Diffusion is a fundamental physical phenomenon that underlies the molecular basis of all biology. For example, virtually all aspects of DNA metabolism require proteins to search through a vast sea of nonspecific DNA to locate specific binding targets, and these searches are governed by diffusion.1 An effective search relies on the ability of a protein to efficiently locate and recognize specific sites or structures along DNA, but details of these activities remain poorly understood largely due to a lack of experimental methods capable of probing such dynamic processes. Recent advances in single molecule imaging have led to a resurgence of experimental studies seeking to understand how diffusion contributes to protein-nucleic acid interactions.2
Post-replicative mismatch repair (MMR) serves as an exceptional model system for investigations into diffusion-based searches, as the initial steps of this conserved pathway depend upon a series of diverse target search and recognition events that are coordinated by multiple proteins.3-5 MMR corrects DNA synthesis errors, thereby increasing replication fidelity. Mutations in the MMR machinery are associated with hereditary nonpolyposis colorectal cancer and other cancers.6,7 MutSα and MutLα are conserved protein complexes that initiate MMR. MutSα recognizes DNA mismatches and small insertion/deletion loops, whereas MutLα is an endonuclease that cleaves the lesion-bearing DNA strand.6,7 The challenges faced during the MMR target searches can be illustrated by considering that S. cerevisiae incurs only on the order of ~2 mismatches per round of cell division.
To initiate repair, MutSα must first find DNA lesions; MutLα must locate lesion-bound MutSα; and the lesion-bound MutSα/MutLα complex must then search the flanking DNA for signals that distinguish the parental and daughter strands.5,6 There are four potential diffusion-based mechanisms that might be involved: (1) three-dimensional (3D) diffusion (or “jumping”), where the protein finds its targets through direct 3D collisions from solution; (2) one-dimensional (1D) “hopping,” where the protein moves along the same molecule of DNA via a series of correlated submicroscopic dissociation and reassociation events; (3) 1D sliding, where the protein undergoes a random walk along the DNA without dissociation; and (4) intersegmental transfer, where the protein moves from one site to another via a looped intermediate.1 1D hopping, sliding and intersegmental transfer are referred to as facilitated diffusion, because the reduction in dimensionality brought about through use of these mechanisms presents the potential for target site association rates that exceed the limits imposed by pure 3D diffusion.1 These mechanisms are not mutually exclusive, and different combinations may contribute to site-specific targeting for a given protein.
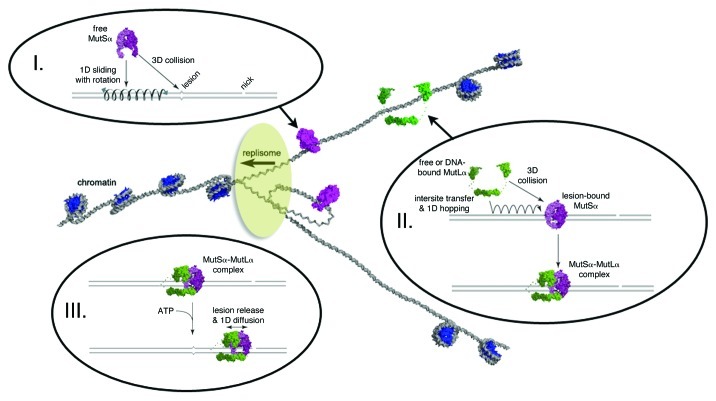
We have developed DNA curtains to allow direct visualization of protein-DNA interactions at the single molecule level.3-5 DNA curtains, in conjunction with lesion bearing substrates, allowed us to observe trajectories of MutSα in real time as it searched for and engaged DNA mismatches, MutLα as it searched for mismatch-bound MutSα and the MutSα/MutLα complex as it scanned the flanking DNA following mismatch recognition.3 These observations suggest that MutSα slid along the DNA while searching for lesions, consistent with a mechanism that required the protein to track the helical phosphate backbone of the DNA (Fig. 1).3,5 In contrast, MutLα hopped on DNA without tracking the DNA helix while searching for lesion-bound MutSα and could also undergo direct intersite transfer between juxtaposed DNA molecules.3,4 MutLα specifically recognized lesion-bound MutSα, and the MutSα/MutLα complex remained stably bound at the lesion.3 Upon binding ATP, the complex was released from the lesion and scanned the flanking DNA through a new mode of more rapid 1D diffusion that appeared to lack any rotational component.3 In addition, the complex became highly resistant to dissociation from the DNA, and the intersite transfer activity of MutLα was suppressed; these unique properties may ensure that MutSα/MutLα remains associated with the damage-bearing strand while scanning the flanking DNA by 1D-diffusion. The released complex no longer recognized mismatches as targets. This finding was especially remarkable given that the redundant nature of 1D diffusion ensures that the MutSα/MutLα complex must encounter the mismatch thousands of times while scanning the flanking DNA for strand discrimination signals.3
Figure 1. Model of the replication fork highlighting the different modes of facilitated diffusion utilized by MutSα and MutLα. (I) MutSα scans newly replicated DNA through 1D sliding, which includes an obligatory rotational component. (II) MutLα searches for and engages lesion-bound MutSα through hopping and/or intersite transfer. (III) Nucleotid binding releases the lesion-bound MutSα/MutLα complex to search for strand discrimination signals through 1D sliding with no rotational requirement in a new state that no longer recognizes DNA lesions. The details by which MMR is coupled to replication, and the impact chromatin imposes on MMR remain open questions.
This work illustrates how transitions between different modes of diffusion are regulated during the early stages of MMR through a combination of mismatch recognition, protein-protein interactions and the binding of nucleotide cofactors. This work also provides insights into structural changes necessary to accommodate the distinct behaviors of the protein complexes. Recent in vivo studies have indicated that MutSα is associated with replication forks in S. cerevisiae, suggesting an intimate coupling between DNA synthesis and DNA repair (Fig. 1).8 This finding prompts the consideration of how DNA replication and mismatch repair are coordinated, and how these processes are influenced by the presence of chromatin. The use of DNA curtains may eventually yield insights into these and other similarly complex biological problems.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23669
References
- 1.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–8. [PubMed] [Google Scholar]
- 2.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–74. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 3.Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, et al. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc Natl Acad Sci USA. 2012;109:E3074–83. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–8. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorman J, Chowdhury A, Surtees JA, Shimada J, Reichman DR, Alani E, et al. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol Cell. 2007;28:359–70. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–9. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 8.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–53. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]