Abstract
Transcription has the capacity to modify mechanically DNA topology, DNA structure, and nucleosome arrangement. Resulting from ongoing transcription, these modifications in turn, may provide instant feedback to the transcription machinery. To substantiate the connection between transcription and DNA dynamics, we charted an ENCODE map of transcription-dependent dynamic supercoiling in human Burkitt lymphoma cells using psoralen photobinding to probe DNA topology in vivo. Dynamic supercoils spread ~1.5 kb upstream of the start sites of active genes. Low and high output promoters handle this torsional stress differently as shown using inhibitors of transcription and topoisomerases, and by chromatin immunoprecipation of RNA polymerase and topoisomerases I and II. Whereas lower outputs are managed adequately by topoisomerase I, high output promoters additionally require topoisomerase II. The genome-wide coupling between transcription and DNA topology emphasizes the importance of dynamic supercoiling for gene regulation.
Chromatin structure, gene regulatory proteins, histones and DNA modification vary temporally with gene expression. DNA structure and topology also may regulate and be modified by nearby genetic activity [1]. Transcribing RNA-polymerase generates supercoils in the DNA template [2] potentially facilitating or impeding DNA-dependent processes [3]. Thus, besides serving as a passive information repository, DNA could actively participate in the real-time regulation of gene activity. Many studies have focused on the dynamics and functions of proteins regulating transcription; in general these studies have not considered the role of DNA structure and topology in gene regulation.
In the “twin domain” theory (twin-supercoiled-domain), threading DNA through the transcription machinery, along a helical path, dynamically drives positive supercoils ahead and negative supercoils behind the translocating RNA polymerase [2]. Negative supercoils untwist while positive supercoils overtwist DNA. Proceeding without pause, RNA polymerase would generate ~7 supercoils per second [4]; unless dissipated this torsional stress would rise to enormous levels disruptive to all genetic processes [1, 5]. DNA topoisomerases transiently break and rejoin the DNA backbone to remove positive and negative supercoils [6]. Depending on the intensity of ongoing transcription and the disposition of topoisomerases, local supercoiling might exceed the relaxation capacity of nearby topoisomerases leaving the residual DNA torsional stress to propagate through the embracing chromatin [7]. This stress might influence the binding of regulatory proteins to DNA, change nucleosome dynamics and reorganize chromatin fibers [3]. Supercoiling may also drive duplex B-DNA into single-stranded or other non-B DNA conformations that alter the ability of DNA and chromatin to loop and twist potentially modifying the function of enhancers and other cis-control elements [8]. Non-B DNA binding proteins maybe require alternative structures, and because non-B DNA is incompatible with nucleosome binding, such structures may sustain nucleosome free regions [9]. Since the magnitude and distribution of supercoiling forces throughout the genome are not known, the extent to which any or all of these potential regulatory mechanisms are realized in vivo has been a matter of speculation.
The accumulation and propagation of torsional stress along a DNA fiber depends on the rate of transcription, the length of the transcribed unit, and the arrangement of promoters (for example, divergent transcription would generate mutually reinforcing upstream negative supercoils) [10–11]. How torsional stress is transmitted through DNA will depend on the topological domains formed by protein-DNA interactions or by the anchoring of DNA to immobile nuclear structures [12]; such domains may concentrate or exclude supercoils. The binding of proteins, nucleosome positioning, and histone modifications might all influence the transmission of torsional stress or the activity of topoisomerases. Fundamental to elucidating the role and the control of torsional stress in gene regulation is the understanding of its disposition within chromosomes.
Whether metazoan chromosomes (like bacteria) are organized into supercoiled domains and whether such supercoils are regulated or regulatory remains controversial [13]. Supercoiling of intracellular DNA has been estimated from the intercalation of psoralen derivatives into DNA; intercalators in general insert between the bases of underwound rather than overwound DNA where the bases are squeezed together [14]. Recent studies in the yeast and fly have provided a coarse-grain view of the distribution of torsional stress along chromosomes, but low resolution has hampered the analysis of the factors governing the generation, relaxation, and transmission of supercoiling at individual genes in vivo [15–16]. Site specific experiments using Southern blots at a handful of genes [14, 17–20] showed that while the genome is generally relaxed, supercoiled DNA exists at a few loci in mammalian and insect cells. This supercoiling remains largely unstudied. Torsional stress has also been measured by monitoring the supercoiling of episomes recovered directly from cells before or after excision from chromosomes, and has been inferred from supercoil-dependent structural transitions in DNA or from the activity of supercoil-dependent recombinases [7, 12, 21]. The low resolution or low throughput of these methods have provided a limited view of the interplay between the factors determining the generation, relaxation, and transmission of DNA supercoiling in vivo.
To address these issues, genomic oligonucleotide microarrays were probed with DNA photo-crosslinked by psoralen in vivo. High resolution mapping revealed dynamic supercoiling transmitted approximately 1.5 kb upstream from transcription start sites (TSSs) to be a characteristic of virtually every transcribed gene. High output promoters sustain higher levels of supercoiling that are counterbalanced by the recruitment of topoisomerases. Topoisomerases I (Topo I) and II (Topo II) are differentially recruited and distinctly deployed illustrating the interconnection between DNA supercoiling and gene regulation.
RESULTS
Overview of the approach
As is well established, psoralen permeates cells, intercalates preferentially into underwound DNA and crosslinks complementary DNA strands upon exposure to UV-light [22] (Fig. 1). Because psoralen intercalation is also favored by high A-T content, but disfavored by nucleosomes and other DNA-protein interactions [16], corrections for sequence and chromatin must be made to estimate supercoiling in vivo. To quantify transcription-generated torsional stress - dynamic supercoiling - psoralen intercalation was compared between actively transcribing cells and cells in which transcription was specifically inhibited just prior to crosslinking. This comparison intrinsically normalized for the effects of sequence and long-lived DNA-protein interactions (such as nucleosomes) to highlight the effect of dynamic supercoiling on psoralen crosslinking.
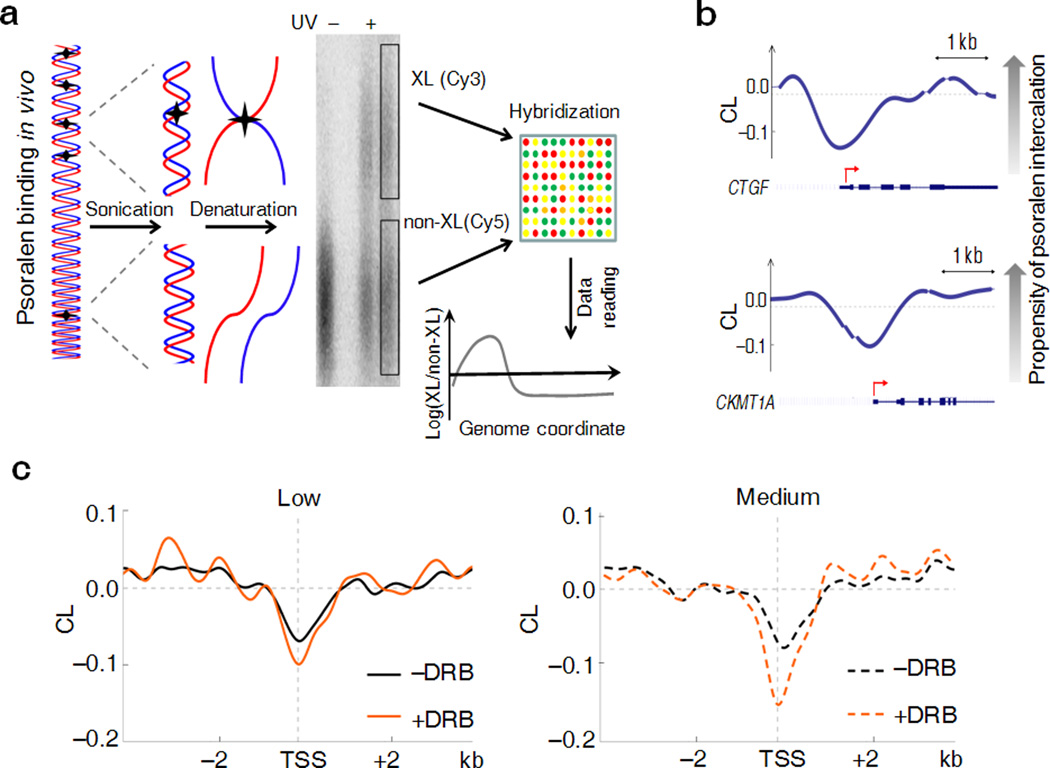
Figure 1. Psoralen photobinding is a genome-probe to detect DNA supercoiling in vivo.
(a) Overview of the approach: treatment of cells with psoralen followed by UV irradiation produces DNA inter-strand crosslinks. Thermal denaturation of genomic DNA fragments results in the formation of two fractions (left). After denaturation, the cross-linked fraction (XL) migrates slowly in gels, while the uncross-linked (non-XL) population is composed of rapidly migrating single-stranded DNA (center). After electrophoretic separation these fractions are purified, fluorochrome labeled and hybridized with densely tiled oligonucleotide arrays (right). The genomic distribution of the ratio of cross-linked and uncross-linked DNA (log 2 scale being 0 at the global mean) represents the efficiency of psoralen intercalation. (b) Representative examples of the psoralen cross-linking map show peculiarities near TSSs. The x axis shows genomic position. The y axis shows the CL level which is the log 2 ratio (crosslinked/un-crosslinked) of the fluorescent signals detected from the DNA microarray. Negative CL values indicate lower propensity of psoralen intercalation. Curves are computationally smoothed. The breaks in the curve correspond to the sequences that were not profiled on the microarray due to uniqueness and conformational issues. Schematic of the genes are represented below each curve with red arrows showing the TSSs. (c) Composite analysis of psoralen CL levels near the transcription start sites of low (from 0 to 20%, left panel) and medium (from 60 to 80% right panel) expressed ENCODE genes before and after treatment of cells with DRB.
To measure crosslinking genome-wide, Raji human B-cells were treated with psoralen and UV-light in G1-phase to minimize the influence of replication and mitosis on DNA topology [7]. Genomic DNA was recovered, sonicated, denatured and electrophoretically fractionated, resolving slowly migrating crosslinked from the faster migrating uncrosslinked DNA. The crosslinked fraction was expected to be enriched for DNA negatively supercoiled at the instant of irradiation (Fig. 1a). The separated fractions labeled with Cy5 or Cy3 were hybridized to high-density oligonucleotide microarrays spanning ENCODE regions [23]. The log ratio (crosslinked/un-crosslinked) of the fluorescent signals defined as the Cross Linking level (CL) provided a continuous picture of psoralen intercalation as a function of genome coordinate. Two computationally smoothed examples are provided (Fig. 1b). The CLs of promoters differed markedly from the CLs of intergenic regions reflecting differences in DNA topology, G-C content or specialized chromatin structures. Therefore the absolute CL from untreated cells was not immediately instructive about the level of dynamic supercoiling.
Because physiologically achievable levels of negative supercoiling increase psoralen intercalation only about two fold relative to relaxed DNA [14], the resulting signal to noise ratio demands experimental replication to achieve statistical significance. Three biological replicates were required for significance of the CL profiles and other maps analyzed here (Supplementary Note, Section 1). The data were averaged across replicates and high frequency noise was filtered by Fourier convolution smoothing [24] (supercoiling levels would be expected to fluctuate on the scale of the torsional (~300 bp) and bending (~150 bp) persistence lengths of DNA, not base pair to base pair). To observe the transcription-dependent supercoiling, the CL maps of cells treated with an inhibitor of transcription elongation were compared with untreated cells. Similarly, to reveal the dynamic character of DNA supercoiling and to examine its regulation, untreated versus topoisomerase-inhibitor treated cells were compared. Because different inhibitors act at different points in the topoisomerase reaction cycle, changes in DNA topology subsequent to treatment should reflect their modes of action [25].
To relate supercoiling with transcription, nuclear RNA was hybridized with these same microarrays. To correlate with psoralen intercalation genes were ranked lowest to highest according to RNA expression. Quintiles of expression were classified: low (0–20%, 20–40%), medium (40–60%, 60–80%) and high (80–100%) (Supplementary Note, Section 2). Because closely spaced TSSs, especially divergent promoters, could confound analysis [7], such promoters were separately analyzed (Supplementary Note, Section 3). This meta-analysis assumed that the ENCODE gene set was representative of the entire genome.
Dynamic supercoils upstream of promoters
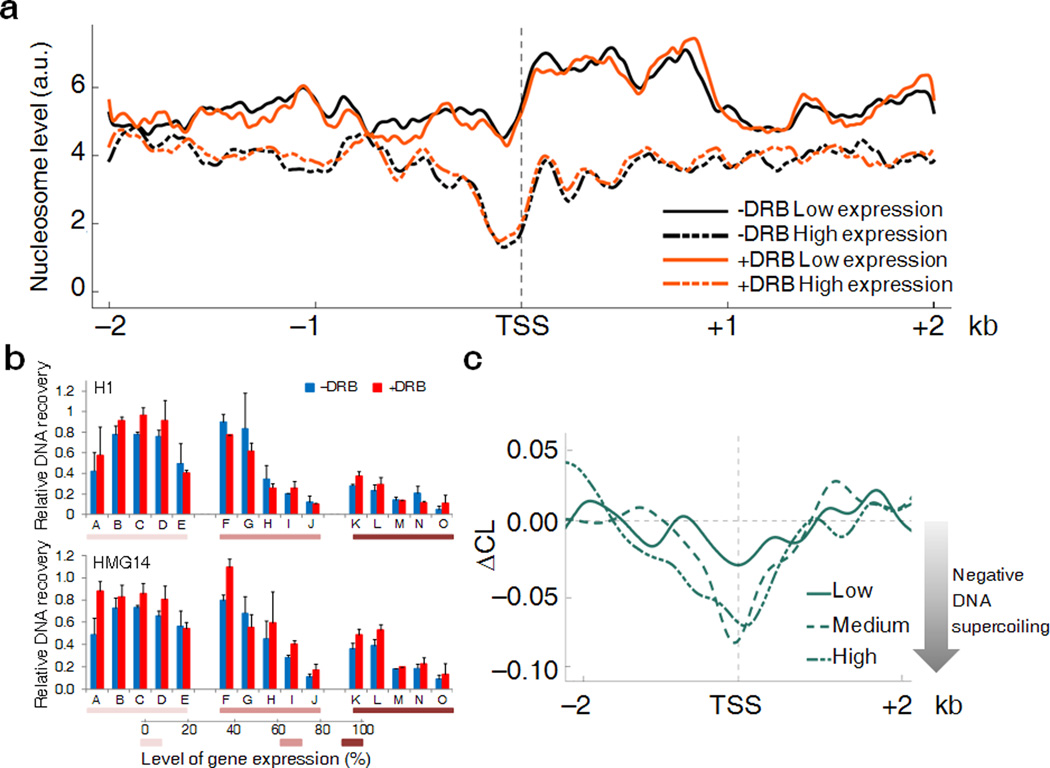
The dynamic range of gene expression is so large, that at the extremes, mechanistic and structural differences might obscure the elastic response of chromatin to applied torque. Therefore, the smoothed CL profiles of 8 kb windows surrounding TSSs of low (0–20%) and medium expressed (60–80%) genes were compared to see if modest differences in torsional stress were detected. Meta-analysis of the data for both sets of promoters revealed troughs of CL (Fig. 1c) at TSSs that largely reside in psoralen unfriendly CpG islands and are laden with transcription and chromatin complexes. The CL profiles were generated also for cells treated with 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB). DRB rapidly enters cells, specifically inhibits CDK9 phosphorylation of RNA polymerase II CTD and blocks transcription pause release [4]. After brief DRB-treatment, the CL-profile should reflect static properties such as sequence and chromatin structure, but not dynamic supercoiling that was expected to be drained by topoisomerases and diffusion of torsional stress away from TSSs. (An alternative drug, α-amanitin permeates cells and inhibits transcriptions too slowly to study dynamic supercoiling [26]). Indeed, the differences in the CL between DRB-treated and untreated cells was maximal near transcriptional start sites and gradually declined up to ~1.5 kb upstream for medium expressed genes (Fig. 1c) while for low expressed genes, the CL remained small and diminished faster upstream of the TSS with or without DRB-inhibition of transcription, just as predicted. To confirm that DRB-elicited CL changes reflected dynamic supercoiling and not large-scale chromatin rearrangements, nucleosome occupancy was monitored across the same gene set analyzed with psoralen crosslinking [27]. Although the nucleosome profiles of active versus inactive genes were distinct (downstream of TSSs active genes harbored fewer, but more well positioned nucleosomes than inactive genes, as previously noted [28]), neither set was affected by DRB-treatment (Fig. 2a). Thus nucleosome rearrangement cannot explain the changes in psoralen cross-linking provoked by DRB. The accessibility of upstream inter-nucleosomal linker DNA to non-nucleosomal chromosome proteins H1 and HMG14 was evaluated by ChIP assay in the presence and absence of DRB across a panel of fifteen genes expressed at different levels (Fig. 2b). As expected, binding of these factors paralleled decreased expression irrespective of DRB-treatment. Dynamic supercoiling seemed likely to explain the difference in psoralen binding upstream regions of active vs. inactive genes. We defined a parameter called CrossLinking Difference (ΔCL) to be a metric of dynamic supercoiling; ΔCL = CL(+DRB) − CL(−DRB) (Supplementary Note, Section 2). ΔCL was defined to give negatively supercoiled regions a negative value; segments devoid of dynamic supercoils have ΔCL near zero. ΔCL reports only transcription-dependent psoralen cross-linking while subtracting the influence of static DNA-protein interactions and DNA sequence. ΔCL profiles in a 4 kb window surrounding TSSs (Fig. 2c) decayed to baseline about 1.5 kb from the TSS of highly expressed genes and were further reduced and absent at low and non-expressed genes respectively. As the twin-supercoiled-domain-theory predicts, ΔCL is diminished within gene bodies where each RNA polymerase is a node between positive and negative supercoils (Supplementary Fig. 1), and so between pairs of elongating polymerase, positive and negative supercoils cancel [2]. In gene bodies nucleosomes are disrupted and reassembled during transcription; for these reasons we focused on upstream regions where interpretation is simpler, reflecting mainly the negative supercoiling emanating from TSSs.
Figure 2. DRB treatment does not affect nucleosome profiles or binding of non-nucleosomal proteins to DNA.
(a) Nucleosome occupancy around the TSSs of low (solid lines) and high (dashed lines) expressed genes in presence (orange lines) or absence (black lines) of DRB. The y axis shows the level of nucleosome binding expressed as tags per million (arbitrary units). The x axis shows the genomic coordinate centered on the TSS. For each position the nucleosome coverage was determined by counting the number of nucleosomes mapping to that position. (b) Enrichment of H1 and HMG14 proteins at promoter regions of selected genes (indicated by alphabetical letter, see Online Methods for details) relative to a reference intergenic region before and after DRB treatment. Promoters are ranked in three groups which have, respectively from left to right, low, medium and high expression. Data are shown as percentage of input (n=3–4, error bars, s.d.). (c) DNA supercoiling around TSSs is determined for low, medium and high expressed genes. The y axis shows the ΔCL which is the computational difference between CL values derived from DRB-treated and DRB-untreated cells. Negative ΔCL values reflect a higher propensity of psoralen to intercalate into the DNA due to transcription-dependent negative supercoiling. The zero point represents absence of transcription-dependent supercoils.
Parameters controlling the level of dynamic supercoiling
In the twin-supercoiled-domain-theory three major factors define the DNA topology of regions upstream of promoters: 1) the rate of supercoil generation by RNA polymerase; 2) how efficiently torsional stress is transported to remote chromatin locations by twist-diffusion or en-bloc rotation of chromatin segments; and 3) the rate of supercoil removal by topoisomerases [3]. The contributions of each of these parameters to upstream supercoiling were examined.
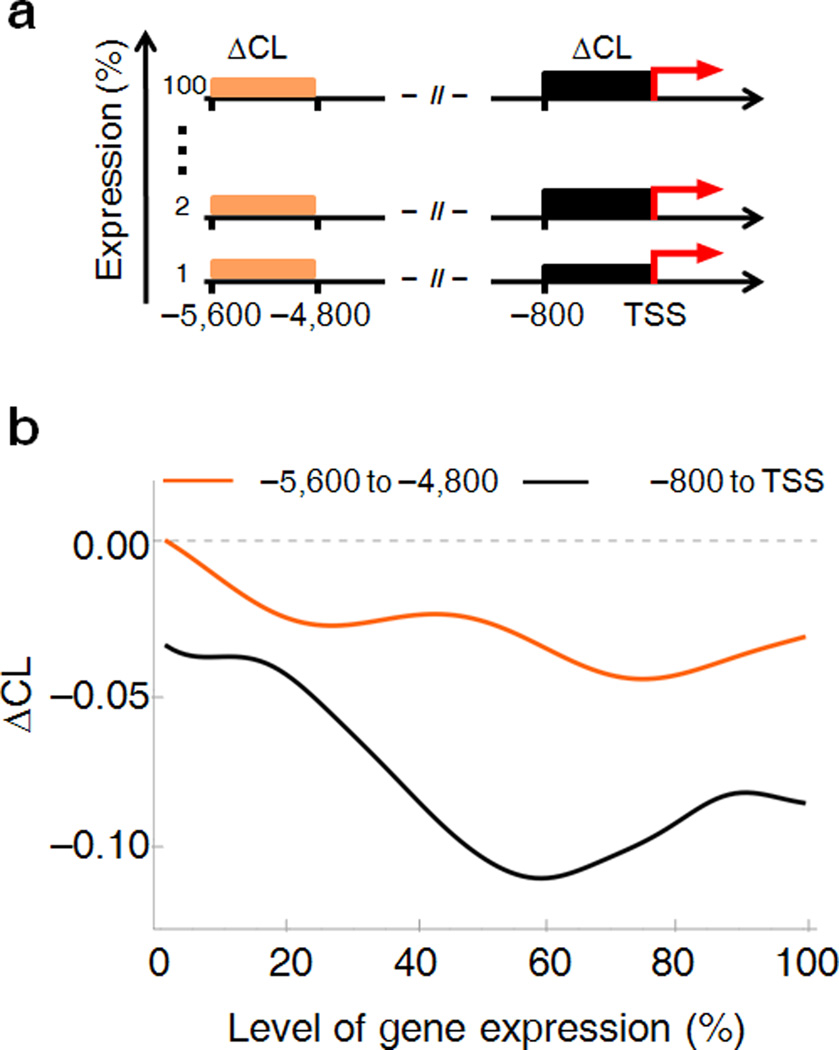
Dynamic supercoiling should increase with transcription; at steady state, the torsional stress generated will be balanced by topoisomerase activity. If torsional stress is freely transmitted through DNA fibers, then increased supercoiling near transcription start sites will be propagated further upstream, unless there are barriers to twist and writhe diffusion. To assess the relationship between dynamic supercoiling and promoter output, ENCODE genes were ranked by expression and the average ΔCL’s from the TSS to −800 bp versus −5,600 to −4,800 bp were compared by sliding-window averaging across the expression spectrum (Fig. 3a, b). ΔCL was near zero in far upstream regions independent of expression, but at TSSs, ΔCL was clearly more negative with high expression. Thus dynamic supercoiling is a local feature of active promoters and not a characteristic of large chromosomal domains.
Figure 3. Differential patterns of supercoil-generation for low-to-medium versus high transcribed genes.
(a) Schematic representation describing the calculation used to determine the relationship between gene expression and DNA topology. (b) The ΔCL signal of upstream promoter-regions was averaged over 800 bp for each single gene and plotted against the level of gene expression (black curve). Smoothing of the curve was done by sliding window average. The ΔCL signal between −5,600 bp and −4,800bp (orange curve) was graphed for comparison.
Dynamic supercoiling is associated with promoters, but not enhancers. Although enhancers bind RNA polymerase, the output of eRNAs transcribed from these regions is meager compared to promoter-sponsored transcription [29]; concordantly ΔCL was at background levels for enhancers irrespective of distance to their associated promoters (Supplementary Fig. 1b). CTCF is a multi-zinc finger protein mediating looping between remote regions of the genome [30]; CTCF-sites were not associated with changes in ΔCL indicating that CTCF-bounded loops do not comprise a capacitor for transcription-generated supercoils (Supplementary Fig. 1c).
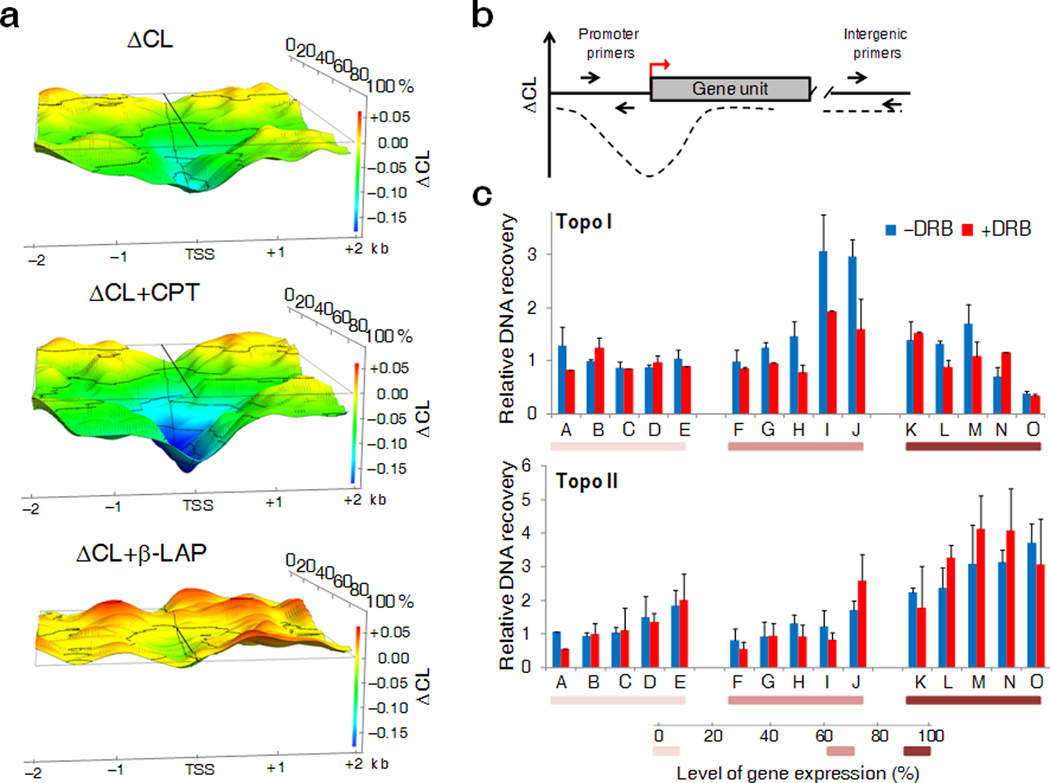
To get a finer view of dynamic supercoiling upstream of TSSs, the ΔCL from −2 kb to +2 kb was graphed across a moving window through the expression spectrum. ΔCL strongly correlated with transcriptional activity as predicted by the twin-domain model. Low expressed genes showed only a small dip in the ΔCL close to the TSS, but as RNA production increased, the ΔCL-signal spread up to 1.5 kb upstream (Fig. 4a, top). ΔCL became increasingly negative as gene expression increased up to 80% of maximal expression. Beyond 80%, DNA supercoiling plateaued or even diminished near TSSs suggesting that special mechanisms are marshaled to contend with the highest levels of torsional stress.
Figure 4. Perturbing the distribution of supercoils with topoisomerase inhibitors reveals the pattern of Topo I and Topo II recruitment to TSSs.
(a) 3-D representation of the ΔCL profiles of genes ranked according to their level of expression in the absence of topoisomerase inhibitors (top panel), in presence of CPT (central panel) or β-LAP (bottom panel). (b) Schematic representation of qPCR design for the ChIP analysis. DNA recovery was determined at promoters and at reference intergenic region. (c) Raji cells were treated with CPT or β-LAP in presence or absence of DRB and Topo I (top panel) or Topo II (bottom panel) occupancy was detected by ChIP. The relative enrichment of the topoisomerases at the promoter area of genes (indicated by alphabetical letter, see Online Methods for details) with different expression levels is shown. Five genes were analyzed in each expression range. Data are normalized to a non-expressed intergenic region (n=3–4, error bars, s.d.).
One way to reduce supercoiling near highest output promoters would be to recruit Topo I or Topo II more effectively. To test this, upstream regions of different output promoters were analyzed by ChIP assay and qPCR (Fig. 4b) with and without DRB. Topo I and Topo II bound to the DNA were trapped using very brief treatments with camptothecin (CPT) and β-lapachone (β-LAP), respectively. CPT highly selectively inhibits strand religation during the Topo I catalytic cycle while β-LAP traps Topo II during the formation of the DNA cleavage complex and inhibits Topo I prior to the strand cleavage [31–33]. For both topoisomerases, the low transcribed genes were hardly enriched relative to a reference intergenic region (Fig. 4c). Whereas Topo II was dramatically recruited to highly active genes, Topo I was most efficiently recruited to the promoter of medium expressed genes. These results confirmed the differential recruitment of Topo I and Topo II according to promoter output. To better relate dynamic supercoiling with topoisomerase activity, topoisomerase recruitment was evaluated with and without transcriptional inhibition. Whereas DRB treatment did not perturb Topo II levels, Topo I was reduced at medium output promoters, those most sensitive to Topo I activity (Fig. 5c). Consistent with Topo I being a supercoil-sensitive enzyme dynamically associating with transcribed loci [34], its recruitment was dependent on transcription activation and supercoil generation. In contrast, Topo II was recruited by features other than dynamic supercoiling.
Figure 5.
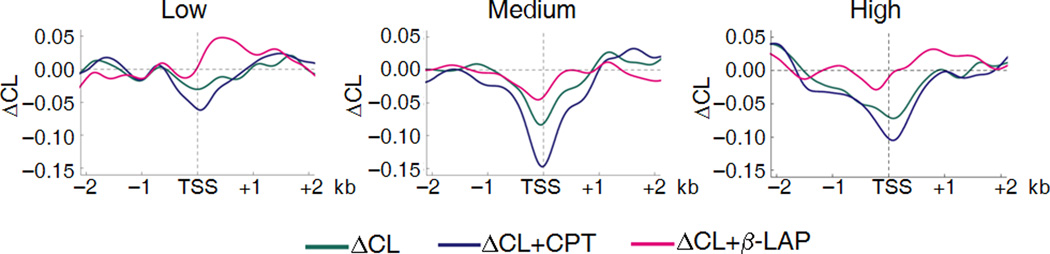
Comparison of ΔCL curves generated in the absence or presence of CPT or β-LAP inhibitors. From left to right respectively low, medium and high expressed genes are shown in each panel.
Dynamic supercoiling appeared sensitive to the distribution and kinetics of topoisomerases. To confirm this, the ΔCLs of promoter regions were compared with and without topoisomerase inhbitor treatment. Topo I nicks a single DNA strand, relaxes supercoils by rotating about the intact DNA strand, then closes the nick. CPT at the DNA-protein interface hinders rotation of the nicked DNA [35]. Consequently, in the presence of CPT, negative supercoiling should intensify transiently upstream of promoters. If the relationship between transcription and supercoiling is as hypothesized, then the ΔCL of upstream of medium expressed genes that depend on Topo I should be more sensitive to CPT than highly expressed genes that recruit Topo II. Indeed, 5 minutes of CPT deepened the ΔCL valley at the TSS and upstream indicating that Topo I activity is broadly applied at promoter regions to control dynamic supercoiling (Fig. 4a, center). The effect of CPT was stronger for medium expressed than for highly expressed genes (Fig. 5). The short treatment insured that the ΔCL reflects changes in DNA topology and not secondary effects [36].
β-LAP, which inhibits Topo I prior to strand nicking, and Topo II in the midst of DNA cleavage [31–32], was selected to infer Top II’s role in resolving topological issues during transcription. Because Topo I and II are functionally partially redundant and Topo I activity increases along with the dynamic supercoiling as demonstrated in Fig. 4c., it was necessary to inhibit Topo I (in order to blunt a confounding compensatory increase in Topo I activity) and Topo II; Topo II function could be inferred from the difference between Topo I and Topo I + II inhibition. To trap a double strand break, both Topo II subunits have to interact simultaneously with the drug on each strand [37]. Thus, with low β-LAP concentration and short treatments, nicks rather than double strand breaks predominate, and diffusion of torsional stress off these nicks should result in the relaxation of regions served by the Topo II. Indeed, 5 minutes of β-LAP treatment uniformly relaxed upstream DNA with the minimization of the ΔCL from the TSS to all upstream points (Fig. 4a, bottom; Fig. 5). Therefore, Topo II acted close to TSSs relaxing negatively supercoiled DNA. Topo II inhibitors that evoked a rapid DNA damage response were not studied (Supplementary Fig. 2).
RNA Polymerase II ChIP-Seq data from Raji cells provided an independent measure of transcriptional activity for the ENCODE genes [38]. The ΔCL profiles of these genes were sorted by pausing index that relates RNA Polymerase II density at promoters versus gene bodies [39]; paused and elongating promoters corresponded to medium and highly expressed genes; genes lacking RNA polymerase were designated silent. The ΔCLs before and after inhibitors, followed the same patterns whether sorted by pausing index or expression (Fig. 5 and Supplementary Fig. 3). Thus, while Topo I predominated at medium, and Topo II at high output promoters, the enzymes relaxed negative supercoils semi-redundantly.
Fine tuning of DNA supercoiling with topoisomerases
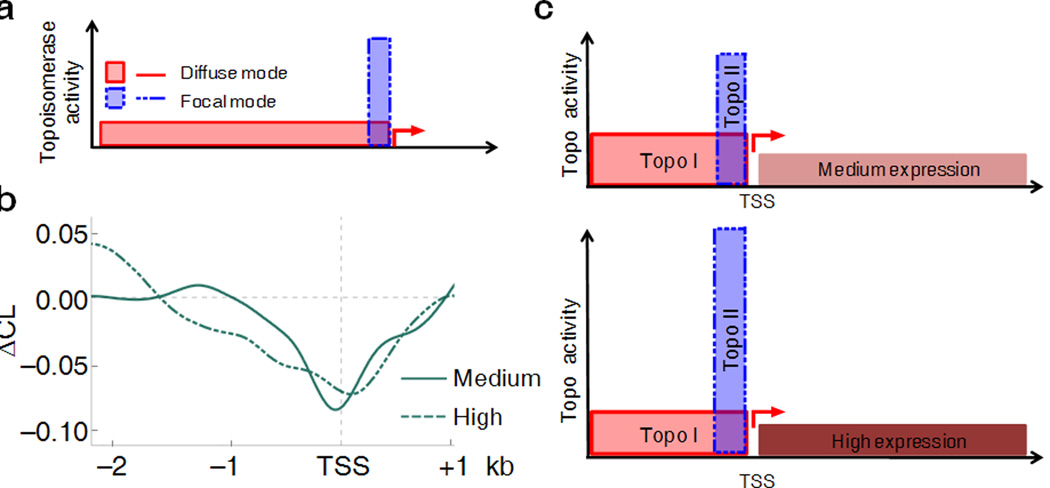
Two scenarios may be hypothesized for the role of topoisomerases in the steady-state regulation of dynamic supercoils. In the first, negative torsional stress generated during transcription spreads into the upstream promoter regions (Fig. 6a, solid red line) where diffusely recruited Topo I and Topo II relieve the stress. Because the probability that a segment upstream from the TSS remains topoisomerase-free falls exponentially with distance, the level of supercoiling is high near TSS but decays sharply upstream with this model. Alternatively, if topoisomerases are recruited focally to the most dynamically stressed DNA, i.e. TSSs (Fig. 6a, dashed blue line), then level of supercoiling would reduced right at the TSS, but beyond this zone, any residual supercoiling would decay only gradually. The ΔCL patterns were compared between sets of genes with different expression to discriminate these possibilities. Whereas the ΔCL upstream of medium transcribed genes decayed rapidly suggestive of diffuse topoisomerase recruitment, at high output promoters, ΔCL diminished at the TSS, but thereafter declined gradually consistent with focal recruitment (Fig. 6b). The observed relationship between transcription, supercoiling, and the response to topoisomerases inhibition suggested that highly active genes recruit Topo II to their TSSs, whereas weakly expressed genes do not.
Figure 6. Differential topoisomerase I and II utilization in the regulation of transcription-induced torsional stress.
(a) Two modes of topoisomerase recruitment at the upstream region of promoters: the diffuse mode (solid line) suggests that topoisomerases are randomly distributed over the upstream promoter regions; the focal mode (dashed line) hypothesizes that topoisomerases work near the TSS. The two modes can be visualized in panel (b) where the ΔCL curves for medium and high expressed genes were generated. (c) Proposed model of supercoiling regulation by topoisomerases. Dynamic supercoiling near medium active genes is managed mainly by Topo I which is distributed over a broad upstream promoter region; whereas highly active promoters recruit Topo II focally near the TSS.
Use of topoisomerase selective inhibitors allowed estimation of the relative relaxation by each enzyme as a function of expression. CPT increased supercoiling more with medium than high expression, but with high expression supercoils diffuse further upstream (Fig. 5, center,and right). With β-LAP, upstream supercoiling was lost for highly active genes (Fig. 5, right) while with medium expression, ΔCL diminished at TSSs and further declined upstream (Fig. 5, center). So topoisomerases acted redundantly upstream of medium expressed genes (Fig. 6c, top), but when transcription intensifies, Topo II was drawn to transcription start sites (Fig. 6c, bottom).
DISCUSSION
The role of dynamic supercoiling in the regulation and execution of genetic transactions has been incompletely described. Though torsional stress has been definitively demonstrated in naked DNA in vitro and in episomes in vivo, the pervasiveness and significance of dynamic supercoiling for most chromosomal genes is not known [7, 40]. Recent studies that used psoralen as a probe for supercoiling in yeast revealed that distinct chromosome compartments confine different levels of DNA helical tension, but lacked sufficient resolution to directly relate DNA topology to gene activity because, 1) yeast genes and the yeast genome are too compact to confine torsional stress to single targets, and 2) the DNAs immobilized on the microarrays were insufficiently short to enable finer mapping [16]. A genome-wide study of psoralen binding to Drosophila polytene chromosomes was limited by the optical resolution of conventional light microscopy [15]. The present work studied ENCODE genes in their normal chromosomal context in the presence of functional topoisomerases. The resolution of high-density oligonucleotide arrays revealed the fine-grain distribution of dynamic supercoiling near promoters and its control by topoisomerases. These experiments showed that transcription-dependent DNA supercoiling was transmitted locally upstream of promoters, but that highly expressed genes relied upon Topo II to dissipate dynamic supercoiling whereas medium expressed genes depend on Topo I.
The level of supercoiling depends on the introduction of torsional stress into DNA, and its removal by topoisomerases or by diffusion to remote regions of the genome [3]. The dynamics of supercoil diffusion depend on the properties of chromatin fibers: the positions of nucleosomes, the interactions between them, inter-nucleosomal linker-binding proteins and the nucleosome modifications could all influence supercoil propagation. Our analysis revealed that torsional stress is dissipated over a short-range (~1.5–2 kb) suggesting that dynamic supercoiling is not usually confined by fixed boundaries in chromatin. Crossing such boundaries, focal high levels of negative supercoiling decrease abruptly [12, 19]. Alternatively, topological domain boundaries for each particular gene may be heteromorphous or transient in nature, resulting in variable domain size between similar cells. The simplest interpretation of our data is that DNA supercoiling upstream of the active promoter is established mostly by frictional restriction to DNA twist diffusion through chromatin. Context dependent supercoiling associated with transgene location supports the idea that chromatin features may modify the distribution of supercoils [17]. Even without fixed boundaries, other architectural features, for example, divergent transcription, could modify the generation and propagation of dynamic supercoils (though the number of divergent promoters expressing both partners was too low to provide more that a hint of mutually reinforcing supercoils, Supplementary Note, Section 3). Meta-analysis of the ΔCL between convergent promoters was further complicated by variable gene length, multiple termination sites, and engaged RNA polymerases downstream of termination sites.
As suggested by transcription defects in cells with mutant topoisomerases, proper supercoil management is required for efficient transcription [41]. Topo I and Topo II, which relax positive and negative supercoils, are fully redundant for transcription in yeast, but double mutants are severely impaired for elongation. However, in mammalian cells the topoisomerases only partially compensate for each other, suggesting specific functions for each in transcription [42]. The topological problems of gene expression may dictate specialized roles for each enzyme since the positive and negative supercoils generated during transcription distort DNA differently and reside in different molecular domains [43]. So recruitment of each topoisomerase to active genes may be context dependent [10, 44]. The results of this study reveal two characteristics of the relaxation of transcription-induced DNA torsional stress by topoisomerases.
First, both Topo I and Topo II prevent the build-up of negative supercoiling in promoter regions. Topo I is a rapid, processive, torque-sensitive enzyme with low activity on nucleosomal templates [45–46] that is well-evolved to act ahead of elongating RNA polymerase where accumulated positive torsional stress and histone modifications mobilize nucleosomes [47]. Inhibiting Topo I slows elongation in vivo [5]. In yeast Topo II binds to nucleosome-free regions near active TSSs [48], and in mammalian cells, the enzyme is enriched near the promoters of Topo II-sensitive genes [49]. In addition, Topo II activity would be favored by the crossed DNA segments [50] as occurs when plectonemes form in negatively supercoiled DNA unbuffered by chromatin rearrangement [1, 51]. Because all elongating transcription complexes impose a 90-degree bend in the template, as downstream DNA is screwed into the RNA polymerase active site, the upstream DNA is translationally rotated generating writhe [52]. Therefore, Topo II would efficiently relax negative supercoiling behind the transcribing RNA polymerase. The twin-supercoiled-domain model predicts that dynamic negative supercoiling is highest at the promoter [2]. Accordingly, the activity of Topo II should be localized near the TSSs of highly active genes as demonstrated here. Some of the changes in ΔCL after β-LAP treatment may arise secondarily from Topo I inhbition. Because Topo I and Topo II are partially compensatory, combinations of inhibitors and rapidly inactivateable mutants will be required to separate the contribution of each topoisomerase to normal transcription.
Second, besides draining supercoils, it may be important to sustain a steady-state level of torsional stress in upstream regions to manage supercoil-driven structural transitions that respond to ongoing transcription [7, 40, 53–54]. The processive, fast but difficult to control Topo I [46] is reduced at promoters of high versus medium expressed genes. DNA relaxation at these genes is accomplished using step-by-step DNA transport by Topo II in which ATP-driven conformational changes accompany very transient DNA breakage [55]. Thus at highly active genes, topological homeostasis could be better enforced by Topo II. Coordinating the rates of transcription and DNA relaxation would adjust supercoiling at different genes; if so, relaxing activity is essential not only to remove topological impediments to transcription, but to establish a sturdy level of negative supercoiling within the regulatory regions of active genes.
DNA supercoiling in regulatory pathways
Much evidence supports the idea that DNA mechanics serves a variety of regulatory functions [3, 8]. In vitro studies functionally couple chromatin structure with DNA topology [47]. DNA supercoiling may assist chromatin remodeling, influence chromatin and DNA structure, and modify DNA-transcription factor interactions [1]. Torsional stress transported through DNA by changing the energy landscape and topology of the chromatin fiber may signal to remote sites [56]. This signal could recruit or restrict binding of DNA conformation-sensitive proteins with regulatory modules [7, 53], or facilitate long-range protein-DNA interactions [47]. The same output states could be achieved only slowly by adjusting the chemical concentrations of transcription factors, whereas supercoiling has the capacity to govern a local transaction in real time. The vast intergenic regions of eukaryotes and the short range of torsional stress propagation insure independent topological regulation of different genes.
Finally, because the structure and mechanics of cellular RNA polymerase is conserved across eukaryotes and prokaryotes, then many of the DNA topology-sensitive regulatory mechanisms in bacteria may also operate in higher organisms. Despite differences in the complexity of the transcription machinery between kingdoms, both are forced to contend with the same polymer physics: the requirement to strongly bend DNA for pre-initiation complex formation and the need to locally melt DNA during initiation [52]. Negative supercoiling facilitates both bending and melting [57–59]; consequently, this fundamental linkage between DNA topology and transcription is conserved [13]. In gyrase-containing bacteria, genomic DNA is globally undertwisted to optimize the transcription of many genes [60], but in higher eukaryotes where genes are often separated by large reaches of inactive DNA, each gene may define its own topology. By coordinating topoisomerase activity with transcription, supercoiling in regulatory regions is dynamically buffered. Supercoil assisted melting might adjust the early rate-limiting steps of transcription and influence RNA production .
As costs decline, this psoralen-based procedure for the analysis of DNA topology may be adapted for NextGen sequencing and will help to uncover other DNA topology-related mechanisms in genome functioning.
Online Methods
Cell culture
Raji cells were synchronized in early G1 phase of the cell cycle by treatment with 1.5% (v/v) DMSO for 96 hours. Cells were released from DMSO in fresh medium and experiments were conducted 6 hours later. When indicated, the gene transcription was inhibited using 40 µM of 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) for 30 minutes. To inhibit topoisomerases, cells were exposed to 10 µM β-lapachone or camptothecin for 5 min.
Psoralen photobinding assay
2×107 cells per 10 ml of media were treated with 140 µL of a 0.9 mg/ml solution of 4,5’,8-trimethylpsoralen in ethanol for 4 min at 37°C. To photocross-link the DNA strands, plates with cells were exposed to 3.6 kJ m−2 of 365 nm light (ultraviolet lamp, model B-100 A, Ultra-Violet Products). Cross-linked genomic DNA was isolated by RNase and Proteinase K treatment in lysis buffer (10 mM Tris-Cl pH 8.0, 100 mM EDTA, 0.5% SDS), followed by repetitive phenol/chloroform extraction and ethanol precipitation. Purified DNA was sonicated (Sonicator, Ultrasonic processor XL, MISONIX Inc. at 15% of power) to produce 250 bp average-size DNA fragments. DNA was then heat-denaturated and incubated at 55°C for 1 hour in glyoxal buffer. Glyoxylated non-cross-linked and cross-linked DNA fragments were separated by electrophoresis (3% agarose gel electrophoresis in 10 mM sodium phosphate buffer (pH 7.0) at 2 volt/cm for 12 hours). With this protocol, the ratio of non-cross-linked DNA to cross-linked fragments is 3 to 1 showing that psoralen concentration inside the cells is in the linear range of DNA binding [7]. After electrophoresis, the gel was incubated with denaturing solution (0.5 M NaOH, 1.5 M NaCl) at 65°C for 3 hours to reverse psoralen crosslinks and stained with SYBR-green [15]. Cross-linked and non-cross-linked DNA fragments were purified by electroelution and hybridized in different combinations to NimbleGen ENCODE arrays (tiled with 12-bp overlapping, 50-mer probes across unique regions of ENCODE as well as 200 kb centered on the c-myc TSS). Three biological replicates, each hybridized to new array were performed. DNA labeling, hybridization, detection, data extraction and quality assessment were performed at NimbleGen.
Gene expression assay
Nuclear RNA was prepared using the Qiagen RNeasy kit. RNA was isolated from the nuclear pellets resuspended in the kit lysis buffer and processed according to the protocol. RNA was converted into double-stranded DNA by using SuperScript Choice System for cDNA synthesis (Invitrogen). cDNAs were sonicated to average fragments of 250 base pairs and hybridized to Nimblegene ENCODE arrays together with genomic DNA sonicated to similar size. In total, three biological replicates with a new array for each were performed. Data were generated at NimbleGen. Expression levels were defined as the average signal at the annotated gene, normalized by the number of probes.
Data Analysis
To smooth the data, the Fourier convolution technique was used [24]. The technique uses a moving window average as a reference, as a result of which the local features are not lost during an unsupervised noise reduction. Microarrays used in this study were designed with each probe having 50 bp and a 12 bp overlap. Thus any given region of the genome is represented by 38 bp non-overlapping sequence between consecutive probes. Fig 1B shows smoothing of two specific regions using 40 probes. During the meta-analysis (Supplementary Note, Section 2) all the transcribed regions were aligned on the transcription start sites (TSS). Since the TSSs are randomly distributed with respect to probes, the data density increases to 1.4 bp per data point (from 38 bp/probe). The meta-analysis presented in this study uses a window size of 400 data points (equivalent to 561 bp, Fig. 2b and subsequent figures). Nimblegen ENCODE (hg18) microarrays had usable data for a total of 855 transcribed regions (Supplementary Note, Section 3). In order to avoid multiple counting of same gene, clusters of those transcripts-genes were identified which were either overlapping or had a TSS within 50 bp of each other. Only the largest “transcribed region” from each of these groups was used. This brought down the total number of transcribed regions to 445 (with 415 unique genes). The complete list of the transcribed regions is provided in the Supplementary Table 1.
Sequence Dependent Background Correction
To remove the sequence bias of psoralen intercalation, the transcribed regions were sorted based on AT content within ±3000 bp of TSS. To reduce systematic errors, these 445 transcribed regions were divided in 10 groups. A correction term, for each of the decades, was calculated by averaging the raw ratios in the flanking regions (−8 kb, −2 kb) and (+2 kb, +8 kb) of each transcribed regions. The data for each of the constituent transcribed regions was then baseline shifted using this base composition correction term to yield the corrected data, which was used for further analysis. See Supplementary Note, Section 2 for a detailed description.
Chromatin Immunoprecipitation (ChIP) and quantitative PCR (qPCR)
ChIP assays were performed with Raji cells as described with minor changes [7, 38]. Briefly, 5×107 cells were cross-linked with 1% formaldehyde and sonicated in TE to produce chromatin fragments of 800 bp on average. Immunoprecipitations with Dynabeads Protein A (Invitrogen) were carried out using 4 µg of antibodies. Immunoprecipitates were washed twice with RIPA buffer (10mM Tris-HCl pH 8.0, 1mM EDTA pH 8.0, 1% Triton X100, 0.1% Na-Deoxycholate, 0.1% SDS, 200mM NaCl); twice with RIPA buffer plus 300mM NaCl; twice with LiCl buffer (10mM Tris-HCl pH 8.0, 1mM EDTA pH 8.0, 250mM LiCl, 0.5% NP40, 0.5% Na-Deoxycholate); and twice with TE. The beads were then resuspended in TE plus 0.25% SDS supplemented with proteinase K (500 µg/ml, Roche) and incubated overnight at 65°C. The DNA was recovered from the eluate by phenol-chloroform extraction followed by ethanol precipitation and dissolved in TE. For qPCR detection performed using the LightCycler 480, the percent of IP enrichment as compared to input was calculated using FastStart DNA Master SYBR Green I kit (Roche Diagnostics) and data are presented as the fold change respect to a reference intergenic region of drug treated cells. Quantification and melting curve analyses were performed using the Roche LightCycler software by the crossing point method as indicated by the supplier. Fifteen genes were analyzed in total; five genes in each group ranked according to the RNA production: 0–20%; 60–80%; 80–100%. All detection primers are listed in Supplementary Table 2. Antibodies used: anti-Topo I (Epitomics, 3552-1); anti-Topo II-β (Epitomics, 3747-1); anti-Histone H1 (Santa Cruz Biotech., sc-8030), and anti-HMG14 (Abcam, ab5212). For all the antibodies used in this study the DNA recovered values were around 100-fold higher than the IgG controls (Santa Cruz Biotechnology: normal mouse IgG, sc-2025; normal rabbit IgG, sc-2027).
Total Protein Extraction and Western Blot
Cells were lysed with RIPA buffer, briefly sonicated, and then pelleted by centrifugation. The supernatant was saved for Western blotting. 40 µg of proteins were subjected to SDS PAGE and then blotted onto a nitrocellulose membrane for incubation with specific antibodies: anti-CHK2 (Cell Signaling, 2662); anti-phospho CHK2 (Cell Signaling, 2661); anti-γ-H2AX (Millipore, 05-636), and anti-Histone H3 (Abcam, ab1791) all used at 1:2000 dilutions.
ChIP-Seq and MNase-Seq data processing
ChIP-seq and MNase-Seq on the formaldehyde cross-linked cells were performed as described [27]. Raw sequencing data from Pol II ChIP-Seq (single-end tags) and MNase-Seq (pair-end tags) libraries were processed using Ilumina Analysis Pipeline and the acquired reads were mapped to the human genome (hg18) using Bowtie software (http://bowtie-bio.sourceforge.net/index.shtml). Only uniquely mapped reads located within the ENCODE regions were retained. For MNase-Seq, we used only those pairs of reads that were between 100 bp and 250 bp apart. The nucleosome coverage map was calculated, for each position, by counting the number of nucleosome-size fragments (centered at the mid-point of each pair of reads) that cover that position.
Supplementary Material
ACKNOWLEDGMENTS
Our research is supported by the Intramural Research Program of the US National Institute of Health, National Cancer Institute, and Center for Cancer Research, the National Library of Medicine and the Food and Drug Adminstration. This study utilized the high-performance computational capabilities of the Helix Systems at the National Institutes of Health, Bethesda, MD (<http://helix.nih.gov>http://helix.nih.gov). We thank R. Casellas and A. Yamane for Illumina sequencing and we thank D. Clark and E. Batchelor for critical comments.
Footnotes
AUTHOR CONTRIBUTIONS
F.K., L.B. A.G. K.B., J.L. and D.L. designed research; F.K. and L.B. performed experiments; A.G. developed analytic and computational tools, A.G., F.K., L.B., D.W. T.M.P and D.L. analyzed data; F.K., A.G., L.B. and D.L. wrote the paper. A.G. and D.L. wrote the supplementary material.
REFERENCES
- 1.Lavelle C. Forces and torques in the nucleus: chromatin under mechanical constraints. Biochem Cell Biol. 2009;87(1):307–322. doi: 10.1139/O08-123. [DOI] [PubMed] [Google Scholar]
- 2.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranello L, et al. The importance of being supercoiled: How DNA mechanics regulate dynamic processes. Biochim Biophys Acta. 2012;1819(7):632–638. doi: 10.1016/j.bbagrm.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darzacq X, et al. In vivo dynamics of RNA polymerase II transcription. Nature Structural & Molecular Biology. 2007;14(9):796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roca J. Transcriptional inhibition by DNA torsional stress. Transcription. 2011;2(2):82–85. doi: 10.4161/trns.2.2.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roca J. The torsional state of DNA within the chromosome. Chromosoma. 2011;120(4):323–334. doi: 10.1007/s00412-011-0324-y. [DOI] [PubMed] [Google Scholar]
- 7.Kouzine F, et al. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15(2):146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 8.Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front Biosci. 2007;12:4409–4423. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 9.Wong B, et al. Characterization of Z-DNA as a nucleosome-boundary element in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104(7):2229–2234. doi: 10.1073/pnas.0611447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durand-Dubief M, et al. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010;29(13):2126–2134. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seila AC, et al. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8(16):2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- 12.Gartenberg MR, Wang JC. Identification of barriers to rotation of DNA segments in yeast from the topology of DNA rings excised by an inducible site-specific recombinase. Proc Natl Acad Sci U S A. 1993;90(22):10514–10518. doi: 10.1073/pnas.90.22.10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Travers A, Muskhelishvili G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 2007;8(2):147–151. doi: 10.1038/sj.embor.7400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer PR, Bat O, Sinden RR. Measurement of localized DNA supercoiling and topological domain size in eukaryotic cells. Methods Enzymol. 1999;304:639–650. doi: 10.1016/s0076-6879(99)04038-0. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto K, Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J Cell Sci. 2004;117(Pt 17):3797–3805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- 16.Bermudez I, et al. A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding. Nucleic Acids Res. 2010;38(19):e182. doi: 10.1093/nar/gkq687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer PR, Sinden RR. Measurement of unrestrained negative supercoiling and topological domain size in living human cells. Biochemistry. 1997;36(11):3151–3158. doi: 10.1021/bi962396q. [DOI] [PubMed] [Google Scholar]
- 18.Jupe ER, Sinden RR, Cartwright IL. Stably maintained microdomain of localized unrestrained supercoiling at a Drosophila heat shock gene locus. EMBO J. 1993;12(3):1067–1075. doi: 10.1002/j.1460-2075.1993.tb05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jupe ER, Sinden RR, Cartwright IL. Specialized chromatin structure domain boundary elements flanking a Drosophila heat shock gene locus are under torsional strain in vivo. Biochemistry. 1995;34(8):2628–2633. doi: 10.1021/bi00008a029. [DOI] [PubMed] [Google Scholar]
- 20.Ljungman M, Hanawalt PC. Presence of negative torsional tension in the promoter region of the transcriptionally poised dihydrofolate reductase gene in vivo. Nucleic Acids Res. 1995;23(10):1782–1789. doi: 10.1093/nar/23.10.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwikardi M, Droge P. Site-specific recombination in mammalian cells catalyzed by gammadelta resolvase mutants: implications for the topology of episomal DNA. FEBS Lett. 2000;471(2–3):147–150. doi: 10.1016/s0014-5793(00)01394-6. [DOI] [PubMed] [Google Scholar]
- 22.Sinden RR, Bat O, Kramer PR. Psoralen cross-linking as probe of torsional tension and topological domain size in vivo. Methods. 1999;17(2):112–124. doi: 10.1006/meth.1998.0723. [DOI] [PubMed] [Google Scholar]
- 23.Myers RM, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghuraman MK, et al. Replication dynamics of the yeast genome. Science. 2001;294(5540):115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 25.Vos SM, et al. All tangled up: how cells direct, manage and exploit topoisomerase function. Nature Reviews Molecular Cell Biology. 2011;12(12):827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen VT, et al. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 1996;24(15):2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei G, et al. Genome-wide mapping of nucleosome occupancy, histone modifications, and gene expression using next-generation sequencing technology. Methods Enzymol. 2012;513:297–313. doi: 10.1016/B978-0-12-391938-0.00013-6. [DOI] [PubMed] [Google Scholar]
- 28.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32(1):37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frydman B, et al. Induction of DNA topoisomerase II-mediated DNA cleavage by beta-lapachone and related naphthoquinones. Cancer Res. 1997;57(4):620–627. [PubMed] [Google Scholar]
- 32.Li CJ, Averboukh L, Pardee AB. beta-Lapachone, a novel DNA topoisomerase I inhibitor with a mode of action different from camptothecin. J Biol Chem. 1993;268(30):22463–22468. [PubMed] [Google Scholar]
- 33.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6(10):789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 34.Zobeck KL, et al. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell. 2010;40(6):965–975. doi: 10.1016/j.molcel.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koster DA, et al. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448(7150):213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 36.Khobta A, et al. Early effects of topoisomerase I inhibition on RNA polymerase II along transcribed genes in human cells. J Mol Biol. 2006;357(1):127–138. doi: 10.1016/j.jmb.2005.12.069. [DOI] [PubMed] [Google Scholar]
- 37.Muslimovic A, et al. Numerical analysis of etoposide induced DNA breaks. PLoS One. 2009;4(6):e5859. doi: 10.1371/journal.pone.0005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouzine F, et al. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol. 2004;11(11):1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 41.Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 42.Baranello L, et al. DNA topoisomerase I inhibition by camptothecin induces escape of RNA polymerase II from promoter-proximal pause site, antisense transcription and histone acetylation at the human HIF-1alpha gene locus. Nucleic Acids Res. 2010;38(1):159–171. doi: 10.1093/nar/gkp817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Timsit Y, Varnai P. Helical chirality: a link between local interactions and global topology in DNA. PLoS One. 2010;5(2):e9326. doi: 10.1371/journal.pone.0009326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.French SL, et al. Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol Cell Biol. 2011;31(3):482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salceda J, Fernandez X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25(11):2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koster DA, et al. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434(7033):671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 47.Zlatanova J, Victor JM. How are nucleosomes disrupted during transcription elongation? HFSP J. 2009;3(6):373–378. doi: 10.2976/1.3249971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sperling AS, et al. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc Natl Acad Sci U S A. 2011;108(31):12693–12698. doi: 10.1073/pnas.1106834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyu YL, et al. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol Cell Biol. 2006;26(21):7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roca J, Wang JC. The probabilities of supercoil removal and decatenation by yeast DNA topoisomerase II. Genes Cells. 1996;1(1):17–27. doi: 10.1046/j.1365-2443.1996.01001.x. [DOI] [PubMed] [Google Scholar]
- 51.Wada H, Netz RR. Plectoneme creation reduces the rotational friction of a polymer. EPL (Europhysics Letters) 2009;87(3):38001. [Google Scholar]
- 52.Cheung AC, Sainsbury S, Cramer P. Structural basis of initial RNA polymerase II transcription. EMBO J. 2011;30(23):4755–4763. doi: 10.1038/emboj.2011.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, et al. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006;25(10):2119–2130. doi: 10.1038/sj.emboj.7601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9(12):849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 55.Roca J. The path of the DNA along the dimer interface of topoisomerase II. J Biol Chem. 2004;279(24):25783–25788. doi: 10.1074/jbc.M402555200. [DOI] [PubMed] [Google Scholar]
- 56.Lesne A, Becavin C, Victor JM. The condensed chromatin fiber: an allosteric chemo-mechanical machine for signal transduction and genome processing. Phys Biol. 2012;9(1):013001. doi: 10.1088/1478-3975/9/1/013001. [DOI] [PubMed] [Google Scholar]
- 57.Unniraman S, Nagaraja V. Axial distortion as a sensor of supercoil changes: a molecular model for the homeostatic regulation of DNA gyrase. J Genet. 2001;80(3):119–124. doi: 10.1007/BF02717907. [DOI] [PubMed] [Google Scholar]
- 58.Revyakin A, Ebright RH, Strick TR. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci U S A. 2004;101(14):4776–4780. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73(3):533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 60.Peter BJ, et al. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5(11):R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.