Abstract
The discovery of small noncoding RNAs (sncRNAs) with regulatory functions is a recent breakthrough in biology. Among sncRNAs, microRNA (miRNA), derived from host or virus, has emerged as elements with high importance in control of viral replication and host responses. However, the expression pattern and functional aspects of other types of sncRNAs, following viral infection, are unexplored. In order to define expression patterns of sncRNAs, as well as to discover novel regulatory sncRNAs in response to viral infection, we applied deep sequencing to cells infected with human respiratory syncytial virus (RSV), the most common cause of bronchiolitis and pneumonia in babies. RSV infection leads to abundant production of transfer RNA (tRNA)-derived RNA Fragments (tRFs) that are ~30 nucleotides (nts) long and correspond to the 5′-half of mature tRNAs. At least one tRF, which is derived from tRNA-Glu-CTC, represses target mRNA in the cytoplasm and promotes RSV replication. This demonstrates that this tRF is not a random by-product of tRNA degradation but a functional molecule. The biogenesis of this tRF is also specific, as it is mediated by the endonuclease angiogenin (ANG), not by other nucleases. In summary, our study presents novel information on the induction of a functional tRF by viral infection.
Introduction
During the last decade, significant attention has been directed towards identification of small noncoding RNAs (sncRNAs) and their biological functions (for review, see ref. 1). sncRNAs are 16–35 nucleotides (nts) long and of several classes including microRNA (miRNA), small-interfering RNA (siRNA), piwi-interacting RNA, etc. Among them, miRNA and siRNA are the most extensively studied, and both suppress gene expression by complementary binding to target mRNAs. The role of sncRNAs in regulating antiviral innate immune responses has just emerged and is largely unexplored.2
Discovery of new types of sncRNAs has been greatly facilitated by the development of ultra-high–throughput sequencing (also called next generation sequencing or deep sequencing) technologies. Numerous sncRNAs have been captured and mapped to various origins, such as mRNA (messenger RNA), rRNA (ribosomal RNA), tRNA (transfer RNA), and snoRNA (small nucleolar RNA), indicating that they are derived from these long RNAs. However, their biological significance remains largely unknown. Although a substantial fraction of sncRNAs might be simply degradation intermediates or by-products during RNA turnover, recent studies have demonstrated that some sncRNAs are actually functional molecules with specific biogenesis (reviewed in ref. 3).
sncRNAs derived from tRNAs, also called tRNA-derived RNA Fragments (tRFs), have been recently identified by several research groups (reviewed in ref. 4). There is increasing evidence that they are not by-products from random degradation, but functional molecules. First, tRFs are produced through specific cleavage of tRNA by endonucleases such as angiogenin (ANG), Dicer, and ELAC2.5,6,7,8 Second, tRF expression is regulated either by cellular stresses5,6 or cell proliferation.8 Third, some tRFs are functional, having trans-silencing activity or being necessary for cell proliferation.8,9
Respiratory tract infections are the second leading cause of death worldwide in children <5 years old. The majority of respiratory tract infections are caused by viruses, among which a paramyxovirus called respiratory syncytial virus (RSV) figures prominently (reviewed in ref. 10,11). Nearly all children have had RSV infection by 2–3 years of age.12 RSV infection also increases the morbidity and mortality rate in immunocompromised patients and the elderly, resulting in a substantial health burden.13 There is currently no specific treatment or vaccine for RSV infection. Although prevention strategy using a humanized monoclonal antibody directed to an epitope of the F protein of RSV has been developed, <3% of the at-risk infant population has access to this kind of prevention.14
The role of sncRNAs, especially miRNAs, in host–virus interaction has been documented.15,16 Upon viral infection, host cells alter their sncRNA expression profile as a defense mechanism. On the other hand, viruses can circumvent host defense and promote their propagation by altering cellular sncRNA expression or by expressing their own sncRNAs.17,18,19,20 Nonetheless, most of these studies have focused on the role of miRNAs in viral infection, while other types of sncRNAs remain largely unexplored.
In this study, we have analyzed the global expression profile of sncRNAs in RSV-infected airway epithelial cells by Illumina ultra-high–throughput sequencing in order to identify functionally important sncRNAs. We found that RSV led to tRNA cleavage, resulting in an abundant induction of tRFs. By further investigating the biogenesis and function of RSV-induced tRFs, we found that tRFs are important molecules controlling RSV replication. This is the first report on the role of tRFs in viral replication. This study also provides a potential therapeutic target to control RSV replication, by regulating tRF induction.
Results
Global sncRNA profile in RSV-infected airway epithelial cells
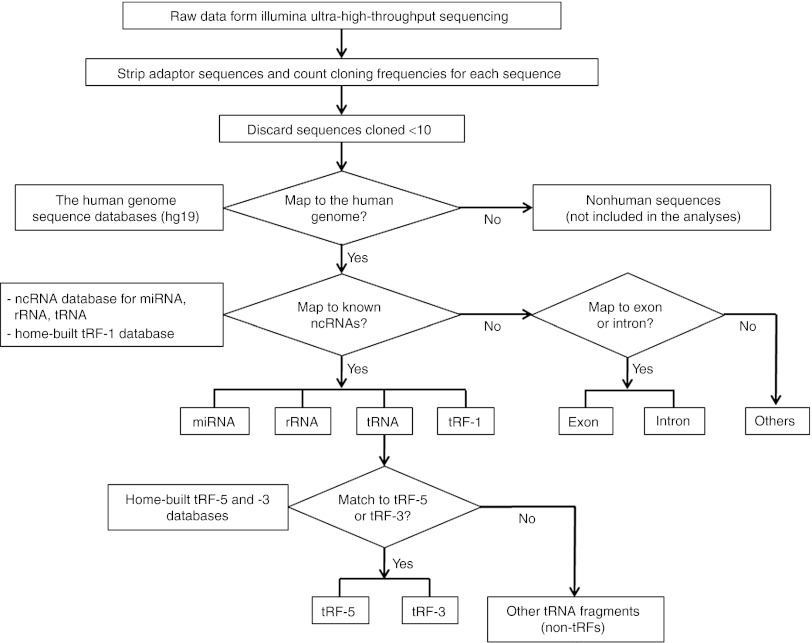
To obtain a global sncRNA expression profile, small RNAs from mock- or RSV-infected A549 cells were subjected to Illumina ultra-high–throughput sequencing. A total of 32,332,590 sequence reads were generated without significant quality issues (Supplementary Figure S1). These raw data were further processed as depicted in Figure 1. After adaptor sequences were stripped, individual sequences with cloning frequency (read numbers) ≥10 were sorted by mapping them to different databases. In brief, the sequences were first mapped to the human genome (hg19; http://hgdownload.cse.ucsc.edu/downloads.html) to eliminate nonhuman sequences, such as those derived from RSV. The filtered human sncRNA sequences were classified by comparing them to the miRNA database (miRBase; http://www.mirbase.org), the rRNA database (RDP; http://rdp.cme.msu.edu/), the tRNA database (GtRNAdb; http://gtrnadb.ucsc.edu/), and the Exon-Intron Database (EID; http://www.utoledo.edu/med/depts/bioinfo/database.html).
Figure 1.
Pipeline of analyses of Illumina high-throughput sequencing data. Flowchart of the sequencing data analyses is depicted. miRNA, microRNA; ncRNA, noncoding RNA; rRNA, ribosomal RNA; tRF, tRNA-derived RNA fragment; tRNA, transfer RNA.
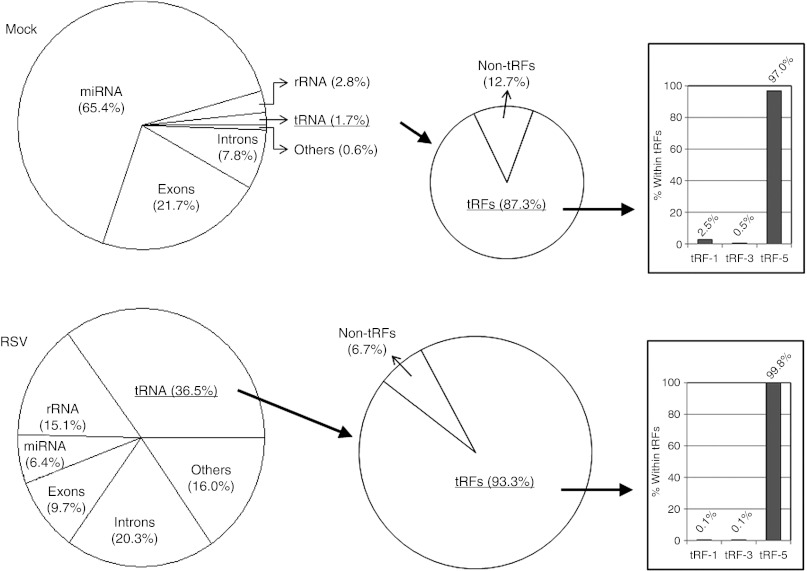
Most of sncRNAs were mapped to miRNAs, tRNAs, rRNAs, or mRNA transcripts, indicating the origin of these sncRNAs (Figure 2). In high-throughput sequencing, the cloning frequency of a sncRNA provides a digital measure of its relative expression level. For fair comparison between mock- and RSV-infected samples, we calculated the relative sequencing frequency of each sncRNA by dividing its raw read numbers by the total read numbers of each experimental group. As shown in Figure 2, a striking difference in sncRNA composition was observed between mock- and RSV-infected samples. In mock-infected cells (those without RSV infection), the majority of sncRNAs (65.4% of total reads) were miRNAs, while sncRNAs mapped to rRNA and tRNA only represented 2.8 and 1.7%, respectively. Following RSV infection, tRNA-derived sncRNAs became the most abundant sncRNAs (36.5% of total reads), while miRNAs were only 6.4%. Except for miRNAs that were processed from their short-lived precursors, other sncRNAs were likely derived from the cleavage of their parental stable RNAs (tRNA, rRNA, etc). However, this cleavage did not seem to be random, as the proportion of individual groups of sncRNA was not equally increased upon RSV infection, and no RNA quality issue was found, as evaluated by 5S rRNA northern hybridization (Figure 3b and other figures shown afterwards). Throughout this paper, we will provide more evidence that sncRNAs from tRNA are not by-products of random RNA degradation.
Figure 2.
Classification of sncRNAs. Human genome-derived sncRNAs were sorted according to their origins, and their relative abundance (calculated from read numbers) was depicted in pie charts (left two charts). tRNA-derived sncRNAs were divided into non-tRFs and tRFs (middle two charts). tRFs were further sorted into tRF-1, 3, and 5 series (bar graphs on the right). The definition of tRFs and the subtypes (-1, -3, and -5 series) are described in the text and depicted in Supplementary Figure S2. miRNA, microRNA; sncRNA, small noncoding RNA; rRNA, ribosomal RNA; RSV, respiratory syncytial virus; tRF, tRNA-derived RNA fragment; tRNA, transfer RNA.
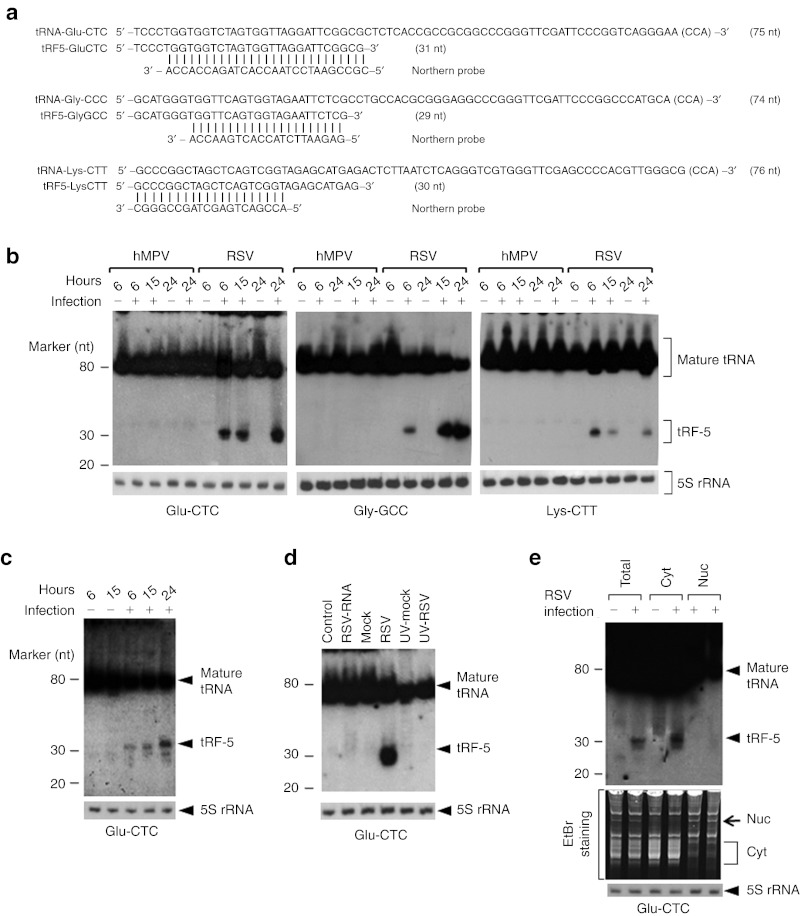
Figure 3.
Experimental validation and characterization of tRF-5s. (a) Sequence alignment of tRF-5s with their parental mature tRNAs and northern probes. The length of each sequence is indicated in parentheses. CCA sequence that is post-transcriptionally added to the 3′-end of tRNA is indicated in brackets. (b) Total RNA from indicated treatments in A549 cells was loaded to a denaturing polyacrylamide gel for northern hybridization using probes indicated in a; 5S rRNA is shown for equal loading. The positions of tRF-5 and mature tRNA are indicated on the right; molecular size markers are indicated on the left. The blot was exposed for ≤8 hours. Data are representative of 2–3 independent experiments. (c) Primary cultured SAE cells were mock- or RSV-infected at MOI of 3 for various lengths of time as indicated. All other experimental conditions were similar to those described in b, except that 4 µg of total RNAs were used for northern hybridization. (d) A549 cells were transfected with purified viral RNAs (lane 2), or infected with naive or UV-inactivated RSV for 24 hours (lanes 4 and 6). Cells treated with Lipofectamine 2000 (lane 1) or mock-infected (lanes 3 and 5) were used as negative controls. Northern hybridization was done as described in b. (e) Total, nuclear, or cytoplasmic RNAs from mock- or RSV-infected A549 cells were subjected to northern hybridization. All other descriptions are the same as in b. Data are representative of 2–3 independent experiments. EtBr, ethidium bromide; hMPV, human metapneumovirus; MOI, multiplicity of infection; nt, nucleotide; rRNA, ribosomal RNA; RSV, respiratory syncytial virus; SAE, small alveolar epithelial cell; tRF, tRNA-derived RNA fragment; tRNA, transfer RNA; UV, ultraviolet.
The enrichment of tRNA-derived sncRNAs following RSV infection
As mentioned earlier, tRNA-derived sncRNAs are an emerging class of sncRNAs that play important biological functions.5,6,8,9 Diverse tRNA-derived sncRNAs have been identified by several research groups, but a consensus nomenclature has not been finalized. Collectively from the literature, two classifications which are not exclusive of each other have been proposed. One is based on their relative location in tRNA (illustrated in Supplementary Figure S2). For those whose sequence precisely matches to the 5′- or 3′-end of mature or precursor tRNAs, they are classified into tRFs. All others are grouped as non-tRFs. Further grouping defines tRF-5 and -3 series depending on which side they are located in the mature tRNA. Another subgroup is the tRF-1 series which corresponds to the 3′-trailer sequences cleaved off from precursor tRNAs during tRNA maturation.8 The other classification is based on their sizes—long tRNA-derived sncRNAs of 30–40 nts (termed as “tRNA halves”) versus short ones less than 30 nts.5,6,21 So far, tRNA halves are known to be induced by cellular stresses and have gained significant attention. They are generated by a single cleavage by an endonuclease from mature tRNA, thus are tRFs.
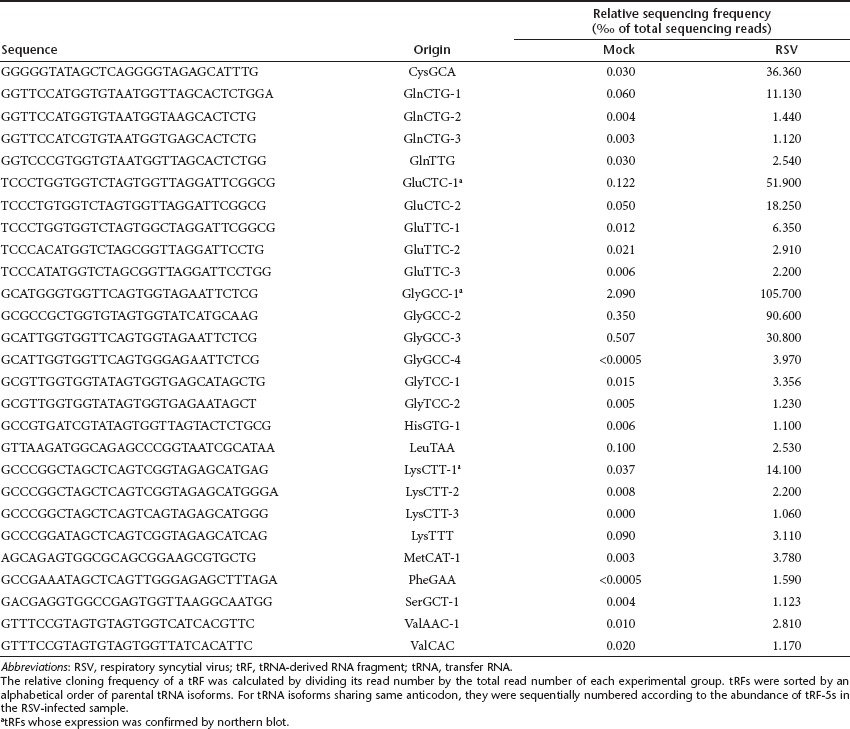
We prefer the “tRF ” nomenclature as this classification is more informative.8 Accordingly, we sorted tRNA-derived sncRNAs by mapping them to our previously built tRF-5, -3, and -1 database.8 As shown in Figure 2, tRFs were dominant over non-tRFs, both in mock- and RSV-infected cells. Notably, the fraction of tRFs increased, while that of non-tRFs significantly decreased (from 12.7 to 6.7%) upon RSV infection. This tendency also argues against the possibility that the induction of tRFs by RSV infection was caused by random RNA degradation. Among tRFs, most of them belonged to the tRF-5 series whereas tRF-3 and -1 series were scarce (Figure 2). Following RSV infection, tRF-5s with a cloned frequency ≥1/‰ (1 per 1,000) are presented in Table 1, and rest of tRF-5s are listed in the Supplementary Table S1.
Table 1. Summary of tRF-5 series with a relative cloning frequency ≥1‰ (reads per mil) in RSV-infected sample.
Expression validation of tRFs by northern blot
We confirmed our sequencing data by northern hybridization which is the most suitable method to validate a newly identified sncRNA. It does not involve any enzymatic treatment and thus visualizes a sncRNA band at its raw size. We chose three tRF-5s that were abundantly cloned after RSV infection (Table 1 and Figure 3a). They were derived from tRNA-Glu-CTC, tRNA-Gly-GCC, and tRNA-Lys-CTT, and thus henceforth named tRF5-GluCTC, tRF5-GlyGCC, and tRF5-LysCTT, respectively.
A band of ~30 nts was readily detectable (Figure 3b), together with a band at ~80 nt representing the corresponding mature tRNA from which each tRF was likely derived. The tRF bands were clearly discrete and prominent along the lane, indicating a specific cleavage event during RSV infection. Most importantly, tRFs of ~30 nts were detected exclusively in the RSV-infected sample. Despite tRNA cleavage, the mature tRNAs were not significantly depleted and their bands kept intense among samples.
Although the band intensity indicated that the tRF-5s were generally much less in quantity than the mature tRNAs, the expression levels of the tRF-5s after RSV infection seemed to be higher than that of the most abundant miRNA, miR-21, based on their cloning frequencies (Supplementary Figure S3a) and the signal intensity in northern hybridization under a similar condition (Supplementary Figure S3b).
Our data also revealed that the induction of tRF-5s was virus specific. Unlike RSV infection, the infection with human metapneumovirus (hMPV), also a pneumovirus of the Paramyxoviridae family, failed to induce tRF5-GluCTC, tRF5-GlyGCC, and tRF5-LysCTT (Figure 3b), further suggesting that tRFs are not random degradation by-products. It is unlikely that the lack of tRF induction by hMPV was due to inefficient hMPV infection, as we and others have demonstrated that A549 cells are permissive to both hMPV and RSV infection, and infected cells exhibit some similar responses including enhanced surface expression of major histocompatibility complex-I.22,23 Collectively, our data indicated that RSV infection induced specific cleavage of tRNA, generating abundant amount of tRF-5s.
Characterization of tRF-5 from tRNA-Glu-CTC
tRF5-GluCTC was chosen for further study, as it was one of the most abundantly cloned tRF-5s (Table 1) and clearly detected by northern hybridization (Figure 3b). We also confirmed that tRF5-GluCTC was inducible by RSV in primary small alveolar epithelial cells (Figure 3c). This observation was in agreement with previous results showing A549 cells as a suitable cell model for investigating the responses of airway epithelial cells to viral infection.24,25
To test whether the induction of tRF5-GluCTC is dependent upon RSV replication, A549 cells were transfected with purified RSV RNA, or infected with live or ultraviolet-inactivated RSV. Untreated- or mock-infected cells were used as controls. Compared with live RSV, neither purified RNA nor ultraviolet-inactivated virus induced tRF5-GluCTC (Figure 3d), suggesting that its induction requires viral replication.
We also determined the subcellular localization of tRF5-GluCTC. Our nuclear/cytoplasmic fractionation indicated that tRF5-GluCTC existed exclusively in the cytoplasm (Figure 3e). The quality of fractionation was assured by western blot of lamin B as a nuclear marker (data not shown)8,26 and ethidium bromide staining of fractionated RNAs. The cytoplasmic RNA was not contaminated by the nuclear RNA and vice versa, as indicated by ethidium bromide-stained bands that were exclusively present in each of the two fractions (designated by arrow and bracket in the bottom panel of Figure 3e).
The molecular function of tRF5-GluCTC
So far, we have demonstrated the specific induction of tRF5-GluCTC (and other tRF-5s) upon RSV infection. A more important question is whether it represents a biologically functional molecule. Given its cytoplasmic localization (Figure 3e), one possibility is that tRF5-GluCTC incorporates into the siRNA/miRNA pathway. It was recently reported that a subset of tRFs exhibit miRNA/siRNA-like trans-silencing capacity.9
To test this possibility, we performed a siRNA/miRNA assay using a sensor plasmid harboring a reverse complementary sequence of tRF5-GluCTC in the 3′-untranslated region of the firefly luciferase gene (Pp) (Figure 4a). If tRF5-GluCTC acts like an siRNA/miRNA, it would recognize the complementary sequence as a target site, therefore leading to suppressed expression of Pp.27 We transfected cells with a cognate sensor plasmid (designated as “Pp-anti_GluCTC_WT”) or a control sensor plasmid lacking the target site (designated as “Pp-vector”), together with a plasmid expressing renilla luciferase (Rr) for normalization. The relative luciferase activity (Pp/Rr values; y-axis in Figure 4b) was calculated and compared between treatments.
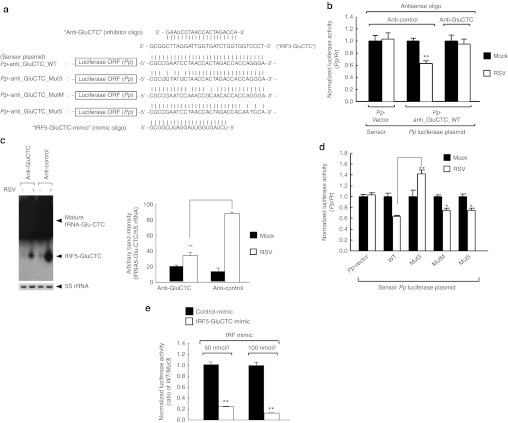
Figure 4.
tRF5-GluCTC exhibits miRNA-/siRNA-like trans-silencing capacity. (a) Sequence alignment of Pp-anti_GluCTC_WT with three mutant luciferase sensor plasmids. Mutated nts are in bold and italic. The sequence of tRF5-GluCTC inhibitor and mimic are also present. (b) A549 cells in hexaplicate were co-transfected with a firefly (Pp) luciferase reporter plasmid (cognate “Pp-anti_GluCTC_WT ” plasmid or “Pp-vector ” plasmid, 0.1 µg/well of 24-well plate), a plasmid expressing renilla (Rr) luciferase, and 120 nmol/l antisense oligos (“anti-GluCTC ” or “anti-control”). After 2 hours post-transfection, cells were mock- or RSV-infected, then harvested at 6 hours post-infection to measure luciferase activities. In all experiments, Pp luciferase values were normalized to renilla (Rr) luciferase values. Values at y-axis (Pp/Rr) are a representative of three independent experiments and are expressed as mean ± SE. **On the second white bar (RSV-infected and Pp-anti_GluCTC transfected) denotes P value <0.01, relative to the second black bar (mock-infected and Pp-anti_GluCTC transfected). (c) A549 cells were treated as described in b and total RNA was subjected to northern hybridization as described in Figure 3b (left panel). Densitometric analysis of tRF5-GluCTC band intensity, quantified using VisionWorksLS image acquisition and analysis software from UVP (Upland, CA) is shown after normalization to 5S rRNA. Data are summarized from three independent experiments (right panel). **P < 0.01, relative to second white bar. (d) A549 cells in hexaplicate were transfected with indicated Pp luciferase reporter plasmids. At 24 hours post-transfection, cells were mock- or RSV-infected, then harvested at 15 hours post-infection to measure luciferase activities. All other descriptions are the same as in b. *P < 0.05, **P < 0.01, respectively, relative to second white bar. (e) A549 cells in hexaplicate were transfected with indicated tRF-mimic oligos. Luciferase plasmids, Pp-anti_GluCTC_WT or _Mut3 (0.1 µg/well), and a plasmid expressing Rr luciferase, were co-transfected. At 40 hours post-transfection, cells were lysed for luciferase assays. Pp values were first normalized by Rr values, and then the Pp/Rr values of “Pp-anti_GluCTC_WT ” were normalized to those of “Pp-anti_GluCTC_Mut3”, yielding relative Pp/Rr values (y-axis). **Denotes P value <0.01, relative to control-mimic (black bars) in respective concentration. All other descriptions are the same as in b. miRNA, microRNA; nts, nucleotides; ORF, open-reading frame; RSV, respiratory syncytial virus; siRNA, small-interfering RNA; tRF, tRNA-derived RNA fragment; tRNA, transfer RNA; WT, wild-type.
Compared with mock infection, RSV infection significantly decreased the relative luciferase activity of “Pp-anti_GluCTC_WT ” but did not affect that of “Pp-vector ” (left two bars versus middle two bars in Figure 4b), suggesting that RSV-induced tRF5-GluCTC could have a trans-silencing activity like miRNA/siRNA. Because RSV infection could lead to pleiotropic effects on the infected cells, we wanted to determine that the observed decrease of luciferase activity was tRF5-GluCTC specific. We inhibited tRF5-GluCTC by transfecting cells with 20-nt antisense oligonucleotides (see Figure 4a for its sequence) which contained a backbone phosphorothioate and 5 nts on each end substituted with 2′-O-methyl ribonucleotides, as described.28,29 Compared with a nontargeting control oligoribonucleotide (designated as “anti-control”), transfection of anti-GluCTC efficiently blocked RSV-induced tRF5-GluCTC (Figure 4c) and accordingly restored the luciferase activity that was decreased by RSV infection (right two bars versus middle two bars in Figure 4b).
Next, we dissected functional domains of tRF-GluCTC for its trans-silencing capacity, by introducing 3–4 nt mutations at the 3′-, middle-, or 5′-region of the tRF-GluCTC target site in the sensor plasmid “Pp-anti_GluCTC ” as depicted in Figure 4a. Contrary to Pp-anti_GluCTC_WT or the other two mutants, the luciferase activity of “Pp-anti_GluCTC_Mut3 ” (“Mut3 ” in Figure 4d) was completely unresponsive to RSV infection, indicating that the 3′-portion of tRF-GluCTC was critical for its trans-silencing activity. The major targeting domain of tRF-GluCTC was different from that of miRNA/siRNA whose 5′-portion (called “seed sequence”) is known to be important for target recognition.1 Thus, the trans-silencing mechanism used by this tRF was distinct from that of canonical miRNA/siRNA.
We also attempted the ectopic expression of tRF5-GluCTC, by designing a synthetic oligoribonucleotide (designated as “tRF5-GluCTC-mimic ” in Figure 4a). Since our data shown in Figure 4d demonstrated that tRF5-GluCTC was distinct from canonical miRNA/siRNA and the 3′-portion of tRF5-GluCTC was critical, we designed a duplex of 20 nts encompassing the 3′- and middle-region of tRF5-GluCTC. The mimic was designed as a duplex form to extend its stability. However, it lacked 2 nt overhangs, the signature end structure of miRNA/siRNA. Upon transfection of this mimic into A549 cells, the luciferase activity of “Pp-anti_GluCTC_WT”, which was normalized by that of “Pp-anti_GluCTC_Mut3”, was significantly suppressed by “tRF5-GluCTC-mimic ” in a dose-dependent manner (Figure 4e). In summary, our data demonstrated that tRF5-GluCTC suppressed target gene expression, through a mechanism distinct from miRNA/siRNA.
tRF5-GluCTC's role in controlling RSV replication
Is there a biological role for tRFs? As we developed methods to suppress or ectopically express tRF5-GluCTC, we could assess cellular phenotype after manipulating this tRF. First, we examined its effect on RSV replication using microplaque immunoperoxidase detection assays to compare the yield of infectious viral particles produced by the cells, which were transfected with anti-control or anti-GluCTC. As shown in Figure 5a, suppression of tRF5-GluCTC led to decreased RSV yield. Two additional experiments, real-time PCR assays for viral gene transcription (Figure 5b) and western blot for viral protein expression (Figure 5c), also supported the result of Figure 5a.
Figure 5.
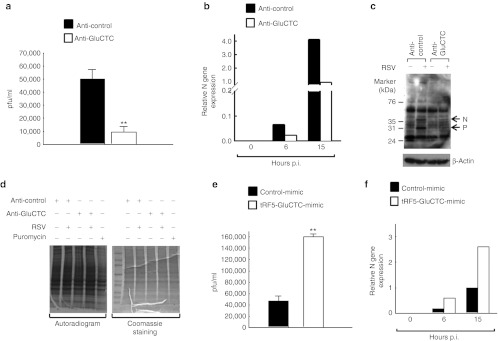
tRF5-GluCTC promotes RSV replication. (a–d). Knockdown of tRF5-GluCTC inhibits RSV replication. Antisense oligos transfection and RSV infection were performed as described in Figure 4b. At 15 hours post-infection (as in a and c) or indicated times post-infection (as in b), viral infectious particles were measured by immunostaining (as shown in a). Viral gene transcription and protein synthesis were investigated by real-time PCR (as shown in b) and western blot (as shown in c), respectively. The total protein synthesis was also assayed for the groups as indicated (as shown in d). (e,f) A549 cells were transfected with 50 nmol/l of indicated mimic oligos. At 6 hours post-transfection, cells were mock- or RSV-infected. At 15 hours post-infection, viral titration and viral gene expression were assayed, respectively. Data are representative of 2–3 independent experiments. **P < 0.01, relative to the black bar. pfu, plaque-forming unit; p.i., post-infection; RSV, respiratory syncytial virus; tRF, tRNA-derived RNA fragment; tRNA, transfer RNA.
Previously, it has been shown that the induction of chemokines and cytokines by RSV infection is viral replication dependent.30 In agreement with the data of Figure 5, the induction of interleukin-8, RANTES, and interferon-β by RSV was significantly decreased, when tRF5-GluCTC was inhibited by anti-GluCTC (Supplementary Figure S4). This result might have been due to side-effects of anti-GluCTC, for instance, possibly targeting the mature tRNA and thereby interfering with cellular global protein synthesis. This possibility was ruled out as we found that global protein synthesis was not affected upon transfection of anti-GluCTC. To validate our assay, we treated cells with puromycin, a known inhibitor for protein synthesis, as a control and observed the inhibition of protein synthesis as expected (Figure 5d, lane 5). Conversely, overexpression of tRF5-GluCTC-mimic significantly enhanced RSV replication, further confirming the stimulatory role of tRF5-GluCTC in RSV replication (Figure 5e,f).
Biogenesis of tRF5-GluCTC
Next, we attempted to investigate how tRF-5s were generated. There are two likely possibilities for the production of tRF-5s; one is that tRF-5s are products of endonuclease cleavage, and the other is that tRF-5s are remnants of 3′-to-5′-exonuclease digestion. As shown in Supplementary Figure S5, the 3′-side fragment was detectable by northern blot using a probe recognizing the 3′-end, supporting the endonuclease mechanism.
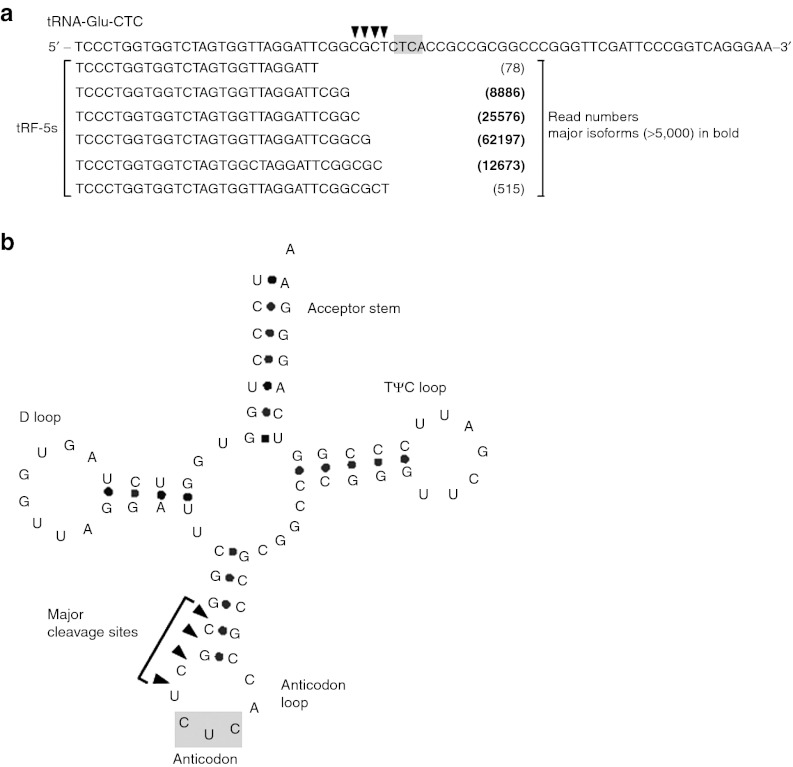
Close inspection of tRF5-GluCTC in the high-throughput data revealed several isoforms with different 3′-ends (Figure 6a and Supplementary Figure S6), which provided a clue to cleavage sites. In the tRNA secondary structure, they were located at the 5′- side of the anticodon loop (Figure 6b). Also, we noticed that the nucleotides before the major cleavage sites were Gs or Cs (Figure 6a and Supplementary Figure S6). These preferences in nucleotide and location further supports the notion that tRFs are not randomly generated, but produced via a specific pathway.
Figure 6.
Cleavage sites in tRNAs to produce tRF5-GluCTC. (a) Alignment of tRF5-GluCTC isoforms to their parental mature tRNA with read numbers listed in parentheses. According to the read numbers of major isoforms (cloned reads >5,000; listed in bold), cleavage sites are designated by downward arrows. The anticodon is highlighted in gray. (b) Major cleavage sites and the anticodon are designated in the tRNA cloverleaf structure. tRF, tRNA-derived RNA fragment; tRNA, transfer RNA.
Ribonucleases targeting the tRNA anticodon loop have been found in both prokaryotes and eukaryotes.31,32,33 Recently, a ribonuclease called ANG has been reported to be responsible for stress-induced cleavage of tRNA at the anticodon loop in mammalian cells.5,6 It is well known that viral infection causes cellular stresses.34,35 To test whether RSV infection produced tRF-5s through ANG induction, we suppressed ANG by a siRNA and measured tRF5-GluCTC in RSV-infected A549 cells. When ANG was suppressed by the siRNA, the induction of tRF5-GluCTC by RSV infection was significantly decreased (>50%) (Figure 7a,b). For comparison, knockdown experiments for other ribonucleases (Dicer, Drosha, ELAC2, and RNaseL) were performed. Dicer and Drosha are required for the generation of miRNAs.1 As mentioned earlier, ELAC2 cleaves precursor tRNA to generate the tRF-1 series (see Supplementary Figure S2). RNaseL is an interferon-induced ribonuclease (reviewed in ref. 36). We found that none of these ribonucleases affected the expression of tRF5-GluCTC upon RSV infection (Figure 7a). Efficient knockdown was confirmed by quantitative reverse transcription (RT)-PCR measurement of each mRNA (Figure 7c) and by western blot selectively for ANG and Dicer (Supplementary Figure S7). Notably, the induction of tRF-5-GluCTC by RSV is independent of Dicer or Drosha, reinforcing that its trans-silencing activity is distinct from that of siRNA/miRNA. Collectively, our data suggest that the production of tRF-5s upon RSV infection specifically required ANG.
Figure 7.
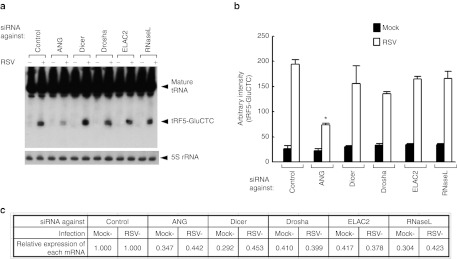
tRF5-GluCTC is generated by angiogenin. (a) A549 cells were transfected with 120 nmol/l of siRNA against indicated proteins or control siRNA as a negative control. At 40 hours post-transfection, the cells were mock- or RSV-infected for 6 hours. Total RNAs were then subjected to northern hybridization as described in Figure 3b (top panel); 5S rRNA expression is shown for equal loading (bottom panel). (b) Densitometric analysis of band intensities of tRF5-GluCTC in a. Band intensity was quantified using VisionWorksLS image acquisition and analysis software from UVP. Band intensity after 5S rRNA normalization are presented as mean ± SE of arbitrary units. *P < 0.05, relative to the first white bar. (c) The suppression of target mRNAs by each siRNA was confirmed by qRT-PCR measurement. The values are the relative expression levels of each mRNA, with the value of control siRNA set as one. ANG, angiogenin; qRT-PCR, quantitative reverse transcription-PCR; RSV, respiratory syncytial virus; siRNA, small-interfering RNA; tRF, tRNA-derived RNA fragment; tRNA, transfer RNA.
Discussion
tRFs are recently identified sncRNAs which have been suggested to regulate cell proliferation, stress-induced translational suppression, and stress granule assembly in mammalian cells.5,6,8 Plants, such as Cucurbita maxima, Arabidopsis thaliana, and Arabidopsis roots, also express tRNA-derived sncRNAs, which are present in the phloem sap or are induced under stress conditions such as oxidative stress and phosphate starvation.37,38 Some microbes such as Streptomyces coelicolor and Tetrahymena thermophila have been reported to produce tRFs in response to starvation,39,40 suggesting universal expression of tRFs among different organisms.
This study is the first report of the induction and function of tRFs in response to virus infection in human airway epithelial cells. We have observed a robust induction of tRNA cleavage upon RSV infection and have provided evidence that tRFs are specifically generated and biologically functional: (i) the induction of tRFs is virus-specific (Figure 3b); (ii) not all tRNAs but a subset of tRNAs are cleaved (Table 1); (iii) cleavage sites are not random but located at the tRNA anticodon loop (Figure 6); and (iv) one tRF has a gene-silencing function, by a mechanism that is distinct from the gene-silencing mechanism of miRNA/siRNA, and promotes RSV replication (Figures 4, 5).
It is known that a miRNA expression profile is cell type-specific and its change in response to viral infections is virus-specific.17 Likewise, we expect differential expression of tRFs in diverse cells/tissues. Previous high-throughput sequencing data captured tRF-5, -3, and -1 series in comparable quantities in prostate cancer cells,8 while tRFs in RSV-infected cells are mostly tRF-5 series. Results shown in Figure 3b also demonstrated that tRF regulation is virus-specific. Given accumulated evidence for the functions of tRFs, future efforts are needed to expand the repertoire of tRFs, which requires accumulation of ultra-high–throughput sequencing data and their proper archive. It should be noted that tRFs were usually filtered away as a routine step in sequencing data analyses. As shown in this manuscript, we are accumulating the knowledge and establishing techniques to explore the functions of tRFs in the context of viral infection, and we expect that these will lead us to define the roles of other tRF-5s, such as tRF5-GlyGCC and tRF5-LysCTT, in response to RSV in the near future.
Recent reports have substantiated that tRNA cleavage is one mechanism of tRF biogenesis. In our data, the majority of tRFs are about 30 nts long and this size range is very similar to that of tRNA halves identified in bacteria, fungi, and plants.39,40,41 In all cases, cleavage sites of these tRFs fall within the anticodon loop. However, we recognized that our tRFs are distinct from previously identified tRNA halves. For example, our cleavage sites are biased towards the 5′-side of the anticodon, whereas the cleavage sites in bacteria, fungi, and plants are distributed throughout the anticodon loop and the TψC arm. In addition, the cleavage enzymes are different. In Saccharomyces cerevisiae, it is Rny1p, an RNase belonging to the RNaseT2 family and induced in response to an oxidative stress.42 In mammalian cells, it is an RNaseA enzyme ANG (Figure 7).5,6
Studies on tRFs are at the very early stage. There are more than 600 tRNA genes in the human genome, and the scale of repertoire of tRFs should be comparable to that of miRNAs. Notably, there are far more tRNA genes than needed for 64 anticodons and 20 amino acids, suggesting that tRNA genes have other functions in addition to their role in protein translation. We speculate that tRFs will be commonly recognized as functional molecules in the future.
In the context of RSV infection, the most important aspect is tRF's biological function. In this study, we have shown that one tRF, tRF5-GluCTC, exhibited trans-silencing capability against target genes (Figure 4). Although such activity by tRF-5s has been reported,9 the tRF-5s therein were ~21 nt thus distinct from our tRF5-GluCTC. How did the 31-nt long tRF5-GluCTC exert a suppressive effect? Given that tRF5-GluCTC was localized in the cytoplasm (Figure 3e), it seemed to recognize complementary sequences in its target mRNAs, but not through the same mechanism used by canonical miRNA/siRNA. First, the 5′-end sequences of its target site (thus the 3′-end of tRF5-GluCTC) was most critical for their interaction (Figure 4a,d), which was opposite to the sequence requirement for miRNA/siRNA. Second, the processing of tRF5-GluCTC did not require Dicer (Figure 7). Third, tRF5-GluCTC was longer than typical miRNA/siRNA. We are currently investigating the molecular mechanism(s) by which tRF5-GluCTC suppressed target gene expression.
Genes involved in host defense to viral infection are functionally important in controlling viral replication.43 Our data demonstrates that tRF5-GluCTC promotes RSV replication. Therefore, it is possible that RSV-induced tRFs (at least tRF5-GluCTC) controls viral replication through suppressing the expression of host defense genes which disfavor virus propagation. Currently, we are searching for tRF5-GluCTC's target genes. This identification will be facilitated by clarifying the molecular mechanism underlying its silencing activity. It is also possible that RSV-induced tRF5-GluCTC regulates RSV replication through targeting mRNAs encoding RSV proteins. However, we think it is unlikely as viral mRNA degradation would inhibit viral replication, which is inconsistent with the results showing the promotion of RSV replication by tRF5-GluCTC (Figure 5). tRF5-GluCTC might interact with the viral genome/antigenome and thereby provide the protection of viral genome/antigenome stability, which might favor the viral replication.
There are a number of other issues to be addressed, including the biogenesis and regulation of tRFs. Is ANG the only nuclease responsible for the generation of tRFs? How is ANG regulated upon RSV infection? In addition to the tRFs identified here, we will expand our investigation to a wider spectrum of tRFs by testing different viruses in diverse host cells. Overall, much more research is needed to understand the role of tRFs in viral infections.
Techniques to express, inhibit, and deliver small RNAs are well developed for siRNA/miRNA-based therapy. For example, nasally inhaled siRNA against RSV genes has been recently shown to offer a fast, potent, and easily administrable antiviral regimen against RSV infection in humans.44,45 Since tRFs, at least for tRF5-GluCTC, promotes RSV infection (Figure 5), coadministration of a siRNA and anti-tRF oligonucleotide may promote stronger antiviral effects, suggesting a potential translational application for this study.
Materials and Methods
Cell lines and virus. HEp-2 cells (ATCC, Manassas, VA) were maintained in minimal essential medium (Invitrogen GIBCO, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and penicillin and streptomycin (100 U/ml). A549, human alveolar type II-like epithelial cells, and 293, a human embryonic kidney epithelial cell line (both from ATCC), were maintained in F12K and minimal essential medium respectively, containing 10% (vol/vol) FBS, 10 mmol/l glutamine, 100 IU/ml penicillin, and 100 µg/ml streptomycin. Primary cultured small alveolar epithelial cells were grown according to the company instructions (ATCC). RSV A2 strain was grown in HEp-2 cells and purified by sucrose gradient as described.46 Viral titer was determined by immunostaining in HEp-2 cells using polyclonal biotin-conjugated goat anti-RSV antibody (Ad direct, Barberton, OH) and streptavidin peroxidase polymer (Sigma, St Louis, MO) sequentially, as described.47
Viral infection. A549 or 293 cells were infected with RSV or hMPV, at multiplicity of infection of 3. An equivalent amount of sucrose solution was added to uninfected cells, as control (mock infection). After initial absorption, viral inoculum was removed and cells were supplied with fresh medium with 2% FBS. Infected cells were scrapped into the medium, followed by sonication and centrifugation. Viruses in the supernatant were harvested and viral titer was determined by immunostaining in HEp-2 cells as described above.
RNA extraction, deep sequencing, RNA mapping, and expression confirmation of tRFs by northern blot. Total cellular RNA was extracted by TRIzol Reagents according to the manufacturer's instruction (Invitrogen, Carlsbad, CA). The RNAs were delivered to Eureka Genomics (Houston, TX) for small RNAs isolation, directional adaptor ligation, cDNA library construction, and sequencing using a Genome Analyzer IIx (Illumina, San Diego, CA). About 485 Mb of sequence data with a total of 32,332,590 sequence reads was generated for mock- and RSV-infected samples, using 36 b single-end sequencing reads. The sequence reads ≥10 after adaptor sequence removal were sent back for further classification. Small RNAs were mapped using Novoalign software (Novocraft Technologies, Selangor, Malaysia) allowing two mismatches.48 After initial alignment, further processing was performed using in-house programs and SAMtools. First, uniquely aligned reads and sequences aligned to more than one genome location (ambiguously aligned reads) were separated. The ambiguously aligned reads were then randomly assigned to one location and combined with uniquely aligned reads for the downstream analysis shown in Figure 1.
Northern hybridization for sncRNAs were performed as described.8 Briefly, RNA was separated in 15% denaturing polyacrylamide gel with 7 mol/l urea and then transferred to a positively charged nylon membrane (Amersham Biosciences, Piscataway, NJ). The membrane was hybridized with 32P-labeled probes in ULTRAhyb-Oligo solution (Life Technologies, Grand Island, NY), followed by washing according to the manufacturer's instruction.
Construction of luciferase sensor plasmids and luciferase assays. The sensor plasmid “Pp-anti_GluCTC_WT ” was constructed by inserting an oligonucleotide, which was complementary to tRF5-GluCTC, into EcoRI/XhoI sites of pcDNA3.1-Zeo(+)-Pp as described.49 Paired primers used for insertion were: 5′-AATTCGCCGAATCCTAACCACTAGACCACCAGGGA-3′ and 5′-TCGATCCCTGGTGGTCTAGTGGTTAGGATTCGGCG-3′ (bold letters represent extra nucleotides to generate EcoRI/XhoI overhangs). Three mutant plasmids were constructed in the same manner (their mutated sequences shown in Figure 4a). An empty pcDNA3.1-Zeo(+)-Pp vector was used as a control (designated as “Pp-vector”).
To investigate the effect of RSV-induced tRF5-GluCTC on mRNA expression, A549 cells were co-transfected with Pp-anti_GluCTC sensor plasmids (firefly plasmids), pRL-CMV plasmids expressing Rr (renilla luciferase), and anti-GluCTC oligonucleotides, using Lipofetamine 2000 according to the manufacturer's instruction (Invitrogen). Pp-vector plasmids and/or anti-control oligonucleotides were used as negative controls. At 2 hours of transfection, the cells were infected with mock or RSV. At 6 hours post-infection, cells were lysed for luciferase assays using a Dual-luciferase kit (Promega, Madison, WI). Data processing and normalization were described in Figure 4. For this experiment, synthetic anti-GluCTC oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). They were 20-nt mixed oligonucleotides which contain a backbone phosphorothioate and have 5 nts on each end substituted with 2′-O-methyl ribonucleotides.28,29 The sequence of “anti-GluCTC ” is shown in Figure 4a and that of “anti-control ” is 5′-CC GCUGAGCTAAAGCCAGCC-3′.
To identify the functional domain(s) of tRF5-GluCTC, A549 cells were co-transfected with Pp-anti_GluCTC sensor plasmids or their mutants, and Rr expressing plasmids for 24 hours, followed by mock or RSV infection (multiplicity of infection of 1). Cells were lysed for luciferase assays at 15 hours post-infection.
For ectopic expression of tRF5-GluCTC, double-stranded RNAs with blunt ends were designed (Figure 4a) and synthesized by Sigma. “Control-mimic ” was purchased also from Sigma. A549 cells were co-transfected with the mimics and luciferase sensor plasmids using Lipofectamine 2000. Cells were harvested for dual-luciferase assays at 40 hours post-transfection. All other descriptions, such as the concentrations of oligonucleotides and plasmids were described in Figures 4e,5e,f.
Protein synthesis assay. Confluent A549 cells, treated with/without antisense oligoribonucleotide and/or RSV infection, were incubated with [35S]-methionine (4 µCi in 2 ml of Dulbecco's modified Eagle's medium containing dialyzed FBS, but no L-glutamine, L-methionine, and L-cystine). Cells were harvested and lysed after 1 hour incubation. The cell lysates were precipitated with 10% trichloroacetic acid, resuspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, and loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel. The gel was stained with Coomassie brilliant blue, dried, and visualized by autoradiography.
RNA interference. siRNAs were purchased from Sigma or Invitrogen; 100 nmol/l of siRNA was transfected into A549 cells, by using Mirus (Madison, WI) according to the manufacturer's recommendations. Forty hours later, A549 cells were mock- or RSV-infected for 6 hours at a multiplicity of infection of 3.
Quantitative real-time PCR. Total cellular RNAs were extracted using TRIzol reagents and viral RNAs released in supernatant were extracted using QIAamp viral RNA kit (Qiagen, Alameda, CA). First strand cDNA was synthesized by using TaqMan RT reagents (ABI, Carlsbad, CA). The RT reaction was performed under the following conditions: 25 °C, 10 minutes; 48 °C, 30 minutes; 95 °C, 5 minutes. Quantitative RT-PCR amplification was performed by using SYBR Sequence information on green-labeled primers for target genes and internal control 18S rRNA is available upon request. Quantitative PCR reactions were performed with the FastStart Universal SYBR Green Master (ROX) (Roche, San Francisco, CA) in the ABI 7500 Sequence Detection System using following conditions: initial steps: 50 °C, 2 minutes and 95 °C, 10 minutes; PCR steps: 95 °C, 15 seconds and 60 °C, 1 minute for 40 cycles. The RT primer to measure the transcription of RSV N gene is: 5′-CTGCGATGAGTGGCAGGCTTTTTTT TTTTTAACTAAAGCTC-3′ primers were designed to incorporate a “tag ” (underlined letters) as part of the assay due to self-priming exhibited by RSV viral RNA.50 The tag sequence was derived from the bacterial chloroamphenicol resistance (Cmr) gene. Sequence with bold letters is complementary to poly A tails of transcribed RSV N gene. Sequence in italics is N gene-specific. At 25 °C annealing temperature, the N-specific sequences would not be sufficient for a stable efficient priming of cDNA from an antigenome of RSV (positive strand). PCR primers for N gene transcription assays are: 5′-ACTACAGTGTATTAGACTTRACAGCAGAAG-3′ (forward) and 5′-CTGCGATGAGTGGCAGGC-3′ (reverse).
Statistical analysis. Statistical significance was analyzed using analysis of variance. P value of <0.05 was considered significant. Mean ± SE is shown.
SUPPLEMENTARY MATERIAL Figure S1. The average sequencing quality score. Figure S2. Diagram of tRFs aligned at a tRNA locus (modified from ref. 8). Figure S3. Northern hybridization of miR-21. Figure S4. Effect of tRF5-GluCTC on cytokine and chemokine secretion. Figure S5. Northern hybridization of 3′-end of GlyGCC tRNA. Figure S6. Cleavage sites in mature tRNAs to produce tRF5 series. Figure S7. Gene silencing of Dicer and ANG. Table S1. Summary of all tRF-5 series.
Acknowledgments
This work was supported by grants from the National Institutes of Health-National Institute of Allergy and Infectious Diseases KAI074829A, the American Lung Association RG232529N, and American Heart Association 12BGIA12060008 to X.B. Authors thank Jonathan Go for the experimental assistance, Betty H Johnson for assistance with manuscript editing, and Cynthia Tribble for manuscript submission. We also thank Antonella Casola and Roberto Garofalo for their comments on the manuscript. The authors declared no conflict of interest.
Supplementary Material
The average sequencing quality score.
Diagram of tRFs aligned at a tRNA locus (modified from ref. 8).
Northern hybridization of miR-21.
Effect of tRF5-GluCTC on cytokine and chemokine secretion.
Northern hybridization of 3′-end of GlyGCC tRNA.
Cleavage sites in mature tRNAs to produce tRF5 series.
Gene silencing of Dicer and ANG.
Summary of all tRF-5 series.
REFERENCES
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Skalsky RL., and, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck AC., and, Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Sobala A., and, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2:853–862. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N.et al. (2010Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly J Biol Chem 28510959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF., and, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW.et al. (2009Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs RNA 152147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A., and, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ., and, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermos CR, Vargas SO., and, McAdam AJ. Human metapneumovirus. Clin Lab Med. 2010;30:131–148. doi: 10.1016/j.cll.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenburg J., and, Boivin G. The distinguishing features of human metapneumovirus and respiratory syncytial virus. Rev Med Virol. 2010;20:245–260. doi: 10.1002/rmv.651. [DOI] [PubMed] [Google Scholar]
- Glezen WP, Taber LH, Frank AL., and, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- Falsey AR, Hennessey PA, Formica MA, Cox C., and, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- Martin JA, Hamilton BE, Sutton PD, Ventura SJ, Menacker F, Kirmeyer S.et al. (2007Births: final data for 2005 Natl Vital Stat Rep 561–103. [PubMed] [Google Scholar]
- Liu X, Wang T, Wakita T., and, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398:57–67. doi: 10.1016/j.virol.2009.11.036. [DOI] [PubMed] [Google Scholar]
- Hess AM, Prasad AN, Ptitsyn A, Ebel GD, Olson KE, Barbacioru C.et al. (2011Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense BMC Microbiol 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss IW., and, Renne R. Viral miRNAs: tools for immune evasion. Curr Opin Microbiol. 2010;13:540–545. doi: 10.1016/j.mib.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölken L, Pfeffer S., and, Koszinowski UH. Cytomegalovirus microRNAs. Virus Genes. 2009;38:355–364. doi: 10.1007/s11262-009-0347-0. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J.et al. (2004Identification of virus-encoded microRNAs Science 304734–736. [DOI] [PubMed] [Google Scholar]
- Reese TA, Xia J, Johnson LS, Zhou X, Zhang W., and, Virgin HW. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol. 2010;84:10344–10353. doi: 10.1128/JVI.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishima K, Sugahara J, Tomita M., and, Kanai A. Sequence evidence in the archaeal genomes that tRNAs emerged through the combination of ancestral genes as 5' and 3' tRNA halves. PLoS ONE. 2008;3:e1622. doi: 10.1371/journal.pone.0001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo RP, Mei F, Manganaro M, Espejo R, Ogra PL., and, Reyes VE. Upregulation of class I major histocompatibility complex (MHC) molecules on respiratory syncytial virus (RSV)-infected airway epithelial cells. Am J Respir Crit Care Med. 1994;149:A987. [Google Scholar]
- Ren J, Kolli D, Liu T, Xu R, Garofalo RP, Casola A.et al. (2011Human metapneumovirus inhibits IFN-ß signaling by downregulating Jak1 and Tyk2 cellular levels PLoS ONE 6e24496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Liu T, Spetch L, Kolli D, Garofalo RP., and, Casola A. Airway epithelial cell response to human metapneumovirus infection. Virology. 2007;368:91–101. doi: 10.1016/j.virol.2007.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Luxon BA, Casola A, Garofalo RP, Jamaluddin M., and, Brasier AR. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J Virol. 2001;75:9044–9058. doi: 10.1128/JVI.75.19.9044-9058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Liu T, Shan Y, Li K, Garofalo RP., and, Casola A. Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog. 2008;4:e1000077. doi: 10.1371/journal.ppat.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ.et al. (2004Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways Cell 11769–81. [DOI] [PubMed] [Google Scholar]
- Yoo BH, Bochkareva E, Bochkarev A, Mou TC., and, Gray DM. 2'-O-methyl-modified phosphorothioate antisense oligonucleotides have reduced non-specific effects in vitro. Nucleic Acids Res. 2004;32:2008–2016. doi: 10.1093/nar/gkh516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideue T, Hino K, Kitao S, Yokoi T., and, Hirose T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15:1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeberle HA, Kuziel WA, Dieterich HJ, Casola A, Gatalica Z., and, Garofalo RP. Inducible expression of inflammatory chemokines in respiratory syncytial virus-infected mice: role of MIP-1alpha in lung pathology. J Virol. 2001;75:878–890. doi: 10.1128/JVI.75.2.878-890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardelt W, Mikulski SM., and, Shogen K. Amino acid sequence of an anti-tumor protein from Rana pipiens oocytes and early embryos. Homology to pancreatic ribonucleases. J Biol Chem. 1991;266:245–251. [PubMed] [Google Scholar]
- Kaufmann G. Anticodon nucleases. Trends Biochem Sci. 2000;25:70–74. doi: 10.1016/s0968-0004(99)01525-x. [DOI] [PubMed] [Google Scholar]
- Suhasini AN., and, Sirdeshmukh R. Transfer RNA cleavages by onconase reveal unusual cleavage sites. J Biol Chem. 2006;281:12201–12209. doi: 10.1074/jbc.M504488200. [DOI] [PubMed] [Google Scholar]
- Akaike T., and, Maeda H. Nitric oxide and virus infection. Immunology. 2000;101:300–308. doi: 10.1046/j.1365-2567.2000.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitko V., and, Barik S. An endoplasmic reticulum-specific stress-activated caspase (caspase-12) is implicated in the apoptosis of A549 epithelial cells by respiratory syncytial virus. J Cell Biochem. 2001;80:441–454. doi: 10.1002/1097-4644(20010301)80:3<441::aid-jcb170>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Castelli J, Wood KA., and, Youle RJ. The 2-5A system in viral infection and apoptosis. Biomed Pharmacother. 1998;52:386–390. doi: 10.1016/s0753-3322(99)80006-7. [DOI] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY.et al. (2009Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing Plant Physiol 1512120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sun L., and, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR., and, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- Haiser HJ, Karginov FV, Hannon GJ., and, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ., and, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM., and, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlier D., and, Lyles DS. Interplay between innate immunity and negative-strand RNA viruses: towards a rational model. Microbiol Mol Biol Rev. 2011;75:468–90, second page of table of contents. doi: 10.1128/MMBR.00007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitko V, Musiyenko A, Shulyayeva O., and, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- Zhang W, Yang H, Kong X, Mohapatra S, San Juan-Vergara H, Hellermann G.et al. (2005Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene Nat Med 1156–62. [DOI] [PubMed] [Google Scholar]
- Guerrero-Plata A, Casola A., and, Garofalo RP. Human metapneumovirus induces a profile of lung cytokines distinct from that of respiratory syncytial virus. J Virol. 2005;79:14992–14997. doi: 10.1128/JVI.79.23.14992-14997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Liu T, Pang L, Li K, Garofalo RP, Casola A.et al. (2011A novel mechanism for the inhibition of interferon regulatory factor-3-dependent gene expression by human respiratory syncytial virus NS1 protein J Gen Virol 92Pt 92153–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N.et al. (2009The Sequence Alignment/Map format and SAMtools Bioinformatics 252078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Kunkeaw N, Jeon SH, Lee I, Johnson BH, Kang GY.et al. (2011Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity RNA 171076–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister R, Rodrigues D, Murray EJ, Laxton C, Westby M., and, Bright H. Use of a highly sensitive strand-specific quantitative PCR to identify abortive replication in the mouse model of respiratory syncytial virus disease. Virol J. 2010;7:250. doi: 10.1186/1743-422X-7-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The average sequencing quality score.
Diagram of tRFs aligned at a tRNA locus (modified from ref. 8).
Northern hybridization of miR-21.
Effect of tRF5-GluCTC on cytokine and chemokine secretion.
Northern hybridization of 3′-end of GlyGCC tRNA.
Cleavage sites in mature tRNAs to produce tRF5 series.
Gene silencing of Dicer and ANG.
Summary of all tRF-5 series.