SUMMARY
RNA binding proteins (RBPs) have emerged as major causative agents of Amyotrophic Lateral Sclerosis (ALS). To investigate the function of TAF15, an RBP recently implicated in ALS, we explored its target RNA repertoire in normal human brain and mouse neurons. Coupling high-throughput sequencing of immunoprecipitated and crosslinked RNA (HITS-CLIP) with RNA-Seq and TAF15 knockdowns, we identified conserved TAF15 RNA targets and we assessed the impact of TAF15 on the neuronal transcriptome. We describe a role of TAF15 in the regulation of splicing for a set of neuronal RNAs encoding proteins with essential roles in synaptic activities. We find that TAF15 is required for a critical alternative splicing event of the zeta-1 subunit of the glutamate N-methyl-D-aspartate (NMDA) receptor (Grin1) that controls the activity and trafficking of NR1. Our study uncovers neuronal RNA networks impacted by TAF15 and sets the stage for investigating the role of TAF15 in ALS pathogenesis.
INTRODUCTION
RBPs, of which heterogeneous ribonucleoprotein particle proteins (hnRNPs) constitute a major class, are multifunctional proteins that are involved in numerous aspects of RNA processing and function (Dreyfuss et al., 2002). ALS is a fatal disease caused by degeneration of motor neurons. In ~20% of cases, ALS is part of a broader neurodegenerative disease spectrum that includes Frontotemporal Lobar Degeneration (FTLD) (Ibrahim et al., 2012; Mackenzie and Neumann, 2011). Dominant mutations in genes coding for four hnRNPs (TDP-43, FUS/TLS, TAF15, and EWS) are found in familial and sporadic cases of ALS (Neumann et al., 2006). RBPs that cause or are associated with ALS and FTLD, have prion-like domains that under pathological conditions promote their aggregation (Ibrahim et al., 2012; Johnson et al., 2008; King et al., 2012).
NMDA receptors (NMDARs) are ionotropic glutamate receptors with central roles in excitatory synaptic activities and neuronal development (Dingledine et al., 1999; Lau and Tymianski, 2010; Paoletti, 2011). NMDARs are heteromeric composed of two obligatory NR1 subunits and two NR2 subunits (NR2A-2D) (Behe et al., 1995; Dingledine et al., 1999). The NR1 subunit, (also known as zeta-1), is encoded by the Grin1 gene and is required for the formation and regulation of functional receptors (Moriyoshi et al., 1991; Paoletti, 2011). Grin1 undergoes differential mRNA splicing to create different isoforms of NR1 subunits that contain distinct regulatory elements within their C-terminal tails including the C1 cassette (Dingledine et al., 1999). The C1 domain, encoded by E19 exon of Grin1, plays critical roles in regulation of NR1 function and trafficking and is subjected to activity-dependent regulation (reviewed in (Li et al., 2007)). Phosphorylation of C1 cassette potentiates NMDA activity and trafficking (Hisatsune et al., 1997). Activation of NMDAR enhances calcium influx and intracellular Ca+2/calmodulin binding (Ehlers et al., 1996; Lan et al., 2001; Okabe et al., 1999; Tingley et al., 1997). CaM binding at the C1 cassette underlies NMDAR-dependent plasticity and protect against excitotoxic cell death (Ehlers et al., 1996).
TAF15 along with FUS and EWS belongs to the FET family of RBPs (Bertolotti et al., 1996). All of the FET members are involved in various aspects of RNA metabolism and are predominantly nuclear. Recently, putative ALS causing mutations were described in the TAF15 gene in familial (Ticozzi et al., 2011) and sporadic cases (Couthouis et al., 2011). Similar to TDP-43 and FUS proteinopathies, the sporadic ALS-associated variants of TAF15 (i.e R408C) caused formation of cytoplasmic aggregates in cultured neurons and neurodegeneration in Drosophila (Couthouis et al., 2011). TAF15 cytoplasmic inclusions are also found in all cases of FUS-FTLD subtypes, further strengthening the notion of a pathogenic role of TAF15 in neurodegeneration (Neumann et al., 2011). TDP-43 and FUS RNA interaction maps have begun to address their impact on neuronal RNA processing (Ishigaki et al., 2012; Lagier-Tourenne et al., 2012; Polymenidou et al., 2011).
TAF15 has been implicated in pre-mRNA splicing (Hoell et al., 2011; Jobert et al., 2009) but the neuronal RNA targets of TAF15 and the impact of TAF15 on the neuronal transcriptome are not known. Here we report the in vivo RNA targets of TAF15 in human brain and mouse neurons and we define conserved groups of neuronal TAF15 targets that implicate TAF15 in the control of mRNAs that code for proteins with essential roles in synaptic activities. We find that TAF15 is required for a critical alternative splicing event of Grin1 E19 exon that controls the trafficking of NMDA glutamate receptor. Our study uncovers neuronal RNA networks impacted by TAF15 and sets the stage for investigating the role of TAF15 in ALS and FTLD pathogenesis.
RESULTS & DISCUSSION
TAF15-RNA Interaction Maps in Human Brain and Mouse Neurons
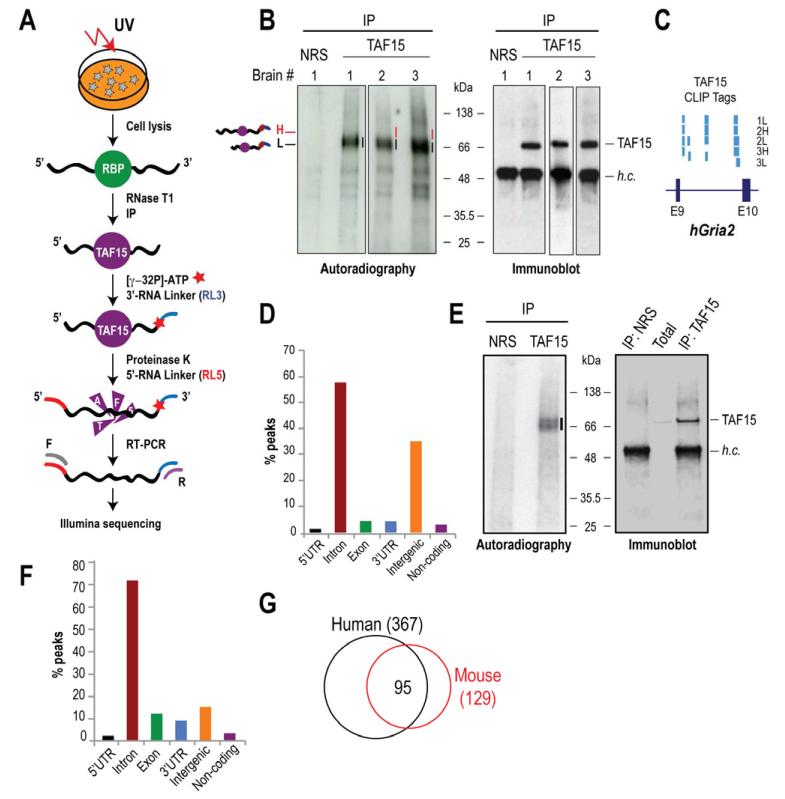
We employed HITS-CLIP (Chi et al., 2009) (Figure 1A) to identify in vivo RNA targets of TAF15 from three unrelated, normal human brains. CLIPs of TAF15 from human brains, resulted in the formation of specific complexes of TAF15 with RNAs, which were absent in the non-immune rabbit serum (NRS) lane (Figure 1B, Figure S1A-C, Supplementary Text). We prepared libraries from the membrane segments containing the main radioactivity signal (1L, 2L and 3L) and from the portions of the membrane just above the main signal in brains 2 and 3 (2H and 3H) (Figure 1B). Attempts to generate cDNA libraries from the NRS negative control failed indicating the stringency of our CLIPs. The five brain TAF15 CLIP libraries generated a total of ~23.9 million reads that mapped to the human genome (hg19). The majority of the reads (~90%) mapped in gene’s sense strands. We did not find any correlation between TAF15 binding and RNA expression levels (Supplementary Text), indicating that peaks containing abundant TAF15 CLIP-tags represent substantial TAF15 RNA binding sites and do not merely correlate with the abundance of the targeted transcripts.
Figure 1. TAF15 HITS-CLIP of human brains and mouse neurons.
A. HITS-CLIP schematic.
B. TAF15 CLIPs from three human brains. Lines (black; 1L, 2L, and 3L, red; 2H and 3H) indicate TAF15-RNA complexes that were excised for library preparation. NRS; non-immune serum (negative control).
C. Screen shot from UCSC Genome Browser of portion of human Gria2 gene (chr4: 158,255,078-158,257,088) with TAF15 peaks.
D. Distribution of human brain CLIP-tag peaks.
E. TAF15 CLIPs from cultured mouse neurons.
F. Distribution of mouse neuron CLIP-tag peaks.
G. Overlap between top human and mouse TAF15 RNA targets.
See also Figures S1, S2 and S3.
To verify the reproducibility and significance of the computational analyses, the bioinformatics were performed for each CLIP sample independently; all conclusions were consistent for all CLIP samples. Genomic positions of TAF15 peaks were highly consistent in all five libraries prepared from the three human brains (Figure 1C, Supplementary Text). We used peaks calling to analyze the CLIP-tag distribution and we found that ~58% of the peaks mapped to introns, ~4% to coding exons, ~4% to 3′-UTRs and ~1% to 5′-UTRs, indicating that TAF15 binds predominantly to pre-mRNAs, which is consistent with the nuclear localization of TAF15 (Figure 1D). A sizable portion of the peaks mapped to intergenic regions (~36%) implying possible TAF15 binding to noncoding RNAs and non-annotated transcripts (Figure 1D).
To uncover transcripts that are likely to be functionally regulated by TAF15, we sought to identify conserved TAF15 binding sites by performing HITS-CLIP in mouse neurons that we differentiated from Embryonic Stem (ES) cells (Figure S1D). We performed two biological replicates of TAF15 HITS-CLIP (Figure 1E, Supplementary Text) and generated a total of ~24.3 million reads that were mapped to the mouse genome (mm9) with the majority of reads (~94%) corresponding to sense strands of genes. Peaks were defined as for human CLIPs. The genomic distribution of mouse TAF15 peaks was similar to that of human CLIP and showed predominant binding of TAF15 to mouse introns (~70%; Figure 1F). Assessment of TAF15 peaks for lincRNA bindings revealed a mean of less than 3% binding in human and mouse libraries. Consistent with our findings, PAR-CLIP of overexpressed FLAG-tagged TAF15 in HEK293 cells, revealed a large fraction of clusters within intronic regions (Hoell et al., 2011). However, our study probes the binding sites of endogenous TAF15 in the context of normal mouse neurons and human brain, which is more relevant to the function of TAF15 in neural tissue and applicable to future studies related to ALS and FTLD.
To examine TAF15 sequence specificity, we extensively searched for enriched Nmers (4mers-7mers) within human and mouse TAF15 CLIP-tags (Table S1). As expected, we observed an increase of Nmer abundance for shorter Nmers. However, consistent with the observation that RBPs bind to degenerate sequences and their binding is influenced by RNP complexes that they associate with (Dreyfuss et al., 2002), we found that even the most enriched Nmer accounts for less than 30% of CLIP-tags. This is consistent with PAR-CLIP of FLAG-tagged TAF15 in HEK293 cells, which uncovered no significant motif (Hoell et al., 2011). Interestingly, clustering of the top 20 6mers using the BliC metric (Habib et al., 2008) revealed a CUG preference suggesting a possible binding specificity.
TAF15 Targets Genes with Synaptic Activities
To identify the most consistently targeted human and mouse RNA transcripts, we ranked genes that contained TAF15 peaks using the Rank Product of the number of CLIP-tags in all CLIP experiments (pfp<0.1). We identified 367 human genes (with mouse homologs) and 129 mouse genes that were consistently targeted by TAF15 (Table S2). The higher number of targets identified in the human samples is expected because of the cellular heterogeneity of human brain that contains various types of neurons and glial cells, compared to the mouse samples, which represent highly pure neurons. Notably, of the highly TAF15 targeted mouse transcripts ~74% are the same in humans (Figure 1G), indicating the presence of a conserved network of neuronal RNAs that are targeted by TAF15. Gene Ontology term analysis (Table 1) revealed enrichment of genes involved in synaptic structure, function and transmission such as subunit zeta-1 of the NMDA receptor (NR1, NMDAR1, Grin1); subunits 2A and 2B of the NMDAR (NR2A, Grin2a; NR2B, Grin2b); potassium voltage-gated channel subfamily D member 2 (Kcnd2); subunits 2 and 4 of the ionotropic AMPA glutamate receptor (Gria2, Gria4); neurexin 1 and 3 (Nrxn1, Nrxn3), neuroligin 1 (Nlgn1) and protocadherin-9 (Pcdh9). GO term analysis of TDP-43 after depletion revealed enrichment of targets with synaptic activities (Polymenidou et al., 2011). Notably, comparing the highly targeted TAF15 mouse RNA transcripts to those of FUS (Lagier-Tourenne et al., 2012) revealed that 146/164 (~89%) of TAF15 targets are also targeted by FUS (Table S3, Figure S3A, Supplementary Text), indicating that the two RBPs possibly regulate common neuronal networks.
Table 1.
Gene Ontology Term analysis for top, conserved, human and mouse TAF15 RNA targets identified by CLIP.
| Gene Ontology Category |
GO Term | Description | P-value |
|---|---|---|---|
|
| |||
|
Molecular
Function |
GO:0008066 | Glutamate receptor activity | 3.51E-007 |
| GO:0005216 | Ion channel activity | 1.12E-006 | |
| GO:0004970 | Ionotropic glutamate receptor activity | 1.24E-006 | |
| GO:0004872 | Receptor activity | 1.90E-006 | |
| GO:0005234 | Extracellular-glutamate-gated ion channel activity | 2.06E-006 | |
|
| |||
|
Cellular
Component |
GO:0005886 | Plasma membrane | 2.03E-013 |
| GO:0030425 | Dendrite | 1.32E-008 | |
| GO:0014069 | Postsynaptic density | 1.88E-008 | |
| GO:0043025 | Neuronal cell body | 9.17E-008 | |
| GO:0030424 | Axon | 1.08E-007 | |
| GO:0045202 | Synapse | 2.12E-007 | |
| GO:0016020 | Membrane | 1.12E-006 | |
| GO:0030288 | Outer membrane-bounded periplasmic space | 1.24E-006 | |
| GO:0045211 | Postsynaptic membrane | 3.43E-006 | |
| GO:0043198 | Dendritic shaft | 3.66E-006 | |
| GO:0031225 | Anchored to membrane | 5.01E-006 | |
| GO:0043195 | Terminal button | 5.57E-006 | |
| GO:0030054 | Cell junction | 8.16E-006 | |
|
| |||
|
Biological
Process |
GO:0007155 | Cell adhesion | 3.53E-008 |
| GO:0007215 | Glutamate signaling pathway | 3.51E-007 | |
| GO:0007158 | Neuron cell-cell adhesion | 6.96E-007 | |
| GO:0007268 | Synaptic transmission | 2.51E-006 | |
| GO:0050804 | Regulation of synaptic transmission | 4.77E-006 | |
| GO:0060079 | Regulation of excitatory postsynaptic membrane potential | 9.50E-006 | |
The Impact of TAF15 in the Transcriptome of Mouse Neurons
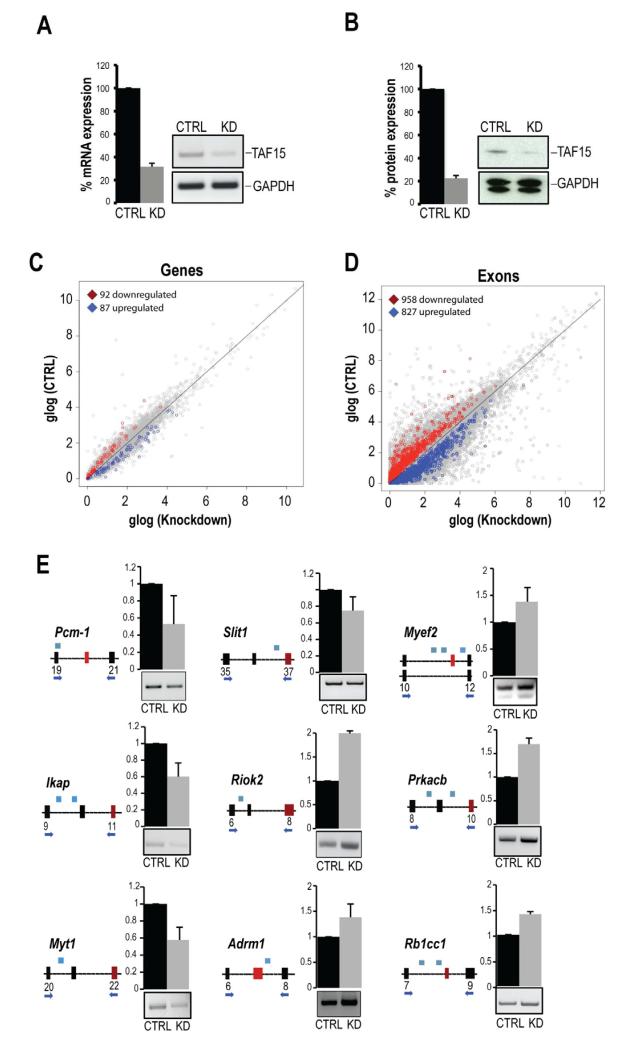
To explore the role of TAF15 in the mouse neuronal transcriptome, we analyzed changes in levels and splicing patterns of RNAs upon depletion of TAF15 by performing control (CTRL) and TAF15 siRNA knockdowns (KD) (Figure S1D) in biological triplicates. We achieved a ~75% reduction of TAF15 mRNA (Figure 2A) and protein levels (Figure 2B). Using RNA-Seq (Vourekas et al., 2012) we generated six libraries (3 CTRL and 3 KD) and obtained a total of ~89.0 and ~72.5 million reads from CTRL and KD libraries, respectively that mapped to the mouse genome. Comparisons of the mRNA expression levels between CTRL and KD RNA-Seq libraries identified 92 protein-coding genes with reduced expression and 87 protein-coding genes with increased expression upon TAF15 knockdown (Figure 2C, Table S4). The few changes in the levels of mRNAs, overall, suggest that TAF15 has no global effect on the stability of mRNAs. At this point it is not clear how TAF15 impacts on the levels of these transcripts.
Figure 2. Impact of TAF15 knockdown on the transcriptome of mouse neurons.
A, B. TAF15 mRNA (A) and protein levels (B) after siRNA knockdown of TAF15 (KD), compared to control knockdown (CTRL), in mouse neurons. Quantitations (A, B) are shown as mean ± s.d., from three biological replicates; changes are significant (P<0.01).
C, D. Scatter plot of genes (C) and exons (D) expression calculated by RNA-Seq from 3 KD and 3 CTRL samples. Differentially expressed genes and exons (N=3; P<0.01) are highlighted, glog; generalized logarithm base 2.
E. RT-PCR validations of differentially included exons in mRNAs whose levels did not change upon TAF15 knockdown. Shown are schematics (constitutive exon: black box; differentially included exon: red box; intron: black line) with primers (arrows) and position of TAF15-peaks (blue boxes). Quantitation (error bars; s.d) and representative gels from RT-PCRs are shown. All detected changes are significant (P<0.01).
See also Figures S2 and S3.
To address the impact of TAF15 in pre-mRNA splicing, we compared exon expression levels between CTRL and TAF15 KD. We identified 958 unique exons of protein-coding genes with significantly (P<0.01) reduced expression (exon exclusion); and 827 unique exons with significantly (P<0.01) increased expression (exon inclusion) upon TAF15 knockdown (Figure 2D, Table S4). We validated by PCR randomly selected exons from Table S4 containing TAF15 CLIP-tags in their vicinity and whose inclusion was either inhibited (pericentriolar material 1 –Pcm-1–; Integrin-linked kinase-associated serine/threonine phosphatase 2C –Ikap–; Myelin transcription factor 1 isoform 4 –Myt1–; and Slit homolog 1 protein precursor –Slit1–) or promoted (serine/threonine-protein kinase RIO2 –Riok2–; proteasomal ubiquitin receptor –Adrm1; myelin expression factor 2 isoform 3 –Myef2–; cAMP-dependent protein kinase catalytic subunit –Prkacb–; and RB1-inducible coiled-coil protein 1 –Rb1cc1–) upon TAF15 knockdown (Figure 2E). We did not detect correlation between the position of TAF15 peaks and the direction of splicing changes (exon inclusion or exclusion). However, we note that the impact of TAF15 in splicing is likely more widespread than the one uncovered by the RNA-Seq experiments because of the stringent criteria that we used to define changed exons and limitations of RNA-Seq (Ozsolak and Milos, 2011).
We tested some of the top targeted TAF15 genes that are associated with neurological disorders including Frmd4a (FERM domain-containing protein 4a isoform 1), Cacna1c (Votlage-dependent L-type calcium channel subunit), Pcdh9 (Protocadherin-9). We also tested Sod1 (superoxide dismutase Cu-Zn), that is mutated in a subset of familial ALS cases. RT-PCR analyses after TAF15 knockdown in mouse neurons revealed marked reduction of Pcdh9, mild reduction of Frmd4a, and Cacna1c but no changes in the levels of Sod1 suggesting a role for TAF15 in the regulation of these transcripts (Figure S3B). Knockdown of TAF15 did not alter the levels of TDP-43 or FUS (Figure S3C); however we note the limitation of this analysis, which is confined to neurons in culture.
As noted from the TAF15 CLIP assays in both human and mouse there was high overlap in TAF15 targets that regulate synaptic activities and functions. We examined a set of targets within this category, which contain differentially regulated exons (P<0.05) and also harbor TAF15-binding sites in a 2Kb distance up- or downstream of the changed exon. Out of these targets, we validated Grin2a, Grin2b, Gria2 and 4, and gamma-aminobutyric acid receptor subunit beta-3 (Gabrb3) and observed reduction of exon inclusion upon depletion of TAF15 among all the genes (Figure S4A). To further explore the effect of TAF15 on those targets, we analyzed mouse cultured neurons expressing FLAG-tagged TAF15 wild-type and TAF15-R408C (a recently identified ALS-variant; Couthouis et al., 2011) (Figure S4B). RT-PCR analyses revealed a reduction of exon inclusion of Grin2b, Gria2, Gabrb3 and a slight increase of exon inclusion of Grin2a and Gria4 in cells expressing TAF15-R408C compared to controls (Figure S4C). These results suggest a role of TAF15 and the ALS-variant (R408C) in regulating targets with synaptic activities and support future investigations to address mechanistically how the levels of TAF15 and the ALS variant affect splicing and the neuronal transcriptome, which currently is unknown.
TAF15 Regulates Inclusion of the C1 Cassette of NR1 Receptor
We noticed TAF15 binding sites near the E19 exon, coding for C1 cassette, of both human and mouse Grin1 of NMDAR (Figure 3A, B) and sought to identify the impact of TAF15 in the regulation of splicing of E19 by performing PCRs in mouse neurons depleted of TAF15. We found an increase of Grin1 transcripts lacking E19 and concomitant decrease of transcripts that harbor E19 (Figure 3C) upon TAF15 knockdown, indicating that TAF15 promotes inclusion of E19. NMDAR play a crucial role in the central nervous system (Dingledine et al., 1999; Petralia et al., 2009) and misregulation of this receptor is relevant to several neurodegenerative disorders including ALS (Plaitakis and Caroscio, 1987; Texido et al., 2011).
Figure 3. TAF15 regulates receptor properties of NR1 via alternative splicing of exon 19 of Grin1.
A. Peptide sequence of C1 cassette of NR1 encoded by E19 of Grin1 with serine 896/897 residues that are subjected to phosphorylation and the calmodulin binding site (aminoacids in blue); the C1 sequence is 100% conserved in humans, mouse and rat.
B. Screen shot from UCSC Genome Browser of portion of human (top) and mouse (bottom) Grin1 gene, coding for NMDAR NR1 subunit zeta-1 (NR1; NMDAR1) with alternative E19 (red box) and location of TAF15 CLIP-tags (blue boxes).
C. TAF15 promotes inclusion of E19/C1 of mouse Grin1. PCR analyses of Grin1 with E18/20 (top) and E19/20 specific primers (bottom). A significant reduction in E19 inclusion was detected in TAF15 knockdowns (KD) compared to control knockdowns (CTRL). Quantitation and representative gels from RT-PCRs are shown.
D – I. Impact of TAF15 on NR1 receptor of rat cortical neurons. All experiments were performed in triplicates from TAF15 knockdowns (KD) and controls (CTRL); error bars in (E-H) represent s.d; all detected changes are significant (P<0.05). (D) TAF15 protein levels after siRNA knockdown of TAF15. (E) NR1 immunoprecipitates were immunoblotted (IB) with an antibody detecting all NR1 protein isoforms (NR1) or detecting specifically the C1 cassette (NR1-C1; left). Quantitation from three experiments. (F) Neuronal lysates were immunoblotted with an antibody detecting phospho-S896/897 NR1 and normalized to GAPDH. (G) Calmodulin-agarose pulldowns were probed by immunoblots for NR1. (H) Cell surface biotinylation (experimental schematic, left panel) of rat cortical neurons after TAF15 CTRL or KD for detection of NR1 that associates with plasma membranes; quantitation from three experiments is shown (right panel). (I) Schematic of TAF15 impact on NR1 function via splicing regulation of E19.
See also Figure S4.
We set out to test the effect of E19 exclusion after TAF15 depletion using both mouse neurons and importantly primary rat cortical neurons that have been used extensively to study NMDAR regulation and function (Williams et al., 1992). We established efficient TAF15 knockdowns in rat cortical neurons (Figure 3D). We used an antibody that recognized all NR1 isoforms to perform immunoprecipitations from CTRL and TAF15 KD and probed the IPs with an antibody that recognizes full-length NR1 or NR1 containing the C1 cassette. The overall levels of NR1 were similar between CTRL and TAF15 KD, however, we detected a ~60% reduction of NR1 receptors containing the C1 cassette in TAF15 KD (Figure 3E) and a ~40% reduction of the phosphorylation level of NR1 subunit in TAF15 depleted neurons (Figure 3F), consistent with the reduction of NR1 receptors containing the C1 cassette that is subject to phosphorylation. Calmodulin-agarose pulldowns and immunoblots for NR1, revealed a ~40% reduction of CaM-bound NR1 in TAF15 KD again consistent with reduction of NR1 receptors containing the C1 cassette that binds to CaM (Figure 3G). To further explore the effect of C1 reduction on targeting of NR1, we used a cell surface biotinylation assay to test the amount of NR1 that associates with plasma membranes. We detected ~25% reduction of the cell surface NR1 in cells depleted of TAF15 compared to CTRL (Figure 3H). Collectively, our observations suggest a role of TAF15 in regulating the splicing of E19, which may alter receptor trafficking (Figure 3I).
Because NMDAR-dependent neurotransmission is affected by changes in receptor trafficking, gating, and alternative RNA splicing of Grin1 isoforms, misregulation of these activities are relevant to several neurodegenerative disorders including ALS (Plaitakis and Caroscio, 1987; Milnerwood et al., 2010; Spalloni et al., 2012). Our findings emphasize the importance of NMDAR regulation and more generally the discovery of human and mouse neuronal RNA targets of TAF15 will facilitate future investigations into the role of TAF15 in ALS and FTLD pathogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture
Mouse embryonic stem cells (ES) were maintained in ES medium on gelatinized plates without feeder cells. Neuronal differentiation was performed as described previously (Wichterle et al., 2009). Cells were maintained into ADFNB+GDNF. Primary rat cortical neurons were prepared and cultured as previously described (Dichter, 1978). Cells were maintained into Neurobasal medium supplemented with B27.
HITS-CLIP and SSD-RNA-Seq
HITS-CLIP from normal human brain and from mouse neurons was performed using anti-TAF15 antibody (Bethyl; A300-308A). HITS-CLIP and Solid Support Directional RNA-Seq were performed as previously described (Chi et al., 2009; Vourekas et al., 2012). The three human brains consisted of fresh temporal cortices containing live cells (neurosurgical specimens). Details are found in Extended Experimental Procedures.
Peak Calling
Peaks were defined by merging overlapping tags into single merged regions and selecting the point with the highest number of aligned reads within each region as the actual peak position. The score of each peak was defined as the total number of tags within the merged region.
Counts of Tags within Genes and Transcripts and Sample Reproducibility
For each annotated gene and transcript, the number of tags, which are contained within defined peaks, was counted. To estimate the reproducibility among samples, the Pearson’s correlation for the gene counts for all pairwise combinations of samples was calculated (Figure S2).
siRNA Silencing
On Days 6 and 8 of neuronal differentiation, TAF15 knockdown was performed using siGENOME SMART-pool (Dharmacon), with lipofectamine RNAiMax Reagent in Opti-MEM + L-Glutamine medium (Invitrogen). Four siRNA duplexes were used for knockdown of TAF15; sense sequences were: (a) GGGGUGAGCAAAGUUA, (b) CCGAGGCCGUGGAGGAUAU, (c) UAGAGGAUAUGGCGGGUCA, and (d) ACAGAAAUGAUCAGCGCAA. Cells were transfected with a final concentration of 10-20 nmol siRNAs. Knockdowns with Silencer negative control siRNA (Dharmacon) served as negative control. On Day 9, cells were harvested for subsequent analyses.
Immunoblots and Immunoprecipitations
Cell lysate was prepared in lysis buffer (20 mM Tris-HCL (pH 7.5), 150 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 0.1% Triton-X100 and complete EDTA-free protease inhibitors (Roche)). Total protein concentration was measured using BioRad reagent per manufacturer’s instructions. Samples were separated in 4-12% NuPAGE gels (Invitrogen) and transferred to nitrocellulose membranes (Invitrogen) for immunblotting. Quantitation of the blots was performed with ImageJ software. Intensity ratio between different conditions was averaged for biological triplicates per group and differences were assessed using Student’s t-test. For immunoprecipitations, cells were washed with cold 1x PBS buffer, collected in RIPA buffer (0.1% SDS, 0.5% Deoxycholate, 1% NP-40, 150 mM NaCl, 50 mM Tris-HCl (pH 8.0)) for 15 min on ice and centrifuged at 10,000 rpm for 10 min to obtain the soluble fractions. The total lysate was incubated with agarose beads (Invitrogen) that were conjugated to appropriate antibodies. Samples were processed for immunoblotting.
Antibodies
The following antibodies were used: rabbit anti-TAF15 (1:10,000; Bethyl Laboratories A300-308A), mouse anti-NMDA NR1 (1:1,000; Millipore 05-432), rabbit anti-NMDAR1 Splice Variant C1 (1:1,000; Millipore AB5046P), rabbit anti-Phospho-NMDAR1 Ser896/897 (1:1,000; EMD 454571), rabbit anti-FUS (1:10,000; Bethyl Laboratories A300-302A-1), rabbit anti-TARDBP (I:10,000; Protein Tech 10782-2-AP), mouse anti-FLAG M2 (1:8,000; Sigma F-3165) and mouse anti-GAPDH (1:10,000; Sigma SAB1405848).
Supplementary Material
HIGHLIGHTS.
Identification of in vivo TAF15 neuronal targets
TAF15 regulates pre-mRNA splicing
TAF15 targets transcripts coding for synaptic proteins
TAF15 regulates alternative splicing of Grin1
ACKNOWLEDGEMENTS
Supported by NIH grants NS072561 and NS056070 to ZM and T32-AG000255 to FI. HITS-CLIP and RNA-Seq libraries deposited in NCBI GEO under accession number GSE43294. FI and ZM conceived and directed experiments. FI performed all experiments and MAM and MAC provided rat cortical neuronal cultures. MM and PA performed all bioinformatical analyses. All authors analyzed data and FI wrote the manuscript with substantial input and editing from MM, PA and ZM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare that they have no conflict of interest.
REFERENCES
- Behe P, Stern P, Wyllie DJ, Nassar M, Schoepfer R, Colquhoun D. Determination of NMDA NR1 subunit copy number in recombinant NMDA receptors. Proc Biol Sci. 1995;262:205–213. doi: 10.1098/rspb.1995.0197. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA- mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichter MA. Rat cortical neurons in cell culture: culture methods, cell morphology, electrophysiology, and synapse formation. Brain Res. 1978;149:279–293. doi: 10.1016/0006-8993(78)90476-6. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Ehlers MD, Zhang S, Bernhadt JP, Huganir RL. Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell. 1996;84:745–755. doi: 10.1016/s0092-8674(00)81052-1. [DOI] [PubMed] [Google Scholar]
- Habib N, Kaplan T, Margalit H, Friedman N. A novel Bayesian DNA motif comparison method for clustering and retrieval. PLoS Comput Biol. 2008;4:e1000010. doi: 10.1371/journal.pcbi.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisatsune C, Umemori H, Inoue T, Michikawa T, Kohda K, Mikoshiba K, Yamamoto T. Phosphorylation-dependent regulation of N-methyl-D-aspartate receptors by calmodulin. J Biol Chem. 1997;272:20805–20810. doi: 10.1074/jbc.272.33.20805. [DOI] [PubMed] [Google Scholar]
- Hoell JI, Larsson E, Runge S, Nusbaum JD, Duggimpudi S, Farazi TA, Hafner M, Borkhardt A, Sander C, Tuschl T. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011;18:1428–1431. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim F, Nakaya T, Mourelatos Z. RNA dysregulation in diseases of motor neurons. Annu Rev Pathol. 2012;7:323–352. doi: 10.1146/annurev-pathol-011110-130307. [DOI] [PubMed] [Google Scholar]
- Ishigaki S, Masuda A, Fujioka Y, Iguchi Y, Katsuno M, Shibata A, Urano F, Sobue G, Ohno K. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci Rep. 2012;2:529. doi: 10.1038/srep00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobert L, Pinzon N, Van Herreweghe E, Jady BE, Guialis A, Kiss T, Tora L. Human U1 snRNA forms a new chromatin-associated snRNP with TAF15. EMBO Rep. 2009;10:494–500. doi: 10.1038/embor.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan JY, Skeberdis VA, Jover T, Grooms SY, Lin Y, Araneda RC, Zheng X, Bennett MV, Zukin RS. Protein kinase C modulates NMDA receptor trafficking and gating. Nat Neurosci. 2001;4:382–390. doi: 10.1038/86028. [DOI] [PubMed] [Google Scholar]
- Lau A, Tymianski M. Glutamate receptors, neurotoxicity and neurodegeneration. Pflugers Arch. 2010;460:525–542. doi: 10.1007/s00424-010-0809-1. [DOI] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M. FET proteins in frontotemporal dementia and amyotrophic lateral sclerosis. Brain Res. 2011 doi: 10.1016/j.brainres.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Milnerwood AJ, Gladding CM, Pouladi MA, Kaufman AM, Hines RM, Boyd JD, Ko RW, Vasuta OC, Graham RK, Hayden MR, et al. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron. 2010;65:178–190. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- Neumann M, Bentmann E, Dormann D, Jawaid A, DeJesus-Hernandez M, Ansorge O, Roeber S, Kretzschmar HA, Munoz DG, Kusaka H, et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Okabe S, Miwa A, Okado H. Alternative splicing of the C-terminal domain regulates cell surface expression of the NMDA receptor NR1 subunit. J Neurosci. 1999;19:7781–7792. doi: 10.1523/JNEUROSCI.19-18-07781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P. Molecular basis of NMDA receptor functional diversity. Eur J Neurosci. 2011;33:1351–1365. doi: 10.1111/j.1460-9568.2011.07628.x. [DOI] [PubMed] [Google Scholar]
- Petralia RS, Al-Hallaq RA, Wenthold RJ. Trafficking and Targeting of NMDA Receptors. 2009 [PubMed] [Google Scholar]
- Plaitakis A, Caroscio JT. Abnormal glutamate metabolism in amyotrophic lateral sclerosis. Ann Neurol. 1987;22:575–579. doi: 10.1002/ana.410220503. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalloni A, Nutini M, Longone P. Role of the N-methyl-D-aspartate receptors complex in Amyotrophic Lateral Sclerosis. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbadis.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, McKenna-Yasek D, Sapp PC, Silani V, Bosco DA, Shaw CE, et al. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:285–290. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- Tingley WG, Ehlers MD, Kameyama K, Doherty C, Ptak JB, Riley CT, Huganir RL. Characterization of protein kinase A and protein kinase C phosphorylation of the N- methyl-D-aspartate receptor NR1 subunit using phosphorylation site-specific antibodies. J Biol Chem. 1997;272:5157–5166. doi: 10.1074/jbc.272.8.5157. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19:773–781. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Peljto M, Nedelec S. Xenotransplantation of embryonic stem cell- derived motor neurons into the developing chick spinal cord. Methods Mol Biol. 2009;482:171–183. doi: 10.1007/978-1-59745-060-7_11. [DOI] [PubMed] [Google Scholar]
- Williams K, Dichter MA, Molinoff PB. Up-regulation of N-methyl-D-aspartate receptors on cultured cortical neurons after exposure to antagonists. Mol Pharmacol. 1992;42:147–151. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.