Abstract
Pulmonary large cell carcinoma - a diagnostically and clinically controversial entity - is defined as a non-small cell carcinoma lacking morphologic differentiation as either adenocarcinoma or squamous cell carcinoma, but suspected to represent an end-stage of poor differentiation of these tumor types. Given the recent advances in immunohistochemistry to distinguish adenocarcinoma and squamous cell carcinoma, and the recent insights that several therapeutically-relevant genetic alterations are distributed differentially in these tumors, we hypothesized that immunophenotyping may stratify large cell carcinomas into subsets with distinct profiles of targetable driver mutations. We therefore analyzed 102 large cell carcinomas by immunohistochemistry for TTF-1 and ΔNp63/p40 as classifiers for adenocarcinoma and squamous cell carcinoma, respectively, and correlated the resulting subtypes with 9 therapeutically-relevant genetic alterations characteristic of adenocarcinoma (EGFR, KRAS, BRAF, MAP2K1/MEK1, NRAS, ERBB2/HER2 mutations and ALK rearrangements) or more common in squamous cell carcinoma (PIK3CA and AKT1 mutations). The immunomarkers classified large cell carcinomas as variants of adenocarcinoma (n=62; 60%), squamous cell carcinoma (n=20; 20%), or marker-null (n=20; 20%). Genetic alterations were found in 38 cases (37%), including EGFR (n=1), KRAS (n=30), BRAF (n=2), MAP2K1 (n=1), ALK (n=3) and PIK3CA (n=1). All molecular alterations characteristic of adenocarcinoma occurred in tumors with immunoprofiles of adenocarcinoma or marker-null, but not in tumors with squamous immunoprofiles (combined mutation rate 50% vs 30% vs 0%, respectively; P<0.001), whereas the sole PIK3CA mutation occurred in a tumor with squamous profile (5%). Furthermore, marker-null large cell carcinomas were associated with significantly inferior disease-free (P<0.001) and overall (P=0.001) survival. In conclusion, the majority (80%) of large cell carcinomas can be classified by immunomarkers as variants of adenocarcinoma or squamous cell carcinoma, which stratifies these tumors into subsets with a distinct distribution of driver mutations and distinct prognoses. These findings have practical implications for diagnosis, predictive molecular testing and therapy selection.
Keywords: large cell carcinoma, TTF-1, ΔNp63/p40, EGFR, KRAS, ALK
INTRODUCTION
Large cell carcinoma is the third most common subtype of non-small cell lung carcinoma after adenocarcinoma and squamous cell carcinoma, representing 3–9% of non-small cell lung carcinomas.1, 2 It is defined in the 2004 World Health Organization classification of lung tumors as an “undifferentiated non-small cell carcinoma that lacks the cytologic and architectural features of small cell carcinoma, glandular or squamous differentiation”.1 In essence, large cell carcinoma is a tumor in which the line of differentiation cannot be identified by light microscopy, as reflected by its alternative designation as “large cell undifferentiated carcinoma”. This designation is reserved for surgically-resected tumors because the lack of morphologic differentiation in small biopsy or cytology samples is usually a reflection of incomplete sampling or poor cell preservation rather than a true lack of differentiating morphology in the entire tumor,1, 3 although this terminology has been applied inconsistently.
Large cell carcinoma has inspired significant controversy over the years, with the main question centered on whether it represents a truly distinct biological entity or an extreme in the poorly-differentiated spectrum of other major types of non-small cell lung carcinoma, namely adeno- and/or squamous carcinomas. The latter concept is supported by the long-known observation that by electron microscopy, large cell carcinomas commonly reveal ultrastructural features of either adeno- or squamous carcinoma.4–6 Similarly, in more recent microarray-based expression profiling studies, these tumors were found to frequently display gene expression patterns resembling either adeno- or squamous carcinoma.7–9 Using the methods employed in diagnostic pathology prior to the era of immunohistochemistry, histochemical stains were found to identify cytoplasmic mucin production in a subset of large cell carcinomas, leading to the recommendation to reclassify such tumors as variants of adenocarcinoma.1, 10 The limitation of mucin stains, however, is that their sensitivity for glandular differentiation is low (~30%),11 and they are therefore variably utilized in routine practice. More recently, it has been noted that by immunohistochemistry, large cell carcinomas commonly express markers typical of adeno- or squamous carcinoma,12–18 raising the prospect that with increasing routine use of immunostains in current pathology practice, large cell carcinoma could become an “endangered species”.13 However, some immunomarkers, previously utilized as “markers of differentiation” in large cell carcinomas, are now known to lack specificity (such as conventional p63 antibody (4A4) and 34βE1211, 15 – the squamous markers, which have a substantial cross-reactivity in lung adenocarcinomas19). Furthermore, no measure of biological accuracy or clinical value of marker-based stratification of large cell carcinoma has been previously demonstrated.
Two recent advances – one in diagnostic pathology and the other in individualized therapy for non-small cell lung carcinomas – make it timely to reassess the feasibility and utility of marker-based reclassification of large cell carcinoma. First, there has been a significant progress in immunomarkers to determine the line of differentiation in non-small cell lung carcinomas. In particular, a notable advance has been recent characterization of ΔN isoform of p63 (p40) as a highly specific squamous marker, unlike the conventional p63 antibody, which in combination with the glandular marker TTF-1 has been shown to reliably distinguish adeno- and squamous carcinomas.20–23 Second, the treatment of patients with non-small cell lung carcinomas has recently undergone a major paradigm shift to a highly individualized approach based on tumor histology and targetable molecular alterations.24 In particular, the recent breakthroughs in targeted therapies have revealed fundamental molecular differences in therapeutically-relevant genetic alterations between adenocarcinoma (e.g. EGFR,25 KRAS,25 ALK26 and BRAF27 mutations) and squamous cell carcinoma (e.g. PIK3CA mutations and several other recently-described genetic alterations),28 which forms the basis for a recommendation to employ predictive molecular tests differentially in patients with these tumors.29 Given the uncertainty with the diagnostic approach and paucity of studies focused on large cell carcinomas, the use of individualized therapies in patients with these tumors is not well-established. In particular, there is little molecular data to inform a strategy for predictive molecular testing in patients with these tumors. While several studies did include a small number of large cell carcinomas, and reported on the presence of EGFR (4%)30 and KRAS (8–30%)31–34 mutations in these tumors, a comprehensive screen for driver mutations in a large series of large cell carcinomas has not been performed. Furthermore, it has not been explored whether the recent improvement in immunomarkers could translate into a more biologically-precise classification of large cell carcinomas, which could inform the selection of predictive molecular tests in patients with these tumors.
Given the above considerations, the goals of this study were to 1) establish the overall rate of targetable mutations in large cell carcinoma, 2) determine whether the distribution of these mutations can be predicted by immunophenotyping, and 3) explore whether immunomarker-defined subsets of large cell carcinoma have distinct clinicopathologic characteristics. We therefore evaluated 102 large cell carcinomas by immunohistochemistry for TTF-1 and ΔNp63 as classifiers for adeno- and squamous carcinoma, respectively, and correlated the resulting subtypes with 9 therapeutically-relevant genetic alterations (EGFR, KRAS, BRAF, MAP2K1, PIK3CA, NRAS, AKT1, ERBB2 and ALK) as well as various clinicopathologic parameters.
MATERIALS AND METHODS
Study design
The study was performed with approval of the Institutional Review Board of the Memorial Sloan-Kettering Cancer Center, New York. A total of 102 large cell carcinomas were identified in the archives during the period of 1999–2011, after exclusion of 11 cases with unavailable or insufficient material for all assays in this study (this represents 2.2% of a total of 5267 resected non-small cell lung carcinomas at our institution during that period). Large cell carcinomas were defined as surgically-resected non-small cell lung carcinomas lacking the morphologic evidence of glandular, squamous or neuroendocrine differentiation by light microscopy. Mucin special stains were not used as part of inclusion or exclusion criteria in this study. Large cell neuroendocrine carcinomas and sarcomatoid (entirely spindle or giant cell) carcinomas were not included. All cases were reviewed by 2 thoracic pathologists (NR, ALM) to confirm the absence of overt morphologic differentiation in all tumors. A representative formalin-fixed paraffin embedded tumor block was selected for each case, and used for immunohistochemistry, molecular and cytogenetic studies, as described below.
Immunohistochemistry
Immunohistochemistry was performed on a Ventana Discovery XT automated stainer (Ventana Medical Systems) as previously described.18,30 Briefly, primary antibodies included ΔNp63/p40 (CalBiochem, 1:2000 dilution) and TTF-1 (SPT24 clone, NovoCastra, 1:100 dilution). Percentage of immunoreactive tumor cells in each tumor was recorded. Based on prior studies, any reactivity for TTF-1 was considered as positive, whereas positivity for ΔNp63 was defined as reactivity in >10% of tumor cells.20, 21 Additional immunostains were performed at the time of diagnosis or as part of this study, as needed, to exclude the possibility of unsuspected metastasis from extra-pulmonary sites, and/or other epithelioid neoplasms, such as melanoma, sarcoma or large cell lymphoma.
Mutation Analysis
DNA extraction
Tumor areas were macrodissected from 10 unstained 5-um thick sections of FFPE tissue to ensure >50% tumor cellularity. Genomic DNA was extracted using the DNeasy Tissue kit (QIAGEN). Extracted DNA was quantified on the NanoDrop 8000 (Thermo Scientific).
Sequenom mass spectrometry genotyping and Sanger sequencing
All cases were genotyped by Sequenom Mass ARRAY system (Sequenom Inc) for 92 hot-spot point mutations in 8 oncogenes: EGFR, KRAS, BRAF, PIK3CA, MAP2K1 (MEK1), NRAS, AKT1, and ERBB2 (HER2), as described in detail previously.25 Samples were tested in duplicate using a series of 6 multiplexed reactions. Briefly, genomic DNA amplification and allele-specific single base extension reactions were performed using primers designed with the Sequenom Assay Designer v3.1 software system (Sequenom Inc). The extension products were quantitatively analyzed using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) on the Sequenom MassArray Spectrometer. Cases with equivocal Sequenom results upon manual review were retested in duplicate by standard sequencing with and without Locked Nucleic Acid oligonucleotide for confirmation.35
EGFR exon 19 fragment analysis
Cases lacking mutations other than PIK3CA by Sequenom were tested in duplicate for EGFR exon 19 deletions/insertions by fragment sizing assay, as previously described.25 Briefly, a 207-bp genomic DNA fragment encompassing the entire exon 19 was amplified using fluorescently-labeled primers, and PCR products were detected by capillary electrophoresis on an ABI 3730 Genetic Analyzer.
Fluorescent in situ hybridization (FISH) for ALK rearrangements
Cases lacking mutations other than PIK3CA by the above methods were further tested for ALK rearrangements by dual color break-apart FISH (Vysis/Abbott Molecular) according to the manufacturer’s recommendations. Briefly, 4um-thick tissue sections were pretreated by deparaffinization in xylene and dehydration in ethanol. FISH analysis and signal capture were performed on fluorescence microscope (AXIO, Zeiss) coupled with ISIS FISH Imaging System (Metasystems). At least 50 interphase nuclei from each tumor were scored, and a sample was considered positive for ALK rearrangement if >15% of tumor cells displayed broken-apart green/red signals and/or single red signals.
Statistical analysis
Mutation frequencies and clinicopathologic parameters were compared using Fisher exact or Kruskal-Wallis test. Disease-free and overall survival was estimated using Kaplan-Meier method with time origin at the time of surgery. Median (range) of available follow-up was 30 (1–120) months. Group comparisons were performed using log-rank test. Statistical analysis was conducted using SAS version 9.2 (SAS Institute Inc) and the clinfun package in R (http://www.r-project.org/).
RESULTS
Tumor and patient characteristics
Clinical characteristics of 102 patients with large cell carcinomas were as follows: age median (range) 63 (37–89), female n=51 (50%), never smoker n=7 (6%), and smoking pack-years median (range) 40 (0–126). Stage distribution was as follows: stage I n=39 (38%), stage II n=35 (34%), and stage III/IV n=28 (27%). Surgical procedures included wedge resection or segmentectomy (n=25), lobectomy or bilobectomy (n=66) and pneumonectomy (n=11). Morphologic review confirmed the lack of overt glandular, squamous or neuroendocrine differentiation in all tumors. Variant morphologies included basaloid (n=7; 1 focally, 6 diffusely), clear cell (n=5; 4 focally, 1 diffusely), rhabdoid (n=3; 2 focally, 1 diffusely), and with focal spindle and/or giant cells (n=14). The rest were classic large cell carcinomas, not otherwise specified (n=73).
Immunomarker-defined subsets of large cell carcinoma
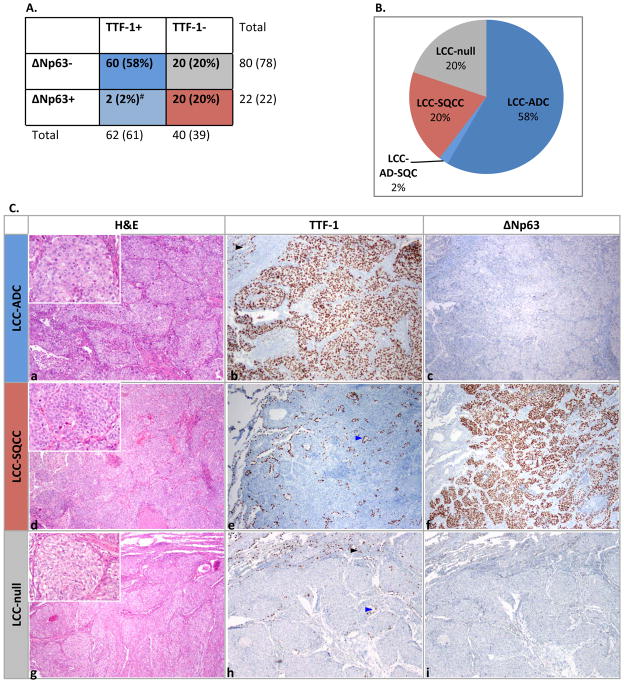
Immunohistochemistry for ΔNp63 and TTF-1 revealed the following immunoprofiles (Figure 1A): 1) ΔNp63−/TTF-1+ (n=60), 2) ΔNp63+/TTF-1− (n=20), 3) ΔNp63+/TTF-1+ (n=2; each markers labeled a distinct cell subpopulation), and 4) ΔNp63−/TTF-1− (n=20). Based on these immunoprofiles, tumors were classified as variants of 1) adenocarcinoma, 2) squamous cell carcinoma, 3) adenosquamous carcinoma, and 4) marker null, respectively (Figure 1B). Expression of TTF-1 in group 1 and ΔNp63 in group 2 was typically seen in the majority of tumor cells: mean ± standard deviation for percentage of tumor cells immunoreactive for TTF-1 or ΔNp63 in those groups was 90±25% (range 10–100%) and 92±14% (range 50–100%), respectively. Examples of microscopic findings are illustrated in Figure 1C. Because of the previously shown similarity of adenosquamous carcinomas to adenocarcinomas in terms of driver mutations and clinicopathologic characteristics,36, 37 the former group was merged with the latter for further analysis.
Figure 1.
Immunohistochemistry-defined subtypes of large cell carcinoma. A. Coexpression profiles of TTF-1 and ΔNp63 (p40). # TTF-1 and ΔNp63 labeled distinct cell populations. B. Pie chart showing TTF-1/ΔNp63 -based subtypes of large cell carcinoma. C. Examples of microscopic findings. H&E shows morphologically-indistinguishable non-small cell carcinomas, all growing as entirely solid nests or sheets of tumor cells with no evidence of either glandular or squamous differentiation. Despite the lack of differentiating morphology, marker profiles provide evidence of submorphologic differentiation as adenocarcinoma (a–c) or squamous cell carcinoma (d–f); g–i illustrates a marker-null large cell carcinomas. Benign pneumocytes (TTF-1+) are seen at the tumor periphery (black arrowheads) or entrapped within the tumor (blue arrowheads). Insets in a, d and g show higher-power images. Abbreviations: ADC adenocarcinoma, AD-SQC adenosquamous carcinoma, LCC large cell carcinoma, SQCC squamous cell carcinoma
Distribution of driver mutations in immunomarker-defined subsets of large cell carcinoma
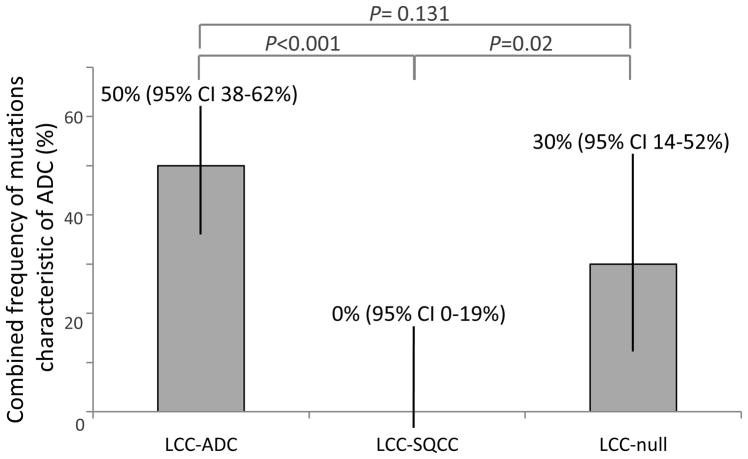
Molecular and cytogenetic analysis of 102 large cell carcinomas revealed that 38 cases (37%) harbored non-overlapping mutations in EGFR (n=1), KRAS (n=25), BRAF (n=2), MAP2K1 (n=1), PIK3CA (n=1) and ALK rearrangements (n=3) (Table 1). All mutations characteristic of adenocarcinoma (EGFR, KRAS, BRAF, MEK1, and ALK) occurred in large cell carcinomas with glandular immunoprofiles or in marker-null tumors but not in tumors with squamous profiles. Combined rate of adenocarcinoma-specific mutations in the above groups was 31/62 (50%) vs 6/20 (30%) vs 0/20 (0%), respectively (P<0.001). The sole PIK3CA mutation occurred in a tumor with a squamous immunoprofile (1/20; 5%). As illustrated in Figure 2, the combined frequency of mutations characteristic of adenocarcinoma was significantly different between large cell carcinomas with adeno- vs squamous (P<0.001) and null vs squamous (P=0.02), but not between adeno- vs null (P=0.13) immunoprofile. KRAS mutations had a 5:1 ratio of smoking-related transversion mutations (G12V, G12C, G13R, Q61H) to transition mutations (G12D, G12S), respectively – a ratio similar to the one found in lung adenocarcinomas in our patient population.38
Table 1.
Distribution of mutations in immunomarker-defined subtypes of large cell carcinoma.
| Immunomarker-defined subtype of LCC | All cases (n=102) | |||
|---|---|---|---|---|
| LCC-ADC† (n=62) | LCC-SQCC (n=20) | LCC-null (n=20) | ||
| EGFR | 1 (2%): Exon 19 18 bp Δ | 0 | 0 | 1 (1%) |
| KRAS | 25 (40%)
|
0 | 5 (25%)
|
30 (29%) |
| BRAF | 1 (2%): G469A | 0 | 1 (5%): V600E | 2 (2%) |
| MEK1 | 1 (2%): K57N | 0 | 0 | 1 (1%) |
| PIK3CA | 0 | 1 (5%): E542K | 0 | 1 (1%) |
| NRAS | 0 | 0 | 0 | 0 |
| AKT1 | 0 | 0 | 0 | 0 |
| HER2 | 0 | 0 | 0 | 0 |
| ALK¥ | 3 (5%) | 0 | 0 | 3 (3%) |
| Any mutation | 31 (50%) | 1 (5%) | 6 (30%) | 38 (37%) |
includes 2 tumors with profiles of adenosquamous carcinoma, of which one harbored a KRAS G12C mutation
no ALK gene rearrangement results available for 4 cases due to FISH failure
Abbreviations: LCC large cell carcinoma, ADC adenocarcinoma, SQCC squamous cell carcinoma
Figure 2.
Distinct distribution of driver mutations in immunomarker-defined subtypes of large cell carcinoma. Shown is a combined frequency of genetic alterations characteristic of adenocarcinoma (EGFR, KRAS, BRAF, MAP2K1, and ALK) ± 95% confidence intervals (CI).
Large cell carcinomas harboring genetic alterations in EGFR or ALK
The sole patient with a tumor harboring an EGFR mutation was a 53 year old woman, whose resected primary lung tumor was morphologically a classic large cell carcinoma - an entirely solid/undifferentiated non-small cell carcinoma with no microscopic evidence of glandular or squamous differentiation. Because of the resemblance of this solid morphology to squamous histology, this tumor was initially interpreted as squamous cell carcinoma, and this patient was therefore also included in our recent series on EGFR mutations in tumors mimicking squamous cell carcinomas [patient ID 12 in ref.25]. By immunohistochemistry, this tumor was TTF-1+/ΔNp63−, supporting adenocarcinoma lineage. Nineteen months after surgery the patient developed brain metastases and was treated with erlotinib. She showed a marked radiologic response with near-complete regression of the brain lesions.
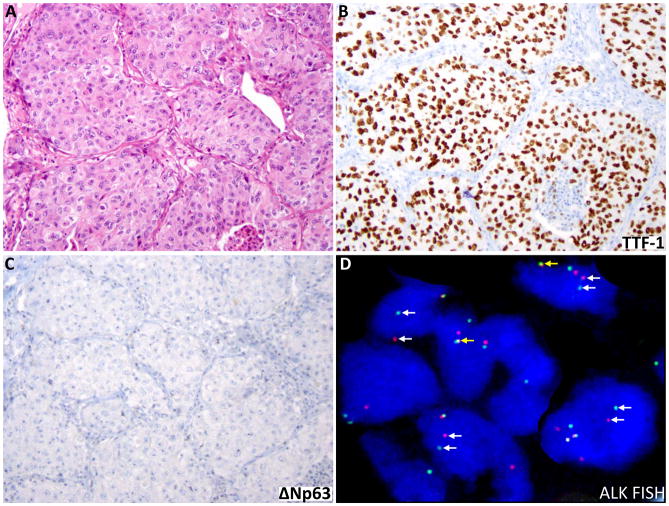
All 3 patients with ALK rearrangements were either never (n=2) or light (n=1; 0.7 pack-years) smokers and were younger (age 60, 52 and 48 years) than the median age of 63 years for patients in this series. The tumors were morphologically classic large cell carcinomas, in which immunoprofiles similarly revealed glandular lineage (TTF-1+/ΔNp63−). Crizotinib response data are not available for these patients. An example of ALK-rearranged large cell carcinoma is illustrated in Figure 3.
Figure 3.
An example of ALK-rearranged large cell carcinoma. A. While there is no evidence of morphologic differentiation by H&E (A), positive TTF-1 (B) and negative ΔNp63 (C) immunostains support glandular lineage. (D) Split red and green signals (white arrows) indicate the presence of ALK rearrangement, whereas the native ALK allele is detected as merged red and green signals yielding a yellow color (yellow arrows).
Clinicopathologic characteristics of immunomarker-defined subsets of large cell carcinoma
As shown in Table 2, a comparison of clinicopathologic characteristics between large cell carcinomas with adeno- vs squamous vs null immunoprofiles did not reveal significant differences in the analyzed parameters (age, gender, smoking, tumor size, and stage), although patients with squamous profiles tended to be older, and had an invariable smoking history in contrast to the occurrence of rare never smokers in the other groups. Of morphologic variants, the only preferential association was between basaloid features and squamous immunoprofile, but this analysis is limited by a small number of cases in each subgroup with variant morphology.
Table 2.
Clinicopathologic features by immunomarker-defined subtype of large cell carcinoma.
| LCC-ADC† (n=62) | LCC-SQCC (n=20) | LCC-null (n=20) | P value | |
|---|---|---|---|---|
|
| ||||
| Age: median (range) | 62 (41–86) | 71 (37–89) | 62 (45–81) | 0.07 |
|
| ||||
| Gender: n (%) | ||||
| Female | 32 (52) | 10 (50) | 9 (45) | 0.96 |
| Male | 30 (48) | 10 (50) | 11 (55) | |
|
| ||||
| Smoking status: n (%) | ||||
| Never | 6 (10) | 0 | 1 (5) | 0.43 |
| Current/former | 56 (90) | 20 (100) | 19 (95) | |
|
| ||||
| Smoking pack-years*: median (range) | 45 (0–110)‡ | 47 (1.5–90) | 40 (0–130) | 0.81 |
|
| ||||
| Tumor size, cm: median (range) | 3.5 (0.4–12) | 3.5 (1.1–9.5) | 3.1 (0.9–9.2) | 0.90 |
|
| ||||
| Stage: n (%) | ||||
| I | 19 (15) | 12 (60) | 8 (40) | 0.18 |
| II | 26 (42) | 6 (30) | 6 (30) | |
| III/IV | 17 (27) | 2 (10) | 6 (30) | |
|
| ||||
| Morphologic variants: n (%) | ||||
| Basaloid | 1 (2) | 4 (20) | 2 (10) | 0.08 |
| Clear cell | 3 (5) | 1 (5) | 1 (5) | |
| Rhabdoid | 1 (2) | 0 | 2 (10) | |
| Focal giant or spindle cells | 9 (15) | 3 (15) | 2 (10) | |
| Not otherwise specified | 48 (77) | 12 (60) | 13 (65) | |
includes 2 tumors with profiles of adenosquamous carcinoma
pack years = number of packs of cigarettes smoked per day X years of smoking
number of pack-years not available for one smoker
Abbreviations: LCC large cell carcinoma, ADC adenocarcinoma, SQCC squamous cell carcinoma
As shown in Figure 4, survival analysis revealed that, remarkably, marker-null group had a dismal 5-year disease-free and overall survival of 9% and 12%, respectively, compared to 55% and 56%, respectively, for tumors with adenocarcinoma profiles, and 46% and 47%, respectively, for tumors with squamous profiles. The inferior survival of marker-null compared to marker-positive groups was statistically significant (P<0.001 for median disease-free and P=0.001 for median overall survival), and remained significant after stage stratification. For stage I tumors with adenocarcinoma profiles, 5-year disease-free survival was 61% (95% confidence intervals 39% – 96%).
Figure 4.
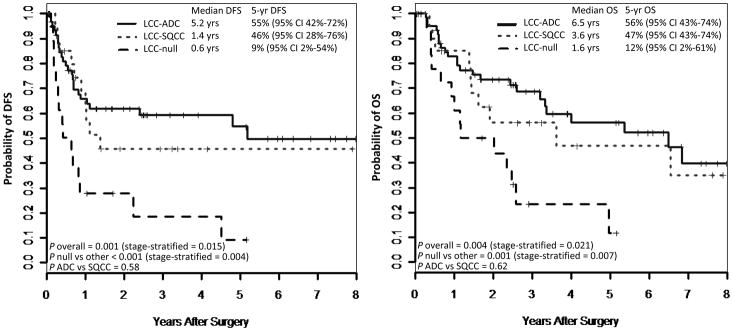
Survival by immunomarker-defined subtype of large cell carcinoma: an adverse prognosis associated with marker-null carcinomas. Abbreviations: DFS disease-free survival, OS overall survival, CI confidence intervals.
DISCUSSION
The present data demonstrate that pulmonary large cell carcinomas – non-small cell carcinomas entirely lacking morphologic differentiation – retain the expression of differentiation markers supporting their histogenetic relationship to poorly-differentiated adeno- or squamous carcinomas in the majority of cases. We show that immunomarker-defined subsets of large cell carcinoma have a distinct spectrum of therapeutically-relevant driver mutations, including EGFR, KRAS and ALK, which parallels their distribution in tumors defined by traditional morphology. Finally, we identify marker-null large cell carcinomas as tumors that have a mutation profile that is similar to adenocarcinoma and a particularly poor prognosis.
Our finding that the expression of TTF-1 and p63 (ΔN isoform) reveals marker profiles akin to adeno- or squamous carcinoma, respectively, in 80% of large cell carcinoma is comparable to the data from prior studies, showing the expression of glandular or squamous immunomarkers in 59–90% of large cell carcinomas.12–18 Similar to this series, prior studies also suggested a more frequent relationship of large cell carcinomas to adeno- than squamous carcinomas, which may reflect the higher overall incidence of adenocarcinomas in the studied patient populations and/or the greater propensity of those tumors for complete loss of morphologic differentiation. However, a direct comparison across studies is limited by differences in the utilized markers, particularly in the studies performed prior to the recent advances in immunohistochemistry. While the use of immunostains, particularly a combination of TTF-1 and p63/p40, is now widely recommended for subtyping of undifferentiated non-small cell carcinomas in small biopsy/cytology samples,3, 39 applying this approach to entirely-undifferentiated non-small cell carcinomas in resections (i.e. large cell carcinomas) has remained controversial. In small biopsy/cytology samples, morphologically-unclassifiable non-small cell carcinomas are in most cases derived from carcinomas with clear evidence of at least focal morphologic differentiation upon resection, and classification of those samples by ancillary studies has gained wide acceptance. Although a similar approach to large cell carcinomas has been suggested by several investigators, the current recommendation is still to retain large cell carcinoma as a morphologically-defined entity,1, 3 at least in part because of the lack of data on accuracy of markers in classifying these tumors and the lack of evidence that such stratification has clinical utility. As discussed below, our data addresses these concerns by providing molecular corroboration for accuracy of immunomarker-based subtyping of large cell carcinomas and by demonstrating a utility of this stratification for the current clinical practice.
The key novel observation in this study is that large cell carcinomas, as a group, have a high frequency (37%) of therapeutically-relevant driver mutations, and that specific mutations are distributed in specific immunomarker-defined tumor subsets, mirroring the mutation profiles expected for morphologically-defined tumors. As such, EGFR, KRAS, BRAF, MAP2K1 and ALK alterations, which are characteristic of adeno- but not squamous carcinomas, were found selectively in tumors with non-squamous immunoprofiles, whereas the only alteration in tumors with squamous profile was a PIK3CA mutation. It may appear aberrant, however, that the frequency of EGFR and KRAS mutations in large cell carcinomas with glandular profiles is 2% and 40%, respectively, whereas the frequency of these mutations in unselected conventional adenocarcinomas in our patient population is ~20% and ~30%, respectively.40 In fact, this mutation frequency is entirely consistent with what is expected for a poorly-differentiated subset of adenocarcinomas. Specifically, it is well established that EGFR mutations occur preferentially in well-to-moderately differentiated adenocarcinomas with non-solid - bronchioloalveolar/lepidic and papillary -patterns,41, 42 while KRAS mutations are enriched in poorly-differentiated adenocarcinomas with solid histology.34, 43 Thus, the lower EGFR and higher KRAS mutation frequency in large cell carcinomas with glandular immunophenotype closely matches the expected frequency of these mutations for tumors in the spectrum of poorly-differentiated adenocarcinomas. Similarly, the frequency of other genetic alterations (ALK, BRAF, MAP2K1, PIK3CA) is comparable to the expected rate of these mutations in conventional adeno- or squamous carcinomas. The lack NRAS, ERBB2, and AKT1 mutations in large cell carcinomas is in line with their overall low expected prevalence (<1%) in lung carcinomas. In addition to establishing the overall frequency of therapeutically-relevant mutations, all of which are linked to either established or investigational targeted agents,44, 45 the mutation data in this study provides a measure of biological accuracy for immunophenotype-based classification of large cell carcinoma by demonstrating a similarity of mutation profiles in immunomarker-defined and morphologically-defined tumors.
To our knowledge, this is the first report of ALK rearrangement in large cell carcinomas. This expands the previously-recognized morphologic spectrum of ALK-positive lung carcinomas, although the propensity of these tumors for solid growth pattern (in addition to their classic association with signet ring cells) has been described.46 Notably, the clinical characteristics of patients with ALK-rearranged large cell carcinomas in this series (never/light smoker, younger age) are similar to what has been described for patient with ALK-positive adenocarcinomas.39,40
The identity of marker-null large cell carcinomas, and whether these represent entirely undifferentiated carcinomas or whether the differentiation lineage can be identified in these tumors by other markers, needs further study. We speculate that at least some of these tumors represent variants of TTF-1-negative adenocarcinomas, since the absence of TTF-1 is known to occur in ~20% of adenocarcinomas, whereas complete absence of p63/p40 expression is unusual for squamous cell carcinomas.19, 47–49 The relationship to adenocarcinoma of at least a subset of marker-null large cell carcinomas is further supported by our finding that they harbor a significant number of KRAS and BRAF mutations (combined rate 30%) – a mutation profile that is more similar to lung adeno- than squamous carcinoma. Identification of a reliable pan-adenocarcinoma marker would be needed to further clarify the nature of marker-null tumors.
Another novel observation in this study is that marker-null large cell carcinomas are associated with a distinctly inferior prognosis compared to differentiation marker-positive tumors. Conversely, the prognosis associated with the latter tumors appears to fall in the lower range of what has been reported for poorly-differentiated/high-grade subset of conventional adeno- and squamous carcinomas. In particular, several recent studies have demonstrated that the presence of solid growth pattern – a hallmark of poor differentiation – is a significant predictor of poor outcome in lung adenocarcinomas, conferring a 60–70% disease-free survival in stage I adenocarcinomas, compared to 80–>90% survival for better-differentiated tumors.50–52 Thus, 61% disease-free survival for stage I large cell carcinomas with glandular immunoprofiles in this study appears to be comparable to what is expected for tumors in the spectrum of poorly-differentiated adenocarcinomas. Although the survival data in this study is limited by a relatively small number of patients in each subgroup, this data in keeping with the concept that large cell carcinomas represent tumors in a continuum of solid growth/poor differentiation with usual types of non-small cell carcinoma. The particularly poor prognosis associated with marker-null large cell carcinomas may reflect the state of poorest differentiation – tumors undifferentiated at both morphologic and biomarker levels. We note that this observation parallels the known adverse prognostic effect of the lack of TTF-1 expression in adenocarcinomas.53–55 The high risk of recurrence suggests that large cell carcinomas overall and particularly marker-null subset could serve as a stage-independent indication for trials evaluating adjuvant chemotherapy, as has been recently suggested for poorly-differentiated adenocarcinomas in general.52
A direct practical utility of the findings in this study is that immunomarker-based stratification of large cell carcinomas could be used to guide the selection of predictive molecular tests in clinical practice. Currently, the standard predictive testing of lung carcinomas includes screening of adenocarcinomas for EGFR mutations (and in some institutions KRAS mutations), as positive and negative predictors, respectively, of sensitivity to EGFR tyrosine kinase inhibitors erlotinib and gefitinib, and for ALK rearrangements as a predictor of sensitivity to crizotinib.29 The standard guidelines, including the National Comprehensive Cancer Network, recommend testing of all large cell carcinomas for genetic alterations characteristic of adenocarcinoma,29 although this recommendation is based on limited data. Our findings support this recommendation for large cell carcinomas with non-squamous immunoprofiles. In contrast, our findings suggest that a biologically-rational triage for large cell carcinomas with squamous marker expression would include testing for genetic events characteristic of squamous rather than adeno-carcinomas, which includes PIK3CA mutations,25 as well as recently-identified DDR2 mutations56 and FGFR1 amplification57 – the markers which are anticipated to become part of routine clinical testing for squamous cell carcinomas in the near future28, 58 (although the degree of tumor-type specificity for the latter alterations still needs further investigation). Identification of tumor lineage in large cell carcinomas by immunoprofiling could thus direct the use of specific molecular and cytogenetic assays appropriate for that tumor type, thus optimizing the use of tissue and resources. The value of identifying targetable alterations in large cell carcinomas is illustrated by a patient with an EGFR mutation in this series, whose metastatic tumor had a marked response to erlotinib.
The second potential practical utility of our findings is for the use of “histology-based” agents -bevacizumab and pemetrexed - which are approved for patients with non-squamous non-small cell carcinomas. In clinical studies, it was suggested that all large cell carcinomas should be regarded as non-squamous based on the superior response of these tumors compared to squamous cell carcinomas,59, 60 although interpretation of this data is limited by the designation of tumors in small samples as large cell carcinoma in those studies. Nevertheless, the data on immunoprofiling of large cell carcinomas in this and other studies shows that a subset of these tumors are variants of squamous cell carcinoma, and therefore lumping all large cell carcinomas as “non-squamous” is biologically imprecise, although clinical responses of the group overall could reflect the predominance of adenocarcinoma variants. Notably, Monica et al18 showed that squamous lineage identified in large cell carcinomas by immunostains correlated with overexpression of thymidylate synthase – a putative target of pemetrexed – a profile that parallels thymidylate synthase expression in usual-type squamous cell carcinomas,18 supporting the concept that immunoprofiling of large cell carcinomas could be useful for the exclusion of non-recommended therapies in patient with these tumors. The actual impact of marker-based stratification on treatment outcomes with “histology-based” agents needs to be determined in clinical studies.
It is important to note that there are several potential limitations to TTF-1/ΔNp63-based classification of large cell carcinomas, despite these markers having been shown to be effective in distinguishing adeno- and squamous carcinomas in several recent studies.20–22 The first limitation is that neither marker is restricted to these tumor types: TTF-1 is also expressed in thyroid carcinomas, high-grade neuroendocrine carcinomas, and occasionally in unexpected settings, such as carcinomas of gynecologic tract, whereas ΔNp63 is expressed in squamous cell carcinomas of any site, urothelial, thymic, trophoblastic and basal cell/myoepithelial tumors (reviewed in refs. 19 and 20) – some of these tumors, when showing predominantly solid growth pattern, can enter in the differential diagnosis with large cell carcinomas. Thus, the interpretation of these markers must be performed in the context of careful morphologic and clinicoradiologic correlation and, if needed, with the use of additional immunostains to exclude the possibility of tumor types other than non-small cell lung carcinoma. This particularly applies to TTF-1/ΔNp63-null tumors, which must also be distinguished from other epithelioid neoplasms, such as melanoma or sarcoma with epitheliod features. The second limitation, mentioned above, is that TTF-1 is not a pan-adenocarcinoma marker, since it recognizes only ~80% of lung adenocarcinomas. The third potential limitation is the uncertainty with the interpretation of focal (<50% but above isolated tumor cells) ΔNp63 reactivity, which, based on prior studies, is not characteristic of either squamous cell carcinoma or adenocarcinoma.20–22 Such reactivity was not observed in this series, and the classification of large cell carcinomas with this uncommon immunoprofile, if encountered, remains to be clarified. While several other glandular and squamous markers are currently available, none are clearly superior in sensitivity and specificity to TTF-1 and ΔNp63, and whether they add value to TTF-1/ΔNp63 panel will be of interest to explore in future studies. Notably, we found that neither of the two additional glandular markers – Napsin A and mucicarmine – was positive in any TTF-1-negative tumors in this series (data not shown). Nevertheless, co-expression of these glandular markers with TTF-1 would enhance its specificity. Conversely, supplementing ΔNp63 with lower-specificity squamous markers, such as p63 (4A4) or 34βE12, is unlikely to be of value.
Also of note is the distinction in the current classification scheme of lung carcinomas between large cell carcinomas and the other class of undifferentiated non-small cell carcinomas that exhibit features of dedifferentiation in the form of spindle or giant/anaplastic cells, which are classified as “sarcomatoid carcinomas”.1 Although there is some morphologic gray-zone between giant cell/anaplastic carcinomas and large cell carcinomas in the higher end of the spectrum of cytologic pleomorphism, the tumors composed entirely of frankly anaplastic or spindle cells were not included in this series, and the ability of immunomarkers to detect residual differentiation and predict driver mutations in this other class of undifferentiated/dedifferentiated carcinomas would be of interest to investigate in a focused study. Further study is also needed to evaluate the potential significance of variant morphologies in large cell carcinomas.
In conclusion, our findings extend the concept that the majorly of large cell carcinomas exhibit immunophenotypic characteristics of either adeno- or squamous carcinomas, and further provide evidence that immunomarker-defined subsets of these tumors have distinct profiles of therapeutically-relevant mutations and distinct prognosis. While the current definition of large cell carcinoma is based on the morphologic criteria (supplemented with low-sensitivity mucin stains),1, 3 this classification groups biologically-heterogeneous tumors in a single category. Our data show that with currently-available markers, stratification of large cell carcinomas would have a utility for triage of tissue for EGFR/KRAS/ALK testing and for prognostication. Furthermore, marker-based stratification is also likely to be important for future clinical and molecular investigations where identification of biologically-precise tumor lineages is increasingly important given a strong trend for tumor type specificity and molecularly-targeting of the emerging therapeutics. We therefore suggest that “large cell carcinomas” with the marker profiles of adeno- or squamous carcinomas should be classified as variants of these respective tumor types, and predictive molecular and cytogenetic tests selected accordingly, whereas the term “large cell (undifferentiated) carcinoma” be reserved for the minority cases lacking differentiation at both morphologic and biomarker levels. Given the increasing use of immunohistochemistry to subtype non-small cell carcinomas in small samples in the current practice, to what degree this approach may already be informally applied in practice at individual institutions, would be of interest to survey.
Acknowledgments
We thank Chyau Lau, Laetitia Borsu, and Angela Marchetti for assistance with Sequenom assays, Joe Dycoco for help with the Thoracic Surgery database, and Marina Asher and Irina Linkov for performing immunohistochemistry. Molecular testing was supported by funding from NIH P01 CA129243 grant to M.L. and M.G.K. The MSKCC Sequenom facility is supported by the Anbinder Fund.
Footnotes
DISCLOSURE/CONFLICT OF INTEREST: none to disclose
References
- 1.Brambilla E, Pugatch B, Geisinger KR, et al. Large cell carcinoma. In: Travis WD, Brambilla E, Müller-Hermelink HK, Harris CC, editors. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. IARC Press; Lyon: 2004. pp. 45–50. [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2008. National Cancer Institute; Bethesda, MD: 2011. http://seer.cancer.gov/csr/1975_2008/, based on November 2010 SEER data submission, posted to the SEER web site. [Google Scholar]
- 3.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Churg A. The fine structure of large cell undifferentiated carcinoma of the lung. Evidence for its relation to squamous cell carcinomas and adenocarcinomas. Hum Pathol. 1978;9:143–56. doi: 10.1016/s0046-8177(78)80105-1. [DOI] [PubMed] [Google Scholar]
- 5.Horie A, Ohta M. Ultrastructural features of large cell carcinoma of the lung with reference to the prognosis of patients. Hum Pathol. 1981;12:423–32. doi: 10.1016/s0046-8177(81)80022-6. [DOI] [PubMed] [Google Scholar]
- 6.Albain KS, True LD, Golomb HM, Hoffman PC, Little AG. Large cell carcinoma of the lung. Ultrastructural differentiation and clinicopathologic correlations. Cancer. 1985;56:1618–23. doi: 10.1002/1097-0142(19851001)56:7<1618::aid-cncr2820560726>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 7.Yamagata N, Shyr Y, Yanagisawa K, et al. A training-testing approach to the molecular classification of resected non-small cell lung cancer. Clin Cancer Res. 2003;9:4695–704. [PubMed] [Google Scholar]
- 8.Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi T, Tomida S, Yatabe Y, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol. 2006;24:1679–88. doi: 10.1200/JCO.2005.03.8224. [DOI] [PubMed] [Google Scholar]
- 10.Yesner R. Large cell carcinoma of the lung. Semin Diagn Pathol. 1985;2:255–69. [PubMed] [Google Scholar]
- 11.Terry J, Leung S, Laskin J, et al. Optimal immunohistochemical markers for distinguishing lung adenocarcinomas from squamous cell carcinomas in small tumor samples. Am J Surg Pathol. 2010;34:1805–11. doi: 10.1097/PAS.0b013e3181f7dae3. [DOI] [PubMed] [Google Scholar]
- 12.Rossi G, Marchioni A, Milani M, et al. TTF-1, cytokeratin 7, 34betaE12, and CD56/NCAM immunostaining in the subclassification of large cell carcinomas of the lung. Am J Clin Pathol. 2004;122:884–93. [PubMed] [Google Scholar]
- 13.Pardo J, Martinez-Penuela AM, Sola JJ, et al. Large cell carcinoma of the lung: an endangered species? Appl Immunohistochem Mol Morphol. 2009;17:383–92. doi: 10.1097/PAI.0b013e31819bfd59. [DOI] [PubMed] [Google Scholar]
- 14.Monica V, Ceppi P, Righi L, et al. Desmocollin-3: a new marker of squamous differentiation in undifferentiated large-cell carcinoma of the lung. Mod Pathol. 2009;22:709–17. doi: 10.1038/modpathol.2009.30. [DOI] [PubMed] [Google Scholar]
- 15.Barbareschi M, Cantaloni C, Del Vescovo V, et al. Heterogeneity of large cell carcinoma of the lung: an immunophenotypic and miRNA-based analysis. Am J Clin Pathol. 2011;136:773–82. doi: 10.1309/AJCPYY79XAGRAYCJ. [DOI] [PubMed] [Google Scholar]
- 16.Conde E, Angulo B, Redondo P, et al. The use of P63 immunohistochemistry for the identification of squamous cell carcinoma of the lung. PLoS One. 2010;5:e12209. doi: 10.1371/journal.pone.0012209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Au NH, Cheang M, Huntsman DG, et al. Evaluation of immunohistochemical markers in non-small cell lung cancer by unsupervised hierarchical clustering analysis: a tissue microarray study of 284 cases and 18 markers. J Pathol. 2004;204:101–9. doi: 10.1002/path.1612. [DOI] [PubMed] [Google Scholar]
- 18.Monica V, Scagliotti GV, Ceppi P, et al. Differential Thymidylate Synthase Expression in Different Variants of Large-Cell Carcinoma of the Lung. Clin Cancer Res. 2009;15:7547–52. doi: 10.1158/1078-0432.CCR-09-1641. [DOI] [PubMed] [Google Scholar]
- 19.Rekhtman N, Ang DC, Sima CS, Travis WD, Moreira AL. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod Pathol. 2011;24:1348–59. doi: 10.1038/modpathol.2011.92. [DOI] [PubMed] [Google Scholar]
- 20.Bishop JA, Teruya-Feldstein J, Westra WH, et al. p40 (ΔNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol. 2012;25:405–15. doi: 10.1038/modpathol.2011.173. [DOI] [PubMed] [Google Scholar]
- 21.Pelosi G, Fabbri A, Bianchi F, et al. D(delta)Np63 (p40) and Thyroid Transcription Factor-1 (TTF1) Immunoreactivity upon Small Biopsies or Cellblocks for Typing Non-small Cell Lung Cancer: A Novel Two-hit, Sparing-Material Approach. J Thor Oncol. 2012;7:281–90. doi: 10.1097/JTO.0b013e31823815d3. [DOI] [PubMed] [Google Scholar]
- 22.Nonaka D. A study of DeltaNp63 expression in lung non-small cell carcinomas. Am J Surg Pathol. 2012;36:895–9. doi: 10.1097/PAS.0b013e3182498f2b. [DOI] [PubMed] [Google Scholar]
- 23.Righi L, Graziano P, Fornari A, et al. Immunohistochemical subtyping of nonsmall cell lung cancer not otherwise specified in fine-needle aspiration cytology: a retrospective study of 103 cases with surgical correlation. Cancer. 2011;117:3416–23. doi: 10.1002/cncr.25830. [DOI] [PubMed] [Google Scholar]
- 24.Langer CJ. Individualized therapy for patients with non-small cell lung cancer: Emerging trends and challenges. Critical reviews in oncology/hematology. 2012;83:130–44. doi: 10.1016/j.critrevonc.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18:1167–76. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol. 2010;17:889–97. doi: 10.1245/s10434-009-0808-7. [DOI] [PubMed] [Google Scholar]
- 27.Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29:3574–9. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 28.Drilon A, Rekhtman N, Ladanyi M, Paik PK. Squamous Cell Carcinomas of the Lung: Emerging Biology, Controversies, and the Promise of Targeted Therapy. Lancet Oncol. 2012 doi: 10.1016/S1470-2045(12)70291-7. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 29.NCCN Guidelines: Non-Small Cell Lung Cancer. Version 2.2012. National Comprehensive Cancer Network FW; PA: [DOI] [PubMed] [Google Scholar]
- 30.Sugio K, Uramoto H, Ono K, et al. Mutations within the tyrosine kinase domain of EGFR gene specifically occur in lung adenocarcinoma patients with a low exposure of tobacco smoking. Br J Cancer. 2006;94:896–903. doi: 10.1038/sj.bjc.6603040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sartori G, Cavazza A, Sgambato A, et al. EGFR and K-ras mutations along the spectrum of pulmonary epithelial tumors of the lung and elaboration of a combined clinicopathologic and molecular scoring system to predict clinical responsiveness to EGFR inhibitors. Am J Clin Pathol. 2009;131:478–89. doi: 10.1309/AJCPH0TRMPXVZW2F. [DOI] [PubMed] [Google Scholar]
- 32.Kwiatkowski DJ, Harpole DH, Jr, Godleski J, et al. Molecular pathologic substaging in 244 stage I non-small-cell lung cancer patients: clinical implications. J Clin Oncol. 1998;16:2468–77. doi: 10.1200/JCO.1998.16.7.2468. [DOI] [PubMed] [Google Scholar]
- 33.Graziano SL, Gamble GP, Newman NB, et al. Prognostic significance of K-ras codon 12 mutations in patients with resected stage I and II non-small-cell lung cancer. J Clin Oncol. 1999;17:668–75. doi: 10.1200/JCO.1999.17.2.668. [DOI] [PubMed] [Google Scholar]
- 34.Rodenhuis S, Slebos RJ. Clinical significance of ras oncogene activation in human lung cancer. Cancer Res. 1992;52:2665s–9s. [PubMed] [Google Scholar]
- 35.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang SM, Kang HJ, Shin JH, et al. Identical epidermal growth factor receptor mutations in adenocarcinomatous and squamous cell carcinomatous components of adenosquamous carcinoma of the lung. Cancer. 2007;109:581–7. doi: 10.1002/cncr.22413. [DOI] [PubMed] [Google Scholar]
- 37.Toyooka S, Yatabe Y, Tokumo M, et al. Mutations of epidermal growth factor receptor and K-ras genes in adenosquamous carcinoma of the lung. Int J Cancer. 2006;118:1588–90. doi: 10.1002/ijc.21500. [DOI] [PubMed] [Google Scholar]
- 38.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–4. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travis WD, Rekhtman N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: strategic management of tissue for molecular testing. Semin Respir Crit Care Med. 2011;32:22–31. doi: 10.1055/s-0031-1272866. [DOI] [PubMed] [Google Scholar]
- 40.Kris MG, Arcila ME, Lau C, et al. Two year results of LC-MAP: an institutional program to routinely profile tumor specimens for the presence of mutations in targetable pathways in all patients with non-small cell lung cancers. J Thorac Oncol. 2011;6:S346–7. [Google Scholar]
- 41.Dacic S, Shuai Y, Yousem S, Ohori P, Nikiforova M. Clinicopathological predictors of EGFR/KRAS mutational status in primary lung adenocarcinomas. Mod Pathol. 2010;23:159–68. doi: 10.1038/modpathol.2009.154. [DOI] [PubMed] [Google Scholar]
- 42.Motoi N, Szoke J, Riely GJ, et al. Lung adenocarcinoma: modification of the 2004 WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. Am J Surg Pathol. 2008;32:810–27. doi: 10.1097/PAS.0b013e31815cb162. [DOI] [PubMed] [Google Scholar]
- 43.Ang DC, Zakowski MF, Ladanyi M, Moreira AL, Rekhtman N. Characteristic Morphology and Immunoprofile of Lung Adenocarcinoma with KRAS Mutations: Propensity for Solid Growth Pattern and Correlation with TTF-1 Expression. Mod Pathol. 2010;23:396A. [Google Scholar]
- 44.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–80. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 45.Vakiani E, Solit DB. KRAS and BRAF: drug targets and predictive biomarkers. J Pathol. 2011;223:219–29. doi: 10.1002/path.2796. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida A, Tsuta K, Nakamura H, et al. Comprehensive Histologic Analysis of ALK-Rearranged Lung Carcinomas. Am J Surg Pathol. 2011;35:1226–34. doi: 10.1097/PAS.0b013e3182233e06. [DOI] [PubMed] [Google Scholar]
- 47.Mukhopadhyay S, Katzenstein AL. Subclassification of non-small cell lung carcinomas lacking morphologic differentiation on biopsy specimens: Utility of an immunohistochemical panel containing TTF-1, napsin A, p63, and CK5/6. Am J Surg Pathol. 2011;35:15–25. doi: 10.1097/PAS.0b013e3182036d05. [DOI] [PubMed] [Google Scholar]
- 48.Pelosi G, Rossi G, Bianchi F, et al. Immunhistochemistry by Means of Widely Agreed-Upon Markers (Cytokeratins 5/6 and 7, p63, Thyroid Transcription Factor-1, and Vimentin) on Small Biopsies of Non-small Cell Lung Cancer Effectively Parallels the Corresponding Profiling and Eventual Diagnoses on Surgical Specimens. J Thorac Oncol. 2011;6:1039–49. doi: 10.1097/JTO.0b013e318211dd16. [DOI] [PubMed] [Google Scholar]
- 49.Tsuta K, Tanabe Y, Yoshida A, et al. Utility of 10 immunohistochemical markers including novel markers (desmocollin-3, glypican 3, S100A2, S100A7, and Sox-2) for differential diagnosis of squamous cell carcinoma from adenocarcinoma of the Lung. J Thorac Oncol. 2011;6:1190–9. doi: 10.1097/JTO.0b013e318219ac78. [DOI] [PubMed] [Google Scholar]
- 50.Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011;24:653–64. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- 51.Solis LM, Behrens C, Raso MG, et al. Histologic patterns and molecular characteristics of lung adenocarcinoma associated with clinical outcome. Cancer. 2012;118:2889–99. doi: 10.1002/cncr.26584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sica G, Yoshizawa A, Sima CS, et al. A grading system of lung adenocarcinomas based on histologic pattern is predictive of disease recurrence in stage I tumors. Am J Surg Pathol. 2010;34:1155–62. doi: 10.1097/PAS.0b013e3181e4ee32. [DOI] [PubMed] [Google Scholar]
- 53.Barlesi F, Pinot D, Legoffic A, et al. Positive thyroid transcription factor 1 staining strongly correlates with survival of patients with adenocarcinoma of the lung. Br J Cancer. 2005;93:450–2. doi: 10.1038/sj.bjc.6602717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anagnostou VK, Syrigos KN, Bepler G, Homer RJ, Rimm DL. Thyroid transcription factor 1 is an independent prognostic factor for patients with stage I lung adenocarcinoma. J Clin Oncol. 2009;27:271–8. doi: 10.1200/JCO.2008.17.0043. [DOI] [PubMed] [Google Scholar]
- 55.Berghmans T, Paesmans M, Mascaux C, et al. Thyroid transcription factor 1--a new prognostic factor in lung cancer: a meta-analysis. Ann Oncol. 2006;17:1673–6. doi: 10.1093/annonc/mdl287. [DOI] [PubMed] [Google Scholar]
- 56.Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 Kinase Gene Identify a Novel Therapeutic Target in Squamous Cell Lung Cancer. Cancer Discovery. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paik PK, Hasanovic A, Wang L, et al. Multiplex Testing for Driver Mutations in Squamous Cell Carcinomas of the Lung. J Clin Oncol. 2012;30(Suppl):abstr 7505. [Google Scholar]
- 59.Johnson DH, Fehrenbacher L, Novotny WF, et al. Randomized phase II trial comparing bevacizumab plus carboplatin and paclitaxel with carboplatin and paclitaxel alone in previously untreated locally advanced or metastatic non-small-cell lung cancer. J Clin Oncol. 2004;22:2184–91. doi: 10.1200/JCO.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 60.Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–51. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]