Abstract
Protein palmitoylation describes the post-translational modification of cysteines by a thioester-linked long chain fatty acid. This modification is critical for membrane association, spatial organization, and the proper activity of hundreds of membrane-associated proteins. Palmitoylation is continuously remodeled, both by spontaneous hydrolysis and enzyme-mediated de-palmitoylation. Bioorthogonal pulse-chase labeling approaches have highlighted the role of protein thioesterases as key regulators of palmitoylation dynamics. Importantly, thioesterases are critical for regulating the spatial organization of key oncogenic proteins, such as Ras GTPases. New inhibitors, probes, and proteomics methods have put a spotlight on this emerging post-translational modification. These tools promise to advance our understanding the enzymatic regulation of dynamic palmitoylation, and present new opportunities for drug development.
Introduction
Protein palmitoylation was first reported just a few months before the classic discovery of tyrosine phosphorylation[1–2], yet more than 30 years later, the importance of palmitoylation is only now gaining significant attention as a widespread, dynamic post-translational modification. This is likely due to a historical lack of robust methods for sensitive analysis of this non-polar, non-antigenic modification. Until recently, the only method to study palmitoylation involved metabolic labeling with [3H]-palmitate, followed by lengthy exposure times ranging from days to weeks. Given the lack of straightforward methods, the dynamics and regulation of protein palmitoylation is largely unexplored.
Protein palmitoylation is clearly important in establishing the spatial localization of many well studied signaling complexes. Cellular transformation by oncogenic v-Hras (H-RasG12V) requires membrane anchoring[3–4], and mutation of a single palmitoylation site eliminates the protein’s oncogenic potential[3]. The rate of palmitate turnover on inactive GDP-bound H-Ras is accelerated > 15 times upon activation[5]. Similarly, activation of G-alpha-s accelerates palmitate turnover nearly 50-fold[6]. Similar findings have been observed for the synaptic scaffolding protein PSD-95, which is rapidly depalmitoylated following glutamate stimulation[7]. Based on these observations, dynamic palmitoylation may be a general regulatory mechanism controlling signal-dependent spatial localization.
The goal of this review is to present recent advances for the detection, annotation, and quantification of dynamic palmitoylation, as well as a discussion of the potential for thioesterase inhibitors to modulate key signaling pathways.
Non-radioactive detection of Palmitoylation
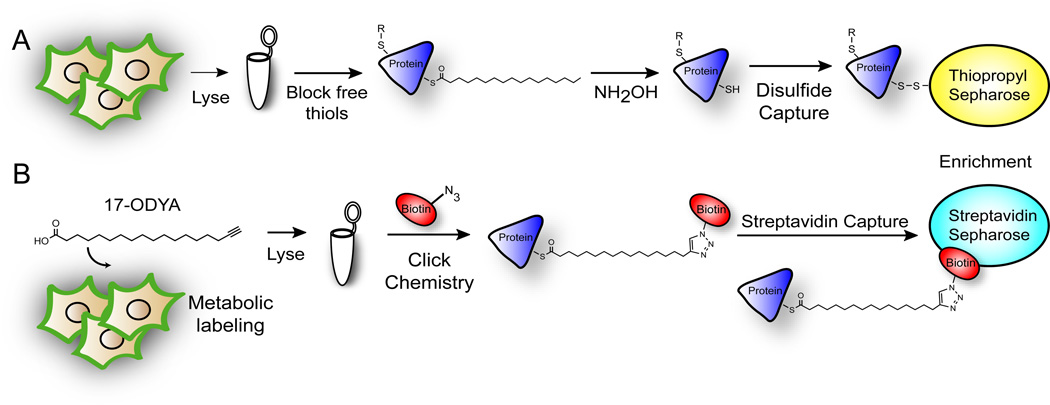
Two complementary methods have been developed in recent years for the non-radioactive detection, enrichment, and mass spectrometry-based annotation of palmitoylated proteins. The first method, termed acyl-biotin exchange, is useful for the static analysis of palmitoylated proteins in native tissues or cells[8–10]. In this method, lysates are first treated with methyl methanethiosulfonate (MMTS) or maleimide to block free thiols. Next, thioesters are hydrolyzed with hydroxylamine, which releasing the acyl chain and exposes new free thiols for disulfide capture[11]. One drawback to this approach is the enrichment of proteins with native thioesters, such as ubiquitin ligases and lipoamide-linked dehydrogenases. New modifications of this approach employ thiol resins for more simplified enrichment[12] (Figure 1A).
Figure 1.
Methods for palmitoylated protein enrichment. (A) Resin-assisted capture of hydroxylamine-sensitive cellular thioesters for static analysis of palmitoylation. After reduction and alkylation, lysates are treated with hydroxylamine to hydrolyze thioesters. Free thiols are captured by disulfide formation using thiopropyl sepharose resin. (B) Bioorthogonal enrichment of 17-ODYA metabolically labeled sites of palmitoylation. Biotin-azide is conjugated by click chemistry to 17-ODYA labeled proteins for streptavidin enrichment.
The second method uses metabolic labeling with the bioorthogonal fatty acid analogue 17-octadecynoic acid. The alkynyl fatty acid analogue is incorporated by the endogenous palmitoylation machinery into native sites palmitoylation. After lysis, labeled proteins are ligated to azide-linked reporter tags by click chemistry[13–14] (Figure 1B). Importantly, all reagents are commercially available and relatively inexpensive. The key advantages are a simplified workflow, high sensitivity, reduced non-specific labeling, and the ability to examine palmitoylation turnover dynamics by classic pulse-chase methods. Unlike ABE, this method only enriches native sites of long-chain fatty acid modification, and not other endogenous thioesters[9–10].
Both enrichment methods have been used to globally annotate palmitoylated proteins by mass spectrometry in a variety of organisms, tissues, and cell lines[9–10,13,15–17]. Altogether, more than 500 palmitoylated proteins have been annotated in mammalian cells. This list contains both integral and membrane-associated proteins, including channels, receptors, and scaffolding proteins. Based on these results, there are likely thousands of palmitoylated cysteine residues in the proteome[15], solidifying protein palmitoylation as pervasive as other widely studied polar post-translational modifications.
Quantitative Analysis of Palmitoylation
Ras is the prototypical palmitoylated protein, and has been used as a model to study the spatial organization, dynamics, and turnover of protein palmitoylation. Upon microinjection of fluorescent, palmitoylated N-Ras, the semi-synthetic protein rapidly distributes to all membranes, and enters a pathway of dynamic palmitoylation and de-palmitoylation[18–19]. N-Ras is quickly de-palmitoylated in the periphery, but re-palmitoylated at the golgi and recycled back to the plasma membrane through the secretory pathway. Complementary live-cell fluorescence imaging with transfected photo-convertible fluorescent protein fusions confirmed these observations. Based on these experiments, palmitoylation is hypothesized to stabilize the membrane attachment and increase the residency time of N-Ras at the plasma membrane[20]. This specific example demonstrates how dynamic palmitoylation can promote spatial organization and function of a key oncogenic signaling protein. Despite these promising observations, there is little evidence studying the dynamics of native proteins to support these findings.
Bioorthogonal metabolic pulse-chase labeling provides a direct approach to profile the global dynamics of palmitoylation on native proteins. Preliminary studies in Jurkat T-cells demonstrated an increase in the rate of Lck de-palmitoylation after pervanadate treatment[21]. Importantly, the rate of palmitoylation turnover was attenuated by treatment with the generic lipase inhibitor methyl arachidonyl fluorophosphonate (MAFP)[22]. This was the first demonstration on native proteins that unspecified serine hydrolases regulate the turnover of palmitoylation on native proteins.
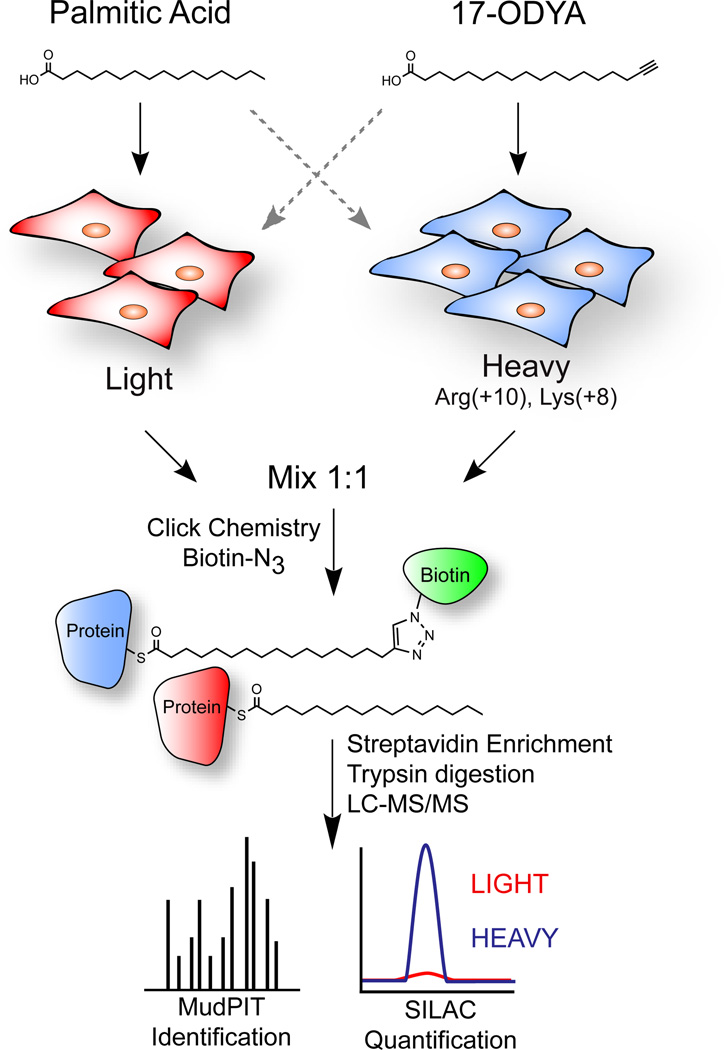
To measure fractional changes in palmitoylation, SILAC quantitative proteomics methods[23] were applied in conjunction with metabolic 17-ODYA and enrichment. BW5147-derived mouse T-cell hybridoma cells were first passaged in isotopic media according to standard SILAC protocols[24]. First, a list of high confidence palmitoylated proteins was established by performing two parallel experiments. In the first experiment, light cells were treated with 17-ODYA for 8 hours, and the heavy cells with palmitic acid, or vice versa (Figure 2). In a second experimental group, heavy and light cells were both treated with 17-ODYA for 8 hours, and one cell lysate was incubated with hydroxylamine to cleave thioesters and release 17-ODYA. In both experimental groups, isotopic paired lysates were mixed at a 1:1 ratio, conjugated to biotinazide, enriched, and digested for high resolution mass spectrometry analysis. The relative enrichment ratio is calculated by comparing the precursor extracted ion chromatograph for each isotopic peptide pair (17-ODYA / Control). A ratio of 1 signifies no specific enrichment, but the majority of peptides were completely absent in the control sample yielding an infinitely high ratio. Approximately 400 proteins displayed specific enrichment in reciprocal labeling experiments in both experimental groups. Similar quantitative proteomics studies in mouse neuronal stem cells identified a partially overlapping set of approximately 300 palmitoylated proteins[17]. These studies highlight the value of reciprocal SILAC approaches for providing high confidence annotation of low abundance palmitoylated proteins.
Figure 2.
Quantitative proteomic analysis of palmitoylated proteins using SILAC. Cells are grown for several passages in media with light or heavy isotopic lysine and arginine. Next, SILAC pairs are labeled with either palmitic acid or 17-ODYA. After metabolic labeling, lysates from isotopic pairs are mixed and conjugated to biotin-azide by click chemistry for high resolution mass spectrometry identification and quantification. Dashed arrows represent replicate experiments switching the order of fatty acid / probe labeling, which is used for combined reciprocal analysis to reduce false positives.
Global dynamics of palmitoylation
The deacylation of Ras is accelerated by protein thioesterases[18–19], yet little is known about the commonality of this mode of regulation, and whether this is specific for small GTPases. Several enzymes have been ascribed protein thioesterase activity, including the lysosomal hydrolase palmitoyl protein thioesterase 1 (PPT1)[25], as well as the soluble lysophospholipases LYPLA1 (APT1)[26–27] and LYPLA2 (APT2)[28–29]. Given the possibility that multiple hydrolases contribute to dynamic palmitoylation, we sought to develop tools for defining the subset of palmitoylated proteins regulated by any combination of lipases.
Any candidate protein thioesterase is likely a member of the serine hydrolase enzyme family[30] with preference towards lipidated substrates. The palmitate analogue hexadecylfluorophosphonate (HDFP) covalently inhibits approximately 20 lipases in T-cell hybridoma cells (including APT1, APT2, and PPT1), without inhibiting serine peptidase, proteases, or small molecule hydrolases[24]. To explore the global dynamics of palmitoylation turnover, SILAC pairs of T-cell hybridoma cells were pulse-labeled with 17-ODYA, and two groups of experiments were performed, one to determine the palmitoylated proteins with rapid turnover, and a second to profile palmitoylated proteins stabilized by HDFP[24]. Using SILAC quantitative analysis, both experiments were performed without over-expression and analyzed globally on native palmitoylated proteins after bioorthogonal enrichment.
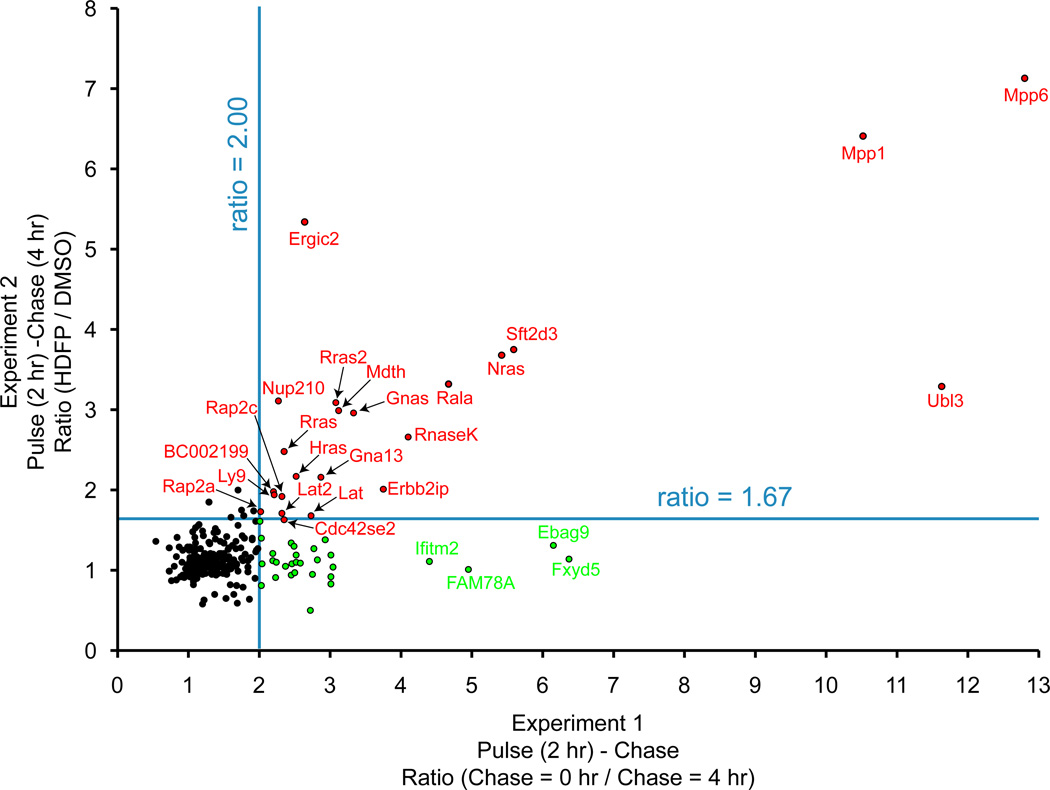
Pulse-chase proteomic analysis revealed several important findings (Figure 3). First, the majority of palmitoylated proteins did not change over the 4 hour chase time period, suggesting dynamic palmitoylation is not a general phenomenon. Secondly, there were several dozen palmitoylated proteins with both rapid turnover dynamics and stabilized by HDFP. This group includes many Ras family GTPases, G-alpha proteins (GNAS and GNA13), and membrane-associated guanylate kinase (MAGUK) proteins (MPP1 and MPP6), which all belong to enzyme families previously characterized as targets of dynamic palmitoylation turnover[31], as well as many other many other palmitoylated proteins implicated in cancer pathogenesis[32–33]. This study does not quantify specific sites of palmitoylation, which likely biases the results towards palmitoylated proteins with fewer, simultaneously regulated sites of palmitoylation. Also, longer pulse-labeling times are required for sufficient 17-ODYA labeling for mass spectrometry detection, making it more difficult to remove 17-ODYA from cellular lipid pools and thus reducing the chase efficiency. Therefore, these findings only highlight the most direct targets of thioesterases with the shortest half-lives. Such global experiments establish a role for protein thioesterases in the regulation of select palmitoylated proteins with established roles in cell growth and cancer.
Figure 3.
Pulse-chase proteomic analysis of dynamic palmitoylation. In experiment 1 (x-axis), SILAC cell pairs were labeled with 17-ODYA for 2 hours, then harvested or chased with 10-fold excess palmitic acid for 4 hours. This experiment highlights proteins with rapid de-palmitoylation or protein turnover. In experiment 2 (y-axis), SILAC cell pairs were labeled with 17-ODYA for 2 hours, and chased with excess palmitic acid with or without the lipase inhibitor HDFP. This experiment highlights palmitoylated proteins where probe labeling is stabilized by lipase inhibition. Blue lines are arbitrary thresholds. Red dots are enzymatically regulated proteins with rapid turnover kinetics. Green dots are proteins with enhanced degradation, as determined by unenriched SILAC proteomics experiments. Adapted with permission from [24]. Copyright (2012) Nature Publishing Group.
Protein thioesterases
Acyl-protein thioesterase 1 (LYPLA1) was first characterized as a lysophospholipase[27], but has several hundred-fold higher activity as a protein thioesterase[26]. Genetic deletion of the yeast homologue of LYPLA1 eliminates the in vitro de-palmitoylation activity from lysates, yet shows a very minor decrease in the turnover of palmitate on GNAS in vivo[34]. Furthermore, LYPLA1 mutant yeast have no defects in growth, mating, or deacylation of other palmitoylated proteins[34]. In mammalian synapses, LYPLA1 mRNA is silenced by the microRNA mi138[35]. After glutamate stimulation, microRNA suppression is relieved by RISC degradation, inducing local synaptic translation of LYPLA1 mRNA[36]. Suppression of LYPLA1 results in a partial reduction in synaptic volume, although there is no direct evidence this is due to the de-palmitoylation activity, and no synaptic protein substrates of LYPLA1 have been identified[35]. Overall, more genetic and biochemical evidence is needed to fully support the hypothesis that endogenous LYPLA1 is responsible for all enzymatic palmitoylation turnover.
Inhibitors of Dynamic Palmitoylation
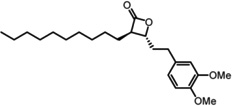
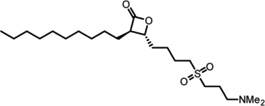
Protein thioesterases regulate the dynamic palmitoylation turnover on Ras and other important proteins involved in the progression of cancer, making LYPLA1 an attractive candidate for inhibitor development. The over-the-counter weight loss drug tetrahydrolipstatin (orlistat) inhibits a diverse subset of lipases to prevent intestinal lipid uptake[37–38]. This natural product contains a reactive β-lactone that is targeted by the nucleophilic serine of a wide range of hydrolases[39]. Careful diversification led to the discovery of a potent LYPLA1-directed, stereo-specific β-lactone inhibitor, named Palmostatin B[40] (Table 1). This compound blocks the acylation cycle of microinjected or transfected Ras in cells, and inhibits the proliferation of myeloid cells transformed with oncogenic H-Ras or N-Ras, but not K-ras4B (which is not palmitoylated)[41]. Competitive activity-based protein profiling using an alkynyl Palmostatin derivative shows inhibition of LYPLA1, LYPLA2, and PPT1 by both Palmostatin B, and the higher potency second generation derivative Palmostatin M[42–43] (Table 1). In vitro, PPT1 has broad thioesterase activity towards many palmitoyl proteins, but is a lysosomal resident hydrolase[44] and unlikely to contribute to dynamic palmitoylation turnover of plasma membrane-bound proteins. Mutations in PPT1 lead to the human neurodegenerative lysosomal storage disease infantile neuronal ceroid lipofuscinosis[45]. Mass spectrometry analysis shows a wide array of other enriched cellular targets[42], highlighting the need for more selective tools to separate the functional roles of each candidate thioesterase.
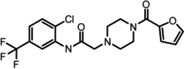
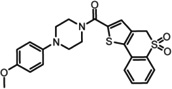
Table 1.
LYPLA1 and LYPLA2 inhibitors.
In order to develop selective inhibitors with more favorable drug-like properties, both LYPLA1 and LYPLA2 were screened in parallel against the NIH Molecular Library of 315,004 compounds using the robust competitive fluorophosphonate-rhodamine fluorescence polarization high-throughput assay (FluoPol-ABPP)[46–47]. Active site occupancy prevents FP-Rhodamine labeling of the enzyme, and hence reduces the fluorescence polarization signal[46]. This time-dependent competitive activity-based assay selects for active site inhibitors with potent binding and long residency times. Lead inhibitors for both LYPLA1 and LYPLA2 converged on a common piperizine amide chemotype[47]. In vitro competitive activity-based protein profiling (ABPP) of initial hits led to the identification of highly selective LYPLA1 and LYPLA2 inhibitors despite their significant homology (68%) (Table 1). Steady-state kinetic analysis reported Ki values of 230 nM (LYPLA2) and 300 nM (LYPLA1)[47]. No other T-cell hybridoma serine hydrolases were inhibited in competitive ABPP-SILAC experiments. Furthermore, these inhibitors maintain potency and selectivity in vivo in mice. These improved reversible probes promise to assign the individual functional roles of both LYPLA1 and LYPLA2, and accelerate the in vivo study of dynamic palmitoylation.
Conclusions
Emerging analytical tools and inhibitors have greatly accelerated our understanding the role of dynamic palmitoylation in the spatial organization of cellular signaling complexes critical for cell growth and membrane polarization. Non-radioactive, bioorthogonal labeling approaches have initiated a new focus on the uncharacterized dynamics of palmitoylation. Exploring the competitive role of palmitoylation with other cysteine modifications may begin to unravel the fundamental role of the palmitoylation cycle and the importance of specifically targeting a subset of palmitoylated proteins for dynamic turnover.
Highlights.
New methods allow direct enrichment of palmitoylated proteins for LC-MS annotation.
SILAC proteomics methods enhance the annotation of low abundance palmitoyl proteins.
Alkynyl fatty acids facilitate pulse-chase labeling for measuring palmitoylation dynamics.
Thioesterase inhibitors are promising new tools to dissect palmitoylation dynamics.
Acknowledgements
Funding is provided by the National Science Foundation Alliance for Graduate Education and the Professorate (J.L.H.), the National Institutes of Health (R00CA151460), and the University of Michigan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jeannie L. Hernandez, Email: jeannieh@umich.edu.
Jaimeen D. Majmudar, Email: jaimeen@umich.edu.
Brent R. Martin, Email: brentrm@umich.edu.
References
- 1.Eckhart W, Hutchinson MA, Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979;18:925–933. doi: 10.1016/0092-8674(79)90205-8. http://dx.doi.org/10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt MFG, Schlesinger MJ. Fatty acid binding to vesicular stomatitis virus glycoprotein: a new type of post-translational modification of the viral glycoprotein. Cell. 1979;17:813–819. doi: 10.1016/0092-8674(79)90321-0. [DOI] [PubMed] [Google Scholar]
- 3.Willumsen BM, Cox AD, Solski PA, Der CJ, Buss JE. Novel determinants of H-Ras plasma membrane localization and transformation. Oncogene. 1996;13:1901–1909. [PubMed] [Google Scholar]
- 4.Buss JE, Sefton BM. Direct identification of palmitic acid as the lipid attached to p21ras. Mol. Cell. Biol. 1986;6:116–122. doi: 10.1128/mcb.6.1.116. http://dx.doi.org/10.1128/MCB.6.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker TL, Zheng H, Walker J, Coloff JL, Buss JE. Distinct Rates of Palmitate Turnover on Membrane-bound Cellular and Oncogenic H-Ras. J. Biol. Chem. 2003;278:19292–19300. doi: 10.1074/jbc.M206956200. http://dx.doi.org/10.1074/jbc.M206956200. [DOI] [PubMed] [Google Scholar]
- 6.Wedegaertner PB, Bourne HR. Activation and depalmitoylation of Gs[alpha] Cell. 1994;77:1063–1070. doi: 10.1016/0092-8674(94)90445-6. http://dx.doi.org/10.1016/0092-8674(94)90445-6. [DOI] [PubMed] [Google Scholar]
- 7.El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. http://dx.doi.org/10.1016/S0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- 8.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 9.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, 3rd, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. http://dx.doi.org/10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. http://dx.doi.org/10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat. Protocols. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. http://dx.doi.org/10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 12. Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. http://dx.doi.org/10.1194/jlr.D011106. This method simplifies the capture and enrichment of native palmitoylation sites on proteins. Mass spectrometry data is presented for both tryptic peptides and specific sites of palmitoylation.
- 13.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. http://dx.doi.org/10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. Robust Fluorescent Detection of Protein Fatty-Acylation with Chemical Reporters. Journal of the American Chemical Society. 2009;131:4967–4975. doi: 10.1021/ja810122f. http://dx.doi.org/10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 15.Wilson JP, Raghavan AS, Yang YY, Charron G, Hang HC. Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M110.001198. http://dx.doi.org/10.1074/mcp.M110.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yount JS, Moltedo B, Yang Y-Y, Charron G, Moran TM, López CB, Hang HC. Palmitoylome profiling reveals S-palmitoylation–dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. http://dx.doi.org/10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, Martin BR, Cravatt BF, Hofmann SL. DHHC5 protein palmitoylates flotillin-2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. J Biol Chem. 2012;287:523–530. doi: 10.1074/jbc.M111.306183. http://dx.doi.org/10.1074/jbc.M111.306183. Using SILAC proteomics methods and 17-ODYA labeling, the authors identify a list of palmitoylated proteins in mouse neruonal stem cells. Cells lacking DHHC5 were also compared to normal cells to annotate DHHC5 substrates.
- 18. Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141:458–471. doi: 10.1016/j.cell.2010.04.007. This report demonstrates the importance of the palmitoylation / de-palmitoylation cycle in the spatial organization of Ras using an array of fluorescent microscopy approaches.
- 19.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. http://dx.doi.org/10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 20.Grecco HE, Schmick M, Bastiaens PI. Signaling from the living plasma membrane. Cell. 2011;144:897–909. doi: 10.1016/j.cell.2011.01.029. http://dx.doi.org/10.1016/j.cell.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 21. Zhang MM, Tsou LK, Charron G, Raghavan AS, Hang HC. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proceedings of the National Academy of Sciences. 2010;107:8627–8632. doi: 10.1073/pnas.0912306107. http://dx.doi.org/10.1073/pnas.0912306107. Using a toolset of bioorthogonal reporters, the authors demonstrate the accelerated turnover of LCK palmitoylation following pervanadate treatment in Jurkat T-cells. This acceleration is attentuated with MAFP treatment, highlighting a specific role for serine hydrolases in de-palmitoylation.
- 22.Deutsch DG, Omeir R, Arreaza G, Salehani D, Prestwich GD, Huang Z, Howlett A. Methyl arachidonyl fluorophosphonate: a potent irreversible inhibitor of anandamide amidase. Biochem Pharmacol. 1997;53:255–260. doi: 10.1016/s0006-2952(96)00830-1. http://dx.doi.org/10.1016/S0006-2952(96)00830-1. [DOI] [PubMed] [Google Scholar]
- 23.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. http://dx.doi.org/10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- 24. Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. http://dx.doi.org/10.1038/nmeth.1769. This study provides the first quantiative proteomic annotation of palmitoylation dynamics using bioothogonal pulse-chase methods. Only a subset of palmitoylated proteins are observed as distinct targets of protein thioesterases.
- 25.Camp L, Hofmann S. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J. Biol. Chem. 1993;268:22566–22574. [PubMed] [Google Scholar]
- 26.Duncan JA, Gilman AG. A Cytoplasmic Acyl-Protein Thioesterase That Removes Palmitate from G Protein alpha Subunits and p21RAS. J. Biol. Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. http://dx.doi.org/10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- 27.Sugimoto H, Hayashi H, Yamashita S. Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J Biol Chem. 1996;271:7705–7711. doi: 10.1074/jbc.271.13.7705. [DOI] [PubMed] [Google Scholar]
- 28.Toyoda T, Sugimoto H, Yamashita S. Sequence, expression in Escherichia coli, and characterization of lysophospholipase II. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1999;1437:182–193. doi: 10.1016/s1388-1981(99)00007-4. http://dx.doi.org/10.1016/S1388-1981(99)00007-4. [DOI] [PubMed] [Google Scholar]
- 29.Tomatis VM, Trenchi A, Gomez GA, Daniotti JL. Acyl-Protein Thioesterase 2 Catalizes the Deacylation of Peripheral Membrane-Associated GAP-43. PLoS One. 2010;5:e15045. doi: 10.1371/journal.pone.0015045. http://dx.doi.org/10.1371/journal.pone.0015045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. http://dx.doi.org/10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smotrys JE, Linder ME. Palmitoylation Of Intracellular Signaling Proteins: Regulation and Function. Annual Review of Biochemistry. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. http://dx.doi.org/10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 32.Yoo BK, Emdad L, Lee SG, Su ZZ, Santhekadur P, Chen D, Gredler R, Fisher PB, Sarkar D. Astrocyte elevated gene-1 (AEG-1): A multifunctional regulator of normal and abnormal physiology. Pharmacol Ther. 2011 doi: 10.1016/j.pharmthera.2011.01.008. http://dx.doi.org/10.1016/j.pharmthera.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. Deregulation of Scribble Promotes Mammary Tumorigenesis and Reveals a Role for Cell Polarity in Carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. http://dx.doi.org/10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duncan JA, Gilman AG. Characterization of Saccharomyces cerevisiae Acyl-protein Thioesterase 1, the Enzyme Responsible for G Protein alpha Subunit Deacylation in Vivo. J. Biol. Chem. 2002;277:31740–31752. doi: 10.1074/jbc.M202505200. http://dx.doi.org/10.1074/jbc.M202505200. [DOI] [PubMed] [Google Scholar]
- 35.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. http://dx.doi.org/10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerjee S, Neveu P, Kosik KS. A Coordinated Local Translational Control Point at the Synapse Involving Relief from Silencing and MOV10 Degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. http://dx.doi.org/10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 37.Borgstrom B. Mode of action of tetrahydrolipstatin: a derivative of the naturally occurring lipase inhibitor lipstatin. Biochim Biophys Acta. 1988;962:308–316. doi: 10.1016/0005-2760(88)90260-3. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y, Burn P. Lipid metabolic enzymes: emerging drug targets for the treatment of obesity. Nat Rev Drug Discov. 2004;3:695–710. doi: 10.1038/nrd1469. http://dx.doi.org/10.1038/nrd1469. [DOI] [PubMed] [Google Scholar]
- 39.Hoover HS, Blankman JL, Niessen S, Cravatt BF. Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorg Med Chem Lett. 2008;18:5838–5841. doi: 10.1016/j.bmcl.2008.06.091. http://dx.doi.org/10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449–456. doi: 10.1038/nchembio.362. http://dx.doi.org/10.1038/nchembio.362. The authors describe an inhibitor or LYPLA1 that blocks Ras dynamics in living cells. This inhibitor also reverses certain maligant characteristics of oncogenic Ras-transformed cells.
- 41. Xu J, Hedberg C, Dekker FJ, Li Q, Haigis KM, Hwang E, Waldmann H, Shannon K. Inhibiting the palmitoylation/depalmitoylation cycle selectively reduces the growth of hematopoietic cells expressing oncogenic Nras. Blood. 2012;119:1032–1035. doi: 10.1182/blood-2011-06-358960. http://dx.doi.org/10.1182/blood-2011-06-358960. Palmostatin B is shown to selectively block the growth of myeloid cells expressing oncogenic N- and H-Ras, but not non-palmitoylated K-Ras. This study highlights the therapeutic potential of LYPLA1 inhibtors for the treatment of certain Ras-dependent cancers.
- 42. Rusch M, Zimmermann TJ, Burger M, Dekker FJ, Gormer K, Triola G, Brockmeyer A, Janning P, Bottcher T, Sieber SA, et al. Identification of Acyl Protein Thioesterases 1 and 2 as the Cellular Targets of the Ras-Signaling Modulators Palmostatin B and M. Angew Chem Int Ed Engl. 2011 doi: 10.1002/anie.201102967. http://dx.doi.org/10.1002/anie.201102967. The authors synthesize an alkynyl Palmostatin M derivative for use as an activity-based probe. Gel and mass spectrometry analysis demonstrate that LYPLA1, LYPLA2, and PPT1 are inhibited by Palmostatin B and M. Kinetic experiments also characerize the inhibition of LYPLA2.
- 43. Hedberg C, Dekker FJ, Rusch M, Renner S, Wetzel S, Vartak N, Gerding-Reimers C, Bon RS, Bastiaens PI, Waldmann H. Development of Highly Potent Inhibitors of the Ras-Targeting Human Acyl Protein Thioesterases Based on Substrate Similarity Design. Angew Chem Int Ed Engl. 2011 doi: 10.1002/anie.201102965. http://dx.doi.org/10.1002/anie.201102965. An optimized β-lactone derivative (Palmostatin M) is presented with enhanced efficacy in cells. Inhibition and kinetic data are shown for LYPLA1.
- 44.Verkruyse LA, Hofmann SL. Lysosomal Targeting of Palmitoyl-protein Thioesterase. Journal of Biological Chemistry. 1996;271:15831–15836. doi: 10.1074/jbc.271.26.15831. http://dx.doi.org/10.1074/jbc.271.26.15831. [DOI] [PubMed] [Google Scholar]
- 45.Vesa J, Hellsten E, Verkruyse LA, Camp LA, Rapola J, Santavuori P, Hofmann SL, Peltonen L. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature. 1995;376:584–587. doi: 10.1038/376584a0. http://dx.doi.org/10.1038/376584a0. [DOI] [PubMed] [Google Scholar]
- 46.Bachovchin DA, Brown SJ, Rosen H, Cravatt BF. Identification of selective inhibitors of uncharacterized enzymes by high-throughput screening with fluorescent activity-based probes. Nat Biotechnol. 2009;27:387–394. doi: 10.1038/nbt.1531. http://dx.doi.org/10.1038/nbt.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J Am Chem Soc. 2012;134:10345–10348. doi: 10.1021/ja303400u. http://dx.doi.org/10.1021/ja303400u. A high-throughput competitive ABPP assay is used to identify novel reversible, selective inhibitors of LYPLA1 and LYPLA2, respectively. Activity-based probes with reduced affinity for LYPLA1 and LYPLA2 are used to profile reversible inhibitor selectivity in lysates, live cells, and in living mice by both fluorescent gels and SILAC-ABPP.