Abstract
Naegleria fowleri is a unicellular eukaryote causing primary amoebic meningoencephalitis, a neuropathic disease killing 99% of those infected, usually within 7–14 days. N. fowleri is found globally in regions including the US and Australia. The genome of the related non-pathogenic species Naegleria gruberi has been sequenced, but the genetic basis for N. fowleri pathogenicity is unclear. To generate such insight, we sequenced and assembled the mitochondrial genome and a 60-kb segment of nuclear genome from N. fowleri. The mitochondrial genome is highly similar to its counterpart in N. gruberi in gene complement and organization, while distinct lack of synteny is observed for the nuclear segments. Even in this short (60-kb) segment, we identified examples of potential factors for pathogenesis, including ten novel N. fowleri-specific genes. We also identified a homologue of cathepsin B; proteases proposed to be involved in the pathogenesis of diverse eukaryotic pathogens, including N. fowleri. Finally, we demonstrate a likely case of horizontal gene transfer between N. fowleri and two unrelated amoebae, one of which causes granulomatous amoebic encephalitis. This initial look into the N. fowleri nuclear genome has revealed several examples of potential pathogenesis factors, improving our understanding of a neglected pathogen of increasing global importance.
Keywords: PAM, meningitis, encephalitis, amoebic infections, infectious disease, genomics, deep sequencing, whole-genome sequencing, amoebic mitochondrial genome
NAEGLERIA fowleri is a deadly human pathogen, and the causative agent of primary amoebic meningoencephalitis (PAM). Cases have been reported from Australia, New Zealand, Africa, Mexico, Venezuela, India, as well as the United States and Europe (Visvesvara and Stehr-Green 1990; Yoder et al. 2010). N. fowleri may be more prevalent than reported, particularly in developing countries. It also exists in temperate regions in association with thermal waters; for example, in hot spring spas in Japan (Izumiyama et al. 2003) or in Yellowstone National Park in the United States (Sheehan et al. 2003). While most of the 130 cases reported in the United States have occurred in the southern-tier states, a single case recently reported from Minnesota indicates that the geographic patterns in the occurrence of N. fowleri infection are changing (Kemble et al. 2012). Consistent with its isolation from thermal waters, N. fowleri is thermotolerant, growing preferentially at 37 °C, but surviving at temperatures up to 45 °C (Kadlec 1975). N. fowleri can exist as a cyst, amoeba, or flagellate. The trophozoites range in size from 10–35 μm and are the primary infective stage of the amoeba, although the cyst form, as carried by wind currents, is associated with cases of PAM (Lawande et al. 1979). Recently, deaths from N. fowleri infection have been reported in the United States in association with the use of tap water in sinus irrigation devices (e.g. neti pots) (Naegleria FAQs 2011).
Infection by N. fowleri occurs when water containing the amoeba enters the nose (e.g. of swimmers), attaches to the olfactory mucosa, and passes through the cribriform plate to reach the olfactory bulb (Martinez and Visvesvara 1997). PAM symptoms include severe headache, nausea, vomiting, fever, stiff neck, and onset of coma and death within approximately ten days (Carter 1972; Martinez and Visvesvara 1997). Disease manifestations are not restricted to immunocompromised patients. Infection in humans is rare, but rapid, with a mortality rate of approximately 99% (Visvesvara and Stehr-Green 1990). The majority of individuals infected with PAM fail to be diagnosed promptly or correctly, and thus most cases are diagnosed postmortem (Heggie 2010). In diagnosed patients, the treatment involves the use of amphotericin B administered both intravenously and intrathecally along with miconazole or fluconazole and rifampin (da Rocha-Azevedo et al. 2009). However, to date only eight cases of successful treatment have been reported (Vargas-Zepeda et al. 2005). A fundamental understanding of N. fowleri at the genomic level would constitute the first step in identifying its mechanisms of pathogenesis, which would guide the development of safer or more effective therapies, and would also facilitate more effective and rapid molecular diagnostics.
N. fowleri is a single-celled microbial eukaryote, in the lineage Heterolobosea and within the supergroup Excavata (Dacks et al. 2008). N. fowleri has a non-pathogenic, non-thermotolerant relative Naegleria gruberi, for which the complete genome sequence has recently been determined (Fritz-Laylin et al. 2010). Several groups have identified differences between N. fowleri and N. gruberi, none of which fully define the pathogenic phenotype of N. fowleri (Marciano-Cabral and Cabral 2007; Serrano-Luna et al. 2007; Marciano-Cabral and Fulford 1986). Due to the unknown and likely multifactorial mechanisms of pathogenesis in N. fowleri, a comparative genomic approach may provide new insights into why N. fowleri causes a severe, quickly fatal disease, whereas N. gruberi is harmless. Specifically, we anticipate that genetic elements enabling pathogenesis that are unique to N. fowleri will be identified, including novel genes (i.e. ORFs with no known homologues), novel paralogues of known gene families, and genes obtained via horizontal gene transfer (HGT). An exploratory genomic approach may identify examples of these, even in the absence of a full genome analysis.
Here we present the use of unbiased next-generation deep sequencing to sequence the 49,530-base pair (bp) mitochondrial genome of N. fowleri to an average of 2,732X coverage. The recently published mitochondrial genome of N. gruberi (Fritz-Laylin et al. 2010) permitted a detailed comparative analysis of the mitochondrial genomes of N. fowleri and N. gruberi. In addition to the entire mitochondrial genome, we sequenced a 60,871-bp segment of the N. fowleri nuclear genome to an average of 501X coverage, and performed parallel analyses to those done for the mitochondrial genome. These studies reveal genes uniquely found in N. fowleri, a cathepsin B homologue and a likely case of HGT, all examples of encoded factors that may play potential roles in N. fowleri pathogenesis.
MATERIALS AND METHODS
Genomic DNA sequencing library preparation
N. fowleri (CDC:V212) was obtained from an existing collection at CDC. It was isolated from the cerebrospinal fluid (CSF) of a PAM patient from Alabama in 1990, for the purpose of diagnosis. All specimens received at CDC for diagnostic purposes are anonymized. Additionally, specimens from the deceased are exempt from IRB. Approximately 0.2 ml of the CSF was inoculated into monolayers of monkey kidney (E6) cell culture. Destruction of the cell culture from invasion of the monolayers by amoebae occurred within three days. Thereafter, amoebae were passaged ~12 times in E6 cell culture to maintain virulence and then established in modified Nelson’s medium with 5% FBS and stored frozen in liquid nitrogen (John and John 1994) Prior to harvesting for DNA extraction, frozen amoebae were thawed and grown to a density of 1×1012 organisms/ml. DNA was extracted with 500:1 of phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v) (Invitrogen Inc., Carlsbad, CA) and purified with the QIAamp DNA mini kit (Qiagen Inc., Valencia, CA) (Zhou et al. 2003). All manipulations of the organism and material extraction performed at the CDC was performed in Biosafety level 2 facilities as specified by CDC Biosafety guidelines, or in Biosafety level 2 facilities which have been specifically certified by the UCSF Biosafety Committee for research laboratories (BUA49187-BU-03-INC) for handling Naegleria fowleri and extracts derived from the organism.
Thirty micrograms of extracted DNA was sent to Macrogen (Seoul, Korea) for 454 GS FLX sequencing (25% 8-kb paired-end, 25% 1-kb paired-end, and 50% random shotgun sequencing), while 10 μg of extracted DNA was prepared for 100 bp paired-end Illumina HiSeq shotgun sequencing with the Illumina Paired-End Genomic DNA sample preparation kit, using nebulization for fragmentation. Genomic assembly was performed using Geneious Assembler (Geneious v. 5.5, Biomatters) using Low Sensitivity/Fast options with no fine tuning (Drummond et al. 2010).
Mitochondrial ORF prediction and annotation
The N. fowleri mitochondrial genomic sequence was used as a BLASTx (Altschul et al. 1990) query to search all annotated N. gruberi mitochondrial proteins at the National Center for Biotechnology Information website (NCBI, http://www.ncbi.nlm.nih.gov/). The BLOSUM62 substitution matrix was used as the default scoring matrix in all BLAST searches. The maximum percent identity of the top hit was used in conjunction with the E-value for ORF prediction and annotation. Sequences retrieved with an acceptable E-value of <5 × 10−2 were considered possible homologues in all BLAST searches.
Nuclear segment ORF prediction and annotation
Open reading frames (ORFs) were predicted using NCBI ORF Finder software (http://www.ncbi.nlm.nih.gov/projects/gorf/) and EMBOSS getorf software (http://emboss.bioinformatics.nl/), in combination with BLAST-searching into the N. gruberi NCBI and/or Joint Genome Institute (JGI, http://www.jgi.doe.gov/) databases, and the NCBI non-redundant (nr) database. The minimum threshold size cut-off for an ORF was 300 bp. ORFs were annotated based on their retrieved candidate orthologues via BLAST searching of the nr database, at an E-value of <5 × 10−2. Those that failed to retrieve any sequence with an acceptable E-value were designated as “putative ORFs”. Candidate N. fowleri ORFs were retrieved from genomic sequence using Sequencher 4.10.1 (Gene Codes Corporation, Ann Arbor, MI), with efforts made to obtain sequences with start and stop codons, as well as being of a similar size to the N. gruberi homologue or the best top BLAST result.
Nuclear segment gene organization
The N. fowleri queries and the retrieved N. gruberi orthologues were used as tBLASTn (Altschul et al. 1990) queries to search the N. gruberi whole genome shotgun database (NCBI). The first criterion for an N. gruberi scaffold to be considered to contain an N. fowleri orthologue was that both the N. fowleri ORF and N. gruberi genes retrieve the same N. gruberi scaffold with an acceptable E-value. The second criterion was that when the N. gruberi protein database downloaded from NCBI was queried against the available N. fowleri nuclear genome sequence using tBLASTn, the N. gruberi gene considered orthologous must retrieve the N. fowleri ORF with an E-value at least ten orders of magnitude smaller (i.e. more significant) than the E-value corresponding to any other query sequence.
Horizontal gene transfer
To investigate possible cases of horizontal gene transfer (HGT), i.e. transfer of genetic material between unrelated organisms, the N. fowleri proteins were used as BLASTp (Altschul et al. 1990) queries to search genomes representing the least divergent and most complete available genome sequences from the major lineages of eukaryotes. Specifically, the following databases were queried: Homo sapiens, Saccharomyces cerevisiae, Arabidopsis thaliana, Dictyostelium discoideum, Entamoeba histolytica, and Tetrahymena thermophila, all hosted by NCBI. For specific reasons having to do with taxonomic sampling points, the nr database was also searched for homologues in Coprinopsis cinerea, Polysphondylium pallidum, and Mastigamoeba balamuthi. The Cyanidioschyzon merolae genome was searched (http://merolae.biol.s.u-tokyo.ac.jp/) as well as the Ostreococcus tauri, Thalassiosira pseudonana, and Emiliania huxleyi genomes hosted by JGI. The Eukaryotic Pathogen Database Resources (EuPathDB, http://eupathdb.org/eupathdb/) were used to search the following organisms: Leishmania major, Trypanosoma brucei, Trypanosoma cruzi, Giardia intestinalis, Trichomonas vaginalis, and Toxoplasma gondii. Additionally, the Acanthamoeba castellanii genome was searched using tBLASTn. The A. castellanii genome is jointly hosted by the Human Genome Sequencing Center and Baylor College of Medicine (http://blast.hgsc.bcm.tmc.edu/bcm/blast/microbialblast.cgi?organism=AcastellaniNeff). Again, the homology criterion was that the sequence must have an E-value of <5×10−2. In the case of A. castellanii, from which only contigs (“contiguous sequences”) were available for searching, genes were manually annotated using Sequencher 4.10.1.
Cathepsin identification
The genomes of T. brucei, T. cruzi, Crithidia fasciculata (www.sanger.ac.uk), L. major, and N. gruberi, were searched for cathepsin sequences using BLASTp, with an E-value cutoff of <5×10−2. These were added to a previously established dataset modified from (Dacks et al. 2008).
Domain identification
ORF domains in the N. fowleri nuclear segment were identified using the Conserved Domain Database (Marchler-Bauer et al. 2010) at NCBI (at an E-value of <5×10−2).
RNA prediction
tRNAscan-SE 1.21 (Schattner et al. 2005) was used to identify tRNAs, RNAweasel (http://megasun.bch.umontreal.ca/RNAweasel/) was used to predict group I and II type introns, tRNAs, and small subunit rRNA, and RNAmmer (Lagesen et al. 2007) was used to predict the large rRNA subunit.
Sequence mapping
The mitochondrial and linear DNA maps of N. gruberi and N. fowleri were created using the program XPlasMap 0.96 (http://www.iayork.com/XPlasMap/).
Signal sequence identification
The program SignalP (Dyrløv Bendtsen et al. 2004) (http://www.cbs.dtu.dk/services/SignalP/) was used to predict possible signal peptides and signal anchors.
Alignments and phylogenetics
Phylogenetic trees were run for the protein homologues of the N. fowleri putative cathepsin B gene (Contig9_16_26041_25267). The dataset of cathepsin B and L homologues had 100 sequences and 156 positions. Sequences were aligned and trimmed to contain only unambiguously homologous positions. Alignment was done using the sequence alignment program MUSCLE 3.5 (Edgar 2004), and masking and trimming were done by eye using MacClade 4.08 (Maddison and Maddison 1989). The alignment is available upon request. ProtTest 1.3 (Abascal et al. 2005) was used to determine that the WAG+G model of protein evolution best fit the data allowing for incorporating correction for a four-category gamma correction for rate variation when appropriate.
MR BAYES v. 3.2.1 (Ronquist and Huelsenbeck 2003) was used to search treespace using 1,000,000 generations. Consensus trees were generated using a burn-in value of 25%. This was validated by plotting likelihood versus generations to ensure that no trees were included prior to the likelihood plateau. Two independent runs, each of four chains, were performed, with convergence of the results confirmed by ensuring a splits frequency of <0.1. PhyML v. 2.4.4 (Guindon and Gascuel 2003) and RAxML-VI-HPC v. 2.2.3 (Stamatakis 2006) were used for maximum-likelihood analyses, and to generate ML-bootstrap values based on 100 pseudo-replicates of each dataset. The tree diagram shown in phylogenetic figures is the best Bayesian topology, with support values listed in the order of Bayesian posterior probability values/PhyML bootstrap values/RAxML bootstrap values.
PCR confirmation of the N. fowleri 60-kb segment from the nuclear genome
For confirmation of organization of the initial contig assembly corresponding to the ~60-kb segment from the N. fowleri nuclear genome, PCR was performed using standard techniques. Briefly, 10 ng of genomic DNA was used as template in a 25 μl PCR reaction containing 1X NEB Standard Taq buffer, 250 μM dNTPs, 10 pmol of forward and reverse primer, and 5 units of purified recombinant Taq polymerase. Forward and reverse primer sequences are shown in Table S1. PCR conditions were 35 cycles of 95°C denaturation for 30s, 50°C annealing for 30s, and a 72°C extension for 90s. 10 μl of amplified material was run out on a 1.5% agarose gel stained with ethidium bromide. Amplified bands of the expected size were visualized under ultraviolet light.
RESULTS
To better understand the genetic makeup of Naegleria fowleri and gain insight into pathogenetic mechanisms, we analyzed extracted amoebic DNA by unbiased deep sequencing using both 454 GS FLX and Illumina HiSeq technology. The goal of 454 pyrosequencing was to provide paired-end scaffolds to facilitate de novo assembly from short 100-bp Illumina reads. This enabled us to assemble the mitochondrial genome and a 60-kb segment from the nuclear genome of N. fowleri. For assembly of the mitochondrial genome of N. fowleri, initial contigs assembled via Geneious Assembler with greater than 100X coverage and >75% nucleotide identity were aligned to the mitochondrial genome of N. gruberi. These initial contigs were then used to assemble the entire mitochondrial genome from 454 data using the Geneious assembler. In total, 393,244 reads assembled into a circular consensus mitochondrial genome of 49,519 nucleotides with an average coverage of 2,732X (range of 766–5,317X). The sequence of the mitochondrial genome of N. fowleri has been deposited into GenBank (accession number JX174181). De novo assembly of additional reads generated a 60-kb segment with an average coverage of 501X (range of 75–8,772X), which we chose for annotation and analysis. This sequence has also been deposited in GenBank (accession number JX827422).
To confirm the organization of the 60-kb assembly reported here, and to rule out the presence of gross indels or translocations, PCR of 11 regions spanning the nuclear genome, each ~1000 bp in length, was performed. Bands of expected size were seen for all 11 PCR amplicons (Fig. 1–2).
Fig. 1–2.
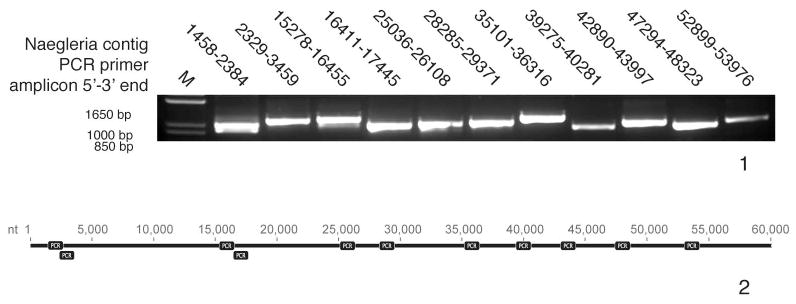
PCR confirmation of assembly. 1. Eleven ~1000 bp amplicons from across the 60-kb segment in the nuclear genome were successfully recovered by PCR amplification. M, ladder. 2. Diagram of where the amplicons were located across the contig.
Statistics
The mitochondrial genome of N. fowleri is 49,519 bp and is AT-rich, having a GC content of only 25.2% (Table 1). Coding sequence comprises 90% of the genome, and no introns are present in the non-coding regions. The N. gruberi mitochondrial genome is slightly larger at 49,842 bp, with a GC content of 22% and coding content of 92%, and also does not contain introns (Table 1). The median exon length for N. fowleri is similar to that of N. gruberi, being shorter by only 32 bp. The N. fowleri mitochondrial DNA contains 70 genes, 46 of which encode proteins and 23 of which encode transfer and ribosomal RNAs.
Table 1.
Mitochondrial genome and nuclear segment statistics from Naegleria gruberi and Naegleria fowleri
| Species | Size (bp) | %GC | %Coding | %Genes with introns | Introns per gene | Median intron length | Exons per gene | Median exon length |
|---|---|---|---|---|---|---|---|---|
| Ng mitochondriaa | 49,842 | 22 | 92 | 0 | 0 | N/Ab | 1 | 795 bp |
| Nf mitochondria | 49,519 | 25.2 | 90 | 0 | 0 | N/Ab | 1 | 766.5 bp |
| Ng nuclear genome | ~41M | 33 | 57.8 | 36 | 0.7 | 60 bp | NDc | NDc |
| Nf nuclear segment | 60,871 | 36.8 | 57.3 | 35 | 0.7 | 87 bp | 1.74 | 432 bp |
The N. fowleri nuclear genome segment is 60,871 bp in length, with a GC content of 36.8%, and a coding content of 57.3% (Table 1). ORF prediction analysis identified 31 putative ORFs in the segment. Table 1 shows that there are 1.74 exons per gene with a median exon length of 432 bp. Thirty-five percent of the ORFs have introns, with 0.7 introns per gene and a median length of 87 bp. These statistics are largely similar to those for the entire N. gruberi genome (Table 1).
Gene complement and organization
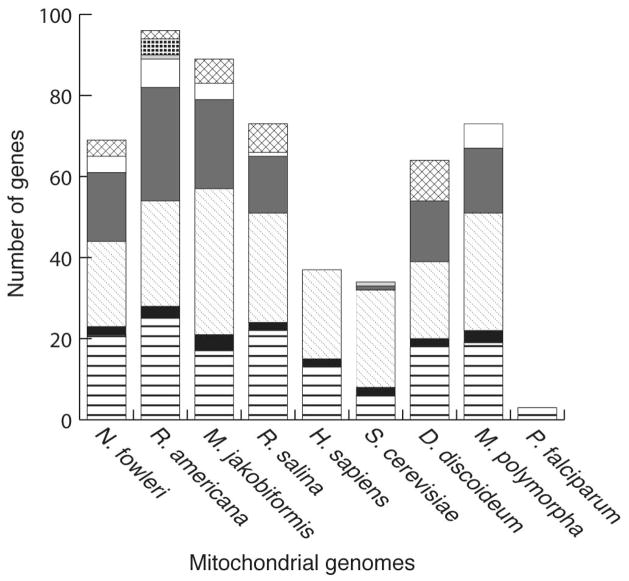
The mitochondrial gene complement of N. fowleri encodes products involved in reductive and oxidative phosphorylation, protein import and maturation, ribosomal proteins, rRNAs, tRNAs, and four ORFs of unknown function (Table S2). The majority of the genes are ribosomal proteins, tRNAs, or are involved in reductive and oxidative phosphorylation. In comparison with the mitochondria of other eukaryotes, as in Fig. 3, the proportions of genes in each category are standard among the diversity observed in other species.
Fig. 3.
Gene content of the Naegleria fowleri and Naegleria gruberi mitochondrial genomes in comparison with other eukaryotes. Genes are included in the classes in the following way, as in (Burger et al. 2003). Reduction and oxidative phosphorylation (horizontal bars): atp1, atp3, atp4, atp6, atp8, atp9; cob; cox1–3; nad1–4, nad4L, nad6–11, sdh2–4. rRNAs (solid black): rnl, rns, rrn5. tRNAs (diagonal dots): trnA-Y. Ribosomal proteins and EF-Tu (solid dark grey): rps1–4, rps7, rps8, rps10–14, rps19; rpl1, rpl5, rpl6, rpl10, rpl11, rpl14, rpl16, rpl18–20, rpl27, rpl31, rpl32, rpl34, rpl36; tufA. Protein import and maturation (solid white): secY, ymf16, tatC, yejR (ccmF), yejU (ccmC), yejV (ccmB), yejW (ccmA); cox11. RNA maturation (solid light grey): rnpB. Transcription (black squares): rpoA-D. Other (hatched): ORFs of unknown function. Data for all organisms except N. fowleri was retrieved from NCBI. In Dictyostelium discoideum, cox1 and cox2 are encoded as a single ORF, but are counted as two genes here. Abbreviations: N. fowleri=Naegleria fowleri; R. americana=Reclinomonas americana; M. jakobiformis=Malawimonas jakobiformis; R. salina=Rhodomonas salina; H. sapiens=Homo sapiens; S. cerevisiae=Saccharomyces cerevisiae; D. discoideum= Dictyostelium discoideum; M. polymorpha=Marchantia polymorpha; P. falciparum=Plasmodium falciparum.
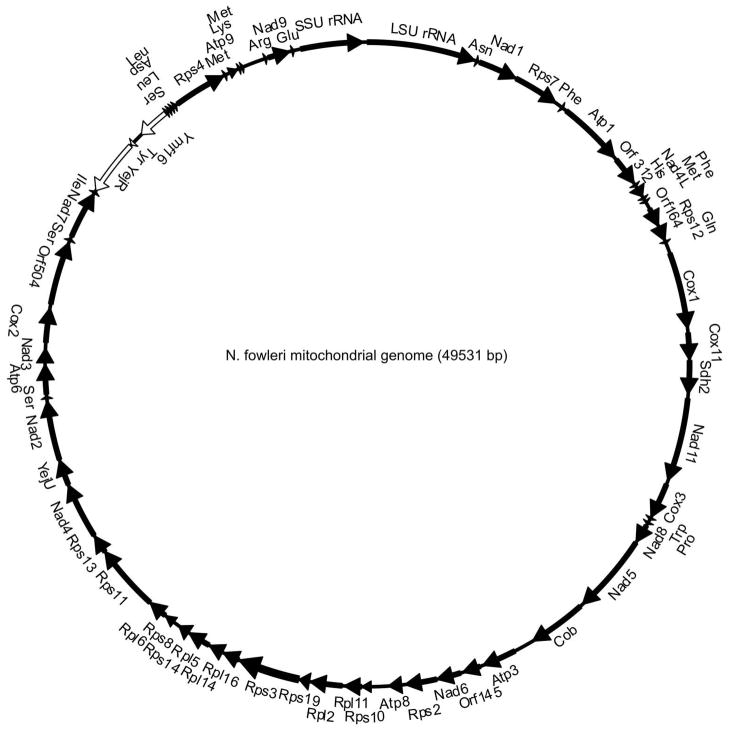
Genes encoded by the N. fowleri mitochondrial genome are tightly packed; only 10% of the genome is non-coding sequence (Fig. 4). ORF prediction software failed to identify any protein-encoding regions in the N. fowleri genome corresponding to genes not present in N. gruberi, and homologues of all annotated N. gruberi genes were found in N. fowleri. The mitochondrial genome of N. fowleri retains the bacteria-like organization represented by a similar gene order of small and large ribosomal proteins to a contiguous alignment of the str, S10, spc, and α operons, seen in N. gruberi and a variety of related single-cell organisms.
Fig. 4.
Circular map of the Naegleria fowleri mitochondrial genome. Genes encoding proteins are annotated based on BLAST results, and genes encoding RNA are annotated based on tRNA and rRNA scanning software predictions (see Materials and Methods section). For the full name of each gene, see Table S2. Black arrows with gene names on the outside of the map represent genes on one strand, and white arrows with gene names on the inside of the map represent genes on the alternate strand. The N. fowleri mitochondrial genome is identical in gene content and organization to the mitochondrial genome of Naegleria gruberi.
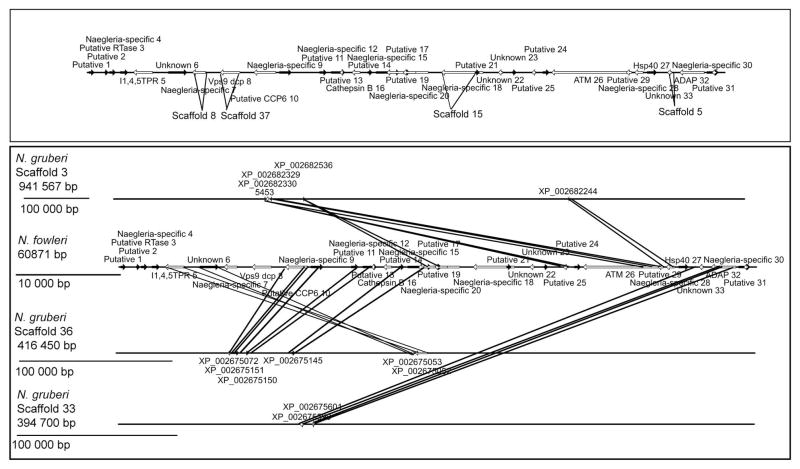
Unlike the collinearity (exact corresponding gene order) observed between the mitochondrial genomes of N. fowleri and N. gruberi, the organization of the N. fowleri ORFs on the 60-kb nuclear segment is not maintained by the N. gruberi orthologues at this arbitrary locus (Fig. 5). Three N. gruberi scaffolds have two or more genes from the N. fowleri nuclear segment, and four genes from the segment are found separately on four additional N. gruberi scaffolds. PCR to amplify the predicted ORFs was used to confirm that this striking result is not due to misassembly of the N. fowleri nuclear segment (Fig. 1–2). Indeed, the organizational dissimilarity between the predicted N. fowleri ORFs and N. gruberi orthologues notably contrasts with the relative similarity in statistics and gene complement between the two organisms.
Fig. 5.
Comparative collinearity analysis of the predicted Naegleria fowleri ORFs on the 60-kb nuclear segment and their homologues in the Naegleria gruberi nuclear genome. Black right arrows indicate ORFs on one strand, while white left arrows indicate ORFs on the other strand. (Top) Genes on the N. fowleri segment and the organization of homologues on individual scaffolds. (Bottom) The N. fowleri segment is in the middle, with the arrangements of homologous genes on N. gruberi scaffolds containing two or more genes above and below. Annotations of N. fowleri genes are based on BLAST results (see Materials and Methods), while those for N. gruberi are the GenBank protein accession for the homologue of each N. fowleri ORF. Scale bars are given for each map at 100,000 bp, with the exception of the N. fowleri segment, for which the scale bar indicates 10,000 bp. There is a distinct lack of collinearity between the two Naegleria species at this region of the N. fowleri genome.
Thirty-one ORFs were identified in the 60-kb segment from the N. fowleri nuclear genome (Table S3; Fig. 5). Of the 31 ORFs, 12 appear to have homologues in multiple eukaryotic genomes, while nine of the 31 ORFs appear to be found only in N. fowleri and N. gruberi. Interestingly, 10 of the 31 ORFs were not identified as having a homologue in any other eukaryote, and therefore might be specific to N. fowleri. Of the ORFs with homologues in other eukaryotic genomes, seven contain recognizable domains, such as Vps9 and ERV1, as identified by searching the conserved domain database (Table 2). Six ORFs with homologues in other eukaryotic genomes match functionally annotated proteins. There is possible evidence for a retrotransposon in the segment, as two adjacent ORFs have a reverse transcriptase or RNase H domain. One additional ORF of unknown identity was identified as having a signal anchor (Contig9_30_59176_60250), and was thus predicted to be a type II membrane protein.
Table 2.
Naegleria fowleri nuclear segment genes retrieving characterized homologues, or with domains in the Conserved Domain Database
| N. fowleri gene | Annotationa | Notesb |
|---|---|---|
| Contig9_3_2113_2703 | Putative reverse transcriptase | Reverse transcriptase domains |
| Contig9_5_6233_4411 | Inositol 1, 4, 5-triphosphate receptor | Calcium channel glycoprotein activated by inositol triphosphate |
| Contig9_10_17987_15908 | Possible cytosolic carboxypeptidase 6 | Possible metallocarboxypeptidase involved in deglutamylation |
| Contig9_16_26041_25267 | Cathepsin B | Cysteine protease |
| Contig9_24_43494_44097 | Erv1/Alr domain containing protein | Erv1/Alr family proteins involved in iron-sulfur cluster biogenesis |
| Contig9_26_51515_44333 | ATM (ataxia telangiectasia mutated) | Cell cycle checkpoint protein, with serine/threonine kinase domains |
| Contig9_27_53454_54366 | Hsp40 | Heat shock protein, with DNAJ and RRM domains |
| Contig9_32_58903_56581 | ADAP | ArfGAP with dual pH domains |
| Contig9_4_3139_3870 | Naegleria-specific | Contains RNAse H domain |
| Contig9_18_37776_33830 | Naegleria-specific | Contains serine/threonine kinase domains |
| Contig9_6_7742_9509 | Unknown | Contains leucine-rich repeats |
| Contig9_8_12703_14563 | Unknown | Contains the Vps9 domain, found in vacuolar protein sorting protein 9 |
| Contig9_33_55908_55399 | Unknown | Contains PUB domain |
Annotations were assigned based on the annotation of homologous genes.
Notes contain additional information about the gene.
Cathepsin identification
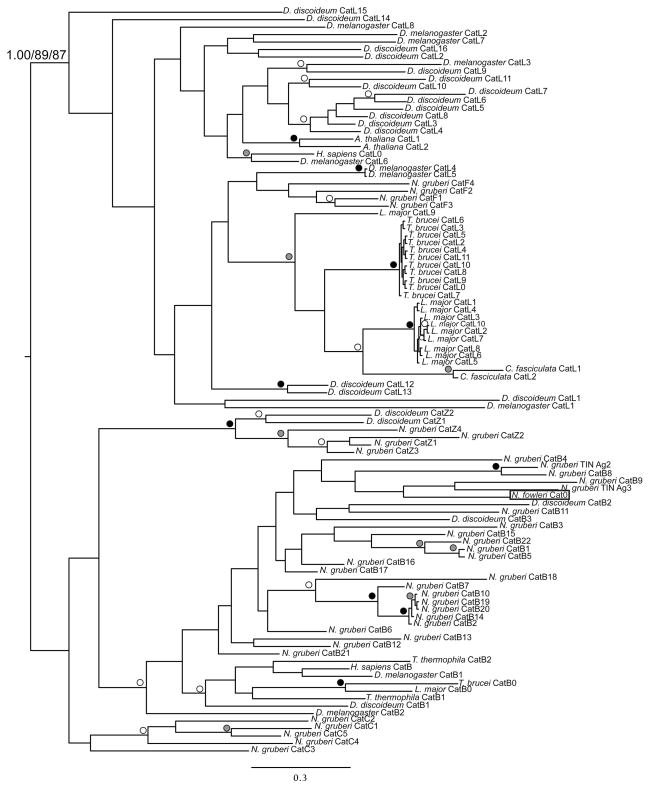
Using N-terminal peptide sequencing, Kim et al. (2009) identified two proteins secreted from N. fowleri that they annotated as cathepsin B and cathepsin B-like, proposing that these cysteine proteases might be involved in the invasion through the blood-brain barrier. Other pathogens, such as the liver fluke Fasciola hepatica, a multicellular parasite, have also been observed to secrete cathepsin B (Law et al. 2003). We identified a cathepsin homologue within the N. fowleri 60-kb nuclear segment (Contig9_16_26041_25267). Interestingly, this ORF was identified as having a signal peptide, raising the possibility of its transport out of the cell via the secretory pathway. Initial BLASTp searches suggested strong similarity to the human cathepsin B, as opposed to cathepsin L. Our phylogenetic analysis (Fig. 6) shows separation of the cathepsin B and L clades with strong statistical support, and groups the N. fowleri cathepsin homologue well within the cathepsin B clade. This analysis also revealed a large expansion of the cathepsin B family in N. gruberi, with multiple independent groups of duplications, two of which producing tubulointerstitial nephritis (TIN) antigen-like genes. While clearly a cathepsin B, the putative N. fowleri homologue is not closely related to any of the N. gruberi sequences.
Fig. 6.
Phylogeny of cathepsin family homologues. This phylogeny is a composite of MrBayes, PhyML, and RAxML results shown on the best Bayesian topology, arbitrarily rooted between the cathepsin B and L clades. The relevant nodes are marked using dots that represent Bayesian posterior probability/PhyML bootstrap/RAxML bootstrap values better than the following thresholds: 1.00/95/95 (black), 0.95/75/75 (grey), and 0.80/50/50 (white). The N. fowleri sequence is boxed. It is found within the cathepsin B clade, supporting its identity as part of the cathepsin B family.
Horizontal gene transfer
HGT events that occurred in N. fowleri, but not in N. gruberi, could contribute to its pathogenic phenotype. In support of this possibility, we identified an ORF encoding a putative 121 amino acid protein (Contig9_23_40599_40961) in the 60-kb segment from the nuclear genome that may have been involved in an HGT event. This sequence did not have a robustly identified N. gruberi homologue. However, upon expanding the search to a diversity of eukaryotes (via the non-redundant, plus organism-specific genomic databases, see Materials and Methods), we did retrieve a corresponding contig of 9,392 bp (high scoring segment pair of 384 amino acids) from the distantly related, amoebozoan, organism A. castellanii (E-value of 4 × 10−5) and one of 311 amino acids from the amoebozoan slime mold P. pallidum (E-value of 3 × 10−5).
DISCUSSION
We have obtained the full mitochondrial genome of N. fowleri and of a 60-kb segment of the N. fowleri nuclear genome, nearly doubling the number of N. fowleri protein entries in GenBank as of November 2012. In addition to providing targets for future molecular parasitological work, these data have provided new insights into the organization and evolution of the Naegleria mitochondrial genome, and a glimpse into the genomic structure of N. fowleri.
The N. fowleri and N. gruberi mitochondrial genomes both reconstitute as circular and encode many of the protein-coding genes, rRNAs, and tRNAs required for mitochondrial function. While the mitochondrial genomic content is variable in eukaryotes (Burger et al. 2003), both N. fowleri and N. gruberi appear to have “classical” collinear sets of mitochondrial genes that are shared among bacteria and many single-celled eukaryotes. Conspicuous in their absence are the genes for RNA maturation. RNA maturation involves the rnpB gene, encoding the ribozyme RNase P, which has been found in the mitochondrial genomes of Reclinomonas americana and S. cerevisiae. It is not known what performs this function in N. fowleri, as an rnpB homologue has not yet been identified in either N. fowleri or N. gruberi, and eukaryotes related to Naegleria lack RNaseP (Burger et al. 2003), with the exception of T. brucei (Salavati et al. 2001).
With the abandonment of the bikont-unikont rooting for the tree of eukaryotes (Roger and Simpson 2009), several features of mitochondrial genomes have been brought into the spotlight. The Heterolobosea (of which Naegleria is a member) are related to two other lineages, most closely to the Euglenozoa and then to the jakobids. The latter prominently have the most bacteria-like mitochondrial genome organization described, both in possession of bacterial RNA polymerase subunits and in operon structure (Lang et al. 1997). No RNA polymerases are encoded in the mitochondrial genomes of Naegleria. Instead, the nuclear genome of N. gruberi encodes a bacteriophage T3/T7-like polymerase (Cermakian et al. 1996), and it is likely that this is the case in N. fowleri, as it is in the vast majority of eukaryotes. However, the N. fowleri and N. gruberi mitochondrial genomes do share a pattern of rps gene organization that is bacteria-like, as is found in at least one member of all six eukaryotic supergroups, which suggests the retention of a plesiomorphic state in these mitochondrial genomes rather than an indication of deeply-branching status for any of the organisms in question (Hauth et al. 2005). Interestingly, no euglenozoans to date are identified as having mitochondrial genomes with bacteria-like organization. Trypanosomatids have maxicircles and minicircles (Westenberger et al. 2006), while the euglenid Diplonema papillatum has a small circular chromosome (Marande et al. 2005).
In general, there is great diversity in observed levels of mitochondrial genome synteny between species (Burger et al. 2003), even between closely related organisms. Within the genus Candida, for instance, there is extensive mitochondrial gene rearrangement between some members, although other members share a conserved gene order (Valach et al. 2011). Thus, it is notable that the N. fowleri and N. gruberi mitochondrial genomes have remarkably similar statistics, gene content, and gene order organization. This is in stark contrast with the gene organization of the 60-kb nuclear segment, which is marked by a conspicuous lack of collinearity between N. fowleri and N. gruberi, as confirmed by our PCR experimental testing of our in silico N. fowleri assembly. Although work with the phylum Apicomplexa has shown extensive genomic rearrangement between species, with little conserved collinearity and synteny between major lineages (DeBarry and Kissinger 2011), the most closely related comparison points to the Heterolobosea, the trypanosomatids, retain highly conserved gene order (Ghedin et al. 2004). While there are several possible genetic mechanisms that lead to genomic re-arrangements, such as transposable elements, or chromosomal breakage, recombination events during meiosis prominently produce this effect. Thus the lack of observed synteny between the Naegleria species may be due to sexual recombination events occurring after the evolutionary split from their last common Naegleria ancestor. N. gruberi has maintained apparently functional copies of genes required for meiosis (Fritz-Laylin et al. 2011). Additionally, there is strong evidence for a sexual cycle in N. lovaniensis (Pernin et al. 1992). Based on these data, a sexual cycle is likely to be operating in Naegleria species, and acting as a source of genetic diversity. If the observed lack of synteny between N. fowleri and N. gruberi is indeed due to meiotic recombination events, this finding would have important implications for the development of drug resistance in N. fowleri.
GC content is widely variable in eukaryotic nuclear genomes. The N. fowleri 60-kb segment and N. gruberi genome are both GC-poor, at 36.8% and 33%, respectively. It is possible that the arbitrary N. fowleri segment that was sequenced might be part of an isochore, a long stretch of DNA homogenous in GC content, as related organisms T. brucei and T. equiperdum have isochore-like organization (Isacchi et al. 1993). On the other hand, assuming that the observed GC content in the 60-kb segment is representative of the overall N. fowleri genome, the slightly elevated GC content relative to N. gruberi might reflect an adaptation to its thermotolerant lifestyle. N. fowleri can grow at 45 °C (Visvesvara et al. 2007), while N. gruberi can only tolerate temperatures up to 37 °C (Griffin 1972). Indeed, the red alga Cyanidioschyzon merolae lives in acidic hot springs at temperatures of 45 °C and has a GC content of 55% (Matsuzaki et al. 2004).
Our analysis of the 60-kb segment of the nuclear genome has furthermore identified examples of several of the anticipated sources of novel factors enabling pathogenesis in N. fowleri. Nine of 31 potential genes in the N. fowleri 60-kb segment were specific to Naegleria species, although the majority of ORFs had well-characterized and likely essential homologues in yeast or mammalian cells. Twenty of the 31 genes in the N. fowleri segment have a homologue in N. gruberi. Previously, the level of divergence between N. fowleri and N. gruberi has been estimated to be comparable to that between humans and frogs based on analyses using 18S ribosomal RNA (Baverstock et al. 1989). Our analyses of the mitochondrial genomes and nuclear genome segments of these two amoebae raise questions not only about the degree of their relatedness, but about how relatedness is best measured. While the lack of collinearity certainly highlights the unexpected diversity in genomes of Naegleria species, the identification of 20 of 31 homologues is evidence that many, if not the majority, of genes may be shared in common between N. fowleri and N. gruberi.
Ten ORFs predicted in the 60-kb nuclear segment were not supported by BLAST searching in either the N. gruberi or the GenBank non-redundant database. Some of these ORFs may be mis-predicted, but others are likely specific to N. fowleri and may constitute a source of genetic novelty. Distinguishing real N. fowleri-specific ORFs is clearly crucial to identifying possible determinants of the pathogenic phenotype, as we expect that pathogenicity is a gain-of-function for N. fowleri (given that most Naegleria species that have been isolated are non-pathogenic). Future molecular biology work on these 10 ORFs and experimental validation on a genome-wide scale is ongoing and will be extremely important.
Another source of genetic novelty, and an example of a potential gain-of-function event through gene duplication, is the cathepsin B homologue (Contig9_16_26041_25267). Cathepsin B is highly expressed in both free-living (Villalobo et al. 2003) and parasitic protists (DuBois et al. 2006) post-excystation. It has been implicated in pathogenicity in several diverse and prominent microbial eukaryotic parasites (Dou et al. 2011, Somanna et al. 2002, Caffrey and Steverding 2009, Kissoon-Singh et al. 2011 inter alia), including in N. fowleri itself (Kim et al. 2009). The cathepsin B homologue identified here is also predicted to have a signal peptide, and therefore has the potential to be secreted.
In the course of classifying Contig9_16_26041_25267, an expansion of the cathepsin B family in N. gruberi was uncovered with 24 cathepsin B members as compared with only four cathepsin L paralogues (Fig. 6). However, the kinetoplastids T. brucei and L. major have expanded their cathepsin L families to contain 11 and nine cathepsin L sequences respectively, while retaining a single cathepsin B homologue. Phylogenetic analysis showed that these expansions occurred independently in kinetoplastids and heteroloboseans (Fig. 6).
The third possible source of genetic novelty identified in N. fowleri is by HGT. The extent and scope of HGT and the role that this has played in adaptation to ecological niches in eukaryotes is controversial. However, examples of transfer across large evolutionary distances have been identified and some have implicated in pathogenesis (de Koning et al. 2000; Fast et al. 2003; Richards et al. 2006; Slot and Rokas 2011). Here we have identified one likely case of inter-supergroup HGT in the N. fowleri nuclear segment, involving N. fowleri and the amoebozoans A. castellanii and P. pallidum, but not N. gruberi. We feel that HGT is the most likely explanation for the observed distribution of this gene, requiring only three evolutionary events, rather than the very high number of independent loss events required in a scenario of evolutionary conservation from a common ancestor. If this is truly HGT, it may be a significant case, as two of these three amoebae are potentially pathogenic to humans. P. pallidum is a non-pathogenic cellular slime mold that feeds on bacteria (Githens and Karnovsky 1973), as do N. fowleri and A. castellanii (Visvesvara et al. 2007) in their free-living stage. Given the common factor of predation of bacteria, it may be that the HGT gene product is involved in some shared response, possibly to phagocytosis of bacteria or an intracellular antibacterial response. However, it may also potentially be involved in pathogenesis, as both N. fowleri and A. castellanii cause amoebic encephalitis, albeit very different forms. The finding of an HGT event in an arbitrary segment comprising <0.15% of the predicted nuclear genome suggests that other HGT events including the one described here may be common and essential macroevolutionary changes to the lifestyle and thus pathogenicity of N. fowleri. This possibility, however, awaits experimental investigation as the homologous ORFs in A. castellanii and P. pallidum have not been characterized and have no identifiable domains.
Overall, this snapshot of the N. fowleri genomic picture has revealed at the same time a highly conserved mitochondrial genome within Naegleria species and widely dissimilar nuclear genomes, with novel genes and candidates for elucidating pathogenic factors. As the field moves into an era of comparative high-throughput genomics, there is great promise that the data generated will yield new insights in the fight against this deadly human pathogen.
Supplementary Material
Table S1. Forward and reverse primers used in PCR amplification of 11 regions of the 60-kb segment
Table S2. N. fowleri mitochondrial genome annotation based on BLAST searching and RNA scanning software
Table S3. Annotation of N. fowleri 60-kb segment based on BLAST results
Acknowledgments
E.K.H is supported by an Alberta Innovates - Health Solutions Graduate Studentship Award. J.B.D. is supported by a Canada Research Chair in Evolutionary Cell Biology, and an Alberta Ingenuity New Faculty Award. C.Y.C. is supported by NIH grant R56-AI08953, a University of California Discovery Grant on the development of novel diagnostics for encephalitis, and an Abbott Pathogen Discovery Award. We would like to thank Dr. Bart Hazes for constructive criticism and suggestions on early versions of this work, and Mike Gray for helpful discussion. We thank Rama Sriram for help in growing amoebae, and Macrogen, Inc. (Korea) for advice on the construction of paired-end libraries for 454 pyrosequencing.
Footnotes
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
LITERATURE CITED
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baverstock PR, Illana S, Christy PE, Robinson BS, Johnson AM. srRNA evolution and phylogenetic relationships of the genus Naegleria (Protista: Rhizopoda) Mol Biol Evol. 1989;6:243–257. doi: 10.1093/oxfordjournals.molbev.a040549. [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Franz Lang B. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Caffrey CR, Steverding D. Kinetoplastid papain-like cysteine peptidases. Mol Biochem Parasitol. 2009;167:12–19. doi: 10.1016/j.molbiopara.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Carter R. Primary amoebic meningo-encephalitis. An appraisal of present knowledge. Trans R Soc Trop Med Hyg. 1972;66:193–213. doi: 10.1016/0035-9203(72)90147-2. [DOI] [PubMed] [Google Scholar]
- Cermakian N, Ikeda TM, Cedergren R, Gray MW. Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res. 1996;24:648–654. doi: 10.1093/nar/24.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha-Azevedo B, Tanowitz HB, Marciano-Cabral F. Diagnosis of infections caused by pathogenic free-living amoebae. Interdiscip Perspect Infect Dis. 2009;2009 doi: 10.1155/2009/251406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks JB, Kuru T, Liapounova NA, Gedamu L. Phylogenetic and primary sequence characterization of cathepsin B cysteine proteases from the oxymonad flagellate Monocercomonoides. J Eukaryot Microbiol. 2008;55:9–17. doi: 10.1111/j.1550-7408.2007.00296.x. [DOI] [PubMed] [Google Scholar]
- Dacks JB, Walker G, Field MC. Implications of the new eukaryotic systematics for parasitologists. Parasitol Int. 2008;57:97–104. doi: 10.1016/j.parint.2007.11.004. [DOI] [PubMed] [Google Scholar]
- de Koning AP, Brinkman FSL, Jones SJM, Keeling PJ. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol Biol Evol. 2000;17:1769–1773. doi: 10.1093/oxfordjournals.molbev.a026275. [DOI] [PubMed] [Google Scholar]
- DeBarry JD, Kissinger JC. Jumbled genomes: Missing apicomplexan synteny. Mol Biol Evol. 2011;28:2855–2871. doi: 10.1093/molbev/msr103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Carruthers VB, Robinson MW, Dalton JP. Cysteine proteases of pathogenic organisms. Vol. 712. Springer; US: 2011. Cathepsin proteases in Toxoplasma gondii; pp. 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious v5.5. 2010 Available from http://www.geneious.com.
- DuBois K, Abodeely M, Sajid M, Engel J, McKerrow J. Giardia lamblia cysteine proteases. Parasitol Res. 2006;99:313–316. doi: 10.1007/s00436-006-0149-4. [DOI] [PubMed] [Google Scholar]
- Dyrløv Bendtsen J, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast NM, Law JS, Williams BAP, Keeling PJ. Bacterial catalase in the microsporidian Nosema locustae: Implications for microsporidian metabolism and genome evolution. Eukaryot Cell. 2003;2:1069–1075. doi: 10.1128/EC.2.5.1069-1075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Ginger ML, Walsh C, Dawson SC, Fulton C. The Naegleria genome: a free-living microbial eukaryote lends unique insights into core eukaryotic cell biology. Res Microbiol. 2011;162:607–618. doi: 10.1016/j.resmic.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, Shu S, Neupane R, Cipriano M, Mancuso J, Tu H, Salamov A, Lindquist E, Shapiro H, Lucas S, Grigoriev IV, Cande WZ, Fulton C, Rokhsar DS, Dawson SC. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Ghedin E, Bringaud F, Peterson J, Myler P, Berriman M, Ivens A, Andersson Br, Bontempi E, Eisen J, Angiuoli S, Wanless D, Von Arx A, Murphy L, Lennard N, Salzberg S, Adams MD, White O, Hall N, Stuart K, Fraser CM, El-Sayed 11NMA. Gene synteny and evolution of genome architecture in trypanosomatids. Mol Biochem Parasitol. 2004;134:183–191. doi: 10.1016/j.molbiopara.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Githens SI, Karnovsky ML. Phagocytosis by the cellular slime mold Polysphondylium pallidum during growth and development. J Cell Biol. 1973;58:536–548. doi: 10.1083/jcb.58.3.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JL. Temperature tolerance of pathogenic and nonpathogenic free-living amoebas. Science. 1972;178:869–870. doi: 10.1126/science.178.4063.869. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hauth AM, Maier UG, Lang BF, Burger G. The Rhodomonas salina mitochondrial genome: bacteria-like operons, compact gene arrangement and complex repeat region. Nucleic Acids Res. 2005;33:4433–4442. doi: 10.1093/nar/gki757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heggie TW. Swimming with death: Naegleria fowleri infections in recreational waters. Travel Med Infect Dis. 2010;8:201–206. doi: 10.1016/j.tmaid.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Isacchi A, Bernardi G, Bernardi G. Compositional compartmentalization of the nuclear genomes of Trypanosoma brucei and Trypanosoma equiperdum. FEBS Lett. 1993;335:181–183. doi: 10.1016/0014-5793(93)80725-a. [DOI] [PubMed] [Google Scholar]
- Izumiyama S, Yagita K, Furushima-Shimogawara R, Asakura T, Karasudani T, Endo T. Occurrence and distribution of Naegleria species in thermal waters in Japan. J Eukaryot Microbiol. 2003;50:514–515. doi: 10.1111/j.1550-7408.2003.tb00614.x. [DOI] [PubMed] [Google Scholar]
- John DT, John RA. Enhancement of virulence in Naegleria fowleri by growth in Vero-cell cultures. J Parasitol. 1994;80:149–151. [PubMed] [Google Scholar]
- Kadlec V. The effects of some factors on the growth and morphology of Naegleria sp and three strains of the genus. Acanthamoeba Folia Parasitol. 1975;22:317–321. [PubMed] [Google Scholar]
- Kemble SK, Lynfield R, DeVries AS, Drehner DM, Pomputius WF, Beach MJ, Visvesvara GS, da Silva AJ, Hill VR, Yoder JS, Xiao L, Smith KE, Danila R. Fatal Naegleria fowleri infection acquired in Minnesota: Possible expanded range of a deadly thermophilic organism. Clin Infect Dis. 2012;54:805–809. doi: 10.1093/cid/cir961. [DOI] [PubMed] [Google Scholar]
- Kim JH, Yang AH, Sohn HJ, Kim D, Song KJ, Shin HJ. Immunodominant antigens in Naegleria fowleri; excretory–secretory proteins were potential pathogenic factors. Parasitol Res. 2009;105:1675–1681. doi: 10.1007/s00436-009-1610-y. [DOI] [PubMed] [Google Scholar]
- Kissoon-Singh V, Mortimer L, Chadee K, Robinson MW, Dalton JP. Cysteine proteases of pathogenic organisms. Vol. 712. Springer; US: 2011. Entamoeba histolytica cathepsin-like enzymes; pp. 62–83. [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BF, Burger G, O’Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- Law RHP, Smooker PM, Irving JA, Piedrafita D, Ponting R, Kennedy NJ, Whisstock JC, Pike RN, Spithill TW. Cloning and expression of the major secreted cathepsin B-like protein from juvenile Fasciola hepatica and analysis of immunogenicity following liver fluke infection. Infect Immun. 2003;71:6921–6932. doi: 10.1128/IAI.71.12.6921-6932.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawande RV, Abraham SN, John I, Egler LJ. Recovery of soil amebas from the nasal passages of children during the dusty Harmattan period in Zaria. Am J Clin Pathol. 1979;71:201–203. doi: 10.1093/ajcp/71.2.201. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Interactive analysis of phylogeny and character evolution using the computer program MacClade. Folia Primatol. 1989;53:190–202. doi: 10.1159/000156416. [DOI] [PubMed] [Google Scholar]
- Marande W, Lukeš J, Burger G. Unique mitochondrial genome structure in diplonemids, the sister group of kinetoplastids. Eukaryot Cell. 2005;4:1137–1146. doi: 10.1128/EC.4.6.1137-1146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2010;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano-Cabral F, Cabral GA. The immune response to Naegleria fowleri amebae and pathogenesis of infection. FEMS Immunol Med Microbiol. 2007;51:243–259. doi: 10.1111/j.1574-695X.2007.00332.x. [DOI] [PubMed] [Google Scholar]
- Marciano-Cabral FM, Fulford DE. Cytopathology of pathogenic and nonpathogenic Naegleria species for cultured rat neuroblastoma cells. Appl Environ Microbiol. 1986;51:1133–1137. doi: 10.1128/aem.51.5.1133-1137.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Visvesvara GS. Free-living, amphizoic and opportunistic amebas. Brain Pathol. 1997;7:583–598. doi: 10.1111/j.1750-3639.1997.tb01076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-i T, Maruyama S, Takahara M, Miyagishima S-y, Mori T, Nishida K, Yagisawa F, Nishida K, Yoshida Y, Nishimura Y, Nakao S, Kobayashi T, Momoyama Y, Higashiyama T, Minoda A, Sano M, Nomoto H, Oishi K, Hayashi H, Ohta F, Nishizaka S, Haga S, Miura S, Morishita T, Kabeya Y, Terasawa K, Suzuki Y, Ishii Y, Asakawa S, Takano H, Ohta N, Kuroiwa H, Tanaka K, Shimizu N, Sugano S, Sato N, Nozaki H, Ogasawara N, Kohara Y, Kuroiwa T. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- Naegleria FAQs. Atlanta, GA: Centers for Disease Control and Prevention; 2011. [Google Scholar]
- Pernin P, Ataya A, Cariou ML. Genetic structure of natural populations of the free-living amoeba, Naegleria lovaniensis Evidence for sexual reproduction. Heredity. 1992;68:173–181. doi: 10.1038/hdy.1992.26. [DOI] [PubMed] [Google Scholar]
- Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. Evolution of filamentous plant pathogens: Gene exchange across eukaryotic kingdoms. Curr Biol. 2006;16:1857–1864. doi: 10.1016/j.cub.2006.07.052. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Simpson AGB. Evolution: revisiting the root of the eukaryotic tree. Curr Biol. 2009;9:R165–R167. doi: 10.1016/j.cub.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Salavati R, Panigrahi AK, Stuart KD. Mitochondrial ribonuclease P activity of Trypanosoma brucei. Mol Biochem Parasitol. 2001;115:109–117. doi: 10.1016/s0166-6851(01)00273-0. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Luna J, Cervantes-Sandoval I, Tsutsumi V, Shibayama M. A biochemical comparison of proteases from pathogenic Naegleria fowleri and non-pathogenic Naegleria gruberi. J Eukaryot Microbiol. 2007;54:411–417. doi: 10.1111/j.1550-7408.2007.00280.x. [DOI] [PubMed] [Google Scholar]
- Sheehan KB, Ferris MJ, Benson JM. Detection of Naegleria sp in a thermal, acidic stream in Yellowstone National Park. J Eukaryot Microbiol. 2003;50:263–265. doi: 10.1111/j.1550-7408.2003.tb00132.x. [DOI] [PubMed] [Google Scholar]
- Slot JC, Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between Fungi. Curr Biol. 2011;21:134–139. doi: 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Somanna A, Mundodi V, Gedamu L. Functional analysis of cathepsin B-like cysteine proteases from Leishmania donovani complex: Evidence for the activation of latent transforming growth factor β. J Biol Chem. 2002;277:25305–25312. doi: 10.1074/jbc.M203034200. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Valach M, Farkas Z, Fricova D, Kovac J, Brejova B, Vinar T, Pfeiffer I, Kucsera J, Tomaska L, Lang BF, Nosek J. Evolution of linear chromosomes and multipartite genomes in yeast mitochondria. Nucleic Acids Res. 2011;39:4202–4219. doi: 10.1093/nar/gkq1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Zepeda J, Gómez-Alcalá AV, Vázquez-Morales JA, Licea-Amaya L, De Jonckheere JF, Lares-Villa F. Successful treatment of Naegleria fowleri meningoencephalitis by using intravenous amphotericin B, fluconazole and rifampicin. Arch Med Res. 2005;36:83–86. doi: 10.1016/j.arcmed.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Villalobo E, Moch C, Fryd-Versavel G, Fleury-Aubusson A, Morin LØ. Cysteine proteases and cell differentiation: Excystment of the ciliated protist Sterkiella histriomuscorum. Eukaryot Cell. 2003;2:1234–1245. doi: 10.1128/EC.2.6.1234-1245.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvesvara GS, Stehr-Green JK. Epidemiology of free-living ameba infections. J Eukaryot Microbiol. 1990;37:25s–33s. doi: 10.1111/j.1550-7408.1990.tb01142.x. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp. Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Mic. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- Westenberger S, Cerqueira G, El-Sayed N, Zingales B, Campbell D, Sturm N. Trypanosoma cruzi mitochondrial maxicircles display species- and strain-specific variation and a conserved element in the non-coding region. BMC Genomics. 2006;7:60. doi: 10.1186/1471-2164-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JS, Eddy BA, Visvesvara GS, Capewell L, Beach MJ. The epidemiology of primary amoebic meningoencephalitis in the USA, 1962–2008. Epidemiol Infect. 2010;138:968–975. doi: 10.1017/S0950268809991014. [DOI] [PubMed] [Google Scholar]
- Zhou L, Sriram R, Visvesvara GS, Xiao L. Genetic variations in the internal transcribed spacers and mitochondrial small subunit rRNA gene of Naegleria spp. J Eukaryot Microbiol. 2003;50:522–526. doi: 10.1111/j.1550-7408.2003.tb00617.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Forward and reverse primers used in PCR amplification of 11 regions of the 60-kb segment
Table S2. N. fowleri mitochondrial genome annotation based on BLAST searching and RNA scanning software
Table S3. Annotation of N. fowleri 60-kb segment based on BLAST results