Abstract
Carbapenem resistant Klebsiella pneumoniae (CRKP) are isolated with increasing frequency, especially from immunocompromized patients. The capsular polysaccharide (CPS) types of CPKP were not determined. Investigation of two CRKP isolates from a 2011 outbreak at the Clinical Center, the National Institutes of Health, identified a new capsular type shared by the two isolates, similar to K. pneumonia K19 and K34 but structurally different than any published K. pneumoniae CPS repeating unit:
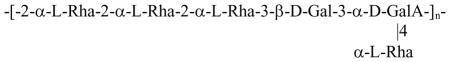
The LPS of the two isolates was found to have no O-specific polysaccharide and the chemical structure of the core oligosaccharides agreed with published data.
If this structure type will be prevalent among CPKP isolates, our findings could facilitate rapid diagnosis and help develop new therapeutic solutions to this antibiotic resistant pathogen.
Keywords: capsular polysaccharide, LPS, structure, KPC, CRKP
Klebsiella pneumoniae is an opportunistic pathogen causing hospital- and community-acquired infections mostly of the respiratory and urinary tracts. Two important virulence factors of K. pneumoniae are capsular polysaccharides (CPS) and lipopolysaccharides (LPS). These two antigens are used for serotyping: K-typing based on the CPS structure, and O-typing based on the LPS’s O-specific polysaccharide (O-SP) structure [1].
Capsular antibodies are protective against capsulated pathogens like pneumococci, meningococci, and Haemophilus influenzae type b [2]. A challenge in developing a Klebsiella CPS-based vaccine is the large number of K antigens (77 K serotypes and many non-typable strains), however only some are known to be associated with human diseases [3]. A 24-valent experimental CPS vaccine was shown to be safe and immunogenic in humans [3;4]. The serotype/s of carbapenem resistant K. pneumoniae (CRKP) isolates are not known. One reason is the lack of commercial anti-capsular sera, another is the occurrence of non-specific serological reactions between sera raised against whole bacteria carrying structurally different K antigens, used for typing in the laboratories [5].
Klebsiella O antigens are currently divided into 8 basic groups. Initially 12 O serotypes were described [6], however detailed chemical analyses revealed that 4 of them are structurally identical [7;8]. New structural types of K. pneumoniae O-antigens are also described [9].
Molecular typing is currently the most common method used in epidemiological studies of K. pneumoniae, yet it does not provide information about the chemical structures of the CPS and LPS essential for developing reagents for active or passive protection from the disease.
In 2011, a CRKP caused an outbreak at the Clinical Center, the National Institutes of Health, infecting 18 patients and causing death of 6 of them [10] The outbreak strain was notable for its transmissibility and persistence in the gastrointestinal tract of patients. We elucidated the CPS and LPS saccharide structures of two isolates from this outbreak.
Based on the colony morphology, we have chosen two outbreak isolates: strain 2796, isolated from patient ID 11 (Table 1 in ref. [10]), with very mucoid colonies, and strain 3264, isolated from patient ID 17 (Table 1 in ref. [10]), with less mucous appearance. The PCR analyses using enterobacterial repetitive intergenic consensus primers (ERIC-PCR) showed high clonal relationship between the two isolates (data not shown), which was in agreement with the results obtained by whole genome sequencing [10]. Both isolates possessed K. pneumoniae carbapenemase (KPC)-1 gene located on their plasmids.
Analysis of the LPS by SDS-PAGE followed by silver staining showed only one low molecular weight band indicating a rough LPS type, with no O-specific polysaccharide chain (Fig. 1). In concordance, this LPS did not react with any O-typing sera. Mild acid hydrolysis of the LPS produced core, consisting of two major variants, differing by the presence of one galacturonic acid, and a disaccharide α-Hep-5-Kdo. Both core and the disaccharide were characterized by 2D NMR and ESI-MS, and the results were in agreement with the published data, Fig. 2 [11]. Two core variants with masses of 1490 Da and 1666 Da were present in similar amounts.
Fig. 1.
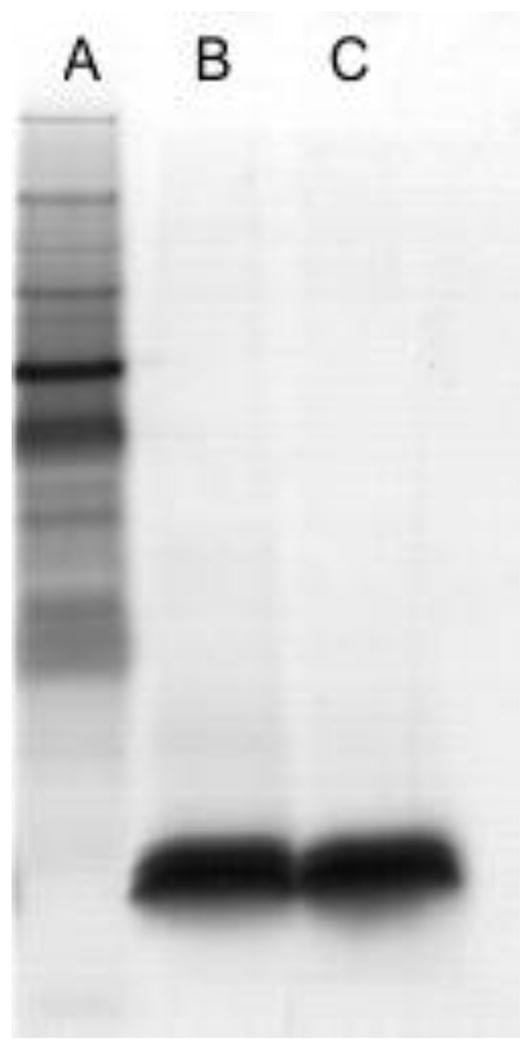
SDS-PAGE analyses of LPS from two isolates of K. pneumoniae: A: marker, B: strain 2796, C: strain 3264.
Fig. 2.
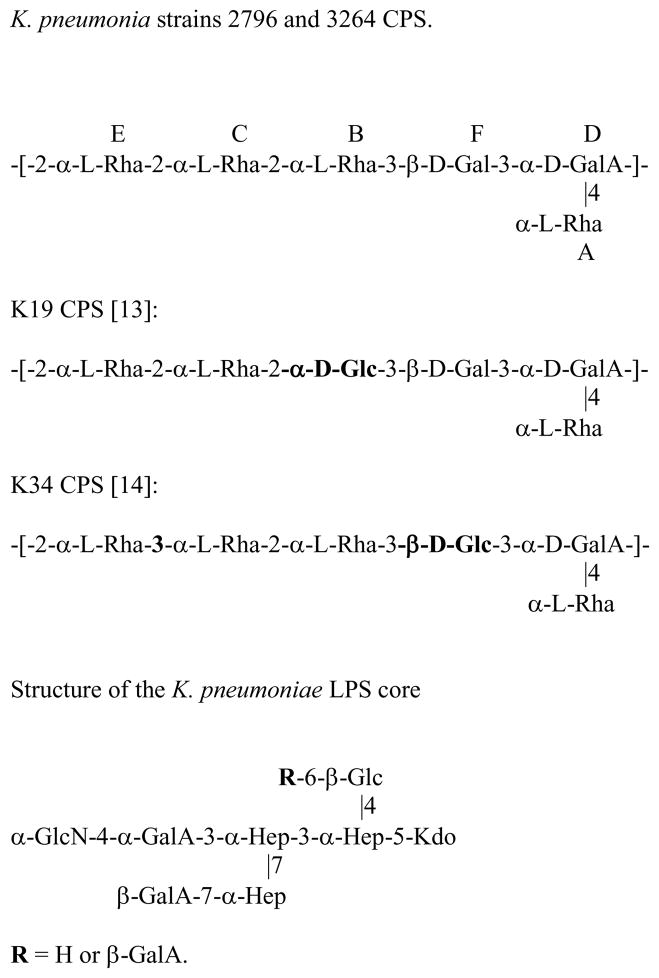
Structures of K. pneumoniae strains 2796 and 3264 CPS analyzed in this study and similar structures of K. pneumoniae K19 and K34 CPSs. Differences between structures are marked in bold type.
Monosaccharide analysis (GC of alditol acetates) of the two isolates revealed Rha and Gal. GC of acetylated methanolysis products showed additionally the presence of GalA. Absolute configurations of the monosaccharides were determined using GC of acetylated or TMS products (for rhamnose) of the reaction with (R)-2-BuOH/AcCl.
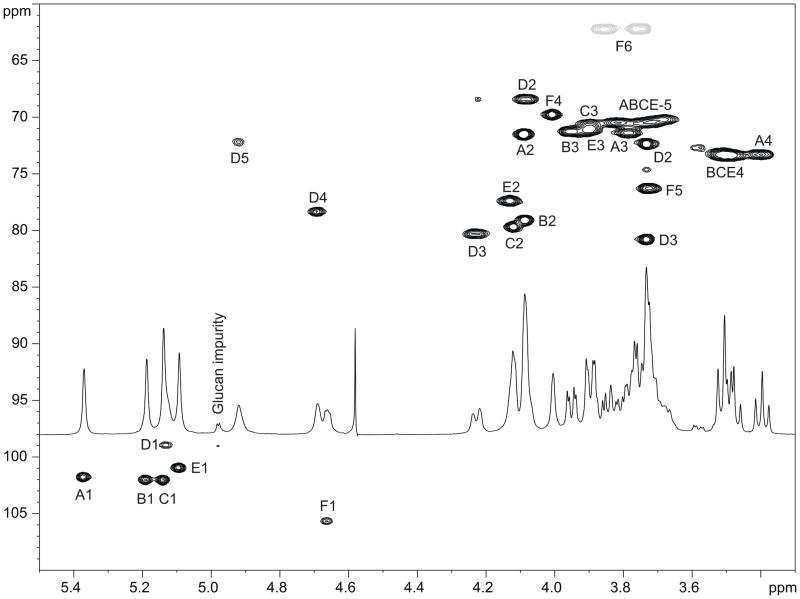
2D NMR spectra of the polysaccharides (gCOSY, TOCSY, NOESY, gHSQC) were recorded at 25 °C and interpreted using Bruker Topspin 2.1 program. Spectra showed four spin systems of α-Rhap, one of β-Galp, and one of α-GalpA, identified by characteristic signal patterns in proton-proton and proton-carbon correlation spectra (Fig. 3). Anomeric configuration for α-rhamnose residues followed from the observation of TOCSY correlations from H-1 to H-2,3,4 (not visible for β-anomers) and position of C-5 signals around 70 ppm (expected at ~73 ppm for β-anomers). Anomeric configuration for α-galacturonic acid residue D followed from J1,2 3.5 Hz and position of C-5 signal at 72.1 ppm. β-Gal F had overlapping H-2 and H-3 signals, distorting J1,2 coupling constant which can not be used for determination of its anomeric configuration. This configuration was deduced from the low-field position of C-1 signal at 105.6 ppm and C-5 signal at 76.2 ppm, typical for β-galactopyranose.
Fig. 3.
HSQC spectrum of the K. pneumoniae CPS (25 °C, 500 MHz). Both strains, 2796 and 3264, gave identical spectra. Grey cross peaks are from CH2 groups, black from CH.
The connection between the monosaccharides was deduced from NOE and methylation data. The following strong NOE were observed: A1:D4,5; A1:F1; B1:F3; C1:B1,2,3; C1:E5; E1:C1,2,3; F1:D3; D1:E1,2, indicating linkage between all monosaccharides except for substitution of Gal F. Due to the overlap of H-2 and H-3 signals of the Gal F one can not determine its substitution position from NOE data. This was done by methylation analysis, which showed the presence of 3-substituted Gal, and terminal and 2-substiuted Rha. Together these data indicate that PS has the structure shown on Fig. 2. 1H and 13C NMR chemical shifts simulated for this structure using the CASPER web service [12] showed good agreement with experimental results.
The structure of this CPS is not identical to any of the published Klebsiella CPS structures, but it resembles K. pneumoniae serotypes K19 [13] and K34 [14] CPSs (Fig. 2), rare human isolates [15]. We observed a cross-reaction with anti-K34 serum; however we did not have anti-K19 serum in our possession.
In summary we present CPS and LPS analyses of two clinical isolates of CRKP, identifying a novel capsular structure and a rough type LPS. The distribution of K-types among multidrug-resistant isolates is unknown. Recently, a first description of a novel capsular gene cluster in a KPC-producing Klebsiella pneumoniae isolated in Brazil was reported [16] and it encourages extending such investigations to other CRKP outbreak strains.
Experimental
1. Characterization of bacteria
K. pneumoniae clinical isolates were obtained from the Clinical Center, the National Institutes of Health. Their identification and spread in the hospital units were reported [10]. Clonal relationship of the genomic DNA of investigated strains was confirmed using the ERIC-PCR method [17]. Briefly, ERIC-PCR conditions were: 7 min/95 °C, 35 cycles consisting of 30 s/95 °C, 1 min/52 °C, 8 min/65 °C and final extension 16 min/65 °C. The following primers were used: ERIC-1R (reverse): ATGTAAGCTCCTGGGGATTCAT; ERIC-1F: AAGTAAGTGACTGGGGTGAGCG. It was further confirmed by the cluster method UPGMA (Quantity One Program, Bio-Rad, Munich, Germany). Both isolates were tested positive by PCR for the presence of KPC-1 using a specific pair of primers as described [18]. Briefly, plasmid DNA was isolated using an A&A DNA isolation kit (A&A Biotechnology, Poland). The presence of plasmid DNA was confirmed electrophoretically, whereas plasmid DNA concentration and purity were assessed using a Picodrop 100 device (Qiagen, Hilden, Germany). PCR conditions were: 5min/95 °C, 35 cycles consisting of 1min/95 °C, 30s/58 °C, 1.5min/72 °C and final extension – 10min/72 °C.
Two isolates were chosen for detailed chemical analysis of their LPS and CPS structures; the selection was based on their colony morphology.
2. Isolation of CPS and LPS
Bacteria were cultured on carbohydrate-rich Worfel–Ferguson agar medium, cells collected and washed with PBS, centrifuged and the pellets washed twice more with PBS. The CPS was extracted by suspending bacterial pellets in PBS and vigorously vortexing for 2h; after centrifugation, the supernatant was dialyzed against water and lyophilized. The CPS preparation contained a viscous material, which was directly subjected to chemical analyses.
LPS was extracted by the hot phenol method and recovered from the water phase [19]. No carbohydrate components were detected in the phenol phase. The LPS was purified by ultracentrifugation. The pellet contained rough LPS as visualized by SDS-PAGE. It was treated with 2 % acetic acid at 100 °C for 3 h to remove lipid A. Soluble products were separated by gel chromatography on a BioGel P-10 column and the oligosaccharide fractions analysed. Both CPS and LPS preparations contained less than 3% of proteins and nucleic acids.
3. NMR spectroscopy
NMR spectra were recorded at 25°C in D2O on a Varian UNITY INOVA 600, instrument, using acetone as reference for proton (2.225 ppm) and carbon (31.5 ppm) spectra. Varian standard programs DQCOSY, NOESY (mixing time of 400ms), TOCSY (spinlock time 120 ms), HSQC, and gHMBC (long-range transfer delay 100 ms) were used. The spectra were processed and analyzed using the Bruker Topspin 2.1 program.
4. Methylation
Methylation was performed by the Ciucanu-Kerek procedure [20]. The products were hydrolyzed with 3 M TFA (120 °C, 3h), dried, reduced with NaBD4, the reagent was destroyed with 0.5 mL of 4 M HCl, and the solution dried under a stream of air, then dried twice with the addition of MeOH (1 mL), then acetylated with 0.4 mL Ac2O – 0.4 mL pyridine for 30 min at 100 °C, dried with air stream and analyzed by GC-MS (Varian Saturn 2000 electron impact ion-trap instrument, capillary column DB-17, 160–260 °C by 4 °/min).
5. Analytical methods
SDS-PAGE used 16% Tricine gels followed by silver staining [21], protein content was measured by Lowry [22], nucleic acid by 260 nm absorption.
6. Serological analyses
Isolates were cultured in Worfel–Ferguson medium, next subcultured 3 times every 24 hours, and incubated for another 4 hours in the same medium. Thereafter, a drop of cells was mixed with a reference K antigen rabbit antiserum on a glass slide and left for 10 min [23]. After application of a coverslip, the preparation was examined for the Quellung reaction under the light microscope using an oil immersion objective. Double immunodiffusion was used to observe reaction of purified LPS with O-typing sera obtained by immunizing rabbits with non-capsulated variants of Klebsiella reference strains [23]. Reference strains with confirmed K and O antigens were from Dr. F. Orskov, The Collaborative Centre of Reference and Research on Escherichia coli and Klebsiella in Copenhagen [23;25].
Table 1.
NMR data for the CPS from K. pneumoniae 3264 (D2O, 25 °C, 500 MHz). The same data were obtained for strain 2796.
| Residue | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| α-Rha A | H | 5.37 | 4.09 | 3.78 | 3.40 | 3.68 | 1.24 |
| C | 101.7 | 71.4 | 71.4 | 73.2 | 70.2 | 17.9 | |
| α-Rha B | H | 5.19 | 4.09 | 3.95 | 3.51 | 3.82 | 1.30 |
| C | 102.0 | 79.1 | 71.2 | 73.4 | 70.1 | 17.9 | |
| α-Rha C | H | 5.14 | 4.12 | 3.90 | 3.48 | 3.72 | 1.28 |
| C | 102.0 | 79.6 | 71.0 | 73.4 | 70.3 | 17.9 | |
| α-Rha E | H | 5.09 | 4.13 | 3.90 | 3.51 | 3.77 | 1.29 |
| C | 100.9 | 77.3 | 71.0 | 73.4 | 70.5 | 17.9 | |
| α-GalA D | H | 5.13 | 4.08 | 4.23 | 4.69 | 4.92 | |
| C | 98.9 | 68.4 | 80.3 | 78.3 | 72.1 | ||
| β-Gal F | H | 4.66 | 3.73 | 3.73 | 4.01 | 3.72 | 3.75; 3.85 |
| C | 105.6 | 72.4 | 80.8 | 69.7 | 76.2 | 62.2 |
Acknowledgments
This work was partially supported by the intramural programs of NICHD and NIAID, National Institutes of Health, Bethesda, MD.
Abbreviations
- CRKP
carbapenem resistant Klebsiella pneumoniae
- KPC
K. pneumoniae carbapenemase
- CPS
capsular polysaccharide
- LPS
lipopolysaccharide
- NOE
Nuclear Overhauser Effect
- gCOSY
gradient Correlation Spectroscopy
- NOESY
Nuclear Overhauser Effect Spectroscopy
- TOCSY
Total Correlation Spectroscopy
- gHSQC
gradient Heteronuclear Single Quantum Coherence
- ERIC
enterobacterial repetitive intergenic consensus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robbins JB, Schneerson R, Szu SC, Fattom A, Yang Y, Lagergard T, Chu C, Sorensen US. Prevention of invasive bacterial diseases by immunization with polysaccharide-protein conjugates. Curr Top Microbiol Immunol. 1989;146:169–180. doi: 10.1007/978-3-642-74529-4_18. [DOI] [PubMed] [Google Scholar]
- 3.Cryz SJ, Jr, Mortimer P, Cross AS, Furer E, Germanier R. Safety and immunogenicity of a polyvalent Klebsiella capsular polysaccharide vaccine in humans. Vaccine. 1986;4:15–20. doi: 10.1016/0264-410x(86)90092-7. [DOI] [PubMed] [Google Scholar]
- 4.Campbell WN, Hendrix E, Cryz S, Jr, Cross AS. Immunogenicity of a 24-valent Klebsiella capsular polysaccharide vaccine and an eight-valent Pseudomonas O-polysaccharide conjugate vaccine administered to victims of acute trauma. Clin Infect Dis. 1996;23:179–181. doi: 10.1093/clinids/23.1.179. [DOI] [PubMed] [Google Scholar]
- 5.Stankiewicz MM. Capsular and somatic antigens of Klebsiella bacilli. Arch Immunol Ther Exp (Warsz) 1990;38:395–406. [PubMed] [Google Scholar]
- 6.Hansen DS, Mestre F, Alberti S, Hernandez-Alles S, Alvarez D, Domenech-Sanchez A, Gil J, Merino S, Tomas JM, Benedi VJ. Klebsiella pneumoniae lipopolysaccharide O typing: revision of prototype strains and O-group distribution among clinical isolates from different sources and countries. J Clin Microbiol. 1999;37:56–62. doi: 10.1128/jcm.37.1.56-62.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorndal H, Lindberg B, Nimmich W. Structural studies on Klebsiella O groups 1 and 6 lipopolysaccharides. Acta Chem Scand. 1971;25:750. doi: 10.3891/acta.chem.scand.25-0750. [DOI] [PubMed] [Google Scholar]
- 8.Kelly RF, MacLean LL, Perry MB, Whitfield C. Structures of the O antigens of Klebsiella serotypes O2(2a,2c), O2(2a,2e,2h) and O2(2a,2f,2g). members of a family of related D-galactan O antigens in Klebsiella spp. J Endotoxin Res. 1995;2:131–140. [Google Scholar]
- 9.Ansaruzzaman M, Albert MJ, Holme T, Jansson PE, Rahman MM, Widmalm G. A Klebsiella pneumoniae strain that shares a type-specific antigen with Shigella flexneri serotype 6. Characterization of the strain and strain and structural studies of the O-antigenic polysaccharide. Eur J Biochem. 1996;237:786–791. doi: 10.1111/j.1432-1033.1996.0786p.x. [DOI] [PubMed] [Google Scholar]
- 10.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vinogradov E, Perry MB. Structural analysis of the core region of the lipopolysaccharides from eight serotypes of Klebsiella pneumoniae. Carbohydr Res. 2001;335:291–296. doi: 10.1016/s0008-6215(01)00216-6. [DOI] [PubMed] [Google Scholar]
- 12.Roslund MU, Säwén E, Landström J, Rönnols J, Jonsson KH, Lundborg M, Svensson MV, Widmalm G. Complete 1H and 13C NMR chemical shift assignments of mono-, di-, and trisaccharides as basis for NMR chemical shift predictions of polysaccharides using the computer program casper. Carbohydr Res. 2011;346:1311–1319. doi: 10.1016/j.carres.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 13.Beurret M, Vignon M, Joseleau JP. Structural investigation of the capsular polysaccharide from Klebsiella K19 by chemical and N.M.R. analyses. Carbohydr Res. 1986;157:13–25. doi: 10.1016/0008-6215(86)85057-1. [DOI] [PubMed] [Google Scholar]
- 14.Joseleau JP, Michon F, Vignon M. Structural investigation of the capsular polysacharide from Klebsilla serotype K-34 and its characterization by N.M.R. spectroscopy. Carbohydr Res. 1982;101:175–185. doi: 10.1016/s0008-6215(00)80998-2. [DOI] [PubMed] [Google Scholar]
- 15.Cryz SJ, Jr, Mortimer PM, Mansfield V, Germanier R. Seroepidemiology of Klebsiella bacteremic isolates and implications for vaccine development. J Clin Microbiol. 1986;23:687–90. doi: 10.1128/jcm.23.4.687-690.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos PI, Picão RC, Vespero EC, Pelisson M, Zuleta LF, Almeida LG, Gerber AL, Vasconcelos AT, Gales AC, Nicolás MF. Pyrosequencing-based analysis reveals a novel capsular gene cluster in a KPC-producing Klebsiella pneumoniae clinical isolate identified in Brazil. BMC Microbiol. 2012;12:173. doi: 10.1186/1471-2180-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westphal O, Jann K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 20.Ciucanu I. Per-O-methylation reaction for structural analysis of carbohydrates by mass spectrometry. Anal chim acta. 2006;576:147–155. doi: 10.1016/j.aca.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 22.Peterson GL. Review of the Folin phenol protein quantitation method of Lowry, Rosebrough, Farr and Randall. Anal Biochem. 1979;100:201–220. doi: 10.1016/0003-2697(79)90222-7. [DOI] [PubMed] [Google Scholar]
- 23.Przondo-Hessek A, Schumacher-Perdreau F, Pulverer G. Cell surface hydrophobicity of Klebsiella strains. Arch Immunol Ther Exp. 1987;35:283–287. [PubMed] [Google Scholar]
- 24.Kauffmann F. On the serology of Klebsiella group. Acta Pathologica Microbiologica Scandinavica. 1949;26:381–406. [Google Scholar]
- 25.Edwards PR, Ewing WH. Identification of Enterobacteriaceae. 3. Burgess Publishing Company; Minneapolis: 1972. [Google Scholar]