Abstract
The Ku heterodimer, composed of Ku70 and Ku80, is the initiating factor of the nonhomologous end joining (NHEJ) double-strand break (DSB) repair pathway. Ku is also thought to impede the homologous recombination (HR) repair pathway via inhibition of DNA end resection. Using the cell-free Xenopus laevis egg extract system, we had previously discovered that Ku80 becomes polyubiquitylated upon binding to DSBs, leading to its removal from DNA and subsequent proteasomal degradation. Here we show that the Skp1-Cul1-F box (SCF) E3 ubiquitin ligase complex is required for Ku80 ubiquitylation and removal from DNA. A screen for DSB-binding F box proteins revealed that the F box protein Fbxl12 was recruited to DNA in a DSB- and Ku-sensitive manner. Immunodepletion of Fbxl12 prevented Cul1 and Skp1 binding to DSBs and Ku80 ubiquitylation, indicating that Fbxl12 is the F box protein responsible for Ku80 substrate recognition. Unlike typical F box proteins, the F box of Fbxl12 was essential for binding to both Skp1 and its substrate Ku80. Besides Fbxl12, six other chromatin-binding F box proteins were identified in our screen of a subset of Xenopus F box proteins: β-TrCP, Fbh1, Fbxl19, Fbxo24, Fbxo28 and Kdm2b. Our study unveils a novel function for the SCF ubiquitin ligase in regulating the dynamic interaction between DNA repair machineries and DSBs.
Keywords: Ku80, Ku86, Ku70, SCF, DNA damage, double-strand break, nonhomologous end joining, Fbxl12, Fbl12, ubiquitin
Introduction
Double-strand breaks (DSBs), one of the most dangerous forms of DNA damage in the cell, can be repaired via two major pathways: homologous recombination (HR) and nonhomologous end joining (NHEJ). NHEJ is initiated by Ku, a heterodimer consisting of Ku70 and Ku80, which recognizes DSBs and recruits additional pathway components to process and repair the damage. Ku recognizes DSBs in a unique manner: the heterodimer forms a toroidal structure with a central channel through which the broken end of the DNA threads.1 It has been suggested that once the DSB is repaired through NHEJ, Ku could potentially become topologically trapped on the DNA.1 Because the central channel of Ku is large enough only to accommodate one duplex of DNA, the protein presumably must be removed from a repaired DSB prior to or at the completion of the next round of DNA replication.2
Ku has an extremely high affinity for DNA ends, with a Kd of about 2 nM,3 and is estimated to be at a concentration of about 300 nM in human cells.2,4 These properties ensure that Ku is among the first, if not the first, factors to bind to a DSB. There is evidence that this affinity for DSBs may present problems for competitive DSB repair pathways. For example, Ku has been shown to regulate DSB repair pathway choice,5-10 likely due to Ku-mediated steric obstruction of DNA end resection, an initial step of the HR pathway.11-13 Successful end resection may therefore require the removal of Ku from DNA ends. In support of this hypothesis, rates of resection have recently been shown to decrease significantly when a bacterial Ku homolog, which is not efficiently removed from DNA in mammalian cells, is substituted for endogenous mammalian protein.14 These data suggest that there may be an active mechanism in cells to remove Ku from DNA, both prior to and following the completion of NHEJ.2
We have used the Xenopus laevis cell-free egg extract system to study the dynamics of Ku on DSBs.15 Using this system, we observed that Ku80 becomes heavily polyubiquitylated via K48-dependent linkages upon binding to DSBs. This ubiquitylation is necessary for the efficient removal of Ku80 from chromatin in egg extract. Surprisingly, although Ku80 is degraded in a ubiquitin-, DSB- and proteasome-dependent manner, proteasome activity is not required per se for its release from DNA. In addition, truncated Ku80 proteins that retained the ability to bind DSBs but not to complete NHEJ were ubiquitylated and degraded with kinetics identical to those of the full-length protein, indicating that it was DSB binding and not NHEJ completion that signaled ubiquitylation.
We showed previously that components of the Skp1-Cul1-F box (SCF) E3 ubiquitin ligase complex interact preferentially with DSBs, and that this DSB interaction is dependent upon the presence of Ku.15 These observations led us to hypothesize that Ku80 is an SCF substrate. Here we report that the SCF complex is, indeed, required for Ku80 ubquitylation, removal from DNA and degradation in egg extracts. The SCF complex generally recognizes its substrates via an interaction between the substrate and the variable F box-containing subunit of the complex. We systematically screened a subset of Xenopus homologs of human F box proteins and identified Fbxl12 as a Ku-sensitive DSB-binding protein. Fbxl12 depletion inhibited both Cul1 and Skp1 binding to DSBs as well as Ku80 ubiquitylation. Besides Fbxl12, we identified six additional F box proteins that interact with chromatin. This study unveils a novel function of the SCF complex in the DNA damage response.
Results
The SCF complex is required for the ubiquitin-dependent removal of Ku80 from DNA
We had previously shown that 35S-labeled Ku80 translated in rabbit reticulocyte lysate and added to Xenopus laevis egg extract was degraded in response to linearized plasmid DNA but not to circular DNA. Ku80 was ubiquitylated upon binding to DSBs leading to its removal from DNA and subsequent degradation by the 26S proteasome. The E3 ubiquitin ligase involved in this process remained unknown, but one tantalizing clue to its identity was our finding that Cul1 and Skp1, components of the SCF E3 ubiquitin ligase complex, bound to DSBs in a Ku-dependent manner.15 This led us to hypothesize that the SCF complex is recruited to DSBs through a substrate interaction with Ku and is responsible for Ku80 ubiquitylation.
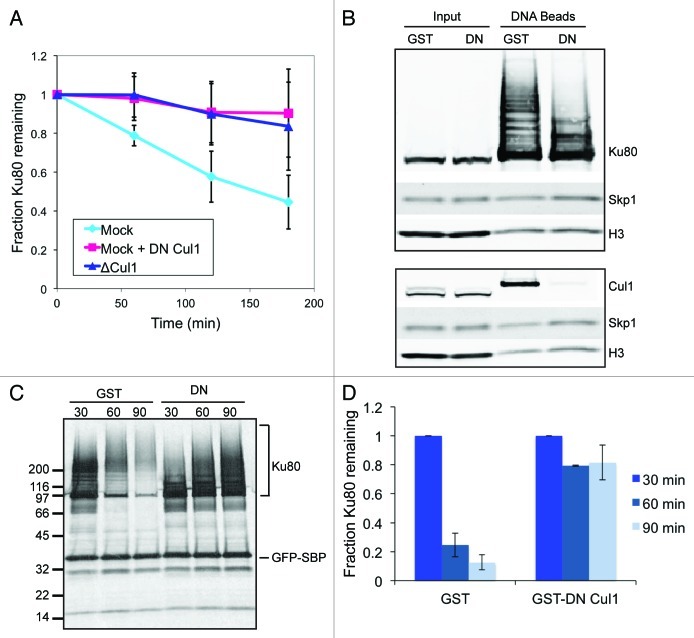
To test whether Ku80 degradation depends on the SCF complex, we generated antibodies against the N terminus of Xenopus laevis Cul1 and used them to immunodeplete Cul1 from egg extracts. Following Cul1 depletion, 35S-labeled Ku80 and linearized DNA were added to extracts, and degradation kinetics monitored. Cul1 depletion clearly led to the stabilization of Ku80 (Fig. 1A), suggesting strongly that the SCF complex is involved in its ubiquitylation.
Figure 1. The SCF complex is required for Ku80 ubiquitylation and degradation in response to DSBs. (A) 35S-labeled Ku80 was added to extract that was either immunodepleted of Cul1, mock-depleted, or mock-depleted and contained GST-tagged dominant-negative Cul1. At one hour intervals, protein was precipitated using trichloro-acetic acid and quantified using a scintillation counter. Average and standard deviation of three independent experiments at each time point are shown. (B) Extracts containing either GST or GST-DN-Cul1 were incubated with SB-DNA beads and inputs and co-purifying proteins were probed with antibodies against Ku80, Skp1, and H3. Beads from a duplicate experiment were probed with antibodies against Cul1, Skp1, and H3 (a loading control). (C) 35S-labeled Ku80 and GFP-SBP, as a loading control, were added to egg extract. After a 45 min pre-incubation, either GST or GST-DN-Cul1 was added. The extract was then incubated with SB-DNA beads. After 30 min, beads were removed from the extract and washed once with and added to extract containing GST or GST-DN-Cul1 but lacking labeled Ku80. Beads were incubated in this extract for an additional 0 (t = 30), 30 (t = 60), or 60 (t = 90) minutes. Bead fractions were visualized by phosphorimager. (D) Ku80 remaining on beads was quantified using a phosphorimager after normalizing to GFP-SBP in three independent experiments. Error bars indicate one standard deviation.
To further confirm this result and ensure that co-depletion of an unknown factor was not responsible for this effect, we used a dominant-negative (DN) version of Cul1 to inactivate the SCF complex. This method takes advantage of the fact that the N-terminal domain of Cul1 binds to Skp1 while the structurally separate C-terminal domain binds to the catalytic subunit Rbx1, and has been previously used in other systems.16 We purified a recombinant protein containing the GST tag and the first 469 residues of the Xenopus laevis Cul1 protein (GST-DN-Cul1). Similar to immunodepletion of Cul1, addition of GST-DN-Cul1 to extract inhibited the DSB-dependent degradation of Ku80 (Fig. 1A).
To test whether the SCF complex is required for Ku80 polyubiquitylation on DNA, DSBs can be modeled in egg extracts using linearized plasmid biotinylated at one end and bound to streptavidin-coated magnetic beads (single biotin DNA beads, or SB-DNA beads). These beads, when incubated in egg extracts, bind to multiple proteins in a DSB-dependent manner.15 Upon binding to SB-DNA beads, Ku80 becomes heavily poly-ubiquitylated. We added SB-DNA beads to extracts containing either GST-DN-Cul1 or GST as a control; DNA beads were purified, and bound proteins were probed by immunoblot. As expected, DN-Cul1 inhibited binding of endogenous Cul1, but not that of Skp1, to SB-DNA beads (Fig. 1B). In addition, DN-Cul1 inhibited much of the Ku80 modification apparent on SB-DNA bead-bound fraction, while it had no effect on Ku80 in the unbound fraction (Fig. 1B).
We had previously noticed a prominent band at about 100 kDa apparent on anti-ubiquitin immunoblots of proteins co-purified with DNA beads (ref. 10 and see Fig. 4, arrow). We assumed that this band represents Cul1 that is modified with the ubiquitin-like peptide Nedd8,17 which cross-reacts with our anti-ubiquitin antibody. In fact, this band can be superimposed with bands that react against both anti-Cul1 and an anti-Nedd8 antibodies, but not with any of the modifications on Ku80 (Fig. S1). This band, like Cul1, disappears in the presence of GST-DN-Cul1. We conclude that this band represents Cul1 on DNA beads that has been modified with Nedd8.

Figure 4. Fbxl12 is required for SCF interaction with DSBs and Ku80 ubiquitylation. Fbxl12 was immunodepleted from egg extracts, which were then incubated with SB-DNA beads. Interacting proteins were analyzed by immunoblot. The arrow points to a Nedd8-conjugated Cul1, which can be detected by anti-ubiquitin antibody. A likely cross-reacting band, which is not depleted by anti-Fbxl12 antibodies, is indicated with an asterisk.
Next, we tested the functional role of SCF on DSBs. We previously showed that ubiquitylation of Ku80 leads to its removal from DNA.15 To test whether the SCF complex is also required for Ku80 removal, we used a technique that we had developed to quantify release of proteins from chromatin. 35S-labeled Ku80 was incubated in extracts and was allowed to bind SB-DNA beads. The beads were then removed from extracts and added to unlabeled, fresh extracts. At various intervals, the beads were isolated, and radioactive Ku80 remaining on the DNA beads was quantified using a phosphorimager. Adding dominant-negative Cul1 to extracts effectively inhibited Ku80 release (Fig. 1C and D). Taken together, these observations strongly indicate that the SCF complex is required for Ku80 ubiquitylation, removal from DNA and proteasomal degradation and, further, that it is likely recruited to DSBs via a substrate interaction with Ku.
The F box protein Fbxl12 binds to DSBs in a Ku-sensitive manner
The SCF complex is generally recruited to its substrates via an interaction between the substrate and the variable F box-containing SCF subunit. Multiple F box proteins within a genome allow the SCF complex to ubiquitylate a variety of distinct substrates in a regulated manner.18 While multiple F box proteins have been identified based on bioinformatic analyses,19 the discovery of substrates recognized by them has been challenging. In fact, the functions of most human F box proteins remain mysterious.20
To identify the F box protein responsible for recruiting the SCF complex to DSBs, we first searched for Xenopus homologs of the 69 known human F box proteins using BLAST. Because the publicly available Xenopus laevis genomic sequence was incomplete at the time these experiments were performed, we also searched the genome of the closely related frog Xenopus tropicalis. In total, we were able to identify clear Xenopus homologs for 58 of the 69 human F box proteins.
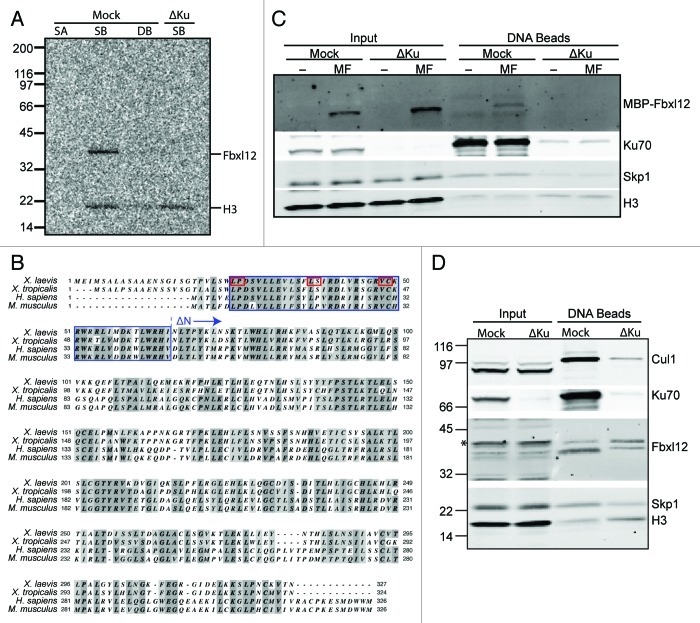
We reasoned that the F box protein required for SCF recruitment to Ku80 on DSBs would, like Cul1 and Skp1, bind to DSBs in a Ku-dependent manner. To search for such an F box protein, we first screened F box proteins that bind to DSBs in egg extracts using SB-DNA beads as described above. To model control undamaged DNA, we used a previously described reagent, linear DNA biotinylated at both ends bound to streptavidin-coated beads (double biotin DNA, or DB-DNA, beads).15 We analyzed 30 of the 58 Xenopus F box protein homologs (Table 1), each of which was 35S-labeled in reticulocyte lysates and allowed to bind DNA beads in egg extracts. Of the 30 F box proteins tested, two did not appear to express in the reticulocyte lysates and 21 were expressed but did not bind to chromatin. In contrast, seven of the tested F box proteins were able to bind to SB-DNA beads (Table 1), four of which showed no obvious preference for DSBs over undamaged DNA (Kdm2b, Fbxl19, Fbxo18, and Fbxo28), and one, Fbxo24, which was not tested for DSB-dependence. In contrast, two (β-TrCP and Fbxl12) showed a clear preference for SB-DNA over DB-DNA bead binding. However, only Fbxl12, expressed from a Xenopus tropicalis cDNA, lost its ability to bind DNA beads following Ku depletion (Fig. 2A).
Table 1. Xenopus F box proteins were screened for Ku sensitivity.
| F-box protein |
IMAGE number |
Xenopus species |
Expressed in RL |
DNA binding |
DSB- dep |
Ku-dep |
|---|---|---|---|---|---|---|
| Ccnf |
5049128 |
laevis |
+ |
– |
N/A |
N/A |
|
Fbh1 |
6316796 |
laevis |
+ |
+ |
– |
N/A |
|
Fbxl12 |
7682433 |
tropicalis |
+ |
+ |
+ |
+ |
| Fbxl14 |
3402730 |
laevis |
– |
N/A |
N/A |
N/A |
| Fbxl16 |
7660186 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxl17 |
6636605 |
laevis |
+ |
– |
N/A |
N/A |
|
Fbxl19 |
6864660 |
laevis |
+ |
+ |
– |
N/A |
| Fbxl20 |
5543149 |
laevis |
– |
N/A |
N/A |
N/A |
| Fbxl22 |
8318195 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxl3 |
5440173 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxl5 |
6317694 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxl8 |
7765941 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxo11 |
6644003 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxo15 |
7856627 |
tropicalis |
+ |
– |
N/A |
N/A |
|
Fbxo24 |
7708113 |
tropicalis |
+ |
+ |
ND |
ND |
| Fbxo25 |
6319137 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxo27 |
7011501 |
laevis |
+ |
– |
N/A |
N/A |
|
Fbxo28 |
7601995 |
tropicalis |
+ |
+ |
– |
N/A |
| Fbxo30 |
5384657 |
tropicalis |
+ |
– |
N/A |
N/A |
| Fbxo36 |
5307822 |
tropicalis |
+ |
– |
N/A |
N/A |
| Fbxo38 |
7391082 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxo41 |
7692156 |
tropicalis |
+ |
– |
N/A |
N/A |
| Fbxo42 |
6953888 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxo44 |
4959662 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxo5-A |
6323527 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxo9 |
4202748 |
laevis |
+ |
– |
N/A |
N/A |
| Fbxw5 |
6980943 |
tropicalis |
+ |
– |
N/A |
N/A |
|
Kdm2b |
6862732 |
laevis |
+ |
+ |
– |
N/A |
| Skp2 |
7393117 |
laevis |
+ |
– |
N/A |
N/A |
| β-TrCP | 5078981 | laevis | + | + | + | – |
cDNAs were screened first for their ability to be translated in rabbit reticulocyte lysate (RL), next their ability to bind SB-DNA beads (DNA binding), their preference for SB-DNA beads over DB-DNA beads (DSB-dep), and, finally, their sensitivity of DSB-binding to the prior depletion of Ku from extracts (Ku-dep). Proteins found to bind to SB-DNA beads are in bold.
Figure 2. The F-box protein Fbxl12 binds to DSBs in a Ku-sensitive manner. (A) 35S-labeled Xenopus tropicalis Fbxl12 and histone H3 were added to egg extract that was either mock- or Ku-depleted. Streptavidin-coated (SA), SB-DNA, or DB-DNA beads were added to the mock-depleted extract and SB-DNA beads were added to the Ku-depleted extract. Bead fractions were visualized by phosphorimager. (B) Protein alignment of Fbxl12 amino acid sequence from Xenopus laevis, Xenopus tropicalis, Homo sapiens and Mus musculus. Indicated are the F box (blue box), conserved F box residues that were mutated (see Fig. 3D and E, red boxes), and the start of the ∆N-Fbxl12 truncation mutant. (C) Egg extracts were either mock- or Ku-depleted, and 1 µM recombinant MBP-Fbxl12 (MF) was then added. These extracts were then incubated with SB-DNA beads, and co-purifying proteins were probed with antibodies against MBP, Ku70, Skp1 or H3. (D) Mock- or Ku-depleted extract was incubated with SB-DNA beads, and bound proteins were probed using antibodies against Xenopus laevis FBXL12. A likely cross-reacting band is indicated with an asterisk.
Fbxl12 is a 37 kDa protein, about which very little is known. The only previously identified substrate for SCFFbxl12 is p57KIP2, a cyclin-dependent kinase inhibitor.21 Interestingly, Fbxl12 has a domain architecture similar to that of the F box protein Skp2, and p57KIP2 is also a SCFSKP2 substrate in mammalian cells.22 However, Skp2, while robustly expressed in reticulocyte lysate, did not bind SB-DNA beads in our assay (Table 1), and we thus discarded it as a candidate for Ku80 recognition.
To identify the Xenopus laevis homolog of Fbxl12, we searched the NCBI expressed sequence tag (EST) database and identified a clone containing sequence with a predicted 82% identity to the C-terminal 78 residues of the Xenopus tropicalis protein. Upon fully sequencing this clone, we were able to conclude that it did, indeed, contain the full-length Xenopus laevis Fbxl12 cDNA, which encodes a polypeptide that is 34% identical to the human protein and 83% identical to the Xenopus tropicalis protein (Fig. 2B). We then generated and purified out of E. coli recombinant MBP-tagged Xenopus laevis Fbxl12 protein, which bound robustly to SB-DNA beads when added to egg extract. This interaction was also dependent upon the presence of Ku (Fig. 2C), indicating that the Xenopus laevis protein had properties similar to its Xenopus tropicalis homolog.
To study endogenous Fbxl12 in egg extract, we generated polyclonal antibodies against a C-terminal peptide of the Xenopus laevis protein. This antibody recognized an endogenous 37 kDa protein in egg extract, although there were several additional recognized bands, some of which are likely to be modified forms of Fbxl12 (Fig. 2D). When used to probe proteins co-purifying with SB-DNA beads, anti-Fbxl12 identified two bands migrating at approximately 37 and 40 kDa. (Fig. 2D). Upon depletion of Ku from the extract, Cul1 and Skp1 binding to SB-DNA beads was abrogated, as was that of the faster migrating anti-Fbxl12 interacting band. The slower migrating band, however, was not negatively affected by Ku depletion. The nature of the slower migrating band is currently unknown (but see discussion).
Fbxl12 interacts with SCF complex components and its binding to chromatin depends on its F box
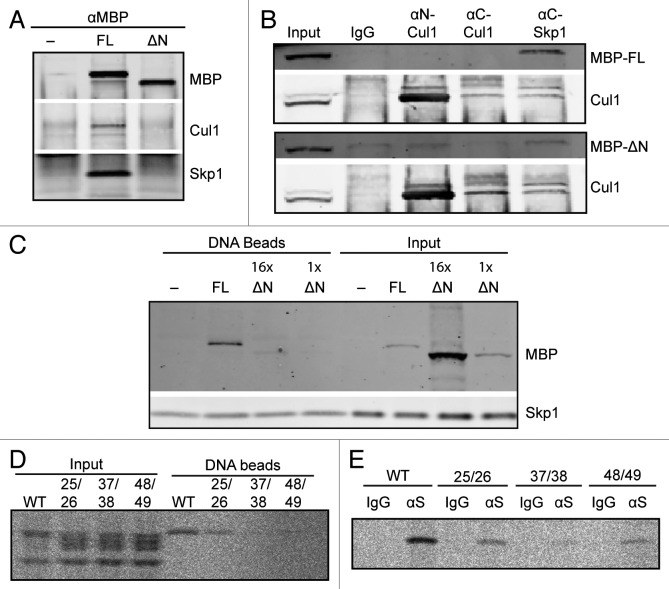
We next sought to confirm the ability of Fbxl2 to interact with other SCF components in Xenopus egg extracts. F box proteins interact with other SCF components via a direct interaction between the F box and Skp1.23 MBP-tagged full-length or ∆N-Fbxl12, which lacks the N-terminus region containing the F box, was incubated in extract and immunoprecipitated using anti-MBP antibodies. An immunoblot of co-purifying proteins revealed that the full-length Fbxl12, but not ∆N-Fbxl12, was able to co-immunoprecipitate both Cul1 and, to a greater extent, Skp1 (Fig. 3A). Similarly, while anti-Skp1 antibodies were able to co-immunprecipitate full-length Fbxl12, they were unable to isolate ∆N-Fbxl12 above background levels (Fig. 3B). In contrast, anti-Cul1 antibodies could not co-isolate Fbxl12: anti-C-terminal Cul1 antibodies did not even efficiently immunoprecipitate Cul1, while anti-N-terminal Cul1 antibodies likely compete with Skp1 for N-terminal domain binding of Fbxl12.
Figure 3. The FBXL12 F-box is required for interaction with SCF components and with DSBs. (A) Extracts containing buffer, MBP-Fbxl12 (FL), or MBP-∆N-Fbxl12 (∆N) were immunopurified using an anti-MBP antibody. Copurified proteins were probed by immunoblot. (B) MBP-Fbxl12 (MBP-FL) or MBP-∆N-Fbxl12 (MBP-∆N) were added to extract, which was then immunpurified using antibodies against N-terminal Cul1, C-terminal Cul1, or C-terminal Skp1. Co-purified proteins were probed using antibodies against MBP and Cul1. (C) MBP-Fbxl12 (FL) was added to egg extract for a final concentration of 1 µM, and MBP-∆N-Fbxl12 (∆N) for a final concentration of 1 ∆M (1x) or 16 ∆M (16x). Extracts were then incubated with SB-DNA beads, and bound proteins were analyzed by immunoblot. (D–E) 35S-labeled Fbxl12 containing the mutations L25A, P26A (25/26), L37A, S38A (37/28), or V48A, C49A (48/49) or wild type Fbxl12 (WT) was added to egg extract and allowed to bind SB-DNA beads (D) or either IgG or anti-Skp1 antibodies (∆S) (E). Bead fractions were analyzed by phosphorimager.
F box proteins typically bind their substrates via domains independent of the F box domain. However, we were surprised to find that MBP-∆N-Fbxl12, unlike the full-length protein, was unable to bind to SB-DNA beads (Fig. 3C). To examine more specific effects of F box mutations, we further generated a series of double point mutations at well-conserved residues in the Fbxl12 F box domain (see Fig. 2B), each of which are known to have important interactions with Skp1.24 Mutant proteins were radioactively labeled and assayed for Skp1 interaction using anti-Skp1 antibodies, and for DNA bead binding. As expected, wild type Fbxl12 both co-purified with Skp1 and bound efficiently to SB-DNA beads. However, the L25A, P26A and V48A, C49A mutants had intermediate, and the L37A, S38A mutant very little, ability to bind to either (Fig. 3D and E; Fig. S2). The correlation between Skp1 binding and chromatin binding suggests that an interaction with Skp1 via the F box may be important for Fbxl12 to interact with chromatin-bound substrate. Such a relationship between Skp1 binding and substrate interaction has been seen previously,25 but is an unusual characteristic for an F box protein.
Fbxl12 recruits the SCF complex to DNA and is required for Ku80 ubiquitylation
Our observations thus far had led us to hypothesize that Fbxl12 was the F box protein responsible for SCF recruitment to DSBs and ubiquitylation of Ku80. To test this, we immundepleted Fbxl12 from egg extract using antibody bound to protein A-coated sepharose beads. Nearly 100% of Fbxl12 was removed from extract, and after depletion both slow- and fast-migrating anti-Fbxl12 bands co-purifying with SB-DNA beads were lost. Consistent with our hypothesis, the binding of both Cul1 and Skp1 was also inhibited in Fbxl12-depleted extract (Fig. 4).
In addition, bulk ubiquitin on SB-DNA beads, most of which is dependent on the presence of Ku80,15 was largely absent following Fbxl12 depletion, closely resembling the result following addition of dominant-negative Cul1 (Fig. 4). The anti-ubiquitin band representing NEDD8-conjugated Cul1 was also missing in Fbxl12-depleted extract.
Finally, the modifications on Ku80 were largely absent in the Fbxl12-depleted extract. In fact, Ku80 modifications in the absence of Fbxl12 closely resemble those from extracts containing dominant-negative Cul1 protein (see Fig. 1B; Fig. S1). These observations strongly suggest that Fbxl12 is, indeed, the F box protein responsible for the recruitment of the SCF complex to DSBs and the ubiquitylation and degradation of Ku80. Therefore, we have shown that Fbxl12 has a previously unrecognized role in the DNA damage response directly at DSBs.
Discussion
The kinetics of protein recruitment to and displacement from chromatin are central to nuclear function, but the latter has been historically largely ignored. In recent years, however, the importance of regulated protein removal from chromatin has gained prominence. A particularly interesting case is that of DNA damage-bound proteins, as these must be removed upon the completion of repair and prior to the return of chromatin to its undamaged state.2 Ku is an exceptionally appealing factor for study as, upon the completion of NHEJ, it is predicted to be topologically trapped on the DNA leading to a potential inhibition of replication and transcription. We previously uncovered a mechanism for the removal of Ku80 from DNA, via what is now becoming recognized as a common and seemingly general process2 in which proteins are removed from chromatin via a mechanism involving their ubiquitylation. Here, we have shown that the SCF complex is required for Ku80 ubiquitylation in Xenopus egg extracts. Moreover, we have identified the F box protein Fbxl12 as being responsible for the recruitment of the SCF complex to DSBs. It has recently been shown that depletion of RNF8, a ubiquitin ligase with an extensively characterized role in the DSB response, inhibits Ku80 ubiquitylation in mammalian cells.26 This observation is seemingly at odds with our data. It is possible, however, that RNF8 ubiquitylates an upstream factor required for the recruitment of the SCF complex to DSBs, or that it is involved in the priming of Ku80 with monoubiquitylation that is then used in subsequent SCF-dependent polyubiquitylations.
Interestingly, of two DSB-binding bands identified using an anti-Fbxl12 antibody, only the faster-migrating one is dependent upon the presence of Ku. Further research will be required to determine whether the slower-migrating band represents a cross-reacting protein or a modified or alternatively spliced form of Fbxl12. However, since both bands are removed by immunodepletion using an anti-Fbxl12 antibody (see Fig. 4) and preliminary results indicate that both bands are competitive with exogenous recobminant Fbxl12 protein (data not shown), it seems likely that these bands represent two isoforms of Fbxl12. If a modified version of Fbxl12 is indeed capable of binding to DSBs independently of Ku, it may be involved in the ability of the SCF complex to distinguish between DSB-bound Ku80 and Ku80 in solution, which is not ubiquitylated even in the presence of DSBs.15 It is unclear at this time, however, why recombinant bacterially expressed and in vitro translated Fbxl12 do not display this Ku-independent ability to bind DSBs.
The SCF complex is known to have multiple roles in DNA damage responses.27 Critical DNA damage factors such as Claspin,28 Chk129 and Cdc25a30,31 are all known SCF substrates. However, a role for the SCF complex in the ubiquitylation of chromatin-bound proteins has not been well established. In contrast, the cullin family member Cul4, which exists in an E3 ubiquitin ligase similar to the SCF complex (CRL4) is known to have many chromatin-bound substrates. For example, the chromatin-bound replication licensing factor Cdt1 and many other proteins involved in regulating re-replication are ubiquitylated by CRL4.32 We suggest that the SCF complex, like CRL4, is active on chromatin-bound proteins. Out of 30 Xenopus F box proteins tested, seven were able to bind strongly to chromatin in Xenopus egg extract. Two of these, Fbxl12 and β-TrCP, were specific for DSBs. Determining how and why β-TrCP is recruited to DSBs will require further investigation, although it is known to be involved in the ubiquitylation of the DNA damage binding protein Claspin28 as well as other factors in the DNA damage response.27 Of the remaining chromatin-binding F box proteins, Fbh1, Fbxl19 and Kdm2b have known DNA- or chromatin-binding motifs (Fbh1 contains a UvrD-like helicase domain and Fbxl19 and Kdm2b both contain zinc and PHD finger domains19). Notably, Kdm2b, a member of the JmjC demethylase family, itself was heavily modified upon binding to chromatin in a pattern resembling polyubiquitylation (data not shown). Intriguingly, ubiquitylation was recently shown to regulate the chromatin localization of a yeast Kdm2b ortholog lacking an F box, Epe1.33 One exciting possibility is that Kdm2b is able to self-regulate its localization using a similar mechanism. The chromatin binding of the remaining two F box proteins, Fbxo24 and Fbxo28, remains mysterious, although Fbxo28 was identified in a screen for proteins interacting with the apoptotic nuclease DNA Fragmentation Factor.34
Our observations indicate that the SCF complex has a previously under-appreciated role in the regulation of chromatin-bound proteins, in particular at sites of DNA damage. Our study establishes a novel molecular function for Fbxl12 in DSB-dependent Ku80 ubiquitylation and will provide a unique tool to further our understanding of how dynamic interactions between repair proteins and DSBs are regulated.
Materials and Methods
Frog egg extract
Meiotic metaphase II-arrested (SCF) Xenopus laevis egg extract was prepared as previously described.35 Protein A-coated Dynabeads (Invitrogen) were bound to antibody according to manufacturer’s recommendations and used to immunodeplete Ku80 and Cul1, as well as for immunoprecipitation experiments. Ku80 was depleted in two rounds of 45 min each on ice, using antibodies against the C-terminal peptide of Ku80. Cul1 was depleted in three rounds of 45 min each on ice, using antibodies against the N-terminus of Cul1. Antibody-bound beads were removed using a magnet.
Xenopus laevis Fbxl12 was depleted in two rounds using protein A-coated sepharose beads bound to anti-Fbxl12 antibody. For each round of depletion, 10 µg antibody was used for every microliter of extract depleted. Extract and antibody-bound beads were incubated together during 40 min of rotation at 4°C for each round. Extract was separated from beads by spinning through a small hole at the bottom of a microcentrifuge tube for 30 sec at 500 x g.
Plasmids
ESTs encoding wild-type Xenopus laevis Ku80 (IMAGE: 4032122), Xenopus laevis Cul1 (IMAGE: 6642922), Xenopus tropicalis Fbxl12 (IMAGE: 7682433), and Xenopus laevis Fbxl12 (IMAGE: 3302221) were purchased from Open Biosystems. A plasmid encoding the H3 gene was obtained from a library of Xenopus laevis cDNAs.36 Point mutations of Xenopus laevis Fbxl12 were generated by site-directed mutagenesis using the oligonucleotides GGA CAC CGG TGT TAT CAT GGG CCG CAG ACA GTG TCC TGC TGG AAG T (L25AP26A), GGA CAC CGG TGT TAT CAT GGG CCG CAG ACA GTG TCC TGC TGG AAG T (L37AS38A), and ACC TCG TCC GCT CCG GAC GAG CTG CCA AGA GAT GGA GAA GGT TGA T (V48AC49A), and their complements. In vitro translation was in rabbit reticulocyte lysate in the presence of 35S-methionine using the SP6 and T7 RNA polymerases according to the manufacturer’s instructions (Promega).
Antibodies and proteins
Antibodies against Ku80, Ku70, Cul1, Skp1, H3, and ubiquitin have been described previously.15 Mouse antibodies against MBP were from New England Biolabs (# E8032). Rabbit polyclonal anti-Fbxl12 antibodies were against the peptide CRGIDELKKSLPNCKVTN and anti-N-terminal Cul1 antibodies were against CMSSRSQNPHGLKQIGLDQIWDD.
GST-DN-Cul1 was generated using the oligonucleotides TCC CCC CGG GAT GTC TTC AAA CAG GAG TCA and AAG GAA AAA AGC GGC CGC TTA CAT CTT TGC ATA GAA CTT CT and cloning into the vector pGEX6P2 (GE Healthcare Life Sciences). Recombinant protein was purified using glutathione sepharose beads (SIGMA) according to manufacturer’s instructions. GST-DN-Cul1 was added to extract at a concentration of 0.75 mM.
MBP- and 6-His-tagged Xenopus laevis Fbxl12 was generated by cloning into the pMAL-c2e vector (NEB), modified by the addition of a PreScission protease site to allow cleavage of the MBP tag, using the oligonucleotides GAT CGT CGA CTT AGT GGT GGT GGT GGT GGT GGT TAG TAA CCT TGC AGT TTG G (reverse, including 6-His tag) and GAT CGG ATC CAT GGA GAT TAT GTC TGC TCT T (forward, full-length) or GAT CGG ATC CAT GAA CCT AAC ACC CTA CAA GCT G (forward, ∆N). Proteins were purified using Ni-NTA-agarose beads (Qiagen) according to manufacturer’s instructions. Protein was added to extract at concentrations indicated.
Chromatin isolation and Ku80 assays
Chromatin isolation and Ku80 release and degradation assays were as described previously.15 To test F box proteins for DNA binding, homologs of human F box proteins were identified by BLAST, in vitro translated in rabbit reticulocyte lysate, and assayed for ability to bind to DNA beads. Candidates were further tested for DSB-dependence and Ku-dependence of chromatin binding.
Immunoprecipitations
Immunoprecipitations were as described previously37 using antibodies bound to protein A-coated magnetic Dynabeads (Invitrogen).
Supplementary Material
Acknowledgments
We thank Cristina Ghenoiu for reagents and Agata Smogorzewska, Elizabeth Garner and members of the Funabiki laboratory for helpful discussions. Christian Zierhut and Simona Giunta assisted with experimental design and John Xue helped generate reagents. L.P. was supported by a postdoctoral fellowship from the Leukemia and Lymphoma Society of America and Rockefeller University training grant T32 CA009673. H.F. is supported by a National Institutes of Health grant (R01 GM075249). The GenBank accession number for Xenopus laevis Fbx112 is KC506729.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here:
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23408
References
- 1.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 2.Postow L. Destroying the ring: Freeing DNA from Ku with ubiquitin. FEBS Lett. 2011;585:2876–82. doi: 10.1016/j.febslet.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blier PR, Griffith AJ, Craft J, Hardin JA. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–601. [PubMed] [Google Scholar]
- 4.Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–9. [PubMed] [Google Scholar]
- 5.Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis J, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med. 2010;207:417–27. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol. 2011;18:75–9. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–42. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38:5706–17. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–4. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/S0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Lee KJ, Davis AJ, Chen DJ. Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. J Biol Chem. 2012;287:4936–45. doi: 10.1074/jbc.M111.306167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu D, Topper LM, Wilson TE. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics. 2008;178:1237–49. doi: 10.1534/genetics.107.083535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao Z, Davis AJ, Fattah KR, So S, Sun J, Lee KJ, et al. Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair (Amst) 2012;11:310–6. doi: 10.1016/j.dnarep.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double-strand break-induced ubiquitylation. J Cell Biol. 2008;182:467–79. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piva R, Liu J, Chiarle R, Podda A, Pagano M, Inghirami G. In vivo interference with Skp1 function leads to genetic instability and neoplastic transformation. Mol Cell Biol. 2002;22:8375–87. doi: 10.1128/MCB.22.23.8375-8387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, et al. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–26. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–70. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 19.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–80. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skaar JR, D’Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell. 2009;137:1358–, 1358, e1. doi: 10.1016/j.cell.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 21.Kim M, Nakamoto T, Nishimori S, Tanaka K, Chiba T. A new ubiquitin ligase involved in p57KIP2 proteolysis regulates osteoblast cell differentiation. EMBO Rep. 2008;9:878–84. doi: 10.1038/embor.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, et al. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc Natl Acad Sci U S A. 2003;100:10231–6. doi: 10.1073/pnas.1831009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–74. doi: 10.1016/S0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 24.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida Y, Murakami A, Tanaka K. Skp1 stabilizes the conformation of F-box proteins. Biochem Biophys Res Commun. 2011;410:24–8. doi: 10.1016/j.bbrc.2011.05.098. [DOI] [PubMed] [Google Scholar]
- 26.Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012;19:201–6. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silverman JS, Skaar JR, Pagano M. SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem Sci. 2012;37:66–73. doi: 10.1016/j.tibs.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mamely I, van Vugt MA, Smits VA, Semple JI, Lemmens B, Perrakis A, et al. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr Biol. 2006;16:1950–5. doi: 10.1016/j.cub.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 29.Zhang YW, Brognard J, Coughlin C, You Z, Dolled-Filhart M, Aslanian A, et al. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol Cell. 2009;35:442–53. doi: 10.1016/j.molcel.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, et al. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- 31.Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, et al. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–74. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 2011;25:1568–82. doi: 10.1101/gad.2068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun S, Garcia JF, Rowley M, Rougemaille M, Shankar S, Madhani HD. The Cul4-Ddb1(Cdt)² ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell. 2011;144:41–54. doi: 10.1016/j.cell.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanus J, Kalinowska-Herok M, Widlak P. Identification of novel putative regulators of the major apoptotic nuclease DNA Fragmentation Factor. Acta Biochim Pol. 2010;57:521–7. [PubMed] [Google Scholar]
- 35.Murray AW. Coordinating cell cycle events. Cold Spring Harb Symp Quant Biol. 1991;56:399–408. doi: 10.1101/SQB.1991.056.01.047. [DOI] [PubMed] [Google Scholar]
- 36.Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–24. doi: 10.1016/S0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- 37.Postow L, Woo EM, Chait BT, Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem. 2009;284:35951–61. doi: 10.1074/jbc.M109.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.