Abstract
Background
To study the molecular characteristics of a long-term, low frequency outbreak of bla KPC-2 in a low prevalence setting involving the hospital environment.
Methodology/Principal Findings
KPC-producing bacteria were screened by selective chromogenic agar and Real-Time PCR. The presence of antibiotic resistance genes was ascribed by PCRs and subsequent sequencing, and the KPC-producing isolates were phylogenetically typed using PFGE and multi-locus sequence typing. Bla KPC-2-plasmids were identified and analysed by S1-nuclease-PFGE hybridization and PCR based replicon typing. A ∼97 kb IncFII plasmid was seen to carry bla KPC-2 in all of the clinical isolates, in one of the isolates recovered from screened patients (1/136), and in the Klebsiella pneumoniae and Enterobacter asburiae isolates recovered from the environment (sinks) in one intensive care unit. The K. pneumoniae strain ST258 was identified in 6 out of 7 patients. An intergenus spread to E. asburiae and an interspecies spread to two different K. pneumoniae clones (ST27 and ST461) of the bla KPC-2 plasmid was discovered. K. pneumoniae ST258 and genetically related E. asburiae strains were found in isolates of both human and environmental origins.
Conclusions/Significance
We document a clonal transmission of the K. pneumoniae ST258 strain, and an intergenus plasmid diffusion of the IncFII plasmid carrying bla KPC-2 in this outbreak. A major reservoir in the patient population could not be unveiled. However, the identification of a persisting environmental reservoir of strains with molecular determinants linked to human isolates, suggests a possible role of the environment in the maintenance of this long-term outbreak.
Introduction
The Klebsiella pneumoniae carbapenemase (KPC) was first identified in the USA in a Klebsiella pneumoniae isolate dated from 1996[1] and has subsequently been reported worldwide.[2] High prevalence rates of KPC have been reported from regions and countries such as USA, Israel, China, and Greece, while a number of countries have reported hospital outbreaks and sporadic cases of KPC-producing K. pneumonia. [2]
KPC has been identified in various species of Enterobacteriaceae and non-fermenters such as Pseudomonas aeruginosa and Acinetobacter baumannii. [2] Further, bla KPC has been identified on plasmids differing in size and structure. This broad species distribution and plasmid diversity is likely due to the location of bla KPC in a functional Tn3-based transposon structure (Tn4401) with a high transposition frequency.[3] With respect to K. pneumoniae, multilocus sequence typing (MLST) has shown that the global dissemination of KPC-producing K. pneumoniae is dominated by isolates belonging to a hyperepidemic clonal complex including sequence type (ST) 258.[4] Although the dissemination and outbreaks of KPC seems to be associated with specific clones several reports are describing outbreaks where several clones and different species are involved [5], [6] as well as individual patients containing different species harbouring bla KPC. [7], [8], [9]
In many countries the emergence of KPC-producing bacteria has been associated with import of isolates from high prevalent areas. In Norway, the first case of K. pneumoniae with KPC was associated with import from Greece in 2007 at a hospital in the southern part of Norway.[10] Subsequently, five additional clinical isolates were identified at the same hospital and a nearby hospital from patients with no recent history of travel or hospitalisation abroad. In this study we describe this long-term outbreak with regards to the molecular characteristics of these isolates, plasmid content and dissemination, and the identification of KPC-producing isolates in the hospital environment.
Materials and Methods
Hospital setting
Sørlandet Hospital (SH) is a 683-bed general hospital enterprise located in three different cities in the southern part of Norway. Due to sharing of some specialized medical functions a certain degree of interchange of patients between the hospitals occurs. Two hospitals belonging to Sørlandet Hospital, SH-Arendal (SH-A), and SH-Kristiansand (SH-K), as well as a tertiary hospital (Oslo University Hospital – Rikshospitalet (OUH-RH)) where involved in this outbreak. A screening programme was implemented during outbreak investigation involving a 12-bed (8 single rooms and one 4-bed room) surgical/medical intensive care unit (ICU-A) at SH-A.
Faecal and environmental screening programme
Faecal screening of ICU-A patients was initiated after the identification of clinical case number 6 (K66-62). Prospective screening was performed on patients (n = 50) on admittance and discharge from the ICU-A during a first screening period from 8th of May to 19th of October 2010, and then only on discharge from the ICU-A during a second screening period from 19th of October to 1st of April 2011 (n = 65). Retrospective screening was attempted on 29 patients that had been hospitalized in the ICU-A at overlapping intervals with patient 6. Seventeen of these were still hospitalized and screened, whereas 4/12 discharged patients were subjected to screening. Patients staying in the ICU-A for more than 14 days were screened repeatedly every second week.
Screening of the environment was undertaken on two occasions, from the 7th to the 9th of June 2010 and on the 27th of December 2010. The screenings involved sink drains in the ICU-A (n = 19), the neighbouring post-operative unit, the coronary unit, and taps for water to dialysis machines in the ICU-A. Environmental samples were obtained with sterile, cotton-tipped swabs (COPAN swabs®). Most areas examined were moist, but the cotton-tip was occasionally moistened when the area examined appeared relatively dry. Screening positive locations were disinfected and then controlled by monthly screening.
Isolation of KPC-producing bacteria and detection of blaKPC during the screening programme
Screening for KPC-producing bacteria was performed using ChromID ESBL agar medium (bioMerieux, la Balme-les-Grottes, France) and an enrichment medium (tryptic-soy broth containing 2 mg/L cefpodoxime). Real-Time PCR of bla KPC. was performed directly on suspensions from the swab, enrichment medium, and colonies from the ChromID ESBL agar[11].
Bacterial strains and clinical data
Clinical data and data on bacterial strains investigated in this study are listed in Table 1 and 2.
Table 1. Molecular characteristics of outbreak strains.
| Isolate | Source (P/E/F) 1 | Month of isolation | Species | PFGE | MLST | bla KPC 2 | bla SHV 2 | bla TEM 2 | Plasmid profiling (kB) 3 | Plasmid hybridization bla KPC 4 |
| K47-25 | P1 | Nov 07 | K. pneumoniae | A2 | 258 | KPC-2 | SHV-11/-12 | TEM-1 | 40, 97, 160 | 97,160 |
| K48-58 | P2 | March 08 | K. pneumoniae | A2 | 258 | KPC-2 | SHV-11/-12 | TEM-1 | 97, 242 | 97 |
| K52-74 | P3 | Oct 08 | K. pneumoniae | A2 | ND | KPC-2 | SHV-11/-12 | TEM-1 | 40, 97, 194 | 97 |
| K54-05 | P4 | Jan 09 | K. pneumoniae | A2 | ND | KPC-2 | SHV-11 | TEM-1 | 40, 97/100 | 97 |
| K57-33 | P5 | March 09 | K. pneumoniae | B | 461 | KPC-2 | SHV-1 | TEM-1 | 48, 97, 225 | 97 |
| K66-62 | P6 | April 10 | K. pneumoniae | A1 | ND | KPC-2 | SHV-11/-12 | TEM-1 | 40, 97, 194 | 97 |
| K66-73 | P7/F | May 10 | K. pneumoniae | A1 | 258 | KPC-2 | SHV-11/-12 | TEM-1 | 40, 97, 194 | 97 |
| K66-74 | P7/F | May 10 | E. asburiae | C1 | - | KPC-2 | NEG | TEM-1 | 97,145 | 97 |
| K67-04 | P1 | Jan 10 | K. pneumoniae | A1 | 258 | NEG | SHV-11/-12 | NEG | 40, 194 | NEG |
| K67-05 | P4 | May 09 | K. pneumoniae | A2 | 258 | NEG | SHV-11/-12 | NEG | 40, 194 | NEG |
| K67-06 | P3 | March 10 | K. pneumoniae | A2 | 258 | NEG | SHV-11/-12 | NEG | 40, 194 | NEG |
| K67-11 | E5 | June 10 | K. pneumoniae | A2 | ND | KPC-2 | SHV-11/-12 | TEM-1 | 40, 97, 194 | 97 |
| K67-12 | E5 | June 10 | E. asburiae | C2 | - | KPC-2 | NEG | TEM-1 | 97,145 | 97 |
| K67-13 | E6 | June 10 | K. pneumoniae | D | 27 | KPC-2 | NEG | TEM-1 | 45, 97, 200 | 97 |
| K67-14 | E6 | June 10 | K. pneumoniae | D | 27 | KPC | NEG | TEM-1 | 45, 97, 200 | 97 |
| K67-15 | E5 | June 10 | K. pneumoniae | A1 | ND | KPC | SHV-11/-12 | TEM-1 | 40, 97, 194 | 97 |
| K67-16 | E5 | June 10 | E. asburiae | C2 | - | KPC | NEG | TEM-1 | 97,145 | 97 |
P1–6 = clinical specimen from patients 1–6, P7/F = fecal screen patient 7, E = specimen from environmental screen (rooms 5, 6).
bla CTX-M was negative in all isolates. bla pAmpC was negative in isolates from patients 1–6, others ND.
Plasmid profiling (S1-nuclease digested DNA (kB)).
Plasmid DNA hybridization with bla KPC specific probes.
Table 2. Clinical data and risk factors for outbreak patients.
| Isolate | Pat-ient | Species | Specimen | Hospita-lization abroad (country) | Month admitted | LOS6/LOS prior to diagnosis | Patient overlap7 | Antibiotic treatment8 | Other risk factors (I/S/R/D) | Role in infection | Discharged to |
| K47-25 | P1 | K. pneumoniae | Expectorate | Greece | Nov 07 | 51/6 | Yes3) | MEM, PTZ | I/R | Uncertain | Home |
| K48-58 | P2 | K. pneumoniae | Urine | No | March 08 | 20/8 | Yes3) | None | None 1) | None | Nursing home |
| K52-74 | P3 | K. pneumoniae | Blood | No | Sep 08 | 36/23 | No5) | MEM, TOB, CTX | I/R/D | Yes | Physical rehabilitation |
| K54-05 | P4 | K. pneumoniae | Expectorate | No | Nov 08 | 106/47 | Yes4, 5) | MEM,TOB | I/R/D | Uncertain | Physical rehabilitation |
| K57-33 | P5 | K. pneumoniae | Urine | No | Nov 08 | 178/131 | Yes4) | MEM, IMI | I/S/R | Uncertain | Nursing home |
| K66-62 | P6 | K. pneumoniae | Expectorate | No | April 10 | 24†/17 | No | MEM | I/S/R | None | Diseased (in hospital) |
| K66-73 K66-74 | P7 | K. pneumoniae E. asburiae | Feces/fecal screening | No | May 10 | 13/5 | No | CTX | I/R | None | Home |
Except urinary catheter.
I = admission to ICU, S = recent surgery (laparotomi), R = artificial ventilator use, D = subjected to haemodialysis.
Patient 1, being readmitted to SH-K, and patient 2 had been hospitalized in the same corridor for two days in March 2008 at SH-K although at separately staffed wards (Med. 2A-K and Med. 2C-K). Measures for contact isolation were not implemented for patient 1 on this occasion.
Patient 4 and patient 5 had been admitted simultaneously to the ICU-A, and had also been referred to a tertiary hospital (OUS-RH) at overlapping intervals.
Patient 3 and patient 4 were hospitalized in the ICU-A two days apart.
LOS = length of stay (days).
Overlap between patients in time and wards.
Anti G-negative antibiotics prior to diagnosis
Species identification
Bacterial identification was performed using the VITEK2 ID-GNB system (bioMérieux, Marcy l'Etoile, France) and/or MALDI-TOF (Microflex LT, Bruker Daltonics) with the MALDI Biotyper 3.0 software version.
Antimicrobial susceptibility testing
Antimicrobial susceptibility of the isolates was determined using Etest (bioMérieux) according to the manufacturer's instruction. The results were interpreted using breakpoints from the European Committee for Antimicrobial Susceptibility Testing.[12]
Molecular characterisation of KPC-producing isolates
PCR and sequencing for bla KPC, bla TEM, bla SHV, bla CTX-M, and a multiplex PCR for the most prevalent plasmid-borne bla AmpC -genes was performed as previously described (Samuelsen JAC 09). Isolates were typed by pulsed-field gel electrophoresis (PFGE) of XbaI-digested total genomic DNA.[10] Strain relatedness was analysed using BioNumerics version 6.0 (Applied Maths, Sint-Martens-Latem, Belgium) using the band-based dice coefficient and the unweighted pairs geometric-matched analysis dendrogram with a position tolerance of 1% for optimization and band comparison. Final comparison of bands for the interpretation of relatedness was performed in accordance with Tenover criteria.[13] Multi-locus sequence typing (MLST) was performed on K. pneumoniae isolates according to the protocol described on the K. pneumoniae MLST web site http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html.
Plasmid analysis
Plasmid profiling was performed using S1-nuclease digested plasmid DNA, separated by PFGE.[10] Subsequently, plasmid DNA was blotted onto nylon membrane and hybridised with bla KPC specific probes to identify carrier plasmids. Furthermore, plasmids were classified into incompatibility groups using the PCR based replicon-typing (PBRT) scheme of Carattoli[14] and hybridisation with replicon FII specific probes as previously described.[10]
Transfer of resistance
Conjugal-transfer experiment was carried out in Luria-Bertani broth at 37°C for clinical strain K47-25 as donor and rifampicin-resistant E. coli J53-2 as recipient, mixed in a 1∶9 ratio respectively. Transconjugants were selected on LB-agar plates (Becton Dickinson, Sparks, MD) supplemented with 100 mg/L rifampicin (Sigma-Aldrich, St. Louis, MO) and 2 mg/L ceftazidime (Sigma-Aldrich).
Ethics statement
This study focuses on the molecular characteristics of bacterial isolates collected as part of the clinical management and microbiology routine work. Faecal screening was performed according to the guidelines from the local hospital and collection of clinical data as part of outbreak investigations for implementation of appropriate infection control measurements. Consequently, ethical approval was not obtained for the study.
Results
Clinical cases
Six KPC-producing K. pneumoniae isolates from clinical samples were recovered in SH from November 2007 to April 2010 (Table 1 and Table 2). The first two isolates had been characterised previously.[10] The first patient had been hospitalized on Crete in September 2007 prior to hospital admission in Norway. The other patients had no history of recent travel abroad. 5 isolates were recovered from SH-A, whereas the isolate from patient 2 was recovered from SH-K. After the identification of KPC-producing K. pneumoniae, measures for contact isolation were initiated for all patients. When patient 1 was readmitted at SH-K in March 2008 measurements for contact isolation was however not implemented. Retrospective analysis of patient histories revealed that all SH-A patients had been hospitalized in the same ICU (ICU-A) as the index patient prior to detection of the KPC-producing isolates. However, during their stay the patients had been admitted to 12 different wards. Further, conventional epidemiological evaluation[15] was able to establish epidemiological links between patient 1 and 2, and 3 and 4, while a possible link was suggested between patient 4 and 5 (Table 2). Time and space overlaps between the clinical cases 1 and 3 as well as 5 and 6 could not be provided for, representing time intervals of 10 and 11 months, respectively.
Several recognized risk factors for ESBL colonization and infection[15], [16], [17], [18] were present in all the patients that had been admitted to the ICU-A prior to isolation of KPC-producing K. pneumoniae (patient 1, 3–6) (Table 2). All patients were above 65 years of age. KPC-producing K. pneumoniae bacteraemia was recognized in one patient, whereas the findings in the other patients were regarded as most likely to be colonisations associated with artificial ventilation or urinary catheter. One patient died, but the KPC-producing K. pneumoniae was not considered to be attributable to infection or death in this patient (Table 2).
ESBL-producing bla KPC-negative K. pneumoniae were also recovered in ambulatory urinary samples due to urinary tract infections (UTI) from three of the of the patients 26, 16, and 4 months after the initial isolation of the KPC-producing K. pneumoniae isolates, respectively (Table 1).
Patient screening
In the patient screening programme only 1/136 patients were positive for KPC-producing Enterobacteriaceae (patient 7). Patient 7, being hospitalized in room 5 for his entire ICU-A stay, was identified with KPC-producing K. pneumoniae (K66-73) and Enterobacter asburiae (K66-74) upon discharge (Table 1), while negative upon admission five days earlier. Room 5 was disinfected using a persulfate based disinfectant after having been occupied by patient 6 and then patient 7 was the first to be admitted two days later.
Environmental screening
KPC-producing bacteria were detected in 4/19 environmental locations in the ICU-A (sink drains in room 5, 6, 9, and the rinsing room). Bla KPC-positive K. pneumoniae were identified in June 2010 from the sink drains in room 5 (K67-11 and K67-15) and room 6 (K67-13 and K63-14), and bla KPC-positive E. asburiae from the sink drain in room 5 (K67-12 and K67-16) (Table 1). The sinks and sink traps were decommissioned and the connecting pipe elbows were disinfected using a chlorine disinfectant before new sinks and sink traps were installed. Monthly environmental screening of these positive locations was then undertaken. Bla KPC-positive K. pneumoniae was again recovered from the sink in room 6 on two occasions in December 2010 triggering a new environmental screening in the ICU-A (data not shown). Additional sink drains in the ICU-A was investigated, and two new locations that were not subjected to investigation in June 2010 were now identified with bla KPC-positive K. pneumoniae (room 9) and bla KPC-positive E. asburiae (room 9 and the rinsing room, data not shown).
Although bla KPC-positive isolates have been identified in the environment no additional clinical cases has been identified since patient 6.
Molecular characterisation of the isolates
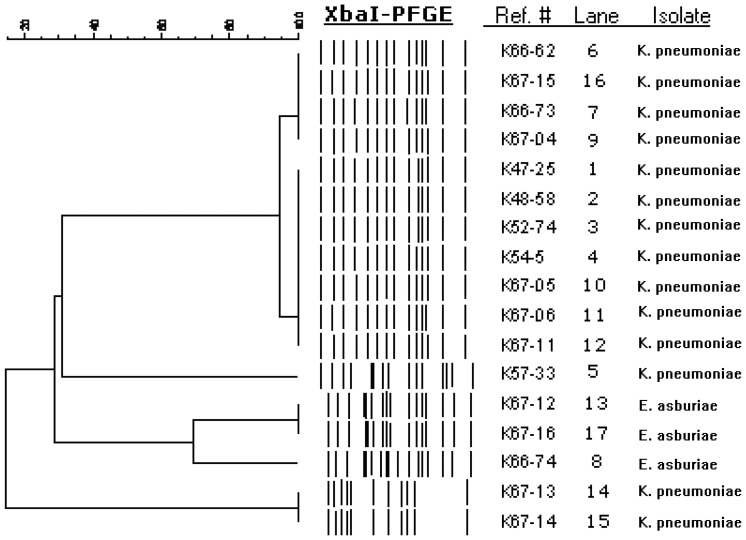
PFGE and MLST typing revealed that 14 K. pneumoniae isolates from both patients and the environment, including the three bla KPC-negative K. pneumoniae UTI-isolates, belonged to two clonally related pulsotypes (A1 and A2), that by MLST were typed to ST258 (Figure 1, Table 1). Distinct K. pneumoniae PFGE/MLST-types were recovered in patient 5 (K57-33; pulsotype B/ST461) and isolates from the sink drain in room 6 (K67-13 and K67-14; pulsotype D/ST27). PFGE of the E. asburiae isolates revealed two closely related pulsotypes corresponding to the E. asburiae (pulsotype C1) isolate from the screening patient (patient 7; K66-47) and the two E. asburiae (pulsotype C2) isolate from room 5 (K67-12 and K67-16). The bla KPC-gene was sequenced in all isolates from patients and from the environmental in June 2010 and found to be bla KPC-2 (Table 1). The bla KPC-positive K. pneumoniae ST258 isolates harboured bla SHV-11, bla SHV-12, and bla TEM-1, whereas the three bla KPC-negative K. pneumoniae ST258 UTI-isolates, harboured bla SHV-12 and bla SHV-11, but were negative for bla TEM. The distinct K. pneumoniae STs in patient 5 (K57-33) and from room 6 (K67-13 and K67-14) harboured bla SHV-1 and lacked the bla SHV-gene, respectively. The bla KPC-positive E. asburiae isolates were devoid of bla SHV but harboured bla TEM-1 (Table 1). PCRs for bla CTX-M and the most prevalent plasmid-borne bla AmpC genes were negative.
Figure 1. Dendrogram of XbaI-digested genomic DNA.
Strains of KPC-producing K. pneumoniae- and E.asburiae-isolates and ESBLA-producing K. pneumonia are shown with percentages of similarity to the right of the dendrogram.
Plasmid analysis
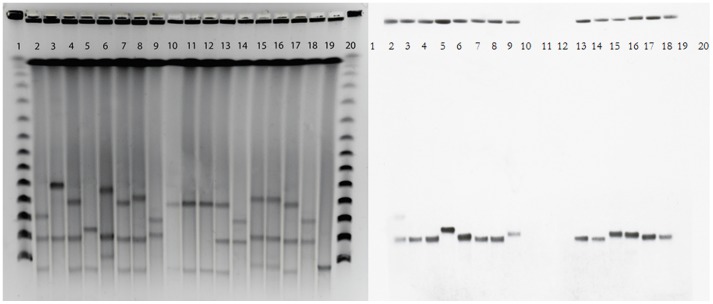
S1-nuclease-PFGE profiles revealed the presence 2-3 plasmids in all isolates ranging from∼40–∼240 kb (Table 1, Figure 2). Hybridization with bla KPC-specific probes identified a bla KPC-carrier plasmid to approximately 97 kb in all bla KPC-positive isolates of K. pneumoniae and E. asburiae. In the bla KPC-positive K. pneumoniae isolate from patient 1 (K47-25) an additional bla KPC-plasmid of ∼160 kb was identified. PBRT and hybridization characterized the ∼97 kb plasmid as an IncFII replicon plasmid. Except for the 97 kb bla KPC-plasmid, separate plasmid profiles were recognized in the other K. pneumoniae pulsotypes/STs and the closely related E. asburiae clones. The plasmid profile among the E. asburiae isolates were identical (Table 1, Figure 2).
Figure 2. PFGE of S1 nuclease-digested total DNA.
Lanes 1 and 20, phage λ DNA ladder (concatemers of 48.5 kb); lane 2, K47-25; lane 3, K48-58; lane 4, K52-74; lane 5, K54-05; lane 6, K57-33; lane 7, K66-62; lane 8, K66-73; lane 9, K66-74; lane 10, K67-04; lane 11, K67-05; lane 12, K67-06; lane 13, K67-11; lane 14, K67-12; lane 15, K67-13; lane 16, K67-14; lane 17, K67-15; lane 18, K67-16; lane 19, K. pneumoniae bla KPC -negative control strain.
Interestingly, the plasmid profile of the pulsotype A/ST258 bla KPC-negative K. pneumoniae clinical UTI isolates was devoid of the ∼97 kb bla KPC-plasmid, indicating loss of this plasmid (Table 1).
Transferability of blaKPC resistance
In vitro plasmid-transfer experiments using K47-25 as the donor strain to E. coli resulted in transconjugants with a frequency of 2.5×10−7 per donor cells.
Antimicrobial susceptibility
The antimicrobial susceptibility profile of the isolates is shown in Table 3. Variable levels of reduced susceptibility to carbapenems were observed in all the bla KPC-positive isolates whereas the three ESBL-positive bla KPC-negative K. pneumoniae ST258 UTI isolates were susceptible to carbapenems (Table 1). A similar susceptibility profile with high-level resistance to penicillins and cephalosporins, resistance to ciprofloxacin, tobramycin, and amikacin, but susceptibility to gentamicin, were common to all K. pneumoniae ST258/pulsotype A isolates. In contrast, the other bla KPC-positive K. pneumoniae pulsotypes were susceptible to ciprofloxacin, tobramycin, amikacin, and the gentamicin MIC was lower. The bla KPC-positive E. asburiae isolates found in the screening positive patient (patient 7) and the environment showed resistance to ciprofloxacin, but were susceptible to the aminoglycosides.
All bla KPC-positive K. pneumoniae ST258 isolates were resistant to colistin. 2 of 3 bla KPC-negative K. pneumoniae UTI isolates were susceptible to colistin, whereas one isolate was resistant (K67-05) displaying a clearly visible double zone (MIC 0.5 mg/L and MIC 16 mg/L).
Discussion
The dissemination of multidrug resistance plasmids among Gram-negative bacteria is one of the major factors contributing to the spread of antimicrobial resistance.[19] However, information about plasmid-dissemination during outbreaks is relatively limited. Often the focus is restricted to specific bacterial species and the dissemination of plasmids into different species might be overlooked. Highly mobile plasmids resulting in ‘plasmid-borne outbreaks’ may delay the recognition of an outbreak, and adds an additional layer of complexity to the molecular investigation of such outbreaks.[20] Plasmids have previously been linked to outbreaks of multi-drug resistant (MDR) Gram-negative bacteria[21], [22], [23], [24], [25], including nosocomial outbreaks of blaKPC-containing plasmids in some studies.[5], [7], [8], [20], [26], [27], [28], [29] Here we describe the dissemination of a KPC-plasmid between strains and species during a small long-term nosocomial outbreak in a low-prevalence setting as well as the possible role of the environment in this context. To the best of our knowledge this is the first nosocomial outbreak report of KPC-producing bacteria from the Nordic countries.
In vivo mobility is indicated by identification of the ∼97 kb IncFII bla KPC-2-plasmid in four distinct MLST/PFGE types of K. pneumoniae and E. asburiae. This observation was supported by successful in vitro conjugation of bla KPC-plasmids. bla KPC –containing Tn4401 has been documented on plasmids of different sizes belonging to different incompatibility groups.[30], [31], [32], [33], [34] While mostly being reported on broad host IncN plasmids[31], bla KPC has recently been reported on narrow host range IncFII plasmids as well, promoting efficient spread within members of Enterobacteriaceae.[33], [35], [36]
It is well known that plasmid-transfer can occur in vivo, but less is known about plasmid-transfer occurring in the hospital environment. We can only speculate were the plasmid-transfer has occurred in this setting. The first clinical cases were associated with the hyperepidemic K. pneumoniae ST258 strain before plasmid-dissemination were observed into other K. pneumoniae STs (ST27 and ST461) and E. asburiae, all of them devoid of non-β-lactam resistances except the latter displaying low-level ciprofloxacin resistance. The KPC-producing E. asburiae remained clinically silent throughout the outbreak and first appeared in faecal screening of patient 7. Faecal screening was not performed on any of the clinical patients, thus whether KPC-producing E. asburiae was carried by any of these patients remains elusive. The finding of a distinct K. pneumoniae (ST27) with the ∼97 kb KPC-plasmid in the hospital environment but not in patients could either be due to the unrecognized presence in patients and in vivo plasmid-transfer or that the plasmid-transfer have occurred in the environment before acquisition by a patient.
Interestingly, loss of bla KPC-plasmids, ∼160 kb plasmid in patient 1 and the ∼97 kb plasmid in the three bla KPC-negative K. pneumoniae UTI isolates, was observed in K. pneumoniae ST258 indicating either a high fitness cost or reduced plasmid stability. Experiments to determine the fitness cost related to the ∼97 kb plasmid and its stability are ongoing.
Reservoirs in the patient or health care worker populations and the environment represent principle modes of spread in nosocomial outbreaks with the patient population being the most important reservoir in high-frequent outbreaks.[37] Introduction of the K. pneumoniae ST258 clone has caused major outbreaks in many hospitals.[38] It is possible that a high level of adherence to standard precautions prevented the establishment of a major gastrointestinal reservoir in our patients, and thus, the prerequisite of a major high-frequent outbreak, were not present.[37], [39], [40], [41] The lack of a patient reservoir and the persistence of the outbreak despite the long interval between known clinical cases could indicate other possible reservoirs contributing to occasional transmissions and maintenance of the outbreak. Health care workers were not screened for faecal or hand carriage partly as no individuals with additional risk factors including dermatitis[37] could be identified and partly due to the fact that the current Norwegian health authority guidelines advocates against screening of health care workers in nosocomial outbreaks of multidrug resistant Gram-negative bacteria.[42] KPC-producing bacteria were detected in as much as 21% of environmental locations in screening samples from sink drains in the ICU-A. Moist surfaces and especially sink drains have been focused in several studies as a possible environmental source of transmission of Enterobacteriaceae, especially Klebsiella spp, to patients.[43], [44], [45], [46], [47], [48] Although the environment were decontaminated and sinks were replaced, KPC-producing E. asburiae and K. pneumoniae were recovered during further environmental screening, suggesting that these strains can survive well in that environment. Contamination of the hands of health care workers due to occasional backsplash during hand washing in a contaminated sink and sink drains[44], [46], [47], [49] or through moist surfaces near sinks and faucets[45] has been suggested as a possible mode of transmission to health care workers and subsequently to patients in the ICU setting, facilitating low-frequent transmissions. The epidemiological link between patient 6 and 7 through their succeeding use of room 5 and the isolation of pulsotype identical KPC-producing K. pneumoniae ST258 from both patients and the sink drain in that room as well as the recovery of closely related KPC-producing E. asburiae from faecal screening in patient 7 and the sink drain could suggest a possible environmental source of colonization of patient 7.
Investigation revealed that several sinks were heavily contaminated by bacteria presumably from waste water washed down the sink after cleaning of patients. A ‘bed bath’-system for cleaning of patients were implemented to shortcut this opportunity. Repetitive disinfection with a biofilm active disinfectant was considered, but disinfection was only performed on two occasions; in June 2010 and January 2011.
In conclusion; the outbreak involved clonal spread of the hyperepidemic K. pneumoniae ST258 stain as well as interspecies plasmid transmission of a ∼97 kb blaKPC-plasmid into two other distinct K. pneumoniae strains and intergenus spread to E. asburiae. The establishment of a local environmental reservoir was documented. The spread among patients has probably occurred partly as a result of transmission between patients in some of the clinical cases, but we infer the possibility that the outbreak was maintained and prolonged with additional clinical cases added due to spread from environmental sources.
Acknowledgments
We acknowledge the contributions from Torill S. Larsen, Erik Bertnes, Astrid Norevik Campbell, Reidun Ørum, Berit Vrålstad, Åsa Melhus, Andre Ingebretsen, Egil Lingaas, Bjørn Iversen, Bjørg Haldorsen, Bettina Aasnæs, and Kine Susann Waade Edvardsen in this work. We thank the Genotyping of Pathogens and Public Health Platform (Institut Pasteur) for coding MLST alleles and profiles available at http://www.pasteur.fr/mlst.
Funding Statement
The study was supported by a fellowship from SÃ ¸rlandet Hospital HF and a research grant from the Northern Norway Regional Health Authority Medical Research Programme. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, et al. (2001) Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrobial Agents and Chemotherapy 45: 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nordmann P, Cuzon G, Naas T (2009) The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9: 228–236. [DOI] [PubMed] [Google Scholar]
- 3. Cuzon G, Naas T, Nordmann P (2011) Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrobial Agents and Chemotherapy 55: 5370–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woodford N, Turton JF, Livermore DM (2011) Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiology Reviews 35: 736–755. [DOI] [PubMed] [Google Scholar]
- 5. Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, et al. (2010) An outbreak of infection due to beta-Lactamase Klebsiella pneumoniae Carbapenemase 2-producing K. pneumoniae in a Greek University Hospital: molecular characterization, epidemiology, and outcomes. Clinical Infectious Diseases 50: 364–373. [DOI] [PubMed] [Google Scholar]
- 6. Wendt C, Schutt S, Dalpke AH, Konrad M, Mieth M, et al. (2010) First outbreak of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae in Germany. Eur J Clin Microbiol Infect Dis 29: 563–570. [DOI] [PubMed] [Google Scholar]
- 7. Sidjabat HE, Silveira FP, Potoski BA, Abu-Elmagd KM, Adams-Haduch JM, et al. (2009) Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clinical Infectious Diseases 49: 1736–1738. [DOI] [PubMed] [Google Scholar]
- 8. Petrella S, Ziental-Gelus N, Mayer C, Renard M, Jarlier V, et al. (2008) Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A beta-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrobial Agents and Chemotherapy 52: 3725–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bennett JW, Herrera ML, Lewis JS, 2nd, Wickes BW, Jorgensen JH (2009) KPC-2-producing Enterobacter cloacae and pseudomonas putida coinfection in a liver transplant recipient. Antimicrobial Agents and Chemotherapy 53: 292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Samuelsen O, Naseer U, Tofteland S, Skutlaberg DH, Onken A, et al. (2009) Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. Journal of Antimicrobial Chemotherapy 63: 654–658. [DOI] [PubMed] [Google Scholar]
- 11. Hindiyeh M, Smollen G, Grossman Z, Ram D, Davidson Y, et al. (2008) Rapid detection of blaKPC carbapenemase genes by real-time PCR. Journal of Clinical Microbiology 46: 2879–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(2012) The European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST Clinical Breakpoint Table v. 2.0, valid from 2012–01–01. Available: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_2.0_120221.pdf Available: 29 June 2012,
- 13. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, et al. (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. Journal of Clinical Microbiology 33: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, et al. (2005) Identification of plasmids by PCR-based replicon typing. Journal of Microbiological Methods 63: 219–228. [DOI] [PubMed] [Google Scholar]
- 15. Skippen I, Shemko M, Turton J, Kaufmann ME, Palmer C, et al. (2006) Epidemiology of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp.: a nested case-control study from a tertiary hospital in London. Journal of Hospital Infection 64: 115–123. [DOI] [PubMed] [Google Scholar]
- 16. Hussein K, Sprecher H, Mashiach T, Oren I, Kassis I, et al. (2009) Carbapenem resistance among Klebsiella pneumoniae isolates: risk factors, molecular characteristics, and susceptibility patterns. Infection Control and Hospital Epidemiology 30: 666–671. [DOI] [PubMed] [Google Scholar]
- 17. Zarkotou O, Pournaras S, Voulgari E, Chrysos G, Prekates A, et al. (2010) Risk factors and outcomes associated with acquisition of colistin-resistant KPC-producing Klebsiella pneumoniae: a matched case-control study. Journal of Clinical Microbiology 48: 2271–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Falagas ME, Rafailidis PI, Kofteridis D, Virtzili S, Chelvatzoglou FC, et al. (2007) Risk factors of carbapenem-resistant Klebsiella pneumoniae infections: a matched case control study. Journal of Antimicrobial Chemotherapy 60: 1124–1130. [DOI] [PubMed] [Google Scholar]
- 19. Carattoli A (2011) Plasmids in Gram negatives: molecular typing of resistance plasmids. International Journal of Medical Microbiology 301: 654–658. [DOI] [PubMed] [Google Scholar]
- 20. Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AK, et al. (2011) Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals Intergenus KPC carbapenemase transmission through a promiscuous plasmid. MBio 2: e00204–00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wiener J, Quinn JP, Bradford PA, Goering RV, Nathan C, et al. (1999) Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA 281: 517–523. [DOI] [PubMed] [Google Scholar]
- 22. Bingen EH, Desjardins P, Arlet G, Bourgeois F, Mariani-Kurkdjian P, et al. (1993) Molecular epidemiology of plasmid spread among extended broad-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates in a pediatric hospital. Journal of Clinical Microbiology 31: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sandegren L, Linkevicius M, Lytsy B, Melhus A, Andersson DI (2012) Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. Journal of Antimicrobial Chemotherapy 67: 74–83. [DOI] [PubMed] [Google Scholar]
- 24. Bagattini M, Crivaro V, Di Popolo A, Gentile F, Scarcella A, et al. (2006) Molecular epidemiology of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit. Journal of Antimicrobial Chemotherapy 57: 979–982. [DOI] [PubMed] [Google Scholar]
- 25. Velasco C, Rodriguez-Bano J, Garcia L, Diaz P, Lupion C, et al. (2009) Eradication of an extensive outbreak in a neonatal unit caused by two sequential Klebsiella pneumoniae clones harbouring related plasmids encoding an extended-spectrum beta-lactamase. Journal of Hospital Infection 73: 157–163. [DOI] [PubMed] [Google Scholar]
- 26. Rasheed JK, Biddle JW, Anderson KF, Washer L, Chenoweth C, et al. (2008) Detection of the Klebsiella pneumoniae carbapenemase type 2 Carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. Journal of Clinical Microbiology 46: 2066–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai JC, Zhou HW, Zhang R, Chen GX (2008) Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli Isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrobial Agents and Chemotherapy 52: 2014–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Curiao T, Morosini MI, Ruiz-Garbajosa P, Robustillo A, Baquero F, et al. (2010) Emergence of bla KPC-3-Tn4401a associated with a pKPN3/4-like plasmid within ST384 and ST388 Klebsiella pneumoniae clones in Spain. Journal of Antimicrobial Chemotherapy 65: 1608–1614. [DOI] [PubMed] [Google Scholar]
- 30. Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, et al. (2010) Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerging Infectious Diseases 16: 1349–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carattoli A (2009) Resistance plasmid families in Enterobacteriaceae. Antimicrobial Agents and Chemotherapy 53: 2227–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carattoli A, Aschbacher R, March A, Larcher C, Livermore DM, et al. (2010) Complete nucleotide sequence of the IncN plasmid pKOX105 encoding VIM-1, QnrS1 and SHV-12 proteins in Enterobacteriaceae from Bolzano, Italy compared with IncN plasmids encoding KPC enzymes in the USA. Journal of Antimicrobial Chemotherapy 65: 2070–2075. [DOI] [PubMed] [Google Scholar]
- 33. Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S (2010) Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. Journal of Antimicrobial Chemotherapy 65: 243–248. [DOI] [PubMed] [Google Scholar]
- 34. Mataseje LF, Boyd DA, Willey BM, Prayitno N, Kreiswirth N, et al. (2011) Plasmid comparison and molecular analysis of Klebsiella pneumoniae harbouring bla(KPC) from New York City and Toronto. Journal of Antimicrobial Chemotherapy 66: 1273–1277. [DOI] [PubMed] [Google Scholar]
- 35. Andrade LN, Curiao T, Ferreira JC, Longo JM, Climaco EC, et al. (2011) Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrobial Agents and Chemotherapy 55: 3579–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, et al. (2011) Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrobial Agents and Chemotherapy 55: 5493–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paterson DL, Bonomo RA (2005) Extended-spectrum beta-lactamases: a clinical update. Clinical Microbiology Reviews 18: 657–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, et al. (2009) Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrobial Agents and Chemotherapy 53: 3365–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pena C, Pujol M, Ardanuy C, Ricart A, Pallares R, et al. (1998) Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended-spectrum beta-lactamases. Antimicrobial Agents and Chemotherapy 42: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lucet JC, Chevret S, Decre D, Vanjak D, Macrez A, et al. (1996) Outbreak of multiply resistant enterobacteriaceae in an intensive care unit: epidemiology and risk factors for acquisition. Clinical Infectious Diseases 22: 430–436. [DOI] [PubMed] [Google Scholar]
- 41. Calfee D, Jenkins SG (2008) Use of active surveillance cultures to detect asymptomatic colonization with carbapenem-resistant Klebsiella pneumoniae in intensive care unit patients. Infection Control and Hospital Epidemiology 29: 966–968. [DOI] [PubMed] [Google Scholar]
- 42.Folkehelseinstituttet. Forebygging og kontroll av spredning av multiresistente gramnegative stavbakterier og ESBL-holdige bakterier i helseinstitusjoner, 2009. http://www.fhi.no/dav/96331178b9.pdf Available: 29 June 2012
- 43. Lowe C, Willey B, O'Shaughnessy A, Lee W, Lum M, et al. (2012) Outbreak of Extended-Spectrum beta-Lactamase-producing Klebsiella oxytoca Infections Associated with Contaminated Handwashing Sinks(1). Emerging Infectious Diseases 18: 1242–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kac G, Podglajen I, Vaupre S, Colardelle N, Buu-Hof A, et al. (2004) Molecular epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae isolated from environmental and clinical specimens in a cardiac surgery intensive care unit. Infection Control and Hospital Epidemiology 25: 852–855. [DOI] [PubMed] [Google Scholar]
- 45. Hobson RP, MacKenzie FM, Gould IM (1996) An outbreak of multiply-resistant Klebsiella pneumoniae in the Grampian region of Scotland. Journal of Hospital Infection 33: 249–262. [DOI] [PubMed] [Google Scholar]
- 46. Hota S, Hirji Z, Stockton K, Lemieux C, Dedier H, et al. (2009) Outbreak of multidrug-resistant Pseudomonas aeruginosa colonization and infection secondary to imperfect intensive care unit room design. Infection Control and Hospital Epidemiology 30: 25–33. [DOI] [PubMed] [Google Scholar]
- 47.de Jong E HJ, Hilkens MGEC, Loeffen FLA, van Leeuwen WB, Melchers WL, Sturm PDJ. (2012) A prolonged outbreak of an extended-spectrum beta-lactamase producing klebsiella pneumonia (EKP) on an ICU due to contamination of sinks. In: Oral presentation of the Twenty-second European Congress of Clinical Microbiology and infectious Diseases, London, E16 1XL, UK. Oral presentation O-124. European Society of Clinical Microbiology and Infectious Diseases, 4002 Basel, Switzerland.
- 48.Domínguez MC V-LS, Conejo MC, Rodríguez-Baño J, Pascual A. (2011) Epidemiological and Environmental Study of an Outbreak Caused by Carbapenemase-Producing Klebsiella oxytoca. In: Posterview of the Fifty-first Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL. Poster K-237. American Society for Microbiology, Washington, DC, USA. [Google Scholar]
- 49. D'Agata EM, Venkataraman L, DeGirolami P, Samore M (1999) Molecular epidemiology of ceftazidime-resistant gram-negative bacilli on inanimate surfaces and their role in cross-transmission during nonoutbreak periods. Journal of Clinical Microbiology 37: 3065–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]