Abstract
Background
The merozoite surface protein-1 (MSP-1) is a candidate target for the development of blood stage vaccines against malaria. Polymorphism in MSP-1 can be useful as a genetic marker for strain differentiation in malarial parasites. Although sequence diversity in the MSP-1 locus has been extensively analyzed in field isolates of Plasmodium falciparum and P. vivax, the extent of variation in its homologues in P. ovale curtisi and P. ovale wallikeri, remains unknown.
Methodology/Principal Findings
Analysis of the mitochondrial cytochrome b sequences of 10 P. ovale isolates from symptomatic malaria patients from diverse endemic areas of Thailand revealed co-existence of P. ovale curtisi (n = 5) and P. ovale wallikeri (n = 5). Direct sequencing of the PCR-amplified products encompassing the entire coding region of MSP-1 of P. ovale curtisi (PocMSP-1) and P. ovale wallikeri (PowMSP-1) has identified 3 imperfect repeated segments in the former and one in the latter. Most amino acid differences between these proteins were located in the interspecies variable domains of malarial MSP-1. Synonymous nucleotide diversity (πS) exceeded nonsynonymous nucleotide diversity (πN) for both PocMSP-1 and PowMSP-1, albeit at a non-significant level. However, when MSP-1 of both these species was considered together, πS was significantly greater than πN (p<0.0001), suggesting that purifying selection has shaped diversity at this locus prior to speciation. Phylogenetic analysis based on conserved domains has placed PocMSP-1 and PowMSP-1 in a distinct bifurcating branch that probably diverged from each other around 4.5 million years ago.
Conclusion/Significance
The MSP-1 sequences support that P. ovale curtisi and P. ovale wallikeri are distinct species. Both species are sympatric in Thailand. The low level of sequence diversity in PocMSP-1 and PowMSP-1 among Thai isolates could stem from persistent low prevalence of these species, limiting the chance of outcrossing at this locus.
Introduction
About half of the world's population resides in areas at risk of contracting malaria, one of the leading causes of morbidity and mortality, accounting for more than 200 million cases and more than 600,000 deaths per annum [1]. Six species in the genus Plasmodium are known to cause human malaria under natural transmission [2]–[4]. However, only malaria caused by Plasmodium falciparum and Plasmodium vivax have been extensively studied, whereas relatively little is known about the less prevalent malaria parasites. Although Plasmodium ovale has been proposed as a valid human malaria species by Stephens in 1922 upon examination of the blood sample from a patient acquiring infection from East Africa [5], it was not until almost a century later that this malaria has been further subdivided into 2 distinct species, i.e. P. ovale curtisi (or classic type) and P. ovale wallikeri (or variant type) based on molecular analysis [4], [6], [7].
P. ovale has a wide geographic distribution across tropical countries, especially Africa, Asia and some Western Pacific islands [8]. In several malaria endemic areas, P. ovale has been found to be sympatric with the major malaria species, P. falciparum and P. vivax. The low parasite densities of P. ovale in infected individuals and its morphological resemblance to P. vivax have hampered efficient microscopy detection, especially when they co-exist with other malaria species in circulation. Therefore, the actual prevalence of this malaria species can be underestimated [8], [9]. Based on limited epidemiological data, it has been estimated that at least 15 million cases occur annually in Sub-Saharan African countries [4]. Because P. ovale possesses a hypnozoite stage in liver cells similar to that found in P. vivax, a relapsing course of infection can ensue.
To date, relatively few molecular markers are available to document the extent of genetic variation and strain differentiation of the sibling species of P. ovale. One of the polymorphic genetic loci that has been well characterized in P. falciparum and P. vivax is the gene encoding the merozoite surface protein-1 (MSP-1) [10], [11]. The precursor of P. falciparum merozoite surface protein-1 (PfMSP-1) is synthesized during schizogony and undergoes primary processing that generates polypeptides of 83, 30, 38 and 42 kDa [12]. The 42-kDa fragment at the C-terminus is further proteolytically cleaved into 33 and 19 kDa fragments by the time of erythrocyte entry. The 19 kDa fragment, containing two epidermal growth factor (EGF)-like motifs, remains attached to the surface of newly invasive merozoite through the ring stage whereas other processed fragments are shed in circulation [12]. Besides being one of the prime asexual erythrocytic vaccine candidates, MSP-1 exhibits extensive sequence divergence within and between different malaria species [13]. Therefore, analysis of this genetic locus will be useful for detailed characterization of the two sibling species of P. ovale.
Recently, two nucleotide sequences of the gene encoding the merozoite surface protein-1 of P. ovale curtisi (PocMSP-1) from Cameroon patients were determined [14]. However, the MSP-1 sequence of P. ovale wallikeri (PowMSP-1) remains unknown. Herein, we have determined the extent of sequence variation in the MSP-1 locus of isolates derived from symptomatic malaria patients in Thailand whose blood samples contained P. ovale based on polymerase chain reaction (PCR)-based detection targeting the small subunit ribosomal RNA gene. Sequence analysis has led to identification of the MSP-1 sequence of P. ovale wallikeri in Thai patients.
Materials and Methods
Human Ethics Statement
The protocol was reviewed and approved by the Institutional Review Board on Human Research of Faculty of Medicine, Chulalongkorn University (IRB259/54). Written informed consent was obtained from participants or from parents/legally guardians enrolled using an approved consent form.
P. ovale isolates
Blood samples with single infection of P. ovale were obtained from 10 symptomatic malaria patients who acquired the infections from diverse endemic areas of Thailand. Background data of each isolate was shown in Table 1. DNA from each isolate was prepared by using Qiagen DNA mini kit (Qiagen, Hilden, Germany) following the protocol provided by the manufacturer and stored at −40 °C until use. Diagnosis of P. ovale was performed by both microscopy and nested PCR targeting the small subunit ribosomal RNA genes (SSU rRNA) of 5 human malaria species as previously described [15], [16]. The diagnostic primers for P. ovale could amplify the SSU rRNA genes of both classic and variant types.
Table 1. Demographic and parasitologic profiles of patients infected with Plasmodium ovale.
| Isolate | Age (Year) | Sex | Year infected | Place acquiring infection | Parasite density (/µl) | SSU rRNA-PCR | mtCYTB sequence* |
| PO-1 | 27 | Male | 1993 | Kanchanaburi Province | 2,400 | P. ovale | P. ovale wallikeri |
| PO-2 | 28 | Male | 1994 | Kanchanaburi Province | 3,200 | P. ovale | P. ovale wallikeri |
| PO-3 | 33 | Male | 1994 | Kanchanaburi Province | 2,520 | P. ovale | P. ovale wallikeri |
| PO-4 | 19 | Female | 1995 | Tak Province | 1,000 | P. ovale | P. ovale curtisi |
| PO-5 | 14 | Female | 2006 | Tak Province | 2,560 | P. ovale | P. ovale wallikeri |
| PO-6 | 35 | Female | 2007 | Tak Province | 600 | P. ovale | P. ovale wallikeri |
| PO-7 | 12 | Male | 2008 | Kanchanaburi Province | 1,280 | P. ovale | P. ovale curtisi |
| PO-8 | 30 | Male | 2010 | Tak Province | 3,520 | P. ovale | P. ovale curtisi |
| PO-9 | 23 | Male | 2010 | Tak Province | 2,560 | P. ovale | P. ovale curtisi |
| PO-10 | 34 | Male | 2010 | Yala Province | 1,760 | P. ovale | P. ovale curtisi |
Nucleotide differences between the mtCYTB fragment occurring at respective positions 175, 187, 205, 316 and 334 were G, C, T, C and C for P. ovale curtisi and T, T, A, T and T for P. ovale wallikeri.
PCR amplification and sequencing of the mitochondrial cytochrome b gene
A fragment of the mitochondrial cytochrome b (mtCYTB) gene of Plasmodium ovale was amplified by PCR using primers PoCytbF (5′-CTTACATTTACATGGTAGAC-3) and PoCytbR (5′-GCCATTTTGAATTGTATAATAG-3′) in a total volume of 25 µL containing 1 µL of template DNA, 2.5 mM each deoxynucleoside triphosphate, 2.5 µL of 10× PCR buffer, 0.3 µM of each primer and 0.5 unit of ExTaq DNA polymerase (Takara, Seta, Japan). The thermal cycling profile included a preamplification denaturation at 94°C, 1 min; 35 cycles of denaturation at 94°C, 40 s, annealing at 53°C, 30 s and extension at 72°C, 30 s; and post amplification extension at 72°C, 5 min. All amplification reactions were done in an Applied Biosystem GeneAmp® PCR System 9700 thermocycler (PE Biosystems, Foster City, CA). PCR products were analyzed by 1% agarose gel electrophoresis. DNA sequencing was performed directly from the purified PCR product using Qiagen PCR purification kit (Qiagen, Hilden, Germany).
PCR amplification and sequencing of the PoMSP-1 gene
The nucleotide sequence of PoMSP-1 was amplified by nested PCR using PoMSP1F0 (5′-AATTCAAAAATGAAGGTGTTC-3′) and PoMSP1R0 (5′-CTTTTGTATTTACCCTCACTC-3′) as outer primers and PoMSP-1F1 (5′-AGGTGTTCGTATTTGCGCTC-3′) and PoMSP-1R1 (5′-CTCTCTCCTTTTAAAGTAAG-3) as inner primers. Two microlitres of the PCR products from primary PCR were used as template for secondary PCR in a total volume of 30 µl. Amplification conditions for primary and secondary PCRs were identical comprising 35 cycles of 96°C for 20 s, 62°C for 5 min with an initial pre-amplification denaturation at 94°C for 1 min and a final post-amplification extension at 72°C for 10 min. DNA sequences were obtained directly from the PCR-amplified products. Sequences have been deposited in the GenBank™ database under the accession numbers KC137340-KC137349.
Data analysis
Alignment of the PoMSP-1 nucleotide sequences was performed using the default option of the CLUSTAL_X program [17] and manually edited. Insertions/deletions (indels) in coding regions were determined from multiple alignments of amino acid sequences to maintain the reading frame. Sequences of the two Cameroon isolates (GenBank accession numbers FJ824670 and FJ824671) were included for comparison [14]. Tandem repeats were detected by scanning the sequence with a small window, determining the distance between exact matches and testing the statistical criteria as implemented in the Tandem Repeats Finder version 4.0 program [18]. Nucleotide diversity (π) was computed from the average number of pairwise sequence differences at synonymous sites (πS) and nonsynonymous sites (πN) in the sample sequences [19]. Standard errors of these parameters were estimated by the bootstrap method with 1,000 pseudoreplicates using the MEGA 5.05 program [20]. Significant differences between πS and πN by Z-tests were considered to provide evidence of selective pressure on tested regions. Nucleotide divergence between pairs of closely related malaria species was calculated from the number of base substitutions per site between sequences using the maximum composite likelihood model and its standard error was obtained by 1,000 bootstrap replicates. All sites with gaps were excluded from analysis. Sequences and their GenBank accession numbers included for analysis were the mtCYTB gene of P. falciparum (XM001348736), P. rechenowi (NC002235), P. fieldi (AB444133), P. simiovale (AY800109), P. ovale curtisi (GU723533) and P. ovale wallikeri (HQ712053); and the SSU rRNA (A-type) locus of these species (M19172, Z25819, AB287283, AB287287, JF894405 and JF894411, respectively). Phylogenetic tree was inferred from amino acid sequences by using the maximum likelihood method based on the Jones-Taylor-Thornton (JTT) model [21] as implemented in the MEGA 5.05 program [20]. Reliability of branching patterns was evaluated by bootstrapping using 1,000 iterations. The MSP-1 sequences of other malaria species and their accession numbers included for comparison were P. falciparum (X03371), P. vivax (AF435593), P. malariae (FJ824669), P. knowlesi (DQ220743), P. fragile (AB444067), P. coatneyi (AB266180), P. inui (AB444062), P. hylobati (AB266182), P. cynomolgi (AB444063), P. fieldi (AB444066), P. simiovale (AB266185), P. gonderi (AB444069), P. chabaudi (L22982), P. berghei (U43521), P. yoelii (XM721164), P. gallinaceum (AJ809338) and P. reichenowi (AJ786604). Estimation of divergence time between PocMSP-1 and PowMSP-1 was inferred from interspecies conserved domains by using the Bayesian Evolutionary Analysis by Sampling Trees (BEAST) package based on Markov Chain Monte Carlo (MCMC) algorithms [22]. The divergence time was calibrated with PfMSP-1 and PrMSP-1 assuming that P. falciparum and P. reichenowi have diverged along with their respective human and chimpanazee hosts since 6±0.5 million years ago (MYA). Analysis was performed by using uncorrelated lognormal relaxed clock, Yule Process for the tree prior, HKY site model with estimated base frequencies and a 4 category gamma site heterogeneity model. Simulations were run for 25,000,000 cycles and logged at every 1,000 cycles.
Results
Amplification and sequencing of mtCYTB of P. ovale
Amplification of the mtCYTB gene of P. ovale encompassing 357 bp fragment has generated single PCR products for all isolates examined. Among the ten isolates described here, sequence analysis identified 5 nucleotide substitutions that segregated perfectly into only two mtCYTB genotypes. The mtCYTB sequences of isolates PO-3, PO-4, PO-5, PO-6 and PO-8 contained G, C, T, C and C at positions 175, 187, 205, 316 and 334 (numbering from the first nucleotide in the forward amplification) consistent with P. ovale curtisi (GenBank accession number HQ712052) whereas the remaining isolates having T, T, A, T and T at these respective sites belonged to P. ovale wallikeri (accession number HQ712053) (Table 1).
Amplification and sequencing of PoMSP-1
PoMSP-1 was successfully amplified by PCR in all isolates, generating single PCR products of expected size (∼5 kb). Direct sequencing of these purified PCR fragments has shown no superimposed signal on electropherograms of PoMSP-1of these samples, suggesting no clonal mixtures in isolates examined. All 5 MSP-1 sequences of P. ovale curtisi contained 5,181 bp whereas 4 of 5 P. ovale wallikeri had 5,016 bp and the remaining isolate (PO-10) from southern Thailand possessed 5,043 bp. In total, 7 distinct PoMSP-1 nucleotide sequences were identified among 10 Thai isolates. Perfectly identical sequences were observed in 3 isolates belonging to P. ovale curtisi (PO-4, PO-8 and PO-9) and 2 isolates identified as P. ovale wallikeri (PO-2 and PO-3).
Comparison of MSP-1 from P. ovale curtisi and P. ovale wallikeri
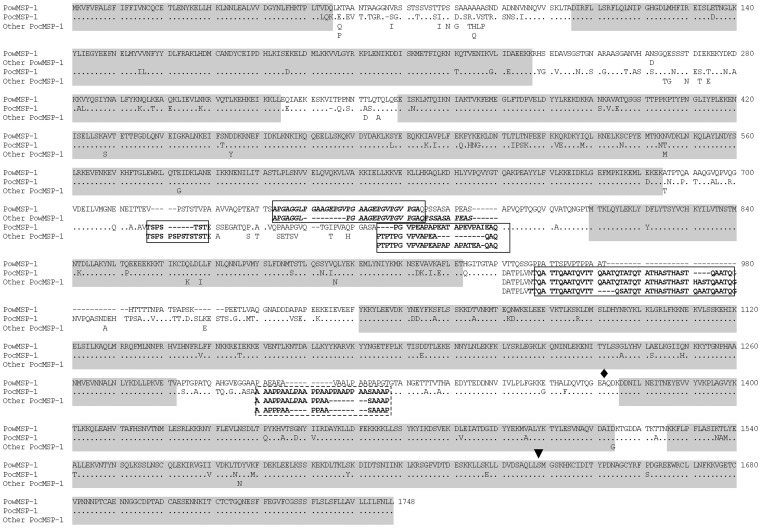
Sequence comparison of MSP-1 derived from human, nonhuman primate, avian and murine malaria in previous studies have shown that malarial MSP-1 could be partitioned into 15 domains comprising 7 variable domains flanked by interspecies conserved sequences [13], [14] (Figure 1). Alignment of the PoMSP-1 sequences from Thai isolates has identified two distinct groups corresponding to each species as determined by the mtCYTB genotypes (Table 1). For PoMSP-1 of P. ovale curtisi (PocMSP-1), 4 repeats-encoding regions were identified as shown in Figure 1. On the other hand, a 9-nucleotide repeat region having the consensus sequence AGGAGTACC was found in PoMSP-1 of P. ovale wallikeri (PowMSP-1) that was conserved among isolates. In non-repeats regions, codon difference between PocMSP-1 and PowMSP-1 as well as insertion or deletion of amino acid residues were more pronounced outside interspecies conserved domains (95/311 codons, 30.55%) than those in interspecies conserved domains (87/1288 codons, 6.75%), consistent with the general pattern of interspecific sequence diversity of malarial MSP-1 locus. The potential cleavage sites generating 42 KDa and 19 KDa fragments were conserved among isolates. Although 38 nucleotide differences were detected in the 42 KDa-encoding fragments of both PocMSP-1 and PowMSP-1 resulting in 18 codon changes, these substitutions were conserved for each species. In the 19 KDa fragment, a single amino acid difference between PocMSP-1 and PowMSP-1 was identified, i.e. Ser1661Pro (position after Figure 1).
Figure 1. Alignment of amino acid sequences of PocMSP-1 and PowMSP-1 among 10 clinical isolates and two Cameroon strains (GenBank accession numbers HQ712052 and HQ712053).
Dots and dashes represent residues identical to PowMSP-1 and deletions, respectively. ‘Other PocMSP-1’ and ‘Other PowMSP-1’ denotes possible substitutions in other isolates. Repeat regions are in bold, italicized and boxed. Interspecies conserved domains are shaded. Potential cleavage sites for generating 42 KDa- and 19 KDa-fragments are shown as diamond and arrow head above the alignment.
Nucleotide diversity
Analysis of nucleotide substitution in PocMSP-1 and PowMSP-1 has shown that synonymous nucleotide diversity (πS) exceeded nonsynonymous nucleotide diversity (πN) (Table 2). However, these differences did not reach statistically significant levels. On the other hand, πS significantly exceeded πN (p<0.001) when both species were considered together, suggesting that purifying selection has shaped variation in the PoMSP-1 locus preceding speciation, presumably from functional or structural constraint on the protein.
Table 2. Synonymous (πS) and nonsynonymous (πN) nucleotide diversity in non-repeat region of MSP-1.
| πS±S.E. | πN±S.E. | |
| P. ovale curtisi (Thai and Cameroon isolates, n = 7) | 0.0027±0.0013 | 0.0017±0.0005 |
| P. ovale curtisi (Thai isolates, n = 5) | 0.0013±0.0009 | 0.0001±0.0001 |
| P. ovale wallikeri (Thai isolates, n = 5) | 0.0005±0.0004 | 0.0000±0.0000 |
| P. ovale (all) | 0.0793±0.0080*** | 0.0160±0.0016 |
Z-tests of the hypothesis that πS = πN: *** p<0.001.
When πS and πN were computed for all P. ovale MSP-1, including both PocMSP-1 and PowMSP-1, the values were much greater than those computed for PocMSP-1 and PowMSP-1 separately (Table 2), as would be expected when combining data from two non-recombining species.
Phylogenetic analysis and nucleotide divergence
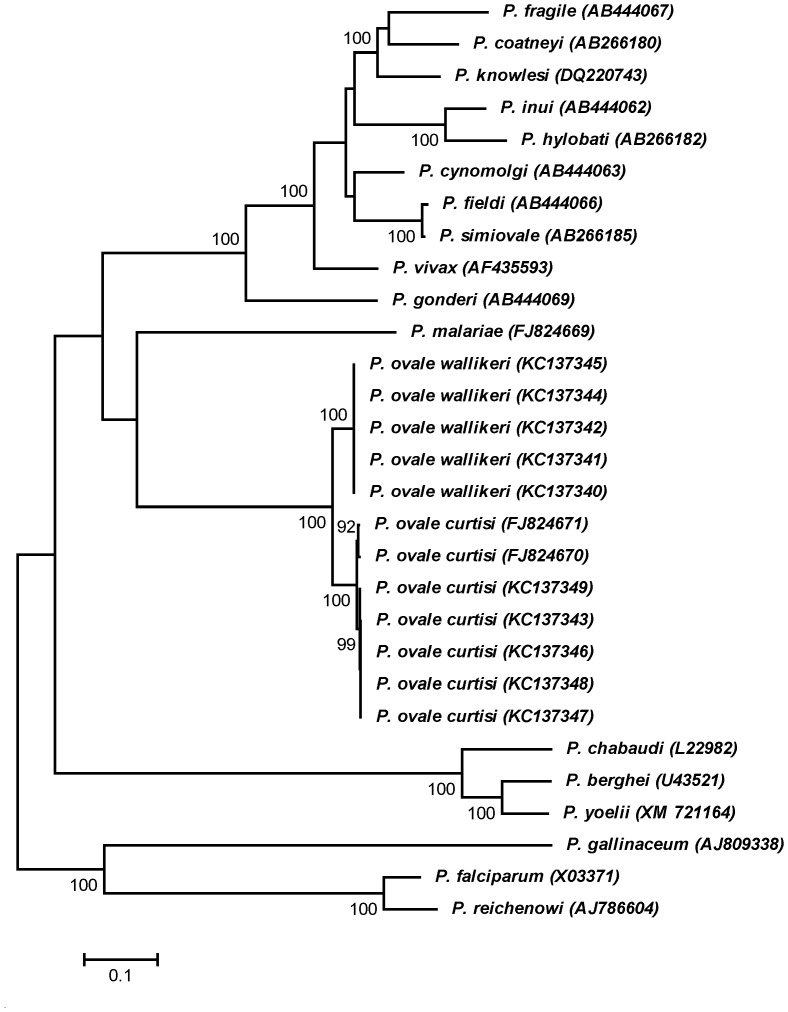
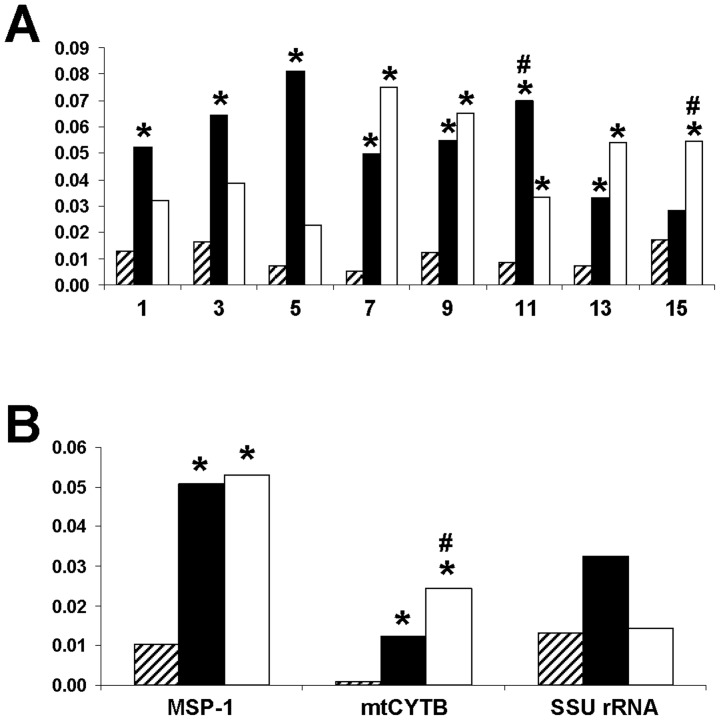
Phylogenetic inference from concatenated amino acid sequences of human, nonhuman primate, murine and avian malarial MSP-1 using the maximum likelihood method has shown that PoMSP-1 and MSP-1 of P. malariae (PmMSP-1) shared a node deep in the tree as previously noted [14]. Importantly, PocMSP-1 and PowMSP-1 occupied a distinct bifurcating branch with 100% bootstrap support (Figure 2). It is noteworthy that the branch length from the node of PocMSP-1 and PowMSP-1 was longer than that separating MSP-1 of P. fieldi (PfiMSP-1) and P. simiovale (PsoMSP-1). It is of note that the evolutionary divergence based on nucleotide sequences between PocMSP-1 and PowMSP-1 was significantly greater than those between PfiMSP-1 and PsoMSP-1 in all but one interspecies conserved domain (Figure 3). Likewise, the nucleotide divergences of concatenated interspecies conserved domains between PfMSP-1 and MSP-1 of P. reichenowi (PrMSP-1), and between PocMSP-1 and PowMSP-1 were significantly more than that of PfiMSP-1 and PsoMSP-1. Meanwhile, comparison of nucleotide divergence at the mtCYTB locus between these closely related malaria species has shown similar findings as those for the MSP-1 locus. Furthermore, the mean divergence at the SSU rRNA locus between P. ovale curtisi and P. ovale wallikeri exceeded that seen for the other species pair (Figure 3). The average divergence time between PocMSP-1 and PowMSP-1 was estimated to be around 4.5±0.07 MYA (95% Highest Posterior Density 0.5 – 7.7 MYA). This would correspond to a rate of 1.6×10−9 to 5.4×10−9 substitutions per site per year, which is close to previous estimates of the range of nucleotide substitution rates of other malarial genes [23]–[25].
Figure 2. Neighbor-joining tree inferred from the MSP-1 intersepecies conserved sequences of human, nonhuman primate, avian and murine malaria.
GenBank accession numbers are in paraentheses. Bootstrap values more than 50% based on 1,000 replicates are shown along the branches. Scale indicates amino acid substitutions per site.
Figure 3. Nucleotide divergence between P. fieldi and P. simiovale (upward diagonal), P. ovale curtisi and P. ovale wallikeri (filled) and P. falciparum and P. reichenowi (unfilled) at each interspecies conserved domain of MSP-1 (A), entire intersepecies conserved domains of MSP-1, mitochondrial cytochrome b (mtCYTB) and small subunit ribosomal RNA (SSU rRNA) (B) based on the number of base substitutions per site between sequences using the maximum composite likelihood model.
Asterisks and pound signs indicate significant differences when compared with P. fieldi/P. simiovale and between P. ovale curtisi/P. ovale wallikeri and P. falciparum/P. reichenowi, respectively.
Discussion
Our recent PCR-based diagnosis of malaria species distribution in Thailand involving 5,044 malaria patients during 2006–2007 and 2008–2009 in major endemic areas have shown that malaria caused by P. ovale contributed to 1.03% and 0.13%, respectively, of all Plasmodium identified [15], [16]. The low prevalence of P. ovale in our studies was not caused by PCR primer-escape detection because our P. ovale-specific primers target conserved sequences in the SSU rRNA genes of both P. ovale curtisi and P. ovale wallikeri [15], [16]. However, our recent study has shown that using a more sensitive PCR target such as the mitochondrial cytochrome b locus has increased the number of P. ovale-positive cases than that using the SSU rRNA target because the copy of mtCYTB per cell outnumbers that of SSU rRNA [26]. This also implies that some P. ovale infections, especially those co-infecting with other malaria species, occurring at a very low parasite density could be undiagnosed by SSU rRNA-based PCR. Therefore, the actual burden of P. ovale infection could be underestimated. Meanwhile, the distribution of P. ovale in Thailand exhibited geographic variation with a higher prevalence in endemic areas bordering Myanmar than those bordering Cambodia and Malaysia [15], [16].
Recent molecular analysis of various genetic loci of P. ovale from diverse geographic origins has purported that the extant P. ovale population contained 2 distinct and non-recombining species designated P. ovale curtisi and P. ovale wallikeri [4], [7]. Phylogenetic analysis has clearly placed both siblings into distinct bifurcating branch with high bootstrap support. Molecular epidemiological studies have further revealed that both P. ovale curtisi and P. ovale wallikeri are sympatric in Angola, Congo, Equatorial Guinea, Uganda, Ghana, Bangladesh and Myanmar whereas only P. ovale curtisi was previously identified in Thailand [4], [27]–[29]. Herein, analyses of the mtCYTB and the MSP-1 loci have further supported co-existence of P. ovale curtisi and P. ovale wallikeri in this country. Although the actual prevalence of these siblings species in Thailand could not be determined due to the low prevalence of these parasites, our analysis has identified equal number of these parasites, suggesting that both species has circulated in this country at a comparable frequency.
The MSP-1 sequences of P. ovale wallikeri newly identified here were structurally differed from those of P. ovale curtisi in terms of number of repeat regions and several nucleotide differences (Figure 1). Like other malarial genes, these repeats could be evolved by slippage-strand mispairing or related mechanisms [30]. However, it is noteworthy that the majority of amino acid differences between PocMSP-1 and PowMSP-1 sequences occurred outside interspecies conserved domains of MSP-1. The extent of nucleotide diversity in MSP-1 of P. ovale curtisi seems to be greater than that of P. ovale wallikeri whereas nucleotide diversity at synonymous sites exceed that of nonsynonymous sites in non-repeat regions of both species although no significant difference was observed. On the other hand, when MSP-1 of both species were considered together, synonymous nucleotide diversity significantly outnumbered nonsynonymous nucleotide diversity, suggesting that purifying selection may shape the pattern of sequence diversity in the MSP-1 locus preceding speciation. Both synonymous and nonsynonymous nucleotide diversity were much greater when both PocMSP-1 and PowMSP-1 were included in the computation than when PocMSP-1 and PowMSP-1 were considered separately (Table 2). This result is consistent with the hypothesis of Sutherland and Polley [4], [31] that Poc and Pow entered the human host lineage separately, being sampled from an ancestral P. ovale population that was more diverse than either P. ovale curtisi or P. ovale wallikeri is today.
A phylogenetic tree inferred from the MSP-1 sequences has placed P. ovale curtisi and P. ovale wallikeri in a distinct bifurcating branch with 100% bootstrap support. It is noteworthy that the branch length from the node separating these sibling species of P. ovale seems to be longer than that separating P. fieldi and P. simiovale. Comparison of sequences in interspecies conserved domains in the MSP-1 locus of some closely related malaria has shown that nucleotide divergence between P. ovale curtisi and P. ovale wallikeri significantly exceeded that between P. fieldi and P. simiovale in all but one domain. The overall nucleotide divergence in interspecies conserved domains between P. ovale curtisi and P. ovale wallikeri was comparable to that between P. falciparum and P. reichenowi. Therefore, the MSP-1 sequence also supports speciation of these sibling species of P. ovale akin to other loci such as mtCYTB and SSU rRNA [4]. Recent analysis using the mtCYTB locus and the gene encoding glyceraldehyde-3-phosphatase has suggested that time to the most recent common ancestor to P. ovale curtisi and P. ovale wallikeri was between 1.0 and 3.5 MYA [4]. Meanwhile, the divergence time in the MSP-1 locus of malaria parasites seems to be much more ancient than house-keeping gene loci. The dimorphic prototypes of PfMSP-1, represented by K1 and MAD20 strains, seem to share the last common ancestor around 27 - 35 MYA [23], [24] whereas the divergence time between P. vivax and P. knowlesi coincides with the time of radiation of Southeast Asian macaques about 5 MYA [32], [33]. However, our analysis has suggested that the split of PocMSP-1and PowMSP-1 seems to be relatively more recent. Therefore, the MSP-1 sequences of P. ovale curtisi and P. ovale wallikeri support ancient divergence times of malaria lineages [34], [35].
One of the major mechanisms generating genetic diversity in the MSP-1 locus is interallelic recombination between distinct alleles during malarial sexual development in anopheleine vectors [10], [11]. Epidemiological studies have shown that the degree of heterologous mating in malaria populations is positively correlated with transmission rates and the prevalence of mixed allele infections [36]. In Thailand, the extent of diversity in PvMSP-1 seems to be highly variable with extensive sequence diversity among isolates from northwestern region where both prevalence and mixed allele infections of P. vivax have been more pronounced than other endemic areas of the country [15], [16]. On the other hand, only few PvMSP-1 alleles were detected in P. vivax populations from southern Thailand as a consequence of preceding population bottlenecks, probably from extensive malaria control measures [37]. By contrast, the extent of diversity in both PocMSP-1 and PowMSP-1 in this study was at least an order of magnitude lower than that for PvMSP-1 whereas perfectly identical PowMSP-1 or PocMSP-1 sequences were identified in isolates collected over a decade apart. The low level of sequence diversity in both PocMSP-1 and PowMSP-1 could stem from repeated genetic bottlenecks, low transmission rate and autogamous breeding. Unfortunately, the limited number of samples in this study precludes a meaningful analysis of genetic recombination at this locus. On the other hand, the prevalence of P. ovale in Thailand has been persistently low despite using molecular detection [15], [16]; thereby, limiting the chance for heterozygous mating comparing with other malaria species.
In conclusion, the nucleotide divergence of the MSP-1 sequences support that P. ovale curtisi and P. ovale wallikeri are distinct species. The low level of within species diversity at the MSP-1 sequences could be a result of low transmission rate and repeated bottleneck effects.
Acknowledgments
We are grateful to all patients who donated their blood samples for this study. We thank Napaporn Kuamsab for technical assistance.
Funding Statement
This study was supported by the Thailand Research Fund (grant nos. RSA5480008) to CP; Government Research Budget (fiscal year 2012) to CP and SJ; and GM43940 from the United States National Institutes of Health to ALH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Healh Organization (WHO) (2011) World malaria report 2011.Geneva, Switzerland: WHO Press. Available: http://www.cdc.gov/malaria/malaria_worldwide/impact.html. Accessed 2012 December 1. [Google Scholar]
- 2. Chin W, Contacos PG, Collins WE, Jeter MH, Alpert E (1968) Experimental mosquito transmission of Plasmodium knowlesi to man and monkey. Am J Trop Med Hyg 17: 355–358. [DOI] [PubMed] [Google Scholar]
- 3.Garhnam PCC (1966) Malaria parasites and other haemosporidia, first ed. Blackwell Scientific Publications, Oxford. [Google Scholar]
- 4. Sutherland CJ, Tanomsing N, Nolder D, Oguike M, Jennison C, et al. (2010) Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J Infect Dis 201: 1544–1550. [DOI] [PubMed] [Google Scholar]
- 5. Stephens JWW (1922) A new malaria parasite of man. Ann Trop Med Parasitol 16: 383–388. [Google Scholar]
- 6. Tachibana M, Tsuboi T, Kaneko O, Khuntirat B, Torii M (2002) Two types of Plasmodium ovale defined by SSU rRNA have distinct sequences for ookinete surface proteins. Mol Biochem Parasitol 122: 223–226. [DOI] [PubMed] [Google Scholar]
- 7. Win TT, Jalloh A, Tantular IS, Tsuboi T, Ferreira MU, et al. (2004) Molecular analysis of Plasmodium ovale variants. Emerg Infect Dis 10: 1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins WE, Jeffery GM (2005) Plasmodium ovale: parasite and disease. Clin Microbiol Rev 18: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dinko B, Oguike MC, Larbi JA, Bousema T, Sutherland CJ (2013) (in press) Persistent detection of Plasmodium falciparum, P. malariae, P. ovale curtisi and P. ovale wallikeri after ACT treatment of asymptomatic Ghanaian school-children. Int J Parasitol: Drugs and Drug Resistance [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanabe K, Mackay M, Goman M, Scaife JG (1987) Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum . J Mol Biol 195: 273–287. [DOI] [PubMed] [Google Scholar]
- 11. Putaporntip C, Jongwutiwes S, Sakihama N, Ferreira MU, Kho WG, et al. (2002) Mosaic organization and heterogeneity in frequency of allelic recombination of the Plasmodium vivax merozoite surface protein-1 locus. Proc Natl Acad Sci USA 99: 16348–16353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holder AA (1988) The precursor to major merozoite surface antigens: structure and role in immunity. Prog Allergy 41: 72–97. [PubMed] [Google Scholar]
- 13. Sawai H, Otani H, Arisue N, Palacpac N, de Oliveira Martins L, et al. (2010) Lineage-specific positive selection at the merozoite surface protein 1 (msp1) locus of Plasmodium vivax and related simian malaria parasites. BMC Evol Biol 10: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Birkenmeyer L, Muerhoff AS, Dawson GJ, Desai SM (2010) Isolation and characterization of the MSP1 genes from Plasmodium malariae and Plasmodium ovale . Am J Trop Med Hyg 82: 996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Putaporntip C, Hongsrimuang T, Seethamchai S, Kobasa T, Limkittikul K, et al. (2009) Differential prevalence of Plasmodium infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J Infect Dis 199: 1143–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jongwutiwes S, Buppan P, Kosuvin R, Seethamchai S, Pattanawong U, et al. (2011) Plasmodium knowlesi malaria in humans and macaques Thailand. Emerg Infect Dis 17: 1799–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nei M (1987) Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- 20. Tamura K, Peterson D, Peterson N, Stecher G, Nei M (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood Evolutionary Distance and Maximum Parsimony Methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282. [DOI] [PubMed] [Google Scholar]
- 22. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hughes AL (1992) Positive selection and interallelic recombination at the merozoite surface antigen-1 (MSA-1) locus of Plasmodium falciparum . Mol Biol Evol 9: 381–393. [DOI] [PubMed] [Google Scholar]
- 24. Polley SD, Weedall GD, Thomas AW, Golightly LM, Conway DJ (2005) Orthologous gene sequences of merozoite surface protein 1 (MSP1) from Plasmodium reichenowi and P. gallinaceum confirm an ancient divergence of P. falciparum alleles. Mol Biochem Parasitol 142: 25–31. [DOI] [PubMed] [Google Scholar]
- 25. Putaporntip C, Jongwutiwes S, Iwasaki T, Kanbara H, Hughes AL (2006) Ancient common ancestry of the merozoite surface protein 1 of Plasmodium vivax as inferred from its homologue in Plasmodium knowlesi . Mol Biochem Parasitol 146: 105–108. [DOI] [PubMed] [Google Scholar]
- 26. Putaporntip C, Buppan P, Jongwutiwes S (2011) Improved performance with saliva and urine as alternative DNA sources for malaria diagnosis by mitochondrial DNA-based PCR assays. Clin Microbiol Infect 17: 1484–1491. [DOI] [PubMed] [Google Scholar]
- 27. Tordrup D, Virenfeldt J, Andersen FF, Petersen E (2011) Variant Plasmodium ovale isolated from a patient infected in Ghana. Malar J 10: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oguike MC, Betson M, Burke M, Nolder D, Stothard JR, et al. (2011) Plasmodium ovale curtisi and Plasmodium ovale wallikeri circulate simultaneously in African communities. Int J Parasitol 41: 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fançony C, Gamboa D, Sebastião Y, Hallett R, Sutherland C, et al. (2012) Various pfcrt and pfmdr1 genotypes of Plasmodium falciparum cocirculate with P. malariae, P. ovale spp., and P. vivax in northern Angola. Antimicrob Agents Chemother 56: 5271–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hughes AL (2004) The evolution of amino acid repeat arrays in Plasmodium and other organisms. J Mol Evol 59: 528–535. [DOI] [PubMed] [Google Scholar]
- 31.Sutherland CJ, Polley SD (2011) Genomic insights into the past, current and future evolution of human parasites of the genus Plasmodium. In: Tibayrenc M, editor. Genetics and Evolution of Infectious Diseases. London: Elsevier. pp. 607–635. [Google Scholar]
- 32. Putaporntip C, Jongwutiwes S, Thongaree S, Seethamchai S, Grynberg P, et al. (2010) Ecology of malaria parasites infecting Southeast Asian macaques: evidence from cytochrome b sequences. Mol Ecol 19: 3466–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Torsi AJ, Morales JC, Melnick DJ (2003) Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution 57: 1419–1435. [DOI] [PubMed] [Google Scholar]
- 34. Silva JC, Egan A, Friedman R, Munro JB, Carlton JM (2011) Genome sequences reveal divergence times of malaria parasite lineages. Parasitology 138: 1737–17349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hughes AL, Verra F (2010) Malaria parasite sequences from chimpanzee support the co-speciation hypothesis for the origin of virulent human malaria (Plasmodium falciparum). Mol Phylogenet Evol 57: 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Conway DJ (2007) Molecular epidemiology of malaria. Clin Microbiol Rev 20: 188–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jongwutiwes S, Putaporntip C, Hughes AL (2010) Bottleneck effects on vaccine-candidate antigen diversity of malaria parasites in Thailand. Vaccine 19: 3112–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]