00056-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Qarn M, Eichler J, Sharon N. Not just for Eukarya anymore: N-glycosylation in Bacteria and Archaea. Curr Opin Struct Biol. 2008;18:544–550. doi: 10.1016/j.sbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Aicart-Ramos C, Valero RA, Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta. 2011;1808:2981–2994. doi: 10.1016/j.bbamem.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Aivaliotis M, Gevaert K, Falb M, et al. Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J Proteome Res. 2007;6:2195–2204. doi: 10.1021/pr0700347. [DOI] [PubMed] [Google Scholar]
- Aivaliotis M, Macek B, Gnad F, Reichelt P, Mann M, Oesterhelt D. Ser/Thr/Tyr protein phosphorylation in the archaeon Halobacterium salinarum --a representative of the third domain of life. PLoS One. 2009;4:e4777. doi: 10.1371/journal.pone.0004777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aka JA, Kim GW, Yang XJ. K-acetylation and its enzymes: overview and new developments. Handb Exp Pharmacol. 2011;206:1–12. doi: 10.1007/978-3-642-21631-2_1. [DOI] [PubMed] [Google Scholar]
- Alam M, Lebert M, Oesterhelt D, Hazelbauer GL. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989;8:631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers SV, Szabo Z, Driessen AJM. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J Bacteriol. 2003;185:3918–3925. doi: 10.1128/JB.185.13.3918-3925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Barak S, Liddell S, Wardell K, Mevarech M. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl Environ Microbiol. 2010;76:1759–1769. doi: 10.1128/AEM.02670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman-Price N, Mevarech M. Genetic evidence for the importance of protein acetylation and protein deacetylation in the halophilic archaeon Haloferax volcanii. J Bacteriol. 2009;191:1610–1617. doi: 10.1128/JB.01252-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfin SM, Kendall RL, Hall L, Weaver LH, Stewart AE, Matthews BW, Bradshaw RA. Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proc Natl Acad Sci USA. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabote RD, Saier MH Jr. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol Mol Biol Rev. 2005;69:608–634. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy SL, Ng SY, Carnegie DS, Jarrell KF. Site-directed mutagenesis analysis of amino acids critical for activity of the type I signal peptidase of the archaeon Methanococcus voltae. J Bacteriol. 2005;187:1188–1191. doi: 10.1128/JB.187.3.1188-1191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Sauer RT. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartig D, Lemkemeier K, Frank J, Lottspeich F, Klink F. The archaebacterial hypusine-containing protein. Structural features suggest common ancestry with eukaryotic translation initiation factor 5A. Eur J Biochem. 1992;204:751–758. doi: 10.1111/j.1432-1033.1992.tb16690.x. [DOI] [PubMed] [Google Scholar]
- Bazan JF, Weaver LH, Roderick SL, Huber R, Matthews BW. Sequence and structure comparison suggest that methionine aminopeptidase, prolidase, aminopeptidase P, and creatinase share a common fold. Proc Natl Acad Sci USA. 1994;91:2473–2477. doi: 10.1073/pnas.91.7.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beja O, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry. 1999;38:14138–14145. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- Brooun A, Zhang W, Alam M. Primary structure and functional analysis of the soluble transducer protein HtrXI in the archaeon Halobacterium salinarium. J Bacteriol. 1997;179:2963–2968. doi: 10.1128/jb.179.9.2963-2968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Iyer LM, Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol Biosyst. 2011;7:2261–2277. doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Aebi M. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta. 1999;1426:239–257. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- Calo D, Eichler J. Crossing the membrane in Archaea, the third domain of life. Biochim Biophys Acta. 2011;1808:885–901. doi: 10.1016/j.bbamem.2010.03.020. [DOI] [PubMed] [Google Scholar]
- Calo D, Kaminski L, Eichler J. Protein glycosylation in Archaea: Sweet and extreme. Glycobiology. 2010a;20:1065–1079. doi: 10.1093/glycob/cwq055. [DOI] [PubMed] [Google Scholar]
- Calo D, Eilam Y, Lichtenstein RG, Eichler J. Towards glyco-engineering in Archaea: Replacing Haloferax volcanii AglD with homologous glycosyltransferases from other halophilic archaea. Appl Environ Microbiol. 2010b;76:5684–5692. doi: 10.1128/AEM.00681-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo D, Guan Z, Eichler J. Glyco-engineering in Archaea: Differential N-glycosylation of the S-layer glycoprotein in a transformed Haloferax volcanii strain. Microb Biotechnol. 2011;4:461–470. doi: 10.1111/j.1751-7915.2011.00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha SS, An YJ, Lee CR, et al. Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber. EMBO J. 2010;29:3520–3530. doi: 10.1038/emboj.2010.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol Microbiol. 2006;61:259–268. doi: 10.1111/j.1365-2958.2006.05226.x. [DOI] [PubMed] [Google Scholar]
- Charlebois RL, Lam WL, Cline SW, Doolittle WF. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc Natl Acad Sci USA. 1997;84:8530–8534. doi: 10.1073/pnas.84.23.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charollais J, Van Der Goot FG. Palmitoylation of membrane proteins. Mol Membr Biol. 2009;26:55–66. doi: 10.1080/09687680802620369. [DOI] [PubMed] [Google Scholar]
- Clarke S. Protein methylation. Curr Opin Cell Biol. 1993;5:977–983. doi: 10.1016/0955-0674(93)90080-a. [DOI] [PubMed] [Google Scholar]
- Cohen-Rosenzweig C, Yurist-Doutsch S, Eichler J. AglS, a novel component of the Haloferax volcanii N-glycosylation pathway, is a dolichol phosphate-mannose mannosyltransferase. J Bacteriol. 2012 doi: 10.1128/JB.01716-12. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–11. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Dalbey RE, von Heijne G. Signal peptidases in prokaryotes and eukaryotes-a new protease family. Trends in Biochem Sci. 1992;17:474–478. doi: 10.1016/0968-0004(92)90492-r. [DOI] [PubMed] [Google Scholar]
- DasSarma S, Fleishmann EM. Archaea: A laboratory manual – Halophiles. NY: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- De Castro RE, Maupin-Furlow JA, Gimenez MI, Herrera Seitz MK, Sanchez JJ. Haloarchaeal proteases and proteolytic systems. FEMS Microbiol Rev. 2006;30:17–35. doi: 10.1111/j.1574-6976.2005.00003.x. [DOI] [PubMed] [Google Scholar]
- Dilks K, Rose RW, Hartmann E, Pohlschroder M. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol. 2003;185:1478–1483. doi: 10.1128/JB.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith M. The Halohandbook: Protocols for Halobacterial Genetics. 2009 http://www.haloarchaea.com/resources/handbook/Halohandbook_2009_v7.1.pdf.
- Eichler J. Novel glycoproteins of the halophilic archaeon Haloferax volcanii. Arch Microbiol. 2000;173:445–448. doi: 10.1007/s002030000152. [DOI] [PubMed] [Google Scholar]
- Eichler J. Post-translational modification of the S-layer glycoprotein occurs following translocation across the plasma membrane of the haloarchaeon Haloferax volcanii. Eur J Biochem. 2001;268:4366–4373. doi: 10.1046/j.1432-1327.2001.02361.x. [DOI] [PubMed] [Google Scholar]
- Eichler J, Adams MW. Posttranslational protein modification in Archaea. Microbiol Mol Biol Rev. 2005;69:393–425. doi: 10.1128/MMBR.69.3.393-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki M, Shinzato N, Sato H, Nakamura K, Kamagata Y. Comparative proteomic analysis of Methanothermobacter themautotrophicus DH in pure culture and in co-culture with a butyrate-oxidizing bacterium. PLoS One. 2011;6:e24309. doi: 10.1371/journal.pone.0024309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EA, Gilmore R, Blobel G. Purification of microsomal signal peptidase as a complex. Proc Natl Acad Sci USA. 1986;83:581–585. doi: 10.1073/pnas.83.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens CA, Kloppsteck P, Forster A, Zhang X, Freemont PS. Structural and functional implications of phosphorylation and acetylation in the regulation of the AAA+ protein p97. Biochem Cell Biol. 2010;88:41–48. doi: 10.1139/o09-128. [DOI] [PubMed] [Google Scholar]
- Facchin S, Lopreiato R, Ruzzene M, Marin O, Sartori G, Gotz C, Montenarh M, Carignani G, Pinna LA. Functional homology between yeast piD261/Bud32 and human PRPK: both phosphorylate p53 and PRPK partially complements piD261/Bud32 deficiency. FEBS Lett. 2003;549:63–66. doi: 10.1016/s0014-5793(03)00770-1. [DOI] [PubMed] [Google Scholar]
- Falb M, Aivaliotis M, Garcia-Rizo C, Bisle B, Tebbe A, Klein C, Konstantinidis K, Siedler F, Pfeiffer F, Oesterhelt D. Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: a large-scale proteomics survey. J Mol Biol. 2006;362:915–924. doi: 10.1016/j.jmb.2006.07.086. [DOI] [PubMed] [Google Scholar]
- Fass D. Disulfide bonding in protein biophysics. Annu Rev Biophys. 2011 doi: 10.1146/annurev-biophys-050511-102321. in press. [DOI] [PubMed] [Google Scholar]
- Fine A, Irihimovitch V, Dahan I, Konrad Z, Eichler J. Cloning, expression and purification of functional Sec11a and Sec11b, type I signal peptidases of the archaeon Haloferax volcanii. J Bacteriol. 2006;188:1911–1919. doi: 10.1128/JB.188.5.1911-1919.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink-Lavi E, Eichler J. Identification of residues essential for the catalytic activity of Sec11b, one of the two type I signal peptidases of Haloferax volcanii. FEMS Microbiol Lett. 2008;278:257–260. doi: 10.1111/j.1574-6968.2007.01000.x. [DOI] [PubMed] [Google Scholar]
- Fischer C, Geourjon C, Bourson C, Deutscher J. Cloning and characterization of the Bacillus subtilis prkA gene encoding a novel serine protein kinase. Gene. 1996;168:55–60. doi: 10.1016/0378-1119(95)00758-x. [DOI] [PubMed] [Google Scholar]
- Forbes AJ, Patrie SM, Taylor GK, Kim YB, Jiang L, Kelleher NL. Targeted analysis and discovery of posttranslational modifications in proteins from methanogenic archaea by top-down MS. Proc Natl Acad Sci USA. 2004;101:2678–2683. doi: 10.1073/pnas.0306575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouzan D, Ammelburg M, Hobel CF, Stroh LJ, Sessler N, Martin J, Lupas AN. The archaeal proteasome is regulated by a network of AAA ATPases. J Biol Chem. 2012 doi: 10.1074/jbc.M112.386458. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil MA, Sherwood KE, Maupin-Furlow JA. Transcriptional linkage of Haloferax volcanii proteasomal genes with non-proteasomal gene neighbours including RNase P, MOSC domain and SAM-methyltransferase homologues. Microbiology. 2007;153:3009–3022. doi: 10.1099/mic.0.2007/008177-0. [DOI] [PubMed] [Google Scholar]
- Giménez MI, Dilks K, Pohlschröder M. Haloferax volcanii twin-arginine translocation substates include secreted soluble, C-terminally anchored and lipoproteins. Mol Microbiol. 2007;66:1597–1606. doi: 10.1111/j.1365-2958.2007.06034.x. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. Inteins: structure, function, and evolution. Annu Rev Microbiol. 2002;56:263–287. doi: 10.1146/annurev.micro.56.012302.160741. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Greaves J, Prescott GR, Gorleku OA, Chamberlain LH. The fat controller: roles of palmitoylation in intracellular protein trafficking and targeting to membrane microdomains. Mol Membr Biol. 2009;26:67–79. doi: 10.1080/09687680802620351. [DOI] [PubMed] [Google Scholar]
- Grote M, O'Malley MA. Enlightening the life sciences: the history of halobacterial and microbial rhodopsin research. FEMS Microbiol Rev. 2011;35:1082–1099. doi: 10.1111/j.1574-6976.2011.00281.x. [DOI] [PubMed] [Google Scholar]
- Guan Z, Naparstek S, Kaminski L, Konrad Z, Eichler J. Distinct glycan-charged phosphodolichol carriers are required for the assembly of the pentasaccharide N-linked to the Haloferax volcanii S-layer glycoprotein. Mol Microbiol. 2010;78:1294–1303. doi: 10.1111/j.1365-2958.2010.07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Naparstek S, Calo D, Eichler J. Protein glycosylation as an adaptive response in Archaea: Growth at different salt concentrations leads to alterations in Haloferax volcanii S-layer glycoprotein N-glycosylation. Environ Microbiol. 2012;14:743–753. doi: 10.1111/j.1462-2920.2011.02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon JM, Mechulam Y, Schmitter JM, Blanquet S, Fayat G. Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Payne SH, Selengut JD. Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J Bacteriol. 2012;194:36–48. doi: 10.1128/JB.06026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile JD, Kennelly PJ. The activity of an ancient atypical protein kinase is stimulated by ADP-ribose in vitro. Arch Biochem Biophys. 2011;511:56–63. doi: 10.1016/j.abb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Harris RA, Popov KM, Zhao Y, Kedishvili NY, Shimomura Y, Crabb DW. A new family of protein kinases--the mitochondrial protein kinases. Adv Enzyme Regul. 1995;35:147–162. doi: 10.1016/0065-2571(94)00020-4. [DOI] [PubMed] [Google Scholar]
- Hartley MD, Imperiali B. At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Arch Biochem Biophys. 2012;517:83–97. doi: 10.1016/j.abb.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Norais C, Badger JH, et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2010;5:e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase T, Wakabayashi S, Matsubara H, Kerscher L, Oesterhelt D, Rao KK, Hall DO. Complete amino acid sequence of Halobacterium halobium ferredoxin containing an Nε-acetyllysine residue. J Biochem. 1978;83:1657–1670. doi: 10.1093/oxfordjournals.jbchem.a132078. [DOI] [PubMed] [Google Scholar]
- Hase T, Wakabayashi S, Matsubara H, Mevarech M, Werber MM. Amino acid sequence of 2Fe-2S ferredoxin from an extreme halophile, Halobacterium of the Dead Sea. Biochim Biophys Acta. 1980;623:139–145. doi: 10.1016/0005-2795(80)90016-1. [DOI] [PubMed] [Google Scholar]
- Hatakeyama T, Hatakeyama T. Amino acid sequences of the ribosomal proteins HL30 and HmaL5 from the archaebacterium Halobacterium marismortui. Biochim Biophys Acta. 1990;1039:343–347. doi: 10.1016/0167-4838(90)90269-l. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Wu HC. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- Hecker A, Lopreiato R, Graille M, Collinet B, Forterre P, Libri D, van Tilbeurgh H. Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS subcomplex. EMBO J. 2008;27:2340–2351. doi: 10.1038/emboj.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci. 2006;31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Helbig AO, Gauci S, Raijmakers R, van Breukelen B, Slijper M, Mohammed S, Heck AJ. Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol Cell Proteomics. 2010;9:928–939. doi: 10.1074/mcp.M900463-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Hu LI, Lima BP, Wolfe AJ. Bacterial protein acetylation: the dawning of a new age. Mol Microbiol. 2010;77:15–21. doi: 10.1111/j.1365-2958.2010.07204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, et al. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010a;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Reuter CJ, Zuobi-Hasona K, Zhou G, Maupin-Furlow JA. Phosphorylation and methylation of proteasomal proteins of the haloarcheon Haloferax volcanii. Archaea. 2010b;2010:481725. doi: 10.1155/2010/481725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Stevens SM Jr, Maupin-Furlow JA. Posttranslational modification of the 20S proteasomal proteins of the archaeon Haloferax volcanii. J Bacteriol. 2006;188:7521–7530. doi: 10.1128/JB.00943-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Zhou G, Maupin-Furlow JA. The N-terminal penultimate residue of 20S proteasome α1 influences its Nα-acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. J Bacteriol. 2009;191:3794–3803. doi: 10.1128/JB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igura M, Maita N, Kamishikiryo J, Yamada M, Obita T, Maenaka K, Kohda D. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 2008;27:234–243. doi: 10.1038/sj.emboj.7601940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KF, Albers SV. The archaellum: an old motility structure with a new name. Trends Microbiol. 2012;20:307–312. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Jones GM, Kandiba L, Nair DB, Eichler J. S-layer glycoproteins and flagellins: Reporters of archaeal post-translational modifications. Archaea. 2010;2010:612948. doi: 10.1155/2010/612948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Robinson C. Mechanisms of protein import and routing in chloroplasts. Curr Biol. 2004;14:R1064–R1077. doi: 10.1016/j.cub.2004.11.049. [DOI] [PubMed] [Google Scholar]
- Jelinska C, Conroy MJ, Craven CJ, Hounslow AM, Bullough PA, Waltho JP, Taylor GL, White MF. Obligate heterodimerization of the archaeal Alba2 protein with Alba1 provides a mechanism for control of DNA packaging. Structure. 2005;13:963–971. doi: 10.1016/j.str.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Jeong YJ, Jeong BC, Song HK. Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii. Biochem Biophys Res Commun. 2011;405:112–117. doi: 10.1016/j.bbrc.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Johnson LN, Barford D. The effects of phosphorylation on the structure and function of proteins. Annu Rev Biophys Biomol Struct. 1993;22:199–232. doi: 10.1146/annurev.bb.22.060193.001215. [DOI] [PubMed] [Google Scholar]
- Jones JD, O'Connor CD. Protein acetylation in prokaryotes. Proteomics. 2011;11:3012–3022. doi: 10.1002/pmic.201000812. [DOI] [PubMed] [Google Scholar]
- Jones GM, Wu J, Ding Y, Uchida K, Aizawa S, Robotham A, Logan SM, Kelly J, Jarrell KF. Identification of genes involved in the acetamidino group modification of the flagellin N-linked glycan of Methanococcus maripaludis. J Bacteriol. 2012;194:2693–2702. doi: 10.1128/JB.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczowka SJ, Maupin-Furlow JA. Subunit topology of two 20S proteasomes from Haloferax volcanii. J Bacteriol. 2003;185:165–174. doi: 10.1128/JB.185.1.165-174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L, Abu-Qarn M, Guan Z, Naparstek S, Ventura VV, Raetz CRH, Hitchen PG, Dell A, Eichler J. AglJ adds the first sugar of the N-linked pentasaccharide decorating the Haloferax volcanii S-layer glycoprotein. J Bacteriol. 2010;192:5572–5579. doi: 10.1128/JB.00705-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L, Guan Z, Abu-Qarn M, Konrad Z, Eichler J. AglR is required for addition of the final mannose residue of the N-linked glycan decorating the Haloferax volcanii S-layer glycoprotein. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagen.2012.06.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L, Aitio O, Helin J, Guan Z, Permi P, Bamford D, Eichler J, Roine E. Diversity in prokaryotic glycosylation: An archaeal-derived N-linked glycan contains legionaminic acid. Mol Microbiol. 2012;84:578–593. doi: 10.1111/j.1365-2958.2012.08045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadzic I, Maupin-Furlow J, Humbard M, Singh P, Goodlett D. Chemical cross-linking, mass spectrometry and in silico modeling of proteasomal 20S core particles of the haloarchaeon Haloferax volcanii. Proteomics. 2012;12:1806–1814. doi: 10.1002/pmic.201100260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ. Archaeal protein kinases and protein phosphatases: insights from genomics and biochemistry. Biochem J. 2003;370:373–389. doi: 10.1042/BJ20021547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ, Potts M. Life among the primitives: protein O -phosphatases in prokaryotes. Front Biosci. 1999;4:D372–D385. doi: 10.2741/kennelly. [DOI] [PubMed] [Google Scholar]
- Kikuchi A, Sagami H, Ogura K. Evidence for covalent attachment of diphytanylglyceryl phosphate to the cell-surface glycoprotein of Halobacterium halobium. J Biol Chem. 1999;274:18011–18016. doi: 10.1074/jbc.274.25.18011. [DOI] [PubMed] [Google Scholar]
- Kim D, Forst S. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology. 2001;147:1197–1212. doi: 10.1099/00221287-147-5-1197. [DOI] [PubMed] [Google Scholar]
- Kimura M, Arndt E, Hatakeyama T, Hatakeyama T, Kimura J. Ribosomal proteins in halobacteria. Can J Microbiol. 1989;35:195–199. doi: 10.1139/m89-030. [DOI] [PubMed] [Google Scholar]
- Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA. Genetic and proteomic analyses of a proteasome-activating nucleotidase A mutant of the haloarchaeon Haloferax volcanii. J Bacteriol. 2008a;190:193–205. doi: 10.1128/JB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland PA, Humbard MA, Daniels CJ, Maupin-Furlow JA. Shotgun proteomics of the haloarchaeon Haloferax volcanii. J Proteome Res. 2008b;7:5033–5039. doi: 10.1021/pr800517a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klussmann S, Franke P, Bergmann U, Kostka S, Wittmann-Liebold B. N-terminal modification and amino-acid sequence of the ribosomal protein HmaS7 from Haloarcula marismortui and homology studies to other ribosomal proteins. Biol Chem Hoppe Seyler. 1993;374:305–312. doi: 10.1515/bchm3.1993.374.1-6.305. [DOI] [PubMed] [Google Scholar]
- Koch MK, Oesterhelt D. MpcT is the transducer for membrane potential changes in Halobacterium salinarum. Mol Microbiol. 2005;55:1681–1694. doi: 10.1111/j.1365-2958.2005.04516.x. [DOI] [PubMed] [Google Scholar]
- Koch MK, Staudinger WF, Siedler F, Oesterhelt D. Physiological sites of deamidation and methyl esterification in sensory transducers of Halobacterium salinarum. J Mol Biol. 2008;380:285–302. doi: 10.1016/j.jmb.2008.04.063. [DOI] [PubMed] [Google Scholar]
- Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. Evolution of two-component signal transduction. Mol Biol Evol. 2000;17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- Konrad Z, Eichler J. Lipid modification of proteins in Archaea: attachment of a mevalonic acid-based lipid moiety to the surface-layer glycoprotein of Haloferax volcanii follows protein translocation. Biochem J. 2002;366:959–964. doi: 10.1042/BJ20020757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. Evolution of two-component signal transduction. Mol Biol Evol. 2000;17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- Kuntz C, Sonnenbichler J, Sonnenbichler I, Sumper M, Zeitler R. Isolation and characterization of dolichol-linked oligosaccharides from Haloferax volcanii. Glycobiology. 1997;7:897–904. doi: 10.1093/glycob/7.7.897. [DOI] [PubMed] [Google Scholar]
- Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wlodawer A. A family portrait of the RIO kinases. J Biol Chem. 2005a;280:37297–37300. doi: 10.1074/jbc.R500013200. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wlodawer A. The RIO kinases: an atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim Biophys Acta. 2005b;1754:14–24. doi: 10.1016/j.bbapap.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Lechner J, Sumper M. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J Biol Chem. 1987;262:9724–9729. [PubMed] [Google Scholar]
- Lechner J, Sumper M. Structure and biosynthesis of prokaryotic glycoproteins. Annu Rev Biochem. 1989;58:173–194. doi: 10.1146/annurev.bi.58.070189.001133. [DOI] [PubMed] [Google Scholar]
- Lechner J, Wieland F, Sumper M. Biosynthesis of sulfated saccharides N-glycosidically linked to the protein via glucose. Purification and identification of sulfated dolichyl monophosphoryl tetrasaccharides from halobacteria. J Biol Chem. 1985a;260:860–866. [PubMed] [Google Scholar]
- Lechner J, Wieland F, Sumper M. Transient methylation of dolichyl oligosaccharides is an obligatory step in halobacterial sulfated glycoprotein biosynthesis. J Biol Chem. 1985b;260:8984–8989. [PubMed] [Google Scholar]
- Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev. 2005;26:147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- Lee HS, Kim YJ, Bae SS, et al. Cloning, expression, and characterization of a methionyl aminopeptidase from a hyperthermophilic archaeon Thermococcus sp. NA1. Mar Biotechnol. 2006;8:425–432. doi: 10.1007/s10126-005-6124-8. [DOI] [PubMed] [Google Scholar]
- Leonard CJ, Aravind L, Koonin EV. Novel families of putative protein kinases in bacteria and archaea: evolution of the "eukaryotic" protein kinase superfamily. Genome Res. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- Lindbeck JC, Goulbourne EA, Jr, Johnson MS, Taylor BL. Aerotaxis in Halobacterium salinarium is methylation-dependent. Microbiology. 1995;141:2945–2953. doi: 10.1099/13500872-141-11-2945. [DOI] [PubMed] [Google Scholar]
- Lowther WT, Matthews BW. Structure and function of the methionine aminopeptidases. Biochim Biophys Acta. 2000;1477:157–167. doi: 10.1016/s0167-4838(99)00271-x. [DOI] [PubMed] [Google Scholar]
- Lowther WT, Matthews BW. Metalloaminopeptidases: common functional themes in disparate structural surroundings. Chem Rev. 2002;102:4581–4608. doi: 10.1021/cr0101757. [DOI] [PubMed] [Google Scholar]
- Lupas A, Flanagan JM, Tamura T, Baumeister W. Self-compartmentalizing proteases. Trends Biochem Sci. 1997;22:399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- Mackay DT, Botting CH, Taylor GL, White MF. An acetylase with relaxed specificity catalyses protein N-terminal acetylation in Sulfolobus solfataricus. Mol Microbiol. 2007;64:1540–1548. doi: 10.1111/j.1365-2958.2007.05752.x. [DOI] [PubMed] [Google Scholar]
- Magidovich H, Yurist-Doutsch S, Konrad Z, Ventura VV, Hitchen PG, Dell A, Eichler J. AglP is a S-adenosyl-L-methionine-dependent methyltransferase that participates in the N-glycosylation pathway of Haloferax volcanii. Mol Microbiol. 2010;76:190–199. doi: 10.1111/j.1365-2958.2010.07090.x. [DOI] [PubMed] [Google Scholar]
- Mann RK, Beachy PA. Novel lipid modifications of secreted protein signals. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- Mao DY, Neculai D, Downey M, et al. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol Cell. 2008;32:259–275. doi: 10.1016/j.molcel.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez V, Frohlich T, Armache JP, Sohmen D, Donhofer A, Mikolajka A, Berninghausen O, Thomm M, Beckmann R, Arnold GJ, Wilson DN. Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. J Mol Biol. 2011;405:1215–1232. doi: 10.1016/j.jmb.2010.11.055. [DOI] [PubMed] [Google Scholar]
- Marsh VL, McGeoch AT, Bell SD. Influence of chromatin and single strand binding proteins on the activity of an archaeal MCM. J Mol Biol. 2006;357:1345–1350. doi: 10.1016/j.jmb.2006.01.074. [DOI] [PubMed] [Google Scholar]
- Marsh VL, Peak-Chew SY, Bell SD. Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. J Biol Chem. 2005;280:21122–21128. doi: 10.1074/jbc.M501280200. [DOI] [PubMed] [Google Scholar]
- Martinez A, Traverso JA, Valot B, Ferro M, Espagne C, Ephritikhine G, Zivy M, Gigilone C, Meinnel T. Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics. 2008;8:2809–2831. doi: 10.1002/pmic.200701191. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Igura M, Nyirenda J, Matsumoto M, Yuzawa S, Noda NN, Inagaki F, Kohda D. Crystal structure of the C-terminal globular domain of oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 Å resolution. Biochemistry. 2012;51:4157–4166. doi: 10.1021/bi300076u. [DOI] [PubMed] [Google Scholar]
- Mattar S, Scharf B, Kent SB, Rodewald K, Oesterhelt D, Engelhard M. The primary structure of halocyanin, an archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J Biol Chem. 1994;269:14939–14945. [PubMed] [Google Scholar]
- Maupin-Furlow J. Proteasomes and protein conjugation across domains of life. Nat Rev Microbiol. 2012;10:100–111. doi: 10.1038/nrmicro2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin-Furlow JA, Gil MA, Humbard MA, Kirkland PA, Li W, Reuter CJ, et al. Archaeal proteasomes and other regulatory proteases. Curr Opin Microbiol. 2005;8:720–728. doi: 10.1016/j.mib.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Maupin-Furlow JA, Humbard MA, Kirkland PA. Extreme challenges and advances in archaeal proteomics. Curr Opin Microbiol. 2012 doi: 10.1016/j.mib.2012.02.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D, Pochet S, Marliere P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–923. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T, Peynot P, Giglione C. Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochimie. 2005;87:701–712. doi: 10.1016/j.biochi.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Meinnel T, Serero A, Giglione C. Impact of the N-terminal amino acid on targeted protein degradation. Biol Chem. 2006;387:839–851. doi: 10.1515/BC.2006.107. [DOI] [PubMed] [Google Scholar]
- Mengele R, Sumper M. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J Biol Chem. 1992;267:8182–8185. [PubMed] [Google Scholar]
- Mescher MF, Strominger JL. Purification and characterization of a prokaryotic glucoprotein from the cell envelope of Halobacterium salinarium. J Biol Chem. 1976;251:2005–2014. [PubMed] [Google Scholar]
- Mescher MF, Strominger JL. Glycosylation of the surface glycoprotein of Halobacterium salinarium via a cyclic pathway of lipid-linked intermediates. FEBS Lett. 1978;89:37–41. doi: 10.1016/0014-5793(78)80517-1. [DOI] [PubMed] [Google Scholar]
- Milligan DL, Koshland DE Jr. The amino terminus of the aspartate chemoreceptor is formylmethionine. J Biol Chem. 1990;265:4455–4460. [PubMed] [Google Scholar]
- Min KT, Hilditch CM, Diederich B, Errington J, Yudkin MD. Sigma F, the first compartment-specific transcription factor of B. subtilis is regulated by an anti-sigma factor that is also a protein kinase. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- Miranda HV, Nembhard N, Su D, Hepowit N, Krause DJ, Pritz JR, Phillips C, Soll D, Maupin-Furlow JA. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc Natl Acad Sci USA. 2011;108:4417–4422. doi: 10.1073/pnas.1018151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muff TJ, Ordal GW. Assays for CheC, FliY & CheX as representatives of response regulator phosphatases. Methods Enzymol. 2007;423:336–348. doi: 10.1016/S0076-6879(07)23015-0. [DOI] [PubMed] [Google Scholar]
- Nadolski MJ, Linder ME. Protein lipidation. FEBS J. 2007;274:5202–5210. doi: 10.1111/j.1742-4658.2007.06056.x. [DOI] [PubMed] [Google Scholar]
- Naor A, Thiaville PC, Altman-Price N, Cohen-Or I, Allers T, Crecy-Lagard V, Gophna U. A genetic investigation of the KEOPS complex in halophilic archaea. PLoS One. 2012;7:e43013. doi: 10.1371/journal.pone.0043013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Chaban B, VanDyke DJ, Jarrell KF. Archaeal signal peptidases. Microbiology. 2007;153:305–314. doi: 10.1099/mic.0.2006/003087-0. [DOI] [PubMed] [Google Scholar]
- Ng WV, Kennedy SP, Mahairas GG, et al. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann B, Lebert MR, Alam M, Nitz S, Kollmannsberger H, Oesterhelt D, Hazelbauer GL. Identification of volatile forms of methyl groups released by Halobacterium salinarium. J Biol Chem. 1994;269:16449–16454. [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Kakuta J, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- Oke M, Carter LG, Johnson KA, et al. The Scottish Structural Proteomics Facility: targets, methods and outputs. J Struct Funct Genomics. 2010;11:167–180. doi: 10.1007/s10969-010-9090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A, Ginzburg M, Ginzburg BZ, Hochstein LI, Volcani BE. Haloarcula marismortui (Volcani) sp. nov., nom. rev., an extremely halophilic bacterium from the Dead Sea. Int J Syst Bacteriol. 1990;40:209–210. doi: 10.1099/00207713-40-2-209. [DOI] [PubMed] [Google Scholar]
- Oxenrider KA, Kennelly PJ. A protein-serine phosphatase from the halophilic archaeon Haloferax volcanii. Biochem Biophys Res Commun. 1993;194:1330–1335. doi: 10.1006/bbrc.1993.1970. [DOI] [PubMed] [Google Scholar]
- Paik WK, Paik DC, Kim S. Historical review: the field of protein methylation. Trends Biochem Sci. 2007;32:146–152. doi: 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci USA. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- Pechlivanis M, Kuhlmann J. Hydrophobic modifications of Ras proteins by isoprenoid groups and fatty acids--More than just membrane anchoring. Biochim Biophys Acta. 2006;1764:1914–1931. doi: 10.1016/j.bbapap.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Perazzona B, Spudich JL. Identification of methylation sites and effects of phototaxis stimuli on transducer methylation in Halobacterium salinarum. J Bacteriol. 1999;181:5676–5683. doi: 10.1128/jb.181.18.5676-5683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer F, Schuster SC, Broicher A, Falb M, Palm P, Rodewald K, Ruepp A, Soppa J, Tittor J, Oesterhelt D. Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics. 2008;91:335–346. doi: 10.1016/j.ygeno.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Pickl A, Johnsen U, Schonheit P. Fructose degradation in the haloarchaeon Haloferax volcanii involves bacterial type phosphoenolpyruvate-dependent phosphotransferase system, fructose-1-phosphate kinase and Class II fructose-1,6-bisphosphate aldolase. J Bacteriol. 2012 doi: 10.1128/JB.00200-12. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroder M, Gimenez MI, Jarrell KF. Protein transport in Archaea: Sec and twin arginine translocation pathways. Curr Opin Microbiol. 2005;8:713–719. doi: 10.1016/j.mib.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J Mol Biol. 1999;289:729–745. doi: 10.1006/jmbi.1999.2827. [DOI] [PubMed] [Google Scholar]
- Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol. 2011;9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- Ramesh V, RajBhandary UL. Importance of the anticodon sequence in the aminoacylation of tRNAs by methionyl-tRNA synthetase and by valyl-tRNA synthetase in an Archaebacterium. J Biol Chem. 2001;276:3660–3665. doi: 10.1074/jbc.M008206200. [DOI] [PubMed] [Google Scholar]
- Ranjan N, Damberger FF, Sutter M, Allain FH, Weber-Ban E. Solution structure and activation mechanism of ubiquitin-like small archaeal modifier proteins. J Mol Biol. 2010;405:1040–1055. doi: 10.1016/j.jmb.2010.11.040. [DOI] [PubMed] [Google Scholar]
- Reuter CJ, Kaczowka SJ, Maupin-Furlow JA. Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J Bacteriol. 2004;186:7763–7772. doi: 10.1128/JB.186.22.7763-7772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderick SL, Matthews BW. Structure of the cobalt-dependent methionine aminopeptidase from Escherichia coli : a new type of proteolytic enzyme. Biochemistry. 1993;32:3907–3912. doi: 10.1021/bi00066a009. [DOI] [PubMed] [Google Scholar]
- Rudolph J, Oesterhelt D. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium. EMBO J. 1995;14:667–673. doi: 10.1002/j.1460-2075.1995.tb07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J, Tolliday N, Schmitt C, Schuster SC, Oesterhelt D. Phosphorylation in halobacterial signal transduction. EMBO J. 1995;14:4249–4257. doi: 10.1002/j.1460-2075.1995.tb00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagami H, Kikuchi A, Ogura K. Novel isoprenoid modified proteins in Halobacteria. Biochem Biophys Res Commun. 1994;203:972–978. doi: 10.1006/bbrc.1994.2277. [DOI] [PubMed] [Google Scholar]
- Sagami H, Kikuchi A, Ogura K. A novel type of protein modification by isoprenoid-derived materials. Diphytanylglycerylated proteins in Halobacteria. J Biol Chem. 1995;270:14851–14854. doi: 10.1074/jbc.270.25.14851. [DOI] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann H, Klink F. Archaebacterial protein contains hypusine, a unique amino-acid characteristic for eukaryotic translation initiation factor-4D. Syst Appl Microbiol. 1989;11:103–107. [Google Scholar]
- Shi L, Bischoff KM, Kennelly PJ. The icfG gene cluster of Synechocystis sp. strain PCC 6803 encodes an Rsb/Spo-like protein kinase, protein phosphatase and two phosphoproteins. J Bacteriol. 1999;181:4761–4767. doi: 10.1128/jb.181.16.4761-4767.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Potts M, Kennelly PJ. The serine, threonine and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol Rev. 1998;22:229–253. doi: 10.1111/j.1574-6976.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- Shigi N. Post-translational modification of cellular proteins by a ubiquitin-like protein in bacteria. J Biol Chem. 2012;287:17568–17577. doi: 10.1074/jbc.M112.359844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinensky M. Recent advances in the study of prenylated proteins. Biochim Biophys Acta. 2000;1484:93–106. doi: 10.1016/s1388-1981(00)00009-3. [DOI] [PubMed] [Google Scholar]
- Smith RF, King KY. Identification of a eukaryotic-like protein kinase gene in Archaebacteria. Protein Sci. 1995;4:126–129. doi: 10.1002/pro.5560040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbiati J, Chapman-Smith A, Miller JL, Miller CG, Cronan JE Jr. Processing of the N termini of nascent polypeptide chains requires deformylation prior to methionine removal. J Mol Biol. 1999;290:607–614. doi: 10.1006/jmbi.1999.2913. [DOI] [PubMed] [Google Scholar]
- Soppa J. From genomes to function: haloarchaea as model organisms. Microbiology. 2006;152:585–590. doi: 10.1099/mic.0.28504-0. [DOI] [PubMed] [Google Scholar]
- Soppa J. Protein acetylation in archaea, bacteria & eukaryotes. Archaea. 2010;2010:820681. doi: 10.1155/2010/820681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JL, Stoeckenius W. Light-regulated retinal-dependent reversible phosphorylation of Halobacterium proteins. J Biol Chem. 1980;255:5501–5503. [PubMed] [Google Scholar]
- Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2011;30:873–881. doi: 10.1038/emboj.2010.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Escalante-Semerena JC. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol. 2004;340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Storch KF, Rudolph J, Oesterhelt D. Car: a cytoplasmic sensor responsible for arginine chemotaxis in the archaeon Halobacterium salinarum. EMBO J. 1999;18:1146–1158. doi: 10.1093/emboj/18.5.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storf S, Pfeiffer F, Dilks K, Chen ZQ, Imam S, Pohlschröder M. Mutational and bioinformatic analysis of haloarchaeal lipobox-containing proteins. Archaea. 2010;2010:410975. doi: 10.1155/2010/410975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streif S, Oesterhelt D, Marwan W. A predictive computational model of the kinetic mechanism of stimulus-induced transducer methylation and feedback regulation through CheY in archaeal phototaxis and chemotaxis. BMC Syst Biol. 2010;4:27. doi: 10.1186/1752-0509-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom MS, Nunn DN, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper M, Berg E, Mengele R, Strobel I. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J Bacteriol. 1990;172:7111–7118. doi: 10.1128/jb.172.12.7111-7118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg SA, Alam M, Lebert M, Spudich JL, Oesterhelt D, Hazelbauer GL. Characterization of Halobacterium halobium mutants defective in taxis. J Bacteriol. 1990;172:2328–2335. doi: 10.1128/jb.172.5.2328-2335.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Z, Stahl AO, Albers SV, Kissinger JC, Driessenr AJM, Pohlschroder M. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol. 2007;189:772–778. doi: 10.1128/JB.01547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Oki H, Tsukihara T, Ogasahara K, Yutani K, Ogata K, et al. Crystal structure of methionine aminopeptidase from hyperthermophile, Pyrococcus furiosus. J Mol Biol. 1998;284:101–124. doi: 10.1006/jmbi.1998.2146. [DOI] [PubMed] [Google Scholar]
- Thao S, Escalante-Semerena JC. Control of protein function by reversible Nε-lysine acetylation in bacteria. Curr Opin Microbiol. 2011;14:200–204. doi: 10.1016/j.mib.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HJ, Allen MD, Löwe J, Bycroft M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry. 2003;42:11460–11465. doi: 10.1021/bi035033g. [DOI] [PubMed] [Google Scholar]
- Tripepi M, Imam S, Pohlschroder M. Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. J Bacteriol. 2010;192:3093–3102. doi: 10.1128/JB.00133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripepi M, You J, Temel S, Onder O, Brisson D, Pohlschröder M. N-glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J Bacteriol. 2012 doi: 10.1128/JB.00731-12. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunasawa S, Izu Y, Miyagi M, Kato I. Methionine aminopeptidase from the hyperthermophilic Archaeon Pyrococcus furiosus : molecular cloning and overexpression in Escherichia coli of the gene and characteristics of the enzyme. J Biochem. 1997;122:843–850. doi: 10.1093/oxfordjournals.jbchem.a021831. [DOI] [PubMed] [Google Scholar]
- Tyagi N, Anamika K, Srinivasan N. A framework for classification of prokaryotic protein kinases. PLoS One. 2010;5:e10608. doi: 10.1371/journal.pone.0010608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uthandi S, Saad B, Humbard MA, Maupin-Furlow JA. LccA, an archaeal laccase secreted as a highly-stable glycoprotein into the extracellular medium of Haloferax volcanii. Appl Environ Microbiol. 2010;76:733–743. doi: 10.1128/AEM.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Valkenburgh C, Chen X, Mullins C, Fang H, Green N. The catalytic mechanism of endoplasmic reticulum signal peptidase appears to be distinct from most eubacterial signal peptidases. J Biol Chem. 1999;274:11519–11525. doi: 10.1074/jbc.274.17.11519. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–5. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Vetting MW, S de Carvalho LP, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Wagner S, Klug G. An archaeal protein with homology to the eukaryotic translation initiation factor 5A shows ribonucleolytic activity. J Biol Chem. 2007;282:13966–13976. doi: 10.1074/jbc.M701166200. [DOI] [PubMed] [Google Scholar]
- Wang F, Liu M, Qiu R, Ji C. The dual role of ubiquitin-like protein Urm1 as a protein modifier and sulfur carrier. Protein Cell. 2011;2:612–619. doi: 10.1007/s13238-011-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardleworth BN, Russell RJ, Bell SD, Taylor GL, White MF. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 2002;21:4654–4662. doi: 10.1093/emboj/cdf465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- Wieland F, Dompert W, Bernhardt G, Sumper M. Halobacterial glycoprotein saccharides contain covalently linked sulphate. FEBS Lett. 1980;120:110–114. doi: 10.1016/0014-5793(80)81058-1. [DOI] [PubMed] [Google Scholar]
- Wieland F, Heitzer R, Schaefer W. Asparaginylglucose: Novel type of carbohydrate linkage. Proc Natl Acad Sci USA. 1983;80:5470–5474. doi: 10.1073/pnas.80.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland F, Paul G, Sumper M. Halobacterial flagellins are sulfated glycoproteins. J Biol Chem. 1985;260:15180–15185. [PubMed] [Google Scholar]
- Wilson HL, Aldrich HC, Maupin-Furlow J. Halophilic 20S proteasomes of the archaeon Haloferax volcanii : purification, characterization and gene sequence analysis. J Bacteriol. 1999;181:5814–5824. doi: 10.1128/jb.181.18.5814-5824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ohta N, Zhao JL, Newton A. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc Natl Acad Sci USA. 1999;96:13068–13073. doi: 10.1073/pnas.96.23.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, King DM, Kennelly PJ. The Phosphorylation Site Database: A guide to the serine-, threonine- and/or tyrosine-phosphorylated proteins in prokaryotic organisms. Proteomics. 2004;4:1562–1570. doi: 10.1002/pmic.200300711. [DOI] [PubMed] [Google Scholar]
- YaDeau JT, Klein C, Blobel G. Yeast signal peptidase contains a glycoprotein and the Sec11 gene product. Prot Natl Acad Sci USA. 1991;88:517–521. doi: 10.1073/pnas.88.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kang CM, Brody MS, Price CW. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yurist-Doutsch S, Eichler J. Manual annotation, transcriptional analysis and protein expression studies reveal novel genes in the agl cluster responsible for N-glycosylation in the halophilic archaeon Haloferax volcanii. J Bacteriol. 2009;191:3068–3075. doi: 10.1128/JB.01838-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurist-Doutsch S, Abu-Qarn M, Battaglia F, Morris HR, Hitchen PG, Dell A, Eichler J. aglF, aglG and aglI novel members of a gene cluster involved in the N-glycosylation of the Haloferax volcanii S-layer glycoprotein. Mol Microbiol. 2008;69:1234–1245. doi: 10.1111/j.1365-2958.2008.06352.x. [DOI] [PubMed] [Google Scholar]
- Yurist-Doutsch S, Magidovich H, Ventura VV, Hitchen PG, Dell A, Eichler J. N-glycosylation in Archaea: On the coordinated actions of Haloferax volcanii AglF and AglM. Mol Microbiol. 2010;75:1047–1058. doi: 10.1111/j.1365-2958.2009.07045.x. [DOI] [PubMed] [Google Scholar]
- Zhang F, Paterson AJ, Huang P, Wang K, Kudlow JE. Metabolic control of proteasome function. Physiology (Bethesda) 2007;22:373–379. doi: 10.1152/physiol.00026.2007. [DOI] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA binding. J Biol Chem. 2003;278:26071–26077. doi: 10.1074/jbc.M303666200. [DOI] [PubMed] [Google Scholar]
- Zhou G, Kowalczyk D, Humbard MA, Rohatgi S, Maupin-Furlow JA. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:8096–8105. doi: 10.1128/JB.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu BC, Drake RR, Schweingruber H, Laine RA. Inhibition of glycosylation by amphomycin and sugar nucleotide analogs PP36 and PP55 indicates that Haloferax volcanii beta-glucosylates both glycoproteins and glycolipids through lipid-linked sugar intermediates: evidence for three novel glycoproteins and a novel sulfated dihexosyl-archaeol glycolipid. Arch Biochem Biophys. 1995;319:355–364. doi: 10.1006/abbi.1995.1305. [DOI] [PubMed] [Google Scholar]
- Zhu W, Reich CI, Olsen GJ, Giometti CS, Yates JR., III Shotgun proteomics of Methanococcus jannaschii and insights into methanogenesis. J Proteome Res. 2004;3:538–548. doi: 10.1021/pr034109s. [DOI] [PubMed] [Google Scholar]