Abstract
CREB3 proteins comprise a set of ER-localised bZip transcription factors defined by the presence of a transmembrane domain. They are regulated by inter-compartmental transport, Golgi cleavage and nuclear transport where they promote appropriate transcriptional responses. Although CREB3 proteins play key roles in differentiation, inflammation and metabolism, a general framework relating their defining features to these diverse activities is lacking. We identify unique features of CREB3 organisation including the ATB domain, which we show is essential for transcriptional activity. This domain is absent in all other human bZip factors, but conserved in Drosophila CREBA, which controls secretory pathway genes (SPGs). Furthermore, each of the five human CREB3 factors was capable of activating SPGs in Drosophila, dependent upon the ATB domain. Expression of the CREB3 protein, CREB-H, in 293 cells, upregulated genes involved in secretory capacity, extracellular matrix formation and lipid metabolism and increased secretion of specific cargos. In liver cells, which normally express CREB-H, the active form specifically induced secretion of apolipoproteins, including ApoA-IV, ApoAI, consistent with data implicating CREB-H in metabolic homeostasis. Based on these data and other recent studies, we propose of a general role for the CREB3 family in regulating secretory capacity, with particular relevance to specialised cargos.
Keywords: CREB-H, CREBA, CREB3L3, secretion, extracellular matrix, apolipoproteins, lipid metabolism
Introduction
Many aspects of cellular and tissue metabolism are regulated at the ER, which is responsible for the synthesis, modification, and transport of a broad range of proteins, lipids and sterols to their appropriate intra- and extracellular destinations. The ER must also accommodate specialized cargos with distinct requirements for secretion e.g., extracellular collagens, chylomicrons, LDL and HDL droplets, and mucigens. With many additional roles, the ER has orchestrated several homeostatic and quality control mechanisms by which it coordinates responses to disruption of these processes that may result from various cellular stresses, including metabolic fluctuation, mutation, or infection. Amongst these homeostatic mechanisms is that of regulated intramembrane proteolysis (RIP) and activation of a distinct class of transmembrane transcription factors. The prototype members of this class of transcription factors are the sterol regulatory element binding proteins, SREBP 1 and 2, which play key roles in fatty acid and cholesterol metabolism (1–6). Following regulated release from the ER in response to cholesterol or fatty acid fluctuations, SREBPs are transported to the Golgi where they are cleaved in a site-specific manner by Golgi-resident proteases S1P and S2P (2, 5, 7). This cleavage releases the cytosolic domain, which is subsequently transported to the nucleus to effect appropriate transcriptional responses of specific target genes in sterol or fatty acid metabolism. Another transcription factor ATF6, a bZip protein involved in quality control of protein folding, was subsequently shown to be an ER-anchored transmembrane factor regulated by the same overall pathway as SREBP (8, 9). In the case of ATF6, transport from the ER and subsequent Golgi cleavage are not regulated by cholesterol levels, but instead in response to the accumulation of unfolded proteins (8, 10–12).
This specialised set of transmembrane transcription factors has now been extended with the identification of a family of bZip-transmembrane factors related to ATF6. The prototype member, CREB3/Luman, was identified via its interaction with the transcriptional co-activator HCF and was shown subsequently to reside in the ER and to be subject to cleavage by S1P and S2P (13, 14). A mammalian family has now been defined consisting of CREB3 together with the factors OASIS/CREB3L1 (15), BBF2H7/CREB3L2 (16), CREB-H/CREB3L3 (17) and CREB4/CREB3L4 (18, 19). Notwithstanding the overall conservation of bZip-transmembrane organisation, CREB3 proteins exhibit certain unique features conserved within the family and not found in ATF6 that may be indicative of functional differences (20, 21). Among these features, we recently demonstrated that unlike ATF6, the ER localisation of CREB-H was determined by a cytosolic domain termed the ERM (ER retention motif) highly conserved in all CREB3 members but absent from ATF6 (22).
CREB3 factors have been shown to play distinct roles in the ER, although the precise regulatory pathways remain unclear. For example, OASIS and BBF2H7 have recently been shown to be essential for normal bone and cartilage development, as well as terminal differentiation of astrocytes and of intestinal goblet cells (23–26). They are required for production of extracellular matrix cargos during osteoblast, chondrocyte and goblet cell differentiation with target genes including the COPII coat components such as Sec 23 and 24 and cargos including collagen IIa and mucins. CREB-H is found mainly in cells of the liver and the small intestine and to a lesser extent in other cell types. Analyses from different laboratories have implicated CREB-H in diverse processes including acute inflammatory responses (27), hepcidin transcription (28), gluconeogenesis (29, 30) and, more recently, lipid and triglyceride metabolism (31, 32).
Here, we highlight a novel feature of the CREB3 family, the ATB domain, which is crucial for CREB-H transcriptional activity. The ATB domain is only present in CREB3 bZip proteins and defines an evolutionary ancient configuration present in three classes, i.e., a transmembrane class present in two distinct subgroups, and a third class lacking the TM domain. This latter class includes the Drosophila CREBA factor, which is known to regulate secretory activity (33, 34). We show that all human CREB3 factors can activate secretory pathway genes in Drosophila, again with a requirement for the ATB domain. We further identify CREB-H target genes induced by ectopic expression, strongly overlapping with those of OASIS and BBF2H7, including Sec23, Sec24, and ECM components, as well as genes involved in lipid metabolism, such as ApoA-IV and LIPH. We demonstrate increased secretory activity as a result of CREB-H induction, and the selective secretion, at the protein level, of specific cargos including ECM components and apolipoproteins. These results are discussed together with recent data on other CREB3 proteins from which we propose a general role of the CREB3 family in secretory capacity with particular relevance to specialised cargos.
Results
A novel domain of the CREB3 family shared with the Drosophila secretory factor CREBA
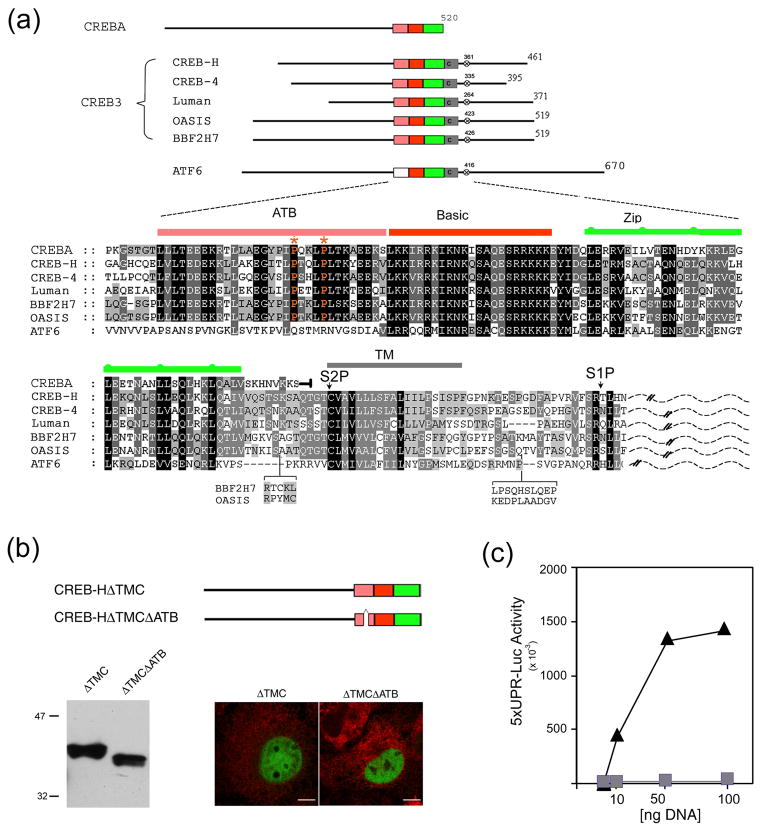
CREB3 proteins exhibit short pockets of limited homology within the N-terminal and C-terminal regions but the most notable feature is the presence of the central highly conserved bZip section. This central region (150 residues) comprises linked sub-domains indicated by coloured boxes and expanded in the corresponding sequence alignment below (Figure 1a). Of particular note for this work is the presence, immediately adjacent to the N-terminal end of the bZip region (red and green boxes), of an additional region of approximately 30 residues (pink box). This region is highly conserved in all CREB3 proteins but lacking in ATF6 and is not part of the typical consensus bZip DNA binding domain (35, 36). It is only found in the CREB3 class of proteins and always immediately adjacent to the bZip domain (see analysis Figure 7). We have therefore named this conserved feature the ATB domain (for adjacent to bZip).
Figure 1. Requirement for the ATB domain in CREB-HΔTMC transcriptional activation.
(a) Organisation of the human CREB3 family members in relation to CREBA and ATF6, showing the ATB domain (pink), the basic region (red), the leucine zipper (green), the transmembrane (TM) domain (grey) and a conserved S1P motif (x). Primary sequence similarity within this central region is shown below. Alignment was performed using ClustalX and annotated with Genedoc. Shading reflects the degree of conservation weighted for chemical similarity. In CREBA, the sequence terminates immediately after the leucine zipper, whereas, in the CREB3 family and in ATF6, homology extends to the TM domain and S1P site. Lumenal sequences C-terminal to the TM domain are indicated by a dashed line. Note the absence of the ATB domain in ATF6, but its virtually complete conservation in CREBA. Optimal alignment based on sequences from additional species is discussed further (see Figure 9). (b) Illustration of CREB-HΔTMC and CREB-HΔTMCΔATB, which lacks 27 residues within the ATB region. COS cells were transfected with the appropriate vectors (1 μg) and expression levels assayed by Western blotting or by immunofluorescence using an antibody to the SV5 epitope tag (CREB-HΔTMC and CREB-HΔTMCΔATB, green channel; calreticulin (an ER marker), red channel). Scale bar, 10 μM. (c) Cells were transfected in triplicate with the target vector 5XATF6-GL3 (1 μg) containing a basal promoter with five copies of the UPR element, and increasing amounts (10 ng, 50 ng or 100 ng) of the expression vectors for CREB-HΔTMC (triangles) or CREB-HΔTMCΔATB (squares). Cells were harvested 24 h post-transfection and luciferase activation was measured.
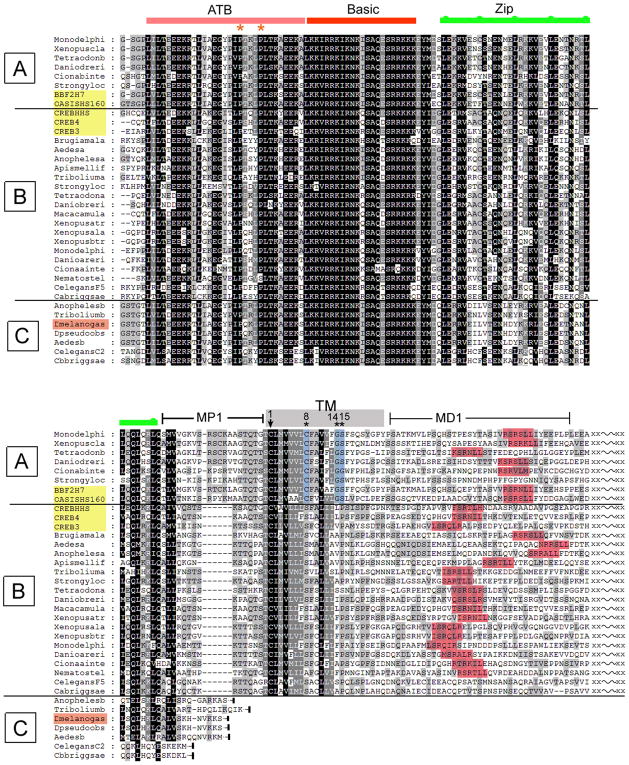
Figure 7. Evolutionary conservation of the CREB3 family and identification of distinct classes.
The figure shows only representative proteins at the amino acid level to allow for key points regarding subgrouping and evolutionary history to be made. For example, numerous CREB3 species from mammals have been omitted, but these all fit with the conclusions and classification discussed in the text. The family of five human CREB3 proteins are shaded in yellow, and thus resolve into the A and B classes. MP1 indicates membrane proximal region 1; TM, transmembrane; MD1, membrane distal region 1. Consensus S1P sites are shaded red. Numbering within the TM domain starts at the completely conserved cysteine. Other features are as described in the text.
To examine the functional relevance of the ATB in CREB-H, we created a version of the nuclear active form, CREB-HΔTMC, containing a short deletion upstream of the bZip domain in the core ATB domain (Figure 1b, CREB-HΔTMCΔATB) and examined transactivation on a test UPR-luciferase target promoter previously shown to be responsive to CREB-H (29). Deletion of the ATB domain affected neither the expression levels nor localisation of the variant which, as expected, localized within the nucleus (Figure 1b). However deletion of the ATB almost completely abrogated transactivation activity (Figure 1c).
One member of the CREB3 family, CREB3L2/BBF2H7, was originally named for its homology within the bZip domain to a Drosophila gene, BBF (16). BBF is identical to and otherwise named as CREBA (37), a factor now known to be expressed in secretory cells and to be critical for transcriptional induction of secretory pathway genes in multiple tissues in Drosophila, especially during high secretory demand (33, 34). We re-evaluated the similarity of CREBA to the CREB3 family and found that although the ATB domain is not present in ATF6, there is strong conservation in amino acid sequence in the ATB regions in CREBA, including a completely conserved PxxLP feature (Figure 1a, ATB domain, asterisks). It was clear that CREB3 proteins and CREBA were ATB-bZip proteins. However the CREBA ATB-bZip configuration is contained at the extreme C-terminus of the protein unlike in the CREB3 family, where the domain is always present in a central location, followed by the transmembrane and lumenal domains (20). These features and observations on an evolutionarily ancient ATB-bZip configuration are discussed further below (Figure 7).
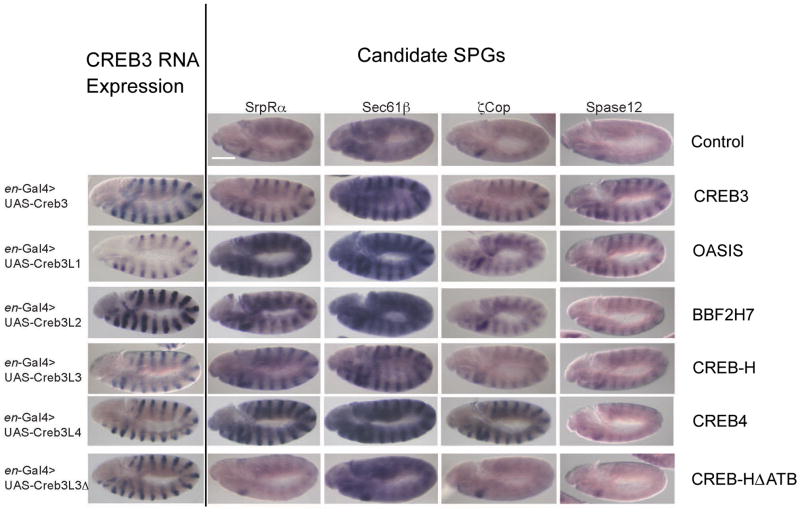
We previously reported that two members of the human CREB3 family were able to activate secretory pathway genes (SPGs) in Drosophila in cells that would otherwise express low or undetectable levels (33). To extend this work, we tested whether this was a generic feature of all human CREB3 proteins. We ectopically expressed each of the CREB3 family members (nuclear forms) in Drosophila epidermal stripes using engrailed promoter-driven Gal4 (epidermal stripe expression) to activate UAS-controlled CREB3 proteins in these tissues. We then examined expression of a series of SPGs in epidermal cells, i.e., SrpRα, Sec61β,ζCop and Spase12. The results (Figure 2) confirmed that both OASIS/CREB3L1 and BBF2H7/CREB3L2 were able to activate SPGs in these cells and that each of the other three members had similar activity. Furthermore, using the ATB deletion variant, we also demonstrated a requirement for the ATB domain, now in the context of physiological activation on these target SPGs (Figure 2).
Figure 2. ATB-dependent activation of SPCGs in Drosophila by CREB3 proteins.
Ectopic expression of the truncated active forms of each CREB3 protein, or the ATB-deletion variant of CREB-H, was achieved using the en-GAL4 driver, which resulted in robust mRNA accumulation of each factor in epidermal stripes (left hand column). Parallel assays were performed for expression of a series of SPCGs. The top row shows wild-type expression of each SPCG tested with little significant background in epidermal stripes, as expected. Expression of the human CREB3 truncated forms induced high-level expression of all SPCGs tested, whereas no significant expression was observed for the ATB-deleted variant. Scale bar, 125 μm.
Thus, human CREB3 proteins and Drosophila CREBA share highly conserved bZip regions that include a unique ATB domain. All of these proteins can activate ectopic expression of SPGs in fly embryos, an activity not found with Drosophila ATF/CrebB, a bZip protein which lacks the ATB (33). Importantly, the conserved ATB domain is required for CREB-H activation of target genes both in human tissue culture cells and in Drosophila embryos.
Establishment of a cell line expressing transcriptionally active CREB-H.ΔTMC
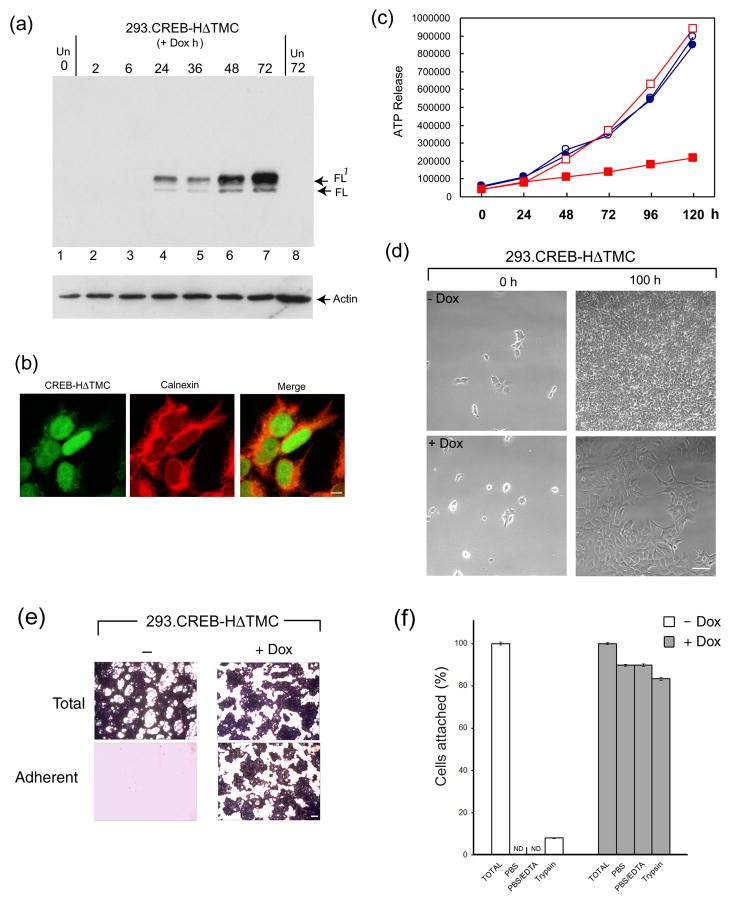
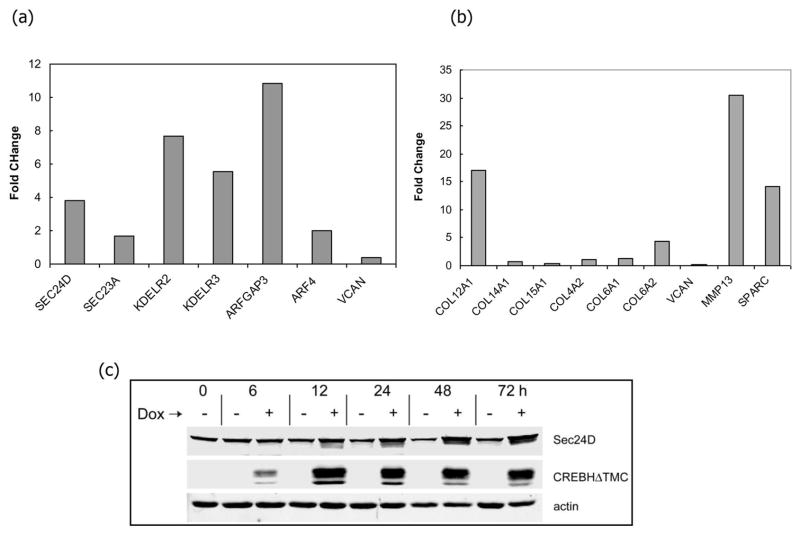
The next goal was to identify the target genes for CREB-H in human cells. We transferred CREB-HΔTMC into a vector for regulated doxycycline (Dox)-induced expression (38). The vector pTRE-CREB-HΔTMC was introduced into 293-Tet On cells and clones containing Dox-inducible expression of CREB-HΔTMC were isolated. CREB-HΔTMC is expressed as a primary product together with a slower migrating species, which we previously showed to be due to efficient phosphorylation (22). Typical results for induction are shown in Figure 3 with the doublet CREB-HΔTMC being readily detectable between 6 and 24 h after Dox addition (Figure 3a). As expected, CREB-HΔTMC was virtually exclusively nuclear with little specific subnuclear localisation (Figure 3b).
Figure 3. Establishment of HEK-293 cells with regulated expression of CREB-HΔTMC.
(a) HEK-293 cells clonally selected for doxycycline regulated expression of CREB-HΔTMC were harvested at different times after induction (0.5 μg/ml of doxycycline) or after control treatment (Un). Tightly regulated expression of CREB-HΔTMC can be seen, from undetectable in untreated cells, increasing between 6 and 24 h after Dox addition. The doublet represents full length CREB-HΔTMC (FL) and a phosphorylated species FL1 as previously demonstrated (22). (b) Induced cells were fixed and stained for the detection of CREB-HΔTMC (green), which localized primarily in the nucleus. Cells were counterstained with the ER marker calnexin (red). Scale bar, 10 μM. (c) 293.CREB-HΔTMC cells ( Dox, open squares: +Dox, filled squares) or a control 293 cell line (filled circles: Dox, open circles: +Dox, filled squares) were seeded at low and incubated in standard growth medium. Dox was added to a duplicate series of each cell line 24 h after seeding. Cells were harvested at intervals thereafter and cell proliferation measured using the Promega luminescent cell viability assay. (d) Phase images of 293.CREB-HΔTMC cells immediately after plating (0 h) or after approximately 4 days (100 h) in the absence or presence of Dox. Scale bar, 100 μM (e) CREB-H induces increased cell adhesion. 293.CREB-HΔTMC cells in cluster dishes (105 cells per well), were induced with Dox or mock treated, incubated for 5 days, then fixed with paraformaldehyde and stained with crystal violet (Total). Duplicate cultures were aspirated, washed with PBS, then fixed and stained (Adherent). Scale bar 1 mm (f) Quantification of cell adhesion as described in the text. Open bars represent cultures grown in the absence of Dox, filled bars represent plus Dox. The total cell recovery was set to 100% in each case. Adherent cells numbers were below the level of significant detection (ND) in the uninduced cultures. The values are averages of three independent experiments, error bars correspond to standard deviations.
Results comparing growth over time after low density seeding of 293.CREB-HΔTMC and control 293 cells are shown in Figure 3c. In the absence of induction, 293.CREB-HΔTMC cells exhibited a similar growth rate to the control cells (open red squares, v. open blue circles). After Dox induction, we observed a reduced growth rate in 293.CREB-HΔTMC (solid red squares), whereas Dox had no effect on control cells (solid blue circles). The effect of Dox induction on 293.CREB-HΔTMC cell growth is also shown in the lower panel of low magnification phase contrast images, showing cells immediately after seeding and after 100 h in the absence and presence of Dox. Notwithstanding the slower growth rate, the 293.CREB-HΔTMC cells were healthy, showed no difference in overall viability from the control cells (data not shown) and also exhibited a flatter morphology (Figure 3d).
Increased cell substratum adhesion induced by CREB-HΔTMC
The flatter cell morphology observed with CREB-H expression may be linked to an increase in cell-substratum attachment of the 293 cells which otherwise readily detach from culture surfaces (39). To examine effects on attachment, we plated 293.CREB-HΔTMC cells at low density, induced CREB-HΔTMC with Dox or left untreated and, when cell monolayers were almost confluent, examined resistance to detachment by washing with PBS containing 1 mM EDTA. Consistent with general observations, 293.CREB-HΔTMC cells (and parental 293 cells, data not shown) in the absence of Dox induction attached weakly and were almost completely removed from the monolayer with washing (Figure 3e). In contrast, upon Dox induction of CREB-HΔTMC, the cell monolayer was quite resistant to detachment (Figure 3e, +Dox). As expected, no such effect of Dox was observed with parental control cells (data not shown). These results are quantitated in Figure 3f showing attached cell numbers after washing with PBS, PBS/EDTA or a brief trypsin treatment (60 s). Any washing of monolayers grown without CREB-HΔTMC induction removed essentially all of the monolayer, with residual cell numbers below the level of sensitivity. In contrast, after Dox induction, more than half the cells remained attached after washing, even after a brief trypsin treatment, though longer trypsin treatment as expected detached the cells. These findings suggest that CREB-H increased cell adhesion to the culture dish, possibly via increased ECM secretion, a conclusion consistent with results in the following sections.
CREB-HΔTMC regulated transcription
To identify CREB-H target genes, we next performed expression profiling by microarray and quantitative RT-PCR in 293.CREB-HΔTMC cells. Based on the time course of the appearance of CREB-HΔTMC (Figure 3), we isolated RNA at either 12 h or 30 h after Dox addition or from mock treated cells. RNA was isolated from triplicate samples for each condition and analysed on Affymetrix Human Gene 1.0 ST whole transcript arrays covering 28,869 genes, each with approximately 26 probes representing each transcript. We found approximately 350 genes with altered expression at a statistically significant value (P<0.001). A complete table of the results is included as Supplementary Table 1. Inspection of this gene set and analysis of distribution within different classification of Gene Ontogeny using the DNAstar suite reveals certain features of this differentially regulated set. Despite sharing the central similarity with ATF6, we found very few CREB-H target genes directly involved in protein folding and chaperone function. This is consistent with previous results indicating that CREB-H does not upregulate classical chaperones such as BiP or GRP94 (27, 29, 40). Perhaps the most salient feature was the relative enrichment of genes involved in three main activities, firstly genes encoding components of the secretory apparatus (e.g., Sec24d, Sec 23a, KDELR3, ARF4, ARFGAP3), secondly, genes linked to the function or composition of extracellular matrix (e.g., Col12a, LOX, SPARC, MMP13), and, thirdly, genes involved in lipoprotein metabolism (e.g., Apo A-IV, Lip H, Lip A, AGPAT9). Of the 350 genes with statistically significant differential regulation, over 30 are classified within the Gene Ontogeny “establishment of localization” subgroup. A selection of the total gene set (Supplementary Table) is summarized under general headings in Table 1. An analysis of the Gene Ontogeny using the David Bioinformatic Resource is given in Supplementary Table 2. We confirmed the induction of a selection of transcripts by qPCR. Within 12 h of Dox addition, Sec24d, KDELR2 and KDELR3, and ARFGAP all showed significant upregulation (Figure 4a), consistent with the microarray results. Similarly, we confirmed the selective upregulation of the Col12a1 gene relative to other collagen genes, as well as induction of MMP13 and the extracellular protein SPARC (Figure 4b). We also analyzed expression of a representative protein of the secretory components, Sec24d, which is known to be involved in coat formation in anterograde ER to Golgi transport (41). We observed significant increases in the level of Sec24d protein within 12–24 h after induction, which was sustained for up to 4 days (Figure 4c). Importantly, we find a very significant degree of overlap in the identification of CREB-H target genes reported here and those identified upon expression in HeLa cells of the active form of OASIS, including as examples Sec24d, Sec23a, KDELR1 and KDELR3, ARFGAP3 (33).
TABLE 1.
Results of microarray analysis in 293.CREB-HΔTMC for genes differentially regulated after the addition of Dox to induce CREB-HΔTMC. Assays were performed in triplicate at 12h and 24h after Dox addition and analysed on Affymetrix Human Gene 1.0 ST arrays. Genes with an FDR-adjusted p-value of less than 0.05 are considered as differentially expressed and the table indicates a selection of these genes with changes in gene expression over 1.5x, grouped on the basis of potential pathways or function. A table of the full complement of differentially regulated genes is attached in Supplementary Table 1. These genes were also analysed using the David Bioinformatic Resource for Gene Ontogeny using a multiple linkage threshold of 0.5, a cut-off EASE score of probability of 0.01. This analysis is presented in Supplementary Table 2
List of oligonucleotides used for Drosophila fly strain construction
| CREB3L3 FGW |
5′ | CACCATGAATACGGATTTAGCTGCTGG | 3′ |
| CREB3L3 RGW |
5′ | TTAACAGGTGCCTGTCTGGGCTGA | 3′ |
| CREB3L4 FGW |
5′ | CACCATGGATCTCGGAATCCCTGACCTG | 3′ |
| CREB3L4 RGW |
5′ | CTAGCTGGTCTGGGCAGCTTTGTTGG | 3′ |
| CREB3 FGW |
5′ | CACCATGGAGCTGGAATTGGATGCTGG | 3′ |
| CREB3 RGW |
5′ | CTATATCTCAATCACCATGGCCTGGAG | 3′ |
Figure 4. Induction of candidate genes by CREB-HΔTMC.
(a,b) Cells were harvested at 12 h or 24 h after Dox addition and total cell RNA isolated, and processed as described in materials and methods. Panel a (12 h) shows changes for a selected set of genes encoding proteins involved in secretory function. Panel b (24 h) shows changes for a selected set of genes encoding extracellular matrix proteins and different isoforms of collagen. Specific selective increases were observed for MMP13, SPARC and Col12a1. (c) Cells plated as in (a) were induced with and without Dox and incubated for different times thereafter. Cell lysates were harvested and analysed by Western blotting for expression levels of Sec24d using Lamin B as a loading control.
Effect of CREB-HΔTMC induction on protein secretion
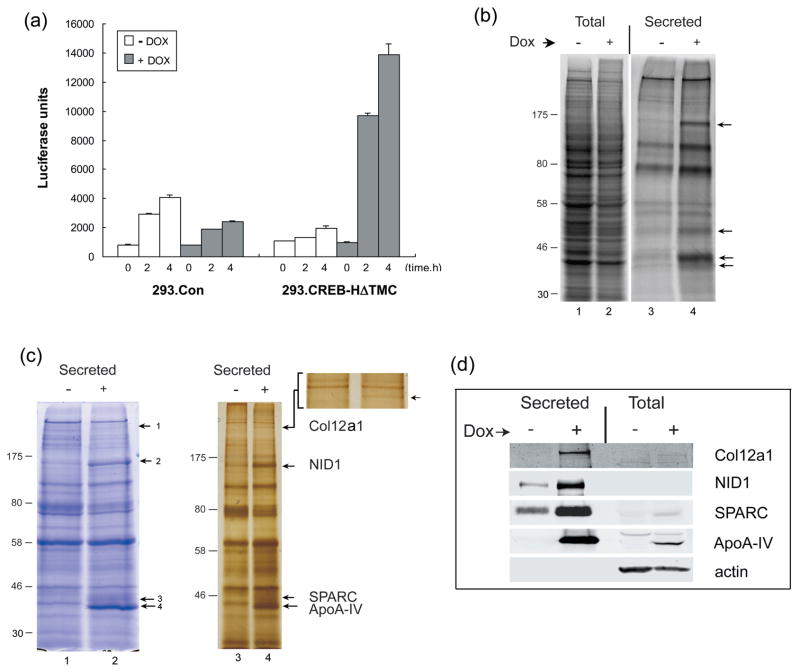
Considering the relative enrichment of genes involved in the secretory pathway, and the identification of secreted cargo components among the potential target genes, we next directly examined whether we could detect any alteration in secretion after CREB-H induction. We first addressed general secretion rates by introducing into 293.CREB-HΔTMC or control cells, a version of luciferase that contains a natural signal sequence for secretion of a stable luciferase. We then measured secretion rates with and without Dox induction. Approximately 24 h after treatment, cells were washed several times to remove any accumulated luciferase, fresh medium was added and the rate of luciferase accumulation in the medium measured over time (Figure 5a). A significant increase in luciferase secretion was observed in the CREB-HΔTMC induced cells and this increase is likely underestimated due to the reduction in overall growth rate upon Dox induction (see Figure 3). We performed identical experiments in control 293 cells not containing CREB-H and, as expected, observed no effect of Dox addition on secretion rates (Figure 5a).
Figure 5. Increased secretion of specific proteins induced by CREB-H.
(a) CREB-H induces increased secretion levels. 293tet-on (Control) and 293.CREB-HΔTMC cell lines were plated at 1.5×105 cells/well in six well cluster dishes coated with collagen and transfected 24 h later with 1 μg of pTKGluc. The cells were induced with Dox or mock treated 24 h after plating and 48 h after induction were, washed three times with cold PBS and fed fresh medium. Supernatant (100 μl) was collected at different time points (0, 2 and 4 h) and analysed for secreted luciferase. (b) Endogenous protein secretion induced by CREB-HΔTMC. Cells were labelled and harvested as described in materials and methods. Total cell samples were separated by SDS-PAGE (lanes 1 and 2). Corresponding media was collected, debris removed by centrifugation, and secreted proteins isolated by TCA precipitation (lanes 3,4). Total samples were loaded at one quarter of the cell equivalents compared to the secreted samples. (c) Identification of cargos exhibiting selective increases in secretion. Scale up of analysis of secreted proteins was as described in materials and methods. Proteins precipitated from the media were separated by SDS-PAGE and subjected to total protein analysis by either Coomassie (lanes 1,2) or silver staining (lanes 3,4). The inset shows an enlargement of the top portion of the gel to aid identification of one of the novel secretd species. Specific bands (numbered in images) were cut from the stained gels and analyzed by mass spectrometry. (d) Confirmation of secreted proteins by western blotting. Samples analysed in (a) by total protein staining were analysed by western blotting using antibodies to each protein as indicated. Total cell samples were loaded at one quarter the cell equivalents compared to secreted samples. The abundant secretion of Apo A-IV is apparent, increasing from an undetectable level in uninduced cells. Increases in the other species were also evident, each migrating with the same mobility to the band identified also by mass spectrometry. Actin is shown as a loading control for the total protein samples and was not detected in the secreted samples.
To pursue the effect of CREB-H on secretion, we next examined its effect on endogenous proteins firstly on an analytical scale after pulsing with 35S-methionine. Cells were induced as before and switched into serum free medium prior to collection, minimizing the large background of serum proteins in the medium. Twenty four hours after induction, cell samples and corresponding media were harvested and total protein or secreted protein profiles examined by autoradiography after SDS-PAGE separation (Figure 5b). Perhaps not surprisingly, we could discern no differences in the complex profile of total synthesized proteins at the resolution of one dimensional gel separation (Figure 5b, lanes 1, 2). However there was a notable difference in the profile of secreted proteins upon Dox induction (Figure 5b, lanes 3, 4). Overall, there was an approximately 3-fold increase in abundance of total secreted proteins when normalized for overall synthetic rate, together with a selective increase in a small number of specific species (lane 4, arrows). To identify these species, the analysis was scaled up and both total and secreted proteins isolated in the absence and presence of CREB-H were analyzed by Coomassie or silver staining (Figure 5c). Consistent with the results from metabolic labelling, we observed a number of bands that were selectively increased in the secreted protein profile after CREB-H induction (Figure 5c, lanes 1, 2; labelled 1–4). These bands were reproducibly observed and represented the only species whose increase was apparent in the total sample by this analysis. Whereas band 4 exhibited a pronounced increase, becoming one of the most prominent species in the total secreted profile, other bands were less discernible and more readily detected on gradient gels with silver staining (right hand panel). Each of the four bands was excised and subject to trypsin digestion and mass spectrometry. Unambiguous identification was made for all of the four bands as Col12A1 (band 1), Nidogen 1 (band 2), SPARC (band 3), and ApoA-IV (band 4). Western analysis (Figure 5d) confirmed the increase in secretion of each of these species though in some cases e.g. Nidogen 1, the increase as judged using this particular antibody in blots did not appear as great as that judged based on total protein staining. Altogether, the data provide convincing evidence for the selective increase in secretion of Col12A1, Nidogen 1, SPARC and ApoA-IV upon induced expression of active CREB-H, with ApoA-IV being the major induced secreted component. Importantly, the genes corresponding to each of these cargos was also identified from the microarray data. These data provide a cohesive series of observations wherein CREB-H induces the transcription of a series of genes in secretory capacity, together with certain cargo genes, thereby promoting increased secretion of cargos with potentially coordinated functions.
CREB-HΔTMC induction of protein secretion in liver cells
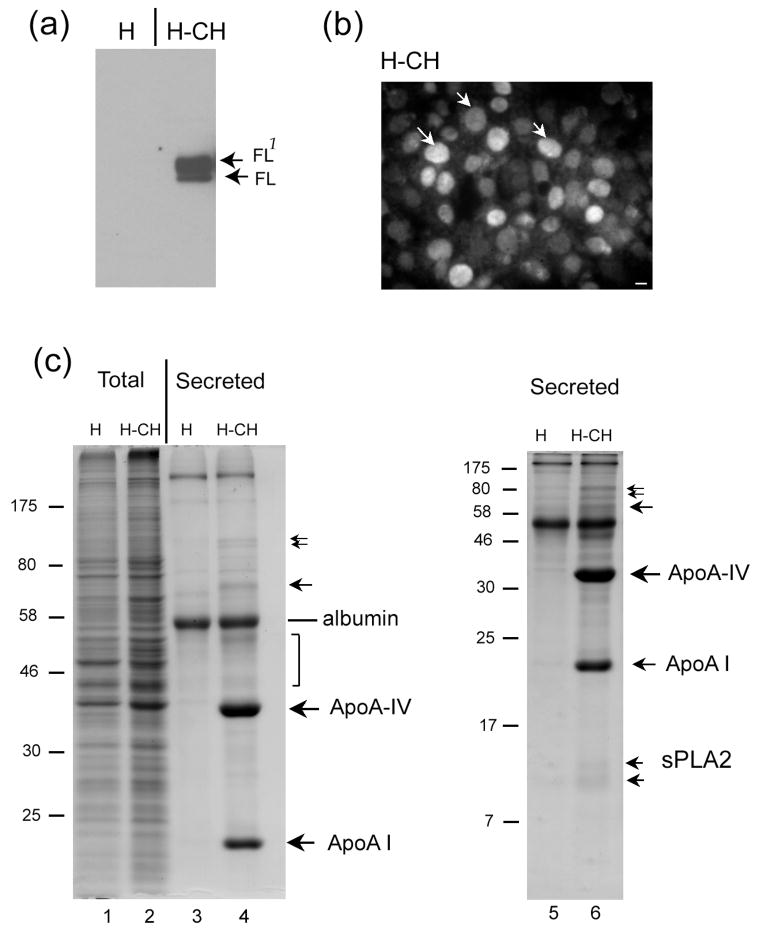
To further establish the effect of CREB-H on protein secretion, we next examined cells in which it is more normally expressed. While present at lower levels in several cell types, CREB-H is detected at high levels in cells of the liver and small intestine. Therefore, we established a liver cell line (HepG2) constitutively expressing the active form. This line (Figure 6a, H-CH) exhibited the typical doublet pattern representing the active form and a phosphorylated form, as seen in the 293 cells, together with pronounced nuclear accumulation (Figure 6b). We then compared total cellular protein content and total secreted proteins from the medium of control HepG2 (H) and H-CH cells (Figure 6c). No significant difference was observed in the total protein profiles (Figure 6c, c.f. lanes 1,2). However there was a striking difference in the secreted protein profiles. Although certain bands remained relatively unaltered, including e.g. albumin, several prominent bands were strongly enriched in the H-CH line (Figure 6c, lanes 3,4 and 5,6, the latter run on a higher percentage gel). Band isolation and mass spectrometry of these bands revealed unequivocal identities for several of these species including ApoA IV, Apo I known components of secreted apolipoprotein complexes, and sPLA2, a secretory phospholipase which acts on phospholipids in cell membranes and on apolipoproteins complexes (42). Our results in liver cells are consistent with those from the inducible 293 cell system, but with differences which likely reflect the fact that CREB-H is mainly expressed in liver and small intestine. The main conclusion here is the demonstration of a consistent theme, in particular in human liver cells, wherein CREB-H activity produces a significant effect on secretory capacity, resulting in selective protein secretion of a distinct class of proteins in this case, apoliproteins and related enzymes. We conclude from these studies that CREB-H increases secretory capacity with selective increases in a distinct class of liver-specific cargos, the apolipoproteins.
Figure 6. Selective protein secretion in liver cells over-expressing nuclear CREB-HΔTMC.
Parental HepG2 cells (H) and HepG2 cells selected for the constitutive expression of CREB-H-ΔTMC (H-CH), were analysed. (a) H-CH cells express the doublet form of CREB-H-ΔTMC identical to that seen with 293Tet-on cells which localized exclusively to nuclei (b). Scale bar, 10 μM. (c) Total cell samples from H or H-CH cells, along with secreted proteins from the corresponding media, were analysed by total protein staining on 10% (lanes 1–4) or 15% (lanes 5,6) SDS PAGE gels. No differences in protein levels could be discerned at this level in the total cell samples. Several proteins, including albumin, remained unchanged in the profile of secreted proteins; however, a number of novel species, including two prominent bands were observed in the medium of H-CH cells. Bands for which unequivocal identifications were made included albumin, Apo A-IV, Apo AI and sPLA2, as indicated.
Evolutionarily conserved configurations of ATB-bZip proteins
Although the existence of five human members of the CREB3 family indicates that there will be distinct variations of a theme, our results defining the ATB domain from sequence conservation and demonstrating function of human CREB3 protein in activation of secretory pathway genes in Drosophila, strongly indicates conserved roles of these proteins. To gain additional insight, we performed sequence analysis across all available databases, searching for potential orthologues based on the presence of the core ATB domain and the invariant defining residues (encompassing the PxxLPL(TS)K indicated above). We identified a number of orthologues with extremely close similarity to the CREB3 proteins, analysis of which allow general principles to be made regarding the family (Figure 7).
Firstly, the ATB domain was found exclusively in a subset of bZip-domain containing proteins and always immediately to the N-terminus of the bZip domain. Secondly, this ATB-bZip configuration was itself found in two major classes, A or B on the one hand, and C. The defining feature of the A/B class (the majority of identified proteins) is the linkage of the ATB-bZip domain to a specific sequence, which conforms in all cases to the transmembrane domain consensus of the CREB3 family (Figure 7, TM, grey bar). Thirdly, those with a TM domain could be divided into two clear subgroupings (A and B classes, Figure 7), dependent upon several co-segregating features (see below). Fourthly, a class of ATB-bZip proteins (class C) that completely conform to the ATB-bZip configuration terminated just adjacent to but upstream of the TM domain. In these cases, it was not that the homology dropped off, but rather that the end of the bZip protein was virtually at the C-terminus of this subclass. This latter class included the Drosophila CREBA species. Finally, with the exception of two Caenorhabditis proteins, all proteins identified in classes A and B, i.e., containing a consensus TM domain, also contain a motif that conforms an S1P cleavage site within approximately 30 residues of the end of the TM region (shaded red).
As indicated, the ATB-bZip-TM class can be divided into groups A and B based on several features as follows. Each of the A class proteins (with fewer representatives in currently available databases) contains an insert between the zipper motif and TM domain. We have labelled this area MP1 (membrane proximal 1) and optimal alignment shows the clear subgrouping dependent on the extended MP1 of class A compared to class B. Strongly reinforcing this subgrouping was the specific co-segregation of features within the TM domain itself. Specifically, in all class A proteins, there was a substitution firstly at TM position 8 (for ease of reference the first conserved cysteine of the TM region is labelled number 1). Position 8 is always a cysteine in class A proteins and always a serine in class B. Coupled to this is the segregation of residues at positions 14 and 15. In all cases, position 15 is a serine in class A proteins and a proline in class B (with a single exception). Residues 14 is always a glycine in class A versus a more bulky hydrophobic residue in class B. This distinction is likely to be particularly relevant in relation to potential qualitative differences in processing of the two groups (see discussion).
With regard to the human CREB3 proteins (shaded yellow) it was now clear that OASIS and BBF2H7 belonged to the A subgroup whereas CREB-H, CREB4 and Luman belong to the B subgroup. Also the class C proteins, which lack the TM region, are somewhat more similar to the class A than the class B proteins in the core ATB-bZip domain. There is generally little homology outside the core ATB-bZip-TM regions, although isolated pockets of homology (data not shown) point to regions worthy of investigation in future comparative examination of family members. We also conclude that the ATB-bZip-TM configuration is a very ancient one, conserved across mammals, to insects, to sea squirt (Ciona), anemones (Nematostella) and present in sponges, the most ancient family in which the ATB-bZip configuration was found. In this latter case, there were two representatives both belonging to the TM group. It is possible therefore that the class C group may have evolved in certain lineages by loss of the TM domain. Features of this analysis taken together with our experimental data provide additional support for our proposals as discussed further below.
Discussion
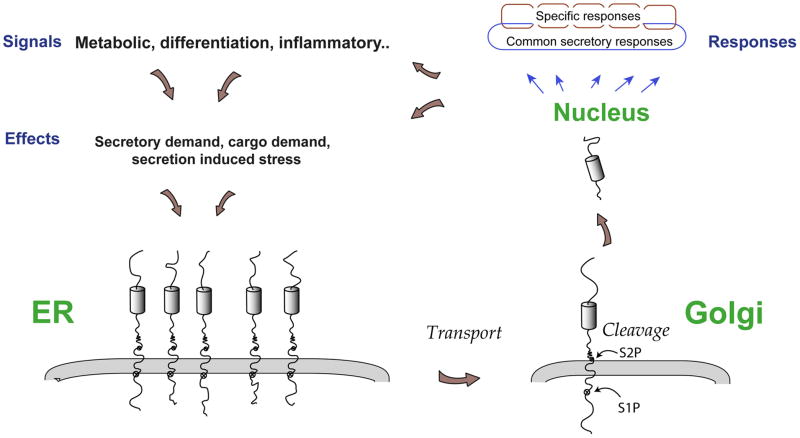
CREB-H is a member of the CREB3 family, a specialised set of ER-anchored bZip transcription factors related to ATF6, a bZip transmembrane factor involved in the unfolded protein response in the ER. Despite this similarity, several lines of evidence indicate that CREB3 family members are regulated in a distinct manner and are likely to be involved in promoting distinct responses to ER stress or demands. However a unifying hypothesis providing a framework for comparative analysis and understanding of the CREB3 family has been lacking. We propose a model (summarised in Figure 8) wherein the core function of these proteins (and their sequence orthologues in other species) is to modulate secretory pathways, coordinating production of specific cargos (which will be distinct in distinct cell types or organisms) with efficient secretory capacity for these cargos and that the ultimate physiological roles of CREB3 proteins are underpinned by these activities. Several lines of evidence provide support for this general hypothesis.
Figure 8. A unifying model proposing a conserved role for CREB3 proteins in responding to potentially diverse signals, leading to increased secretory capacity and cargo secretion.
With regard to secretory functions, responses promoted by CREB3 proteins can be either general and controlled by all CREB3 members, e.g., regulating general flux or supplying limiting factors or be specific for different CREB3 members e.g., increasing distinct limited secretory components required for distinct types of cargos or in different cells. Similarly, apart from genes for secretory components, CREB3 proteins can regulate other targets such as the cargos themselves and again these could be common or distinct. In the response output (top right of diagram), common responses promoted by all family members are indicated by the single blue outline, with responses promoted by individual members superimposed in smaller brown squares. Whereas to date most CREB3 target gene encode either secretory pathway components, secreted cargos or modifying enzymes, additional classes of targets catering to distinct stimuli are likely to also be regulated by CREB3 associated pathways.
ATB-bZip Conservation
CREB3 DNA binding domains are more closely related to the Drosophila melanogaster factor CREBA than to ATF6 or any other bZip factor. We classify a distinct domain, termed the ATB domain shared exclusively between CREBA and the CREB3 family and show it to be critical for transactivation by CREB-H. With respect to specific ATB-bZip organisation, there is no homologue of CREBA in mammalian genomes other than the CREB3 family, increasing further the significance of this feature. This relationship is reinforced by our results indicating that CREB-H target genes are enriched for secretory pathway components. Indeed several individual genes regulated by CREBA in Drosophila (33), such as Sec31a, Sec13, KDELR, ARFGAP3, were also identified in our microarray and qPCR analysis of CREB-H target genes in human cells.
It is likely, notwithstanding the similarity between the CREB3 ATB-bZip domains and the overall themes in target gene induction, that specificity will operate either directly, via differential DNA binding and/or indirectly via selective function of additional cooperative transcription factors. On the other hand, the conservation of the ATB domain, in a region which in other bZip factors is not required for DNA binding (36, 43–45), is strongly indicative of conservation of some aspect of gene targeting or the mechanism of activation. There is an interesting parallel to the ATB domain in the Maf subset of bZip factors, which contain a region termed the EH, flanking the bZip domain (but with no similarity to the ATB domain identified here). The EH domain contributes to DNA binding, probably in a stabilising role rather than conferring specificity (46–48). The precise role of the ATB domain in DNA binding and/or transactivation will be the subject of separate biochemical investigation and side-by-side comparisons with other family members.
CREB3 proteins in secretory activity and cargos in metabolism and ECM
In previous work, certain individual target genes of CREB-H have been reported but with different general conclusions. It was reported that CREB-H could activate certain gluconeogenic target genes (29, 30) though this was not supported in other studies which instead reported activation of a subset of acute phase response genes (27), or upregulation of the iron-binding hormone hepcidin (49). More recent studies of knock-out mice, have revealed CREB-H to be a key factor in metabolic homeostasis of lipids and triglycerides (31, 32). Our results are consistent with these latter studies. In particular, in CREB-H knock-out animals, there is a significant defect in liver expression of several genes which we identify as being upregulated by overexpression of the active form of CREB-H, in particular ApoA-IV and ApoAI. Moreover, in addition to a main phenotype of imbalances in metabolism, several secretory pathway genes including, for example, ADP-ribosylation factor 4a, RAB, RABGAP1l, and Secs including Sec24d, Sec23a and Sec22a were also significantly down-regulated as a consequence of CREB-H loss (GEO GSE29643) (31). Our data add to these previous findings with the demonstration that CREB-H can induce and orchestrate the secretion of high levels of specific cargos in human liver cells, in this case a series of apolipoproteins, and is capable of inducing secretion of at least some of these cargos in cells that otherwise never secrete such proteins. Although not necessary for our general conclusions, it will be very interesting to examine the composition and density profile of apolipoprotein secretion induced by CREB-H in liver cells and to examine any role for CREB-H in secretion in the small intestine, its other main site of expression. For example it may be that CREB-H is involved in chylomicron secretion, whose large particles make unusual demands on secretory capacity.
Our proposal for a general secretory role of CREB3 proteins is also supported by data on other family members including the phenotype of knock-out animals lacking OASIS or BBF2H7 (23, 24) and of mutations in the BBF2H7 homologue in zebrafish (50). Although expressed widely in adult mammalian tissues, the effect of BBF2H7 knock-out in mice was most obvious in cartilage and bone tissue. Particularly in chondrocytes, the ER was abnormally expanded with accumulations of ECM components and defects in collagen secretion and ECM composition (24). Loss of BBF2H7 was accompanied by down regulation of a number of secretory pathway genes, again including e.g., Sec23a, Sec24d and KDELR3. Intriguingly in cell culture, the effects of BBF2H7 loss on ER function and collagen secretion were rescued by Sec23a indicating that a prime function was in remodelling the secretory pathway for ECM cargo secretion and that at least some phenotypic defects were pleiotropic to this role. This conclusion is reinforced by identification of the feelgood mutation in zebrafish, which results in defects in collagen secretion and skeletal formation, as being due to mutation in the zebrafish BBF2H7 homologue (50). Again Sec23a and Sec24d were found to be targets of the zebrafish factor. The main defect reported for OASIS knock-out animals was in bone formation and ECM secretion, in this case reflecting an abnormal ER and reduced ECM and collagen secretion in osteoblasts (23). It is also likely that both these factors function in additional tissues and indeed OASIS has been shown to induce secretory pathway genes such as KDLER3 and Copζ2 and ECM remodelling in pancreatic cells where it is also highly expressed (15, 51, 52). Recent data also indicate that OASIS plays a role in secretion and differentiation of intestinal goblet cells (25). Transcription profiling of CREB3L4, whose expression is high in secretory tissues such as prostate, pancreas and small intestine (18, 19) again identified target genes with a common theme including Sec24d, KDLR2, KDLR3, and ECM components with significant overlap to those identified by us for CREB-H (53). In addition to a strong weighting in secretory pathway genes, there is a notable trend where ECM components are also targets of all of the CREB3 family members, including in this work our demonstration at the protein level of secretion of Col12a, Nidogen 1 and SPARC. These components are known to play a role in cell adhesion and presumably help explain the increased adhesion we observed after CREB-H induction in 293 cells.
Co-segregating features indicative of two subclasses with potential differential processing
With increased numbers of CREB3-like proteins in our analysis, it was clear that the TM class could be segregated into two subclasses. With regard to the MP1 region between the end of the leucine zipper and start of the TM domain, the difference in the two subclasses was not a continuum; rather CREB3 species with a TM domain were either one class or the other. We have shown that this precise region encompasses major determinants of ER retention in CREB-H (21, 22). Furthermore, within TM itself, at position 15, class A proteins always had a serine and class B always a proline (with a single exception). In CREB-H, a class B protein, the proline at this position is critical for cleavage and release of the active form to the nucleus (22). Although the serine presumably permits cleavage in class A proteins, nevertheless the strict distinction strongly suggests potential differences in cleavage mechanism, whether in the enzymes involved or presentation to S2P. Taken together, the distinct subgrouping, firstly in a region shown to influence ER retention/transport and secondly in the TM region, indicate potential differences in regulation and processing between the two classes. Although not the subject of the current work, specific predictions based on these proposals will be tested in future analysis.
In certain species including Drosophila, the class C proteins, which lack a TM domain, activity must be regulated in a distinct manner. Thus, although CREBA, the only class C protein for which we have any information, does indeed regulate secretory pathway genes, it cannot be regulated by pathways dependent upon integral membrane localisation in the ER. It is also possible that CREBA plays multiple roles, not only in responsive, homeostatic controls, but also in other pathways such as differentiation, involving SPGs, or other pathways not limited to SPGs. While the reason for the lack of a TM domain is unclear, the most evolutionary ancient family which we found to contain ATB-BZip proteins was the sponges; in this case the two sponge proteins both contained TM domains. Presumably certain lineages lost the TM domain and although the Drosophila CREBA protein clearly controls secretory function it may have acquired additional roles as a consequence of distinct nuclear localisation.
There is pronounced overlap in the spectrum of genes in the secretion pathway under CREB3 protein control; nonetheless there are also differences in subsets of genes targeted by individual members. It is likely that distinct signals and mechanisms are involved in relaying demand or stress to distinct CREB3 members, though there is currently limited information in this regard. Also whereas most target genes identified to date are either secretory pathway components, secreted cargos or modifying enzymes, CREB3 proteins including TM-containing members may play additional roles outside these activities.
This possibility notwithstanding, taking the following considerations together: 1, that CREB3 factors are anchored in the ER; 2, that they share with CREBA a unique feature of their DNA binding domains, only seen in CREB3/CREBA; 3, that they all function in SPG induction in Drosophila; 4, that independent studies of different family members reveal a consistent theme on target gene involvement in the secretory apparatus and extra-cellular matrix formation; 5, that CREB-H directly induces increased secretion rates and specialised cargo secretion, it is reasonable to propose a general framework for the CREB3 family where their primary role is to orchestrate the secretory apparatus for increased flux though the secretory pathway, potentially with specific regard to specialised cargos or modifying components. We propose that this function underpins their roles in diverse physiological pathways such as promoting differentiation, responding to metabolic fluctuation or potential inflammatory signalling.
Materials and Methods
Isolation of cell lines with regulated CREB-H.ΔTMC expression
293 Tet-On cells (Clontech) were grown in DMEM medium containing 10% Tet-approved foetal calf serum in the presence of G418 at 400 μg/ml. For the inducible expression of the active nuclear form of CREB-H, (CREB-HΔTMC), we constructed a vector based on the doxycycline (Dox) regulated system in pTRE-Tight (Clontech). The vector, pTRE-CREB-HΔTMC (pDJB152), was constructed by inserting a Nhe1-Xba1 fragment containing CREB-HΔTMC from pDJB125 (20) into pTRE-Tight. 293 Tet-on cells were plated in 10 cm dishes in medium containing G418 and transfected with pDJB152 (4 μg/dish), together with the linearized plasmid for puromycin selection pPURO-KAN (0.4 μg/dish). Two days later cells were trypsinised and re-plated in medium containing G418. Puromycin (1 μg/ml) was added 48 hours later. Selection medium was replaced every 4 days and individual G418 and puromycin resistant colonies were isolated and expanded. Cells were screened for Dox-regulated expression of CREB-HΔTMC and selected clonal isolates were amplified for future experiments. In parallel, a control cell line, (293.Con) was selected under the same conditions but not expressing CREB-HΔTMC. Initial titration experiments determined that Dox concentrations of 0.5–1.0 μg/ml yielded optimal expression.
Isolation of a HepG2 cell line constitutively expressing active CREB-H
HepG2 cells were grown in be MEM, 0.1 mM NEAA, 10% FBS, 2 mM L-glutamine and 1 mM sodium pyruvate. We constructed a vector expressing CREB-HΔTMC under the control of the SV40 promoter containing the puromycin resistance marker. The vector, pSVIRES-CREB-HΔTMC (pIF003) was made by inserting the Nhe1-Xba 1 fragment containing CREB-HΔTMC from pDJB125 (20) into the plasmid pIRES-P (provided by Ian Goodfellow). Cells were transfected with 2 μg of DNA/dish. One day later puromycin was added 1 μg/ml, and increased to 1.5 μg/ml subsequently. Individual colonies were then isolated and screened for constitutive expression of CREB-HΔTMC. A clonal isolate (HepG2-CREB-HΔTMC) was used for all experiments.
Cell growth and adhesion assays
Cell number and viability was determined using either an Automated Cell Counter (Countess, Invitrogen) or the Promega Titer-Glo Luminescent Cell Viability Assay Kit measuring ATP release. The direct linear relationship between ATP release and cell number was established in preliminary assays and relative cell numbers were then determined according to the manufacturer’s instructions. For adhesion assays, cells were seeded, induced with Dox and incubated as standard. Cells were then subject to washes with PBS, PBS plus 1 mM EDTA, or a brief trypsin treatment (60 s). Cells remaining after washing were imaged, or detached with trypsin and counted. Quantitations were based in three independent experiments.
Secretion assays
Assays were based on a system for detection of a secreted form of Gaussia luciferase (NEB). We constructed a plasmid expressing the Gaussia luciferase under the control of the herpes simplex virus thymidine kinase promoter. The plasmid pCMVGluc (NEB) was digested with BamHI/XbaI and the appropriate fragment cloned under the control of the TK promoter in the backbone of pDJB125 similarly digested with BamHI/XbaI to create the plasmid pTKGLuc. Control 293 Tet-On cells or 293.CREB-HΔTMC cells were plated (1.5 × 105 cells/well) in six well dishes coated with collagen and transfected 24 h later with 1 μg of pTKGluc. After 24 h, cells were induced with Dox or left untreated. Following a 24 h incubation, cells were washed three times with cold PBS and incubated in fresh medium. Medium was then collected at different time points (0, 2 and 4 h) and analysed for secreted luciferase according to the manufacturer instructions using the Gaussia kit (NEB). Experiments were performed in triplicate for each measure and repeated at least three times.
Analysis of endogenous protein secretion induced by CREB-HΔTMC
293.CREB-HΔTMC cells were plated in poly-l-lysine coated 100 mm dishes. After 24 h cells were either induced with doxycycline (1 μg/μl) or left untreated and allowed to grow for more 48 h. For analytical scale radiolabelling, cells (−/+ Dox induction) were washed (3 x warm PBS, 1x DMEM media lacking methionine and cysteine) and then labelled for 24 h in the same medium containing 50 μCi/ml of EasyTag™ EXPRESS35S Protein Labelling Mix (Perkin Elmer). Media was collected, debris removed by centrifugation, and secreted proteins isolated by TCA precipitation. For total cell samples, the cells were washed with PBS, harvested, and extracted in buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP40, 0.5% DOC) for 30 min at 4°C. Samples were clarified (10,000×g for 10 min) and SDS sample buffer was added to 1X concentration. For larger scale identification of secreted proteins by total protein staining, cells in normal medium containing 10% serum (10 × 100 mm dishes; −/+ Dox) were incubated for 48 h and the media then removed and replaced with serum free media (−/+ Dox) for an additional 24 h to allow the secretion of proteins into the media. The medium was collected (total of 90 ml of media from each sample) and subject to TCA precipitation. Precipitated proteins were resuspended in 400 μl of 2xSDS sample buffer and equal cell equivalents separated by SDS-PAGE. Specific bands were cut from the stained gels and analyzed by mass spectrometry. Total cell samples were prepared as above.
Protein Identification by MALDI mass spectrometry
Following SDS-PAGE, proteins were identified by the PNAC Facility, Department of Biochemistry, University of Cambridge, UK. Briefly, gel bands were excised and subjected to the following treatment (30 min per step, 20°C, in 200 μl 100 mM ammonium bicarbonate/50% acetonitrile): 1, Reduction with 5 mM tris(2-carboxyethyl)phosphine; 2, Alkylation by addition of iodoacetamide (25 mM); 3, Removal of liquid and washing. Gel slices were dried under vacuum for 10 min and 25 μl 100 mM ammonium bicarbonate containing 10 μg/ml modified trypsin (Promega) was added. Digestion was for 17 h at 32°C. Peptides were recovered and desalted using μC18 ZipTip (Millipore) and eluted to a Maldi target plate using 2 μl alpha-cyano-4-hydroxycinnamic acid matrix (Sigma) in 50% acetonitrile/0.1% trifluoroacetic acid. Peptide masses were determined using a MALDI micro MX mass spectrometer (Waters) and analysed with Masslynx software. Database searches of the mass fingerprint data were performed using Mascot (www.matrixscience.com). Where required, Maldi post-source decay analysis was also performed to generate peptide fragment information.
Immunofluorescence studies
Cells were plated on poly-l-lysine coated glass slides placed in plastic tissue culture dishes. Routine immunofluorescence was performed exactly as described previously (21, 22). Primary antibodies were diluted in PBS-10% NCS and applied for 20 min. Primary antibodies used were anti-V5 (1:500, Invitrogen) for the SV5-tag; anti-calnexin polyclonal (1:200, Calbiochem) as a marker for the ER. Fluorochrome (Alexa 488, Molecular Probes or Alexa 543, Pierce) conjugated secondary antibodies of appropriate specificity were used at 1:200.
Western blot analysis
Proteins were analysed by separation either on standard 7.5 %, 10 or 15% SDS-PAGE gels. Proteins were transferred to nitrocellulose membranes which were then blocked with PBS-0.05% Tween20 (PBST) containing 5% non-fat dried milk. After blocking, membranes were incubated with primary antibodies as follows: anti-SV5 (1:10000, Invitrogen), Sec24d (1:1000, gift of Randy Schekman), anti-ApoA-IV (1:1000, 1D6B6, Cell Signalling), anti-SPARC (1:200, 1B2, Sigma-Aldrich), rabbit anti-NID1 (1:500, SAB452006, Sigma-Aldrich), anti-Col12A1(1:200, A-11, Santa Cruz Biotechnology), anti-actin (1:2500, 20–33, A5060 Sigma) or anti-actin (AC-40, A4700 Sigma). Appropriate HP-coupled secondary antibodies were used and blots processed using chemiluminescence detection reagents (Pierce). Alternatively, fluorescently labelled secondary antibodies conjugated to Dylight680 or Dylight800 were used. In the latter case, membranes were blocked in PBS containing 0.5x blocking solution (Licor Biosciences) and target proteins visualized using a LiCor Bioscience Odyssey Infrared Imaging System.
RNA extraction and microarray
Cells were plated in triplicate in six well dishes (2 × 105 cells/well) and incubated with or without Dox (0.5 μg/ml) 24 h after plating. Cells were harvested at different time points and total cell RNA isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The quality of total RNA was assessed on an Agilent Bioanalyser. All samples showed RNA Integrity Numbers greater than 8. RNA (100 ng) was then reverse transcribed, amplified and biotin labelled using random priming (Affymetrix Whole Transcript Kit). Samples were hybridised to Affymetrix Human Gene 1.0 ST arrays, which contain approximately 26 probes per gene. The arrays were washed and stained with streptavidin-phycoerythrin using Affymetrix FS450 Fluidics Stations and imaged on the Affymetrix 3000 7G scanner. Alternatively, RNA levels were analysed using Illumina arrays. Total RNA (100 ng) was in vitro transcribed, amplified and biotin labelled using T7 oligo(dT) (Ambion TotalPrep RNA Amplification Kit) and hybridised to Illumina HT-12 arrays. The arrays were washed and stained with streptavidin-Cy3 and imaged on the Illumina iScan scanner. Data sets of triplicates were analyzed using the Bioconductor package LIMMA with the Benjamini-Hochberg (FDR) test for multiple correction. Genes with an FDR-adjusted p-value of less than 0.05 were considered as differentially expressed. Datasets were also analysed using the ArrayStar software package from DNAStar using the Students t-test and FDR correction with concordant results.
Real Time PCR
Real time PCR was performed with customised Taqman Array Plates (ABI) containing gene specific primers to assess induction of selected genes. cDNA was prepared by reverse transcription of RNA samples (2 μg) using Omniscript Reverse Transcriptase according to the manufacturer’s instructions (Qiagen). The cDNA was adjusted to 100 ng/μl and 100 ng incubated with Taqman Gene Expression Master Mix containing AmpliTAq Gold Ultrapure DNA polymerase in a final volume of 20 μl per reaction. Plates were then analysed on a RealPlex QPCR machine (Eppendorf). Relative increases in RNA were then analysed using the Delta Delta Ct method (54) using GAPDH as the reference calibration gene.
Deletion of the ATB domain
The region flanking the core bZip domain, termed the ATB domain (see text) was deleted by first amplifying a fragment by PCR from pDJB125 (containing CREB-HΔTMC). Primers were designed to extend a fragment from the NheI site at the N-terminus of CREB-HΔTMC and terminate at residue E229 followed by nucleotides corresponding to a BspEI site. Once amplified and digested with NheI and BspEI, the fragment was inserted back into pDJB125 similarly digested with NheI and BspEI, where the natural BspEI site encompasses E257. The result is deletion of residues 230 to 257 within CREB-HΔTMC, resulting in the plasmid pCB10 encoding CREB-HΔTMC.ΔATB. A version of this expressing CREB-HΔTMC.ΔATB under the control of the CMV promoter was also constructed by digesting pCB10 with BamHI and XbaI, and inserting the appropriate fragment into similarly digested pDJB150 (20, 22) to yield the vector pML28.
Promoter activation assays
COS cells were plated in 24-well dishes and transfected with the reporter plasmid (5XATF6-GL3) containing five repeats of the CRE-like UPR/ATF6 element (11). Cell lysates were prepared 24 or 48 h after transfection by addition of “Glo Lysis Buffer” according to the manufacturer’s instructions (Promega). Luciferase activity was determined using the “Bright-Glo” luciferase assay system as described by the manufacturer. For comparison with the active form of CREB-H, we used a vector expressing the active form of ATF6 p3XFLAG-CMV-ATF6. Results of luciferase output were obtained using a Perkin Elmer Victor ELISA plate reader.
Fly Strains
UAS-CREB3L 1 and U A S-CREB3L2 were generated as described previously (33). For the additional UAS- constructs, cDNA sequences for CREB3, CREB-H, CREB-HΔATB and CREB4 were amplified from vectors pJS13, pDJB150, pML28, and pJS85, respectively. The sequence CACC was added to the 5 prime end of the forward PCR primers to facilitate cloning into the pENTRD Gateway cloning entry vector. Following the Gateway LR recombination reaction, cDNA sequences were inserted into the pTW untagged UAS- destination vector (Drosophila Gateway Collection, Carnegie Institution, Baltimore, MD). UAS- constructs were then injected into w1118 (white-) flies (Rainbow Transgenics, Inc., Camarillo, CA) and the white+ eye color insertions from transgenic animals were mapped and balanced to establish stable stocks. engrailed (en)-Gal4 was used to drive expression of all CREB3/CREB3L constructs in epidermal stripes (Weiss et. al., 2001). All Gal4-UAS crosses were performed at 25° C.
In situ hybridizations
In situ hybridizations were performed as previously described (55). Images were obtained using a Zeiss Axiophot microscope (Carl Zeiss, Inc.) configured with a Coolpix 4500 digital camera (Nikon). Images were taken using a Plan-Neofluor 20X, 0.50 NA objective. Images were rotated and cropped so that anterior is always to the left, and ventral is down, using Adobe Photoshop.
Supplementary Material
Acknowledgments
We are grateful to Ron Prywes, University of Columbia for plasmids, Dan Bailey (Health Protection Agency UK) for plasmids used in the early part of this work, Tony Brooks and Sonia Shah, University College London for microarray and bioinformatics analysis, Kathy May for technical assistance and Randy Schekman for antibodies. This work was supported by Marie Curie Cancer Care. Additional support was provided by NIH grants RO1 DE013899 to D.A. and K99 DE021461 to R.F.
References
- 1.Sato R, Yang J, Wang X, Evans MJ, Ho YK, Goldstein JL, Brown MS. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1) J Biol Chem. 1994;269(25):17267–17273. [PubMed] [Google Scholar]
- 2.Duncan EA, Brown MS, Goldstein JL, Sakai J. Cleavage site for sterol-regulated protease localized to a leu-Ser bond in the lumenal loop of sterol regulatory element-binding protein-2. J Biol Chem. 1997;272(19):12778–12785. doi: 10.1074/jbc.272.19.12778. [DOI] [PubMed] [Google Scholar]
- 3.Duncan EA, Dave UP, Sakai J, Goldstein JL, Brown MS. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J Biol Chem. 1998;273(28):17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 4.Hua X, Sakai J, Brown MS, Goldstein JL. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J Biol Chem. 1996;271(17):10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- 5.Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1(1):47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85(7):1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 8.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273(50):33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. JBiol Chem. 2000;275(35):27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Baumeister P, Roy B, Phan T, Foti D, Luo S, Lee AS. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20:5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu R, Yang P, O’Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17(9):5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raggo C, Rapin N, Stirling J, Gobeil P, Smith-Windsor E, O’Hare P, Misra V. Luman, the cellular counterpart of herpes simplex virus VP16, is processed by regulated intramembrane proteolysis. Mol Cell Biol. 2002;22(16):5639–5649. doi: 10.1128/MCB.22.16.5639-5649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honma Y, Kanazawa K, Mori T, Tanno Y, Tojo M, Kiyosawa H, Takeda J, Nikaido T, Tsukamoto T, Yokoya S, Wanaka A. Identification of a novel gene, OASIS, which encodes for a putative CREB/ATF family transcription factor in the long-term cultured astrocytes and gliotic tissue. Brain Res Mol Brain Res. 1999;69:93–103. doi: 10.1016/s0169-328x(99)00102-3. [DOI] [PubMed] [Google Scholar]
- 16.Storlazzi CT, Mertens F, Nascimento A, Isaksson M, Wejde J, Brosjo O, Mandahl N, Panagopoulos I. Fusion of the FUS and BBF2H7 genes in low grade fibromyxoid sarcoma. Hum Mol Genet. 2003;12(18):2349–2358. doi: 10.1093/hmg/ddg237. [DOI] [PubMed] [Google Scholar]
- 17.Omori Y, Imai J, Watanabe M, Komatsu T, Suzuki Y, Kataoka K, Watanabe S, Tanigami A, Sugano S. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver- specific expression. Nucleic Acids Res. 2001;29(10):2154–2162. doi: 10.1093/nar/29.10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi H, Fillion C, Labrie Y, Grenier J, Fournier A, Berger L, El-Alfy M, Labrie C. AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up- regulated by androgens in LNCaP human prostate cancer cells. Cancer Res. 2002;62(3):721–733. [PubMed] [Google Scholar]
- 19.Cao G, Ni X, Jiang M, Ma Y, Cheng H, Guo L, Ji C, Gu S, Xie Y, Mao Y. Molecular cloning and characterization of a novel human cAMP response element-binding ( CREB) gene ( CREB4) J Hum Genet. 2002;47(7):373–376. doi: 10.1007/s100380200053. [DOI] [PubMed] [Google Scholar]
- 20.Bailey D, O’Hare P. Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid Redox Signal. 2007;9:2305–2321. doi: 10.1089/ars.2007.1796. [DOI] [PubMed] [Google Scholar]
- 21.Bailey D, Barreca C, O’Hare P. Trafficking of the bZIP transmembrane transcription factor CREB-H into alternate pathways of ERAD and stress-regulated intramembrane proteolysis. Traffic. 2007;8:1796–1814. doi: 10.1111/j.1600-0854.2007.00654.x. [DOI] [PubMed] [Google Scholar]
- 22.Llarena M, Bailey D, Curtis H, O’Hare P. Different mechanisms of recognition and ER retention by transmembrane transcription factors CREB-H and ATF6. Traffic. 2010;11:48–69. doi: 10.1111/j.1600-0854.2009.00997.x. [DOI] [PubMed] [Google Scholar]
- 23.Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, Saitoh M, Nishimura R, Yoneda T, Kou I, Furuichi T, et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nature cell biology. 2009;11:1205–1211. doi: 10.1038/ncb1963. [DOI] [PubMed] [Google Scholar]
- 24.Saito A, Hino S, Murakami T, Kanemoto S, Kondo S, Saitoh M, Nishimura R, Yoneda T, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Imaizumi K. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nature cell biology. 2009;11:1197–1204. doi: 10.1038/ncb1962. [DOI] [PubMed] [Google Scholar]
- 25.Asada R, Saito A, Kawasaki N, Kanemoto S, Iwamoto H, Oki M, Miyagi H, Izumi S, Imaizumi K. The endoplasmic reticulum stress transducer OASIS is involved in the terminal differentiation of goblet cells in the large intestine. The Journal of biological chemistry. 2012;287(11):8144–8153. doi: 10.1074/jbc.M111.332593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito A, Kanemoto S, Kawasaki N, Asada R, Iwamoto H, Oki M, Miyagi H, Izumi S, Sanosaka T, Nakashima K, Imaizumi K. Unfolded protein response, activated by OASIS family transcription factors, promotes astrocyte differentiation. Nature communications. 2012;3:967. doi: 10.1038/ncomms1971. [DOI] [PubMed] [Google Scholar]
- 27.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic Reticulum Stress Activates Cleavage of CREBH to Induce a Systemic Inflammatory Response. Cell. 2006;124(3):587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 28.Vecchi C, Montosi G, Zhang K, Lamberti I, Duncan SA, Kaufman RJ, Pietrangelo A. ER stress controls iron metabolism through induction of hepcidin. Science. 2009;325:877–880. doi: 10.1126/science.1176639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chin KT, Zhou HJ, Wong CM, Lee JM, Chan CP, Qiang BQ, Yuan JG, Ng IO, Jin DY. The liver-enriched transcription factor CREB-H is a growth suppressor protein underexpressed in hepatocellular carcinoma. Nucleic Acids Res. 2005;33:1859–1873. doi: 10.1093/nar/gki332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MW, Chanda D, Yang J, Oh H, Kim SS, Yoon YS, Hong S, Park KG, Lee IK, Choi CS, Hanson RW, Choi HS, Koo SH. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell metabolism. 2010;11:331–339. doi: 10.1016/j.cmet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Giannikopoulos P, Duncan SA, Wang J, Johansen CT, Brown JD, Plutzky J, Hegele RA, Glimcher LH, Lee AH. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nature medicine. 2010;17:812–815. doi: 10.1038/nm.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Wang G, Zheng Z, Maddipati KR, Zhang X, Dyson G, Williams P, Duncan SA, Kaufman RJ, Zhang K. Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology. 2012;55(4):1070–1082. doi: 10.1002/hep.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fox RM, Hanlon CD, Andrew DJ. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. The Journal of cell biology. 2010;191(3):479–492. doi: 10.1083/jcb.201004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abrams E, Andrew D. CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development. 2005;132:2743–2758. doi: 10.1242/dev.01863. [DOI] [PubMed] [Google Scholar]
- 35.Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 36.Fujii Y, Shimizu T, Toda T, Yanagida M, Hakoshima T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat Struct Biol. 2000;7:889–893. doi: 10.1038/82822. [DOI] [PubMed] [Google Scholar]
- 37.Smolik S, Rose R, Goodman R. A cyclic-amp responsive element-binding transcriptional activator in Drosophila melanogaster, dCREBa, is a member of the leucine zipper family. MolCell Biol. 1992;12:4123–4131. doi: 10.1128/mcb.12.9.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papp S, Fadel M, Michalak M, Opas M. Analysis of the suitability of calreticulin inducible HEK cells for adhesion studies: microscopical and biochemical comparisons. Mol Cell Biochem. 2008;307:237–248. doi: 10.1007/s11010-007-9602-7. [DOI] [PubMed] [Google Scholar]
- 40.DenBoer LM, Hardy-Smith PW, Hogan MR, Cockram GP, Audas TE, Lu R. Luman is capable of binding and activating transcription from the unfolded protein response element. Biochem Biophys Res Commun. 2005;331:113–119. doi: 10.1016/j.bbrc.2005.03.141. [DOI] [PubMed] [Google Scholar]
- 41.Jensen D, Schekman R. COPII-mediated vesicle formation at a glance. J Cell Sci. 2011;124:1–4. doi: 10.1242/jcs.069773. [DOI] [PubMed] [Google Scholar]
- 42.Murakami M, Taketomi Y, Sato H, Yamamoto K. Secreted phospholipase A2 revisited. Journal of biochemistry. 2011;150(3):233–255. doi: 10.1093/jb/mvr088. [DOI] [PubMed] [Google Scholar]
- 43.Landschulz WH, Johnson PF, McKnight SL. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243(4899):1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- 44.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 45.Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 46.Kerppola TK, Curran T. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene. 1994;9:3149–3158. [PubMed] [Google Scholar]
- 47.Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 48.Kurokawa H, Motohashi H, Sueno S, Kimura M, Takagawa H, Kanno Y, Yamamoto M, Tanaka T. Structural basis of alternative DNA recognition by Maf transcription factors. Molecular and cellular biology. 2009;29:6232–6244. doi: 10.1128/MCB.00708-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vecchi C, Montosi G, Zhang K, Lamberti I, Duncan SA, Kaufman RJ, Pietrangelo A. ER stress controls iron metabolism through induction of hepcidin. Science. 2009;325:877–880. doi: 10.1126/science.1176639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melville DB, Montero-Balaguer M, Levic DS, Bradley K, Smith JR, Hatzopoulos AK, Knapik EW. The feelgood mutation in zebrafish dysregulates COPII-dependent secretion of select extracellular matrix proteins in skeletal morphogenesis. Disease models & mechanisms. 2011;4(6):763–776. doi: 10.1242/dmm.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vellanki RN, Zhang L, Guney MA, Rocheleau JV, Gannon M, Volchuk A. OASIS/CREB3L1 induces expression of genes involved in extracellular matrix production but not classical endoplasmic reticulum stress response genes in pancreatic beta-cells. Endocrinology. 2010;151:4146–4157. doi: 10.1210/en.2010-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Omori Y, Imai J, Suzuki Y, Watanabe S, Tanigami A, Sugano S. OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem Biophys Res Commun. 2002;293(1):470–477. doi: 10.1016/S0006-291X(02)00253-X. [DOI] [PubMed] [Google Scholar]
- 53.Ben Aicha S, Lessard J, Pelletier M, Fournier A, Calvo E, Labrie C. Transcriptional profiling of genes that are regulated by the endoplasmic reticulum-bound transcription factor AIbZIP/CREB3L4 in prostate cells. Physiol Genomics. 2007;31:295–305. doi: 10.1152/physiolgenomics.00097.2007. [DOI] [PubMed] [Google Scholar]
- 54.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Lehmann R, Tautz D. In situ hybridization to RNA. Methods in cell biology. 1994;44:575–598. doi: 10.1016/s0091-679x(08)60933-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.