Abstract
Cyclin E1 is expressed at the G₁/S phase transition of the cell cycle to drive the initiation of DNA replication and is degraded during S/G₂M. Deregulation of its periodic degradation is observed in cancer and is associated with increased proliferation and genomic instability. We identify that in cancer cells, unlike normal cells, the closely related protein cyclin E2 is expressed predominantly in S phase, concurrent with DNA replication. This occurs at least in part because the ubiquitin ligase component that is responsible for cyclin E1 downregulation in S phase, Fbw7, fails to effectively target cyclin E2 for proteosomal degradation. The distinct cell cycle expression of the two E-type cyclins in cancer cells has implications for their roles in genomic instability and proliferation and may explain their associations with different signatures of disease.
Keywords: cyclin E2, cell cycle, cyclin E1, proliferation, Fbw7
Introduction
Cyclin E1 drives the transition from G1 to S phase through the assembly of pre-replication complexes and activation of the kinase CDK2, leading to the initiation of DNA synthesis. The fundamental role of cyclin E1 in promoting proliferation has led to its identification as an important oncogene in many cancers, and as a downstream target of other oncogenic pathways.1
After induction by mitogens, cyclin E1 peaks in expression at G1/S phase of the cell cycle. This periodicity in expression results from regulation of both mRNA abundance and protein degradation. Upon S phase entry CDK2 phosphorylation of cyclin E1 primes it for phosphorylation by GSK-3β and other kinases.2 These phosphorylation events create short regions in cyclin E1 that are recognized by the F-box protein Fbw7, which is part of the Skp1-Cul1-Rbx1-Fbw7 ubiquitin ligase complex (SCFFbw7).3 SCFFbw7-mediated ubiquitination of cyclin E1 leads to its rapid proteosomal degradation during S phase and G2/M 2.
Failure to degrade cyclin E1 in late S phase promotes overall proliferation4 but also disrupts the initiation of DNA replication5 and inhibits progression through mitosis,6 ultimately resulting in chromosome instability.5,7 Fbw7 deletion or mutation in cancer is a major cause of loss of periodicity in expression of cyclin E1,8 and Fbw7 mutation is itself highly correlated with chromosome instability.9 The disruption of cyclin E1 phosphorylation sites that are recognized by Fbw7 leads to increased aneuploidy, hyperproliferation and failure of differentiation in knock-in mice,10 while overexpression of hyperstable cyclin E1 (T380A) promotes mammary carcinogenesis.8
Cyclins E1 and E2 are redundant during murine development,11 and in most studies and reviews they are collectively referred to as cyclin E. During the cell cycle both cyclin E1 and E2 mRNA transcription is activated by E2F factors to peak at mid-G1 to early S phase,12-15 and transcription of both cyclin E1 and cyclin E2 mRNAs is co-regulated by methyltransferases such as CARM1/PRMT416 and PRMT5.17,18 Consequently cyclins E1 and E2 are thought to have identical temporal regulation, including periodic degradation via Fbw7 during S phase and G2/M.19 Despite these reports of overlapping function and regulation, cyclin E1 and E2 usually have unique relationships with outcome in cancer cohorts and feature in different signatures of disease.15 For example, we have recently found that some breast cancer subtypes overexpress cyclin E2 more strongly than cyclin E1.20 This led us to re-examine the patterns of cyclin E1 and cyclin E2 expression in cancer cells, particularly during the cell cycle. In this manuscript we provide the first evidence that cyclin E1 and E2 are differently regulated during the S phase of cancer cells.
Results
Cyclin E2 expression is maintained in the S phase of cancer cells, whereas cyclin E1 expression peaks at the G1/S transition of the cell cycle
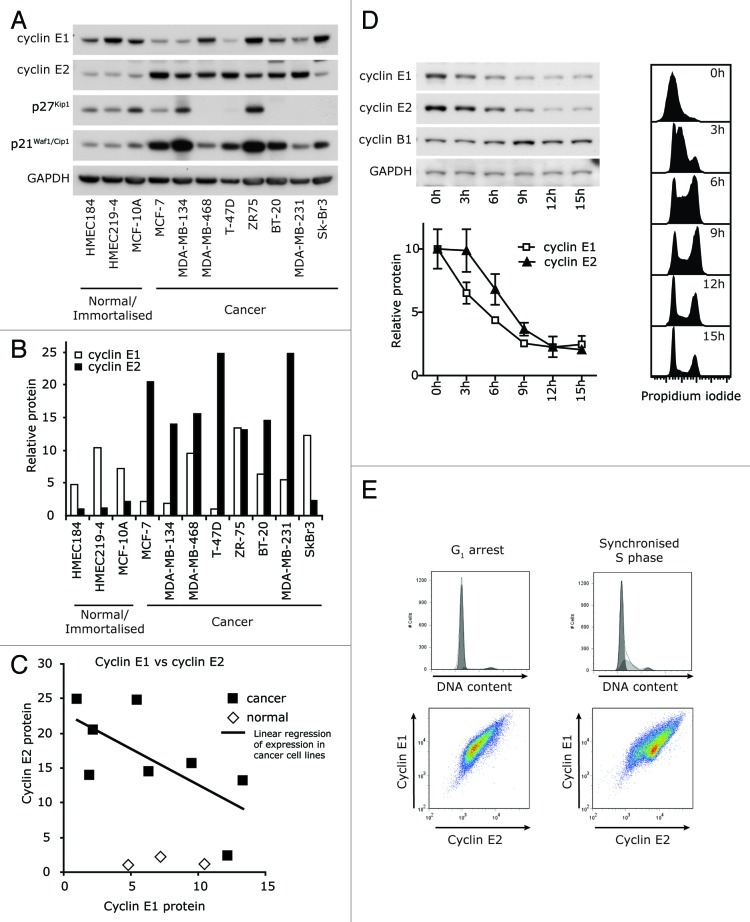
We first compared the absolute levels of cyclin E1 and cyclin E2 in a panel of breast cancer cell lines and their normal/immortalized counterparts using western blotting. Cyclin E2 protein was consistently expressed at high levels in breast cancer cell lines compared with normal or immortalized cell lines, whereas cyclin E1 was not (Fig. 1A and B). Rather than showing concordant expression, the expression of cyclin E1 and cyclin E2 tended to be inversely correlated in cancer cell lines (Fig. 1C).
Figure 1. Cyclin E1 and E2 have discordant cell cycle expression in cancer cells. (A) Breast cancer and normal/immortalized cell line lysates were immunoblotted for cyclins E1 and E2, p21Waf1/Cip1, p27Kip1 and GAPDH. (B) Densitometry was performed on the levels of cyclins E1 and E2, and normalized to expression of GAPDH. (C) Linear regression identified a negative correlation between the expression of cyclin E1 and cyclin E2 (y = -1.0426x + 22.999, R2 = 0.4591; p < 0.0648). (D) T-47D cells were synchronized with HU, released into the cell cycle, and matched lysates and flow cytometry samples collected 0–15h post release. Lysates were immunoblotted as indicated, and densitometry performed on cyclin E1 and E2 expression normalized to GAPDH expression. Cell cycle distribution was analyzed by propidium iodide staining. Data are from duplicate experiments and error bars represent range between replicates. (E) MCF-7 cells in G0/G1 phase (ICI 182780 arrest) and early S phase (estrogen stimulated) were analyzed for DNA content by propidium iodide staining (PI) and expression of cyclins E1 and E2 by flow cytometry.
In order to determine whether the differences in overall cyclin E expression were associated with different patterns of expression during the cell cycle, we used two models of cell cycle synchrony. First, T-47D breast cancer cells were synchronized at G1/S phase by hydroxyurea treatment and then released into the cell cycle. Progress through S phase was apparent within 3 h and the cells reached G2/M at 9 h (Fig. 1D). Cyclin E2 protein was expressed at higher relative levels than cyclin E1 from 3–9 h, and this difference was particularly pronounced in early-mid S phase (Fig. 1D). In MCF-7 breast cancer cells synchronized at G0/G1 by antiestrogen treatment, we observed a constant ratio of expression between cyclin E1 and E2 using flow cytometry (Fig. 1E). Estrogen stimulation into early S phase resulted in the appearance of a second population expressing higher levels of cyclin E2 (Fig. 1E). This suggested a loss of co-ordination between the expression of cyclins E1 and E2 upon entry into S phase.
Cyclin E2 is expressed in S phase in a broad range of breast cancer cell lines, but not normal or immortalized breast cells
In order to confirm that the apparent loss of co-ordination between the expression of cyclins E1 and E2 during entry into S phase was not an artifact of cell synchronization, we next analyzed cyclin E1 and E2 expression vs. DNA content by flow cytometry in a broader range of immortalized mammary cells and breast cancer cell lines. As expected, the highest levels of cyclin E1 were observed during G1, and its expression decreased through S phase in both immortalized (MCF-10A) and cancer cell lines (MCF-7 and BT-20; Fig. 2A). In the immortalized cell line, MCF-10A, the pattern of cyclin E2 expression was similar to that of cyclin E1. However in the cancer cell lines, MCF-7 and BT-20, cyclin E2 expression did not peak until early S phase, and high expression was maintained until mid S phase (Fig. 2A). Across a broader panel of breast cancer cell lines, cyclin E2 expression was also higher in S phase than G1 phase (~2.5-fold) but not in three normal or immortalized cell lines (Fig. 2B), when quantitated from flow cytometry plots. By contrast, cyclin E1 did not have higher relative S phase expression in any of the cancer or immortalized cell lines (Fig. 2B).
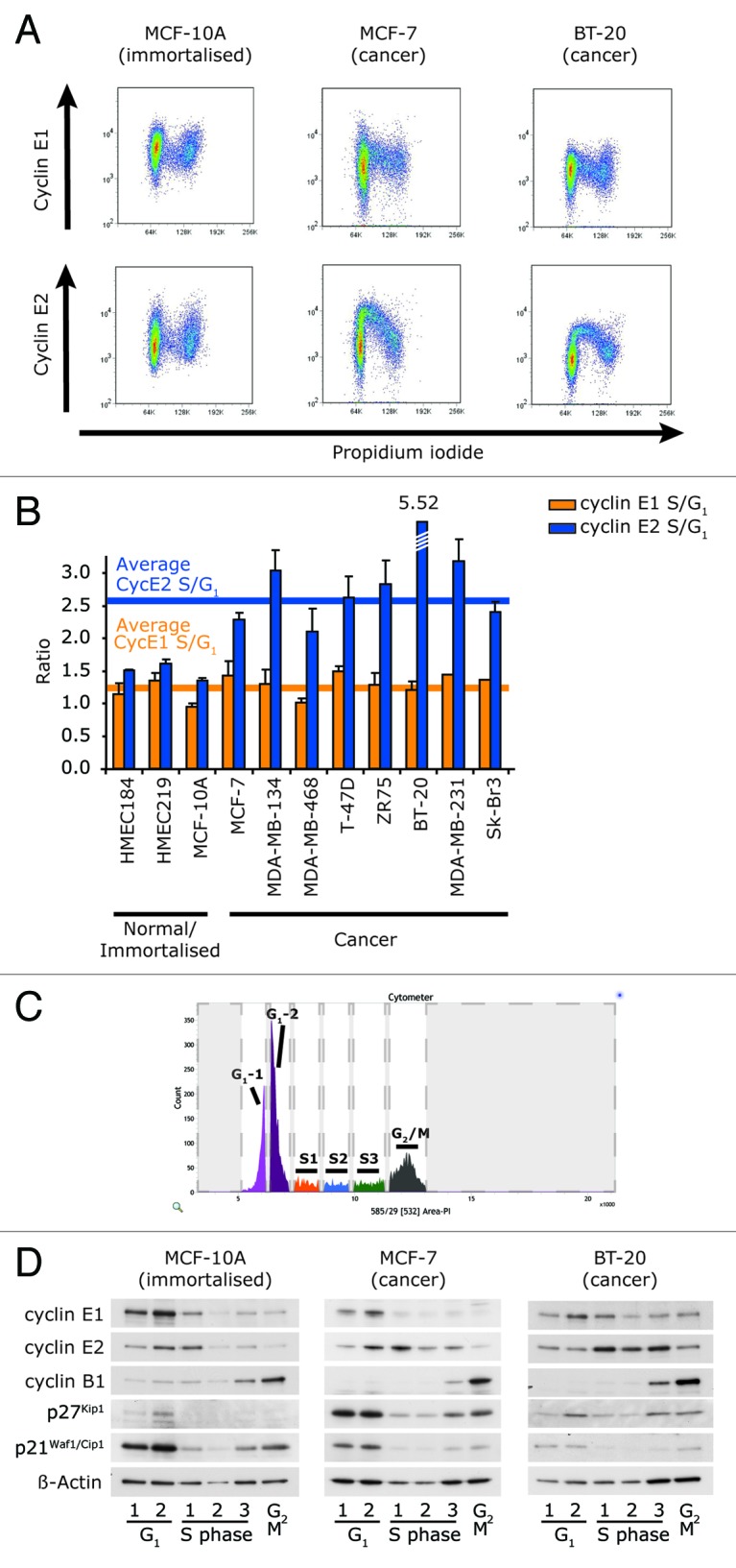
Figure 2. Cyclin E2 is expressed in S phase in a panel of cancer cell lines, but not normal cell lines. (A) Fixed cells were immunoprobed for either cyclin E1 or E2, counterstained with PI and analyzed by flow cytometry. Representative examples of three cell lines are shown. (B) S/G1 phase ratio of expression of cyclin E1 and E2 was quantitated by calculating the ratio of intensity of the geometric mean of expression of each cyclin in S phase to the geometric mean of intensity of expression in G1 phase and determined across a panel of immortalized and breast cancer cell lines. Error bars represent range between replicates. Data are pooled from, or are representative examples of, duplicate data sets. (C) Exponentially growing cells were stained with PI and sorted into six contiguous fractions of the cell cycle according to DNA content, representative example of BT-20 cells shown. (D) MCF-10A, MCF-7 and BT-20 cells were sorted as in (C), and 25,000 cells from each fraction was prepared in sample buffer and separated by SDS-PAGE, followed by western blotting with the antibodies shown.
We further validated our findings by western blotting of cells isolated from distinct stages of the cell cycle, as defined by DNA content. The cell lines were sorted into six fractions of graded intensity of propidium iodide staining, resulting in two G1, three S phase and a G2/M fraction (Fig. 2C). Cells from each fraction were western blotted for cyclin E1 and cyclin E2, as well as cyclin B1, p21Waf1/Cip1 and p27Kip1 to mark cell cycle progression. This confirmed that, unlike cyclin E1, cyclin E2 was expressed predominantly in the S phase of MCF-7 and BT-20 cells, but during the G1/S phase transition of MCF-10A cells (Fig. 2D).
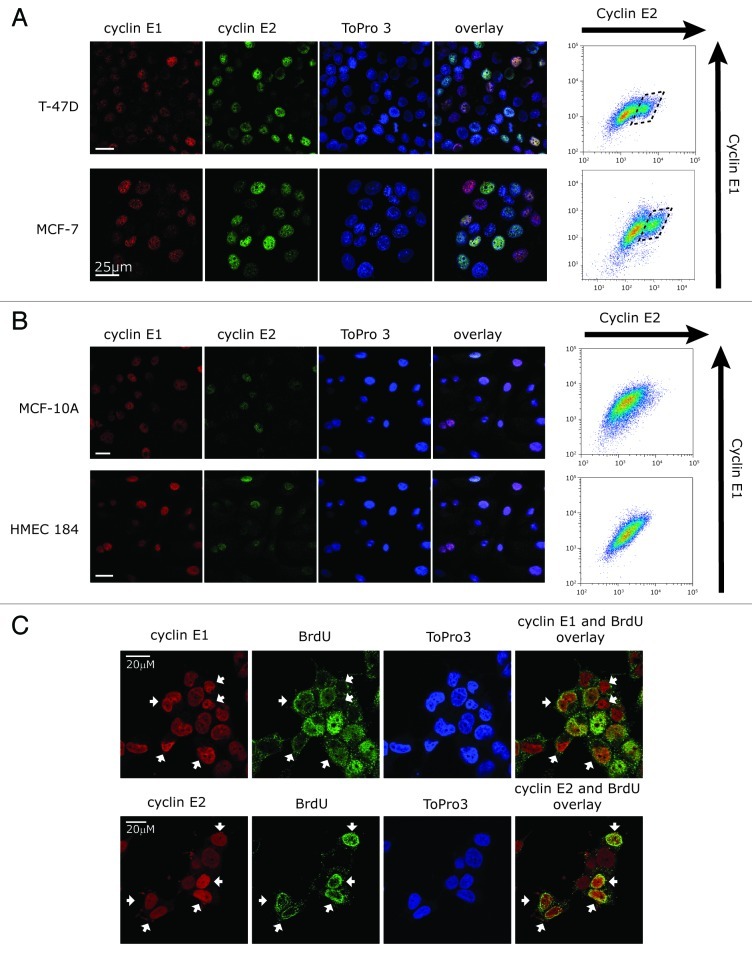
Using both immunofluorescence and flow cytometry to examine the relationship between cyclin E1 and E2 expression in individual cells, we found that the two E-cyclins were expressed in different subsets of cells within breast cancer cell lines (Fig. 3A), with a distinct population of cells expressing high cyclin E2 (Fig. 3A, box). Consistent with our observations of synchronous expression of cyclins E1 and E2 in normal cells, cyclins E1 and E2 were expressed in the same individual normal (HMEC184) and immortalized (MCF-10A) cells (Fig. 3B). Using BrdU staining of T-47D cells, we identified the subset of cells actively undergoing DNA replication (i.e., in S phase). These cells expressed moderate-high levels of cyclin E2 but low levels of cyclin E1 (Fig. 3C), consistent with our observations in Figures 1 and 2.
Figure 3. Cyclin E1 and E2 are expressed in different cell subsets in breast cancer cells. (A and B) Confocal images of (A) breast cancer cells (MCF-7 and T-47D) and (B) immortalized (MCF-10A) or normal (HMEC184) immunoprobed with cyclin E1 (red) or cyclin E2 (green), and counterstained with ToPro3 (blue, nuclei). Fixed cells were analyzed in parallel by flow cytometry for cyclin E1 and E2 levels (right). (C) T-47D cells were pulsed with 20μM BrdU, immunostained for BrdU, cyclin E1 and cyclin E2, and counterstained with ToPro3. White arrows indicate cyclin E1 or cyclin E2 high cells, where high cyclin E2 expression coincides with high BrdU, but high cyclin E1 expression coincides with low BrdU. Scale bars are 20 μm.
Cyclin E1 and cyclin E2 have distinct susceptibility to Fbw7-mediated ubiquitination in cancer cells
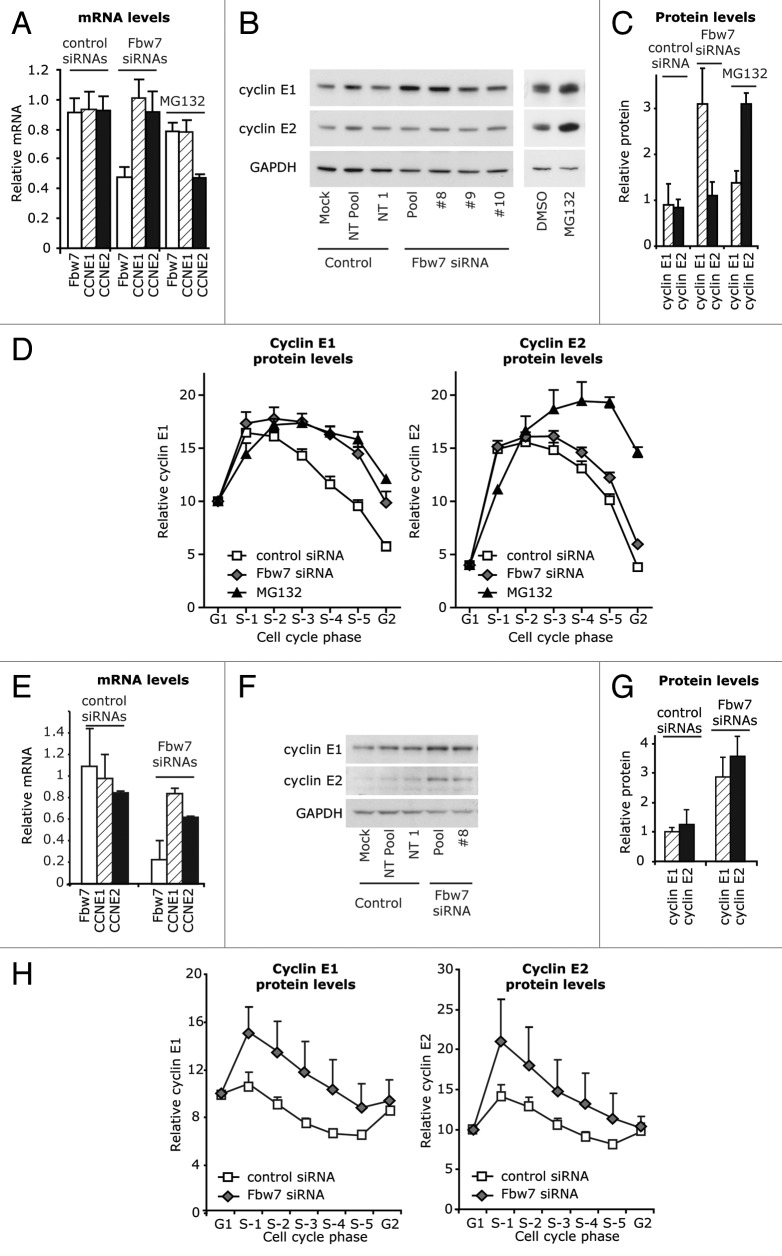
Both cyclin E1 and cyclin E2 have high expression at G1/S (Figs. 1–2), and this is likely due to similar E2F-mediated transcription of cyclin E1 and E2 mRNA during G1 . Since the S phase expression of cyclin E1 is regulated via SCFFbw7-mediated degradation, we hypothesized that differences in cyclin E1 and E2 expression during S phase might result from differences in protein degradation. When T-47D breast cancer cells were treated with with Fbw7 siRNAs, Fbw7 mRNA decreased by 50% (Fig. 4A). This increased cyclin E1 protein levels (3.5x), but did not significantly change cyclin E2 protein levels (1.3x; Fig. 4B and C). Cells were treated in parallel for 24h with MG132, a proteosomal inhibitor. This induced an increase in both cyclin E1 and cyclin E2 protein despite decreased cyclin E2 mRNA (Fig. 4A–C), indicating that cyclin E2 abundance is at least in part regulated by proteosomal degradation. Cells treated with Fbw7 siRNA and MG132 were further analyzed by flow cytometry for cyclin E1 and E2 levels throughout the cell cycle. Fbw7 siRNA treatment stabilized cyclin E1 in mid-late S phase, a pattern that was comparable to the accumulation seen after MG132 treatment (Fig. 4D). By contrast while cyclin E2 significantly accumulated during mid-late S phase after MG132 treatment, this did not occur with Fbw7 siRNA (Fig. 4D).
Figure 4. Cyclin E1 and E2 are differentially regulated by Fbw7 in cancer cell lines, but not normal cell lines. (A) T-47D cells were transfected with Fbw7 and control siRNAs for 48 h, or treated with 8 μM MG132. Lysates were analyzed for Fbw7, CCNE1 and CCNE2 mRNA expression, and (B and C) immunoblotted for cyclin E1 and E2 expression and quantitated by densitometry with normalization to GAPDH. (D) Fixed cells were further immunoprobed for either cyclin E1 or E2, counterstained with PI, and analyzed by flow cytometry. The geometric mean of intensity of expression of cyclin E1 and E2 was quantitated at distinct positions in S phase as identified by PI staining, as indicated in Figure 2C. Data were pooled for control (Mock, NT Pool, NT1) and Fbw7 siRNA (Pool, #8, #9, #10) from quadruplicate experiments and error bars represent S.E.M.. (E–H) HMEC184 cells were transfected with pooled Fbw7 siRNA, Fbw7 siRNA #8, and control treatments (Mock, Non-targeting Pool, Non-targeting #1). (E) Lysates were analyzed by qRT-PCR for Fbw7, CCNE1 or CCNE2 mRNA. (F and G) Lysates were immunoblotted for cyclin E1 and E2 expression and quantitated by densitometry with normalization to GAPDH. (H) Fixed cells were further immunoprobed with antibodies to either cyclin E1 or cyclin E2, counterstained with PI and analyzed by flow cytometry. The geometric mean of intensity of expression of cyclin E1 and E2 was quantitated at distinct positions in S phase as identified by PI staining. Data are pooled from duplicate experiments and error bars represent range between replicates.
Since we had observed downregulation of both cyclin E1 and cyclin E2 during the S phase of normal cells, we performed a similar analysis in the normal cell line, HMEC184. Treatment with Fbw7 siRNAs led to an overall increase in the levels of cyclin E1 and cyclin E2 protein, as observed by western blotting (Fig. 4E–G), despite leading to a slight decrease in cyclin mRNA levels. An analysis of cyclin protein expression through the cell cycle showed that the siRNA treatment led to consistent stabilization during S phase for both cyclin E1 and cyclin E2 (Fig. 4H). This suggested that Fbw7-mediated degradation of cyclin E2 is specifically disrupted in cancer cells.
We confirmed these results in cancer cell lines with naturally occurring dominant-negative (DN) Fbw7 mutations, the ovarian cancer cell line SkOV3 and colon cancer cell line LoVo.21 In SkOV3 cells, cyclin E1 was generally expressed at higher levels during S phase than in G1 (Fig. 5A). However, in other ovarian cancer cell lines, Igrov-1 or A2780, cyclin E1 was highest during G1 phase and decreased in expression during S phase (Fig. 5A and B, representative plots of A2780 shown). The highest levels of cyclin E2 expression were observed during S phase irrespective of Fbw7 mutation (Fig. 5A and B). We observed similar expression patterns when comparing Fbw7 mutant LoVo cells to other colon cancer cell lines HCT116 and SW480 (Fig. 5B): the expression of cyclin E2 was higher in S phase in all the cell lines whether or not they expressed active Fbw7. Thus while Fbw7 drives cyclin E1 turnover during S phase, it is not responsible for the majority of proteosome-mediated turnover of cyclin E2 in cancer cells.
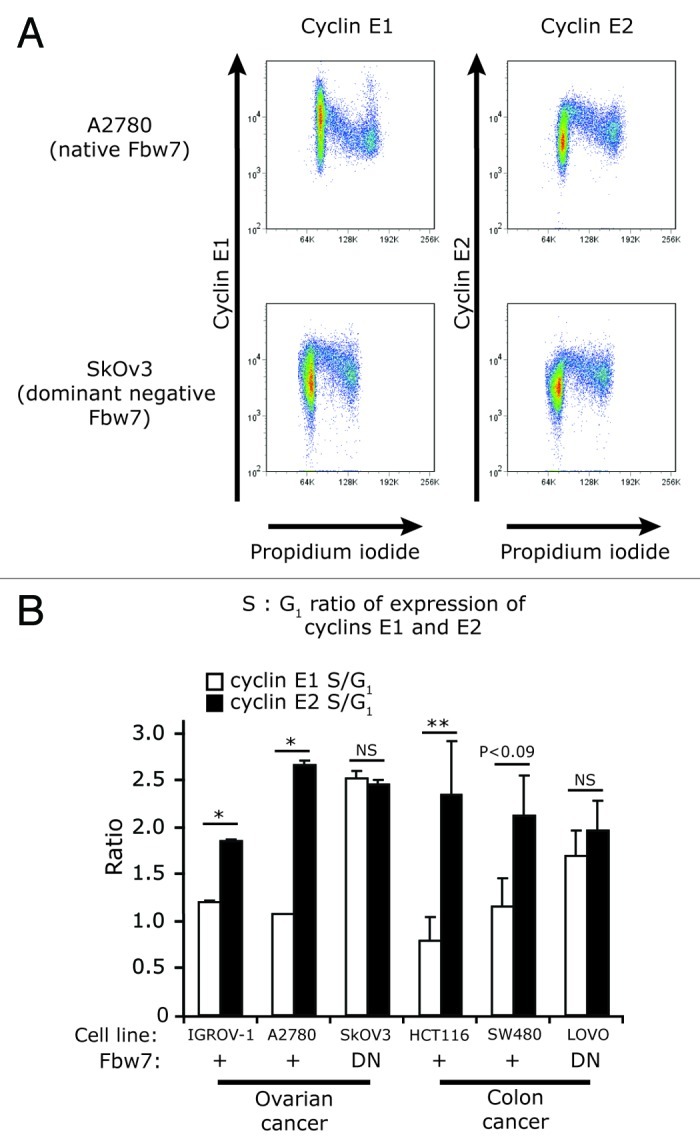
Figure 5. Cyclin E2 stability is not affected by mutation of Fbw7 in ovarian and colon cancer cell lines. (A) Fixed cells from ovarian cancer cell lines A2780 (Fbw7 +/+) and SkOV3 (Fbw7 dominant-negative - DN) were immunoprobed for either cyclin E1 or E2, counterstained with PI, and analyzed by flow cytometry. (B) S/G1 phase ratio of expression of cyclin E1 and E2 in cell lines with wild-type (+) or dominant-negative (DN) Fbw7. SkOv3 (ovarian) and LoVo (colon) cell lines were compared with other ovarian (A2780, IGROV-1) and colon (HCT116, SW480) cancer cell lines. Data are pooled from duplicate experiments and error bars represent range between replicates. Statistical analysis was performed using two-tailed t-tests, ** = p < 0.005, * = p < 0.05.
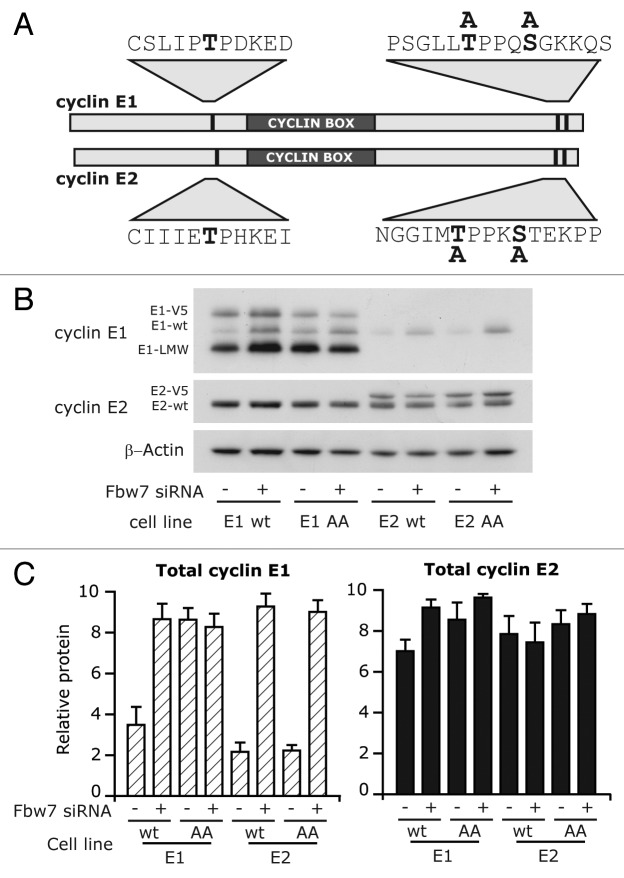
Both cyclin E1 and E2 are E2F target genes, which promote peak mRNA transcription at G1/S. However there are some reports of independent regulation of cyclin E2 mRNA by other transcription factors,15 and these could possibly contribute to cyclin E2 protein expression in S phase. We therefore investigated whether cyclin E1 and E2 expressed under a constitutive promoter also displayed differences in regulation of protein abundance. Fbw7 recognizes cyclin E1 once it is phosphorylated at T74, T395 and S399, the mutation of these sites prevents Fbw7 mediated degradation, and these sites are conserved in cyclin E2.22 Consequently we also investigated the role of these residues in the turnover of exogenous protein. Using the pMSCV vector, we stably overexpressed native cyclins and non-phosphorylatable mutants: T395A/S399A of cyclin E1 (designated cyclin E1 AA), and T392A/S396A of cyclin E2 (designated cyclin E2 AA) (Fig. 6A). Cells expressing wild-type and mutant proteins were treated with Fbw7 siRNA. Cyclin E1 wild-type protein was stabilized by Fbw7 siRNA, whereas cyclin E1 AA protein was not stabilized, consistent with Fbw7-dependent targeting of cyclin E1 (Fig. 6B and C). In contrast neither cyclin E2 wild-type nor E2 AA protein were upregulated by Fbw7 siRNA, and neither protein varied in expression with siRNA treatment. This confirmed that Fbw7 is not efficiently targeting cyclin E2 in cancer cells, and demonstrated that our previous results were not due to differences in endogenous transcription.
Figure 6. Cyclin E2 stability is not affected by mutation of Fbw7-recognition sites in cancer cell lines. (A) Schematic of cyclin E1 and cyclin E2, and position of phosphorylation sites and serine/threonine to alanine substitutions. (B) T-47D cells overexpressing cyclin E1 (wt), cyclin E1 T395A/S399A (AA), cyclin E2 (wt) or cyclin E2 T392A/S396A (AA) were transfected with Fbw7 and control siRNAs. Lysates collected at 72 h were immunoblotted for cyclin E1 and E2, and β-actin. (C) Levels of cyclins E1 and E2 were quantitated by densitometry normalized to β-actin.
Discussion
Distinct regulation of cyclins E1 and E2 in cancer cells
Here we present the first evidence for distinct post-transcriptional regulation of cyclin E1 and E2 in cancer cells. Cyclin E2 expression is sustained during the S phase of cancer cells, and altered targeting by SCFFbw7 contributes to this increased S phase expression. This ultimately results in the discordant expression of cyclins E1 and E2, and expression in distinct subsets of cells. This observation is of broad significance, as it occurs in cell lines derived from breast, ovarian and colon cancers.
Deregulation of the periodic expression of cyclin E1 is well-established as a potent inducer of numerous cellular defects, including proliferative abnormalities10 and genomic instability.7 This deregulation frequently lies downstream of alteration or mutation of Fbw7, but may also be driven by aberrant CDK2 activity23 and through the activity of other proteins such as Ras,24 p21Waf1/Cip1 25, Pin126 and Artemis.27 We observed that the inactivation of cyclin E2 phosphorylation sites via alanine substitution does not increase its stability in cancer cells, unlike similarly mutated cyclin E1. Thus altered phosphorylation is not likely to be the cause of failed targeting by Fbw7. Unfortunately, suitable antibodies are not available to investigate the relative phosphorylation of cyclin E1 and cyclin E2 in cancer cell lines, and this remains an unresolved question.
Cyclin E1 is expressed at high levels in multiple cancer types and drives tumorigenesis autonomously in mouse models, albeit with long latency.1 Cyclin E2, while less thoroughly characterized than cyclin E1 in the context of cancer, has been identified as a candidate mammary oncogene in an MMTV insertional screen,28 and cyclin E2 mRNA is detected at high levels, independently of cyclin E1 mRNA, in various malignancies.15 Cyclin E1 and cyclin E2 mRNAs act as an independent prognostic indicator for overall and metastasis-free survival in breast cancer,29 and cyclin E2 repeatedly features in signatures of poor prognosis of breast cancer that do not include cyclin E1.30-32 Overall these data imply a difference in mRNA expression of cyclins E1 and E2 that is particular to cancer cells, and point to differences in function.
Cyclin E1 protein is overexpressed in about 30% of all breast cancers,33 but our data indicate that this will not be representative of cyclin E2 expression. Not only may cyclin E2 be under distinct transcriptional control,15 we have shown that it is under different degradative control in cancer cells, and total cyclin E expression will not be represented by cyclin E1 alone. These data have implications for the use of cyclin E1 as a biomarker for “cyclin E” activity, and indicate that “cyclin E” is high in a greater proportion of cancers than previously determined. “Cyclin E” expression is also a potential therapeutic target in cancer cells through the use of CDK inhibitors and cyclin E1 siRNA therapy.20,34-36 Applying CDK inhibitors to only cyclin E1 high tumors may limit their potential application, and the use of cyclin E1 specific therapy such as siRNAs would limit the efficacy of anti-cyclin E therapy and could potentially provide a pathway to resistance.
Materials and Methods
Cell culture
Cell lines were obtained and cultured as previously described,37 and HEK293 cells cultured in DMEM F-12 (10% fetal calf serum). T-47D/EcoR cells were derived as described.38 A2780, IGROV-1, HCT116 and SW480 cell lines were obtained from ATCC and cultured in RPMI 1640, 5–10% fetal calf serum (FCS) and insulin (10 μg/ml). LoVo cells were cultured in F-12 Kaighn’s (Gibco) with 10% FCS.
Cyclin E1 (CycE1:IOH27850) and cyclin E2 (CycE2: IOH43526) (Invitrogen) were recombined into pMIG-GW-V539 and retrovirus generated and infected into T-47D/EcoR cells as described.38 Site-directed mutagenesis was performed as previously described.39 Subpopulations with graded expression of GFP and cyclin proteins were separated by sterile FACS.
Synchronization/drug treatments
Cells were synchronized with 1 mM HU (Merck) treatment for 40 h, washed twice with PBS, and fresh media added. Seventy min post removal of HU, cells were pulsed with 20 μM BrdU (Sigma) for 15 min, followed by washing twice with PBS before adding fresh media.
MCF-7 cells were synchronized at G0/G1 with the anti-estrogen ICI 182780 1 x 10−8 M (Selleck Chemicals) for 48 h as described.40 Proliferation was induced with 1 x 10−7 M 17β-estradiol (3,17K-Dihydroxy-1,3,5(10)-estratriene).
Proteosomal inhibition was performed using 8 μM MG132 (carbobenzoxy-L-leucyl-L-leucyl-L-leucinal, Calbiochem), CDK2 inhibitor Roscovitine (Sigma) at 7 μM final concentration, and DMSO was applied to control cultures at the same concentration.
Immunoblotting
Protein lysates were harvested as described,41 and 10–30 μg of lysate were separated using NuPage polyacrylamide gels (Invitrogen) prior to transfer to PVDF membranes. Antibodies, chemiluminescence and densitometry are as described.42
qRT-PCR analysis
Total RNA was extracted using the RNeasy Minikit (Qiagen) and processed with the Reverse Transcription System (Promega). qRT-PCR was performed on an ABI Prism 7900HT (Applied Biosystems, Invitrogen) using gene expression assays cyclin E1 – Hs00180319_m1, cyclin E2 – Hs00180319_m1, Fbw7 - Hs00217794_m1, p107 - Hs00765713_m1 and human RPLP0 – 4326314E (Applied Biosystems). All reactions were performed as described.42
Flow cytometry
Expression of cyclins E1 and E2 was assessed by flow cytometry as described.43 Cells were incubated overnight at 4°C with antibodies to the E-cyclins (E1: EP435E or E2: EP454Y, Epitomics) followed by 1 h incubation at room temperature with secondary antibodies (allophycocyanin conjugated goat anti-mouse, fluorescein 5-isothiocyanate conjugated goat anti-rabbit, Jackson Immunoresearch), co-stained with 10μg/mL PI (Sigma) for 2–5h, and incubated with 50μg/mL RNase A (Sigma). Flow cytometry was performed on a FACSCanto (BD Biosciences). Specific staining of cyclin E1 and E2 was validated using alternative antibodies (HE-12, Santa Cruz; #4132, Cell Signaling) and isotype specific controls. Data were analyzed using FlowJo,44 where signal intensity was calculated by obtaining the geometric mean signal per cell in gated regions.45,46
Cell sorting
Exponentially proliferating cells (1.5 x 107) were fixed and stained as described for flow cytometry analysis. Cells were separated using a 0.45μM filter and sorted into six contiguous windows of propidium iodide fluorescence using an Influx flow cytometer (BD Biosciences), as described.47 25,000 cells from each sorted fraction were solubilized with 1x PAGE sample buffer before analysis by western blotting as described.48
Immunofluorescence and microscopy
Cells were fixed with 4% PFA/PBS for 20 min at room temperature, and post-fixed with methanol at -20°C for 20 min. Alternatively cells were fixed in methanol at -20°C for 20 min. Samples were blocked with 1% BSA/PBS, incubated with primary antibodies at 1:100 for 1h, followed by 1h with secondary antibodies and ToPro3/DAPI as DNA counterstains (Jackson ImmunoResearch). For BrdU co-staining, cells were pulsed with BrdU for 30 min followed by methanol fixation. Slides were fixed for 5 min in 4% PFA/PBS after staining with primary and secondary antibodies, denatured with 1.5M HCl for 10 min at 37°C, and blocked and incubated with anti-BrdU (BD Biosciences).
Confocal microscopy was performed on a Leica DMRBE or DMIRE2 microscope (63x/100x PL APO oil objectives). Images were processed with Adobe Photoshop, and adjusted for optimal brightness/contrast.
siRNA Transfection
siRNAs were purchased from Dharmacon and transfected at 2–100nM using Lipofectamine 2000 (Invitrogen) for 24–72h. The following siRNAs were used: Fbxw7 (J-004264–07-10), siControl Pool (D-001810–10), siControl individual siRNAs (D-001810–1-4) and mock transfection.
Acknowledgments
The authors thank Dr Will Hughes for his assistance with live cell microscopy, and the MLC Flow Cytometry Facility for assistance with cell sorting. This research was supported by the National Health and Medical Research Council of Australia (481307, 535903, 427601), the Cancer Institute NSW (07/CDF/1–28, 11/CDF/3–26, 09/RIG/1–18), the Australian Cancer Research Foundation (ACRF Unit for the Molecular Genetics of Cancer), the Petre Foundation, and the RT Hall Trust. C.E.C. is a National Breast Cancer Foundation and Cure Cancer Australia Fellow and is supported by the Young Garvan Foundation. R.L.S. was a NHMRC Senior Principal Research Fellow. E.A.M. is a Cancer Institute NSW Career Development Fellow.
Glossary
Abbreviations:
- HU
hydroxyurea
- PI
propidium iodide
- BrdU
bromodeoxyuridine
- DNA
deoxyribonucleic acid
- CDK
cyclin dependent kinase
- SCFFbw7
Skp1/Cul1/Rbx1/Fbw7 ubiquitin ligase complex
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflicts of interest and do not need to make any financial disclosures.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23409
References
- 1.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–86. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 2.Welcker M, Singer J, Loeb KR, Grim J, Bloecher A, Gurien-West M, et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell. 2003;12:381–92. doi: 10.1016/S1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 3.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–7. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 4.Juan G, Cordon-Cardo C. Intranuclear compartmentalization of cyclin E during the cell cycle: disruption of the nucleoplasm-nucleolar shuttling of cyclin E in bladder cancer. Cancer Res. 2001;61:1220–6. [PubMed] [Google Scholar]
- 5.Ekholm-Reed S, Méndez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–81. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keck JM, Summers MK, Tedesco D, Ekholm-Reed S, Chuang L-C, Jackson PK, et al. Cyclin E overexpression impairs progression through mitosis by inhibiting APC(Cdh1) J Cell Biol. 2007;178:371–85. doi: 10.1083/jcb.200703202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 9.Ekholm-Reed S, Spruck CH, Sangfelt O, van Drogen F, Mueller-Holzner E, Widschwendter M, et al. Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer. Cancer Res. 2004;64:795–800. doi: 10.1158/0008-5472.CAN-03-3417. [DOI] [PubMed] [Google Scholar]
- 10.Minella AC, Loeb KR, Knecht A, Welcker M, Varnum-Finney BJ, Bernstein ID, et al. Cyclin E phosphorylation regulates cell proliferation in hematopoietic and epithelial lineages in vivo. Genes Dev. 2008;22:1677–89. doi: 10.1101/gad.1650208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, et al. Cyclin E ablation in the mouse. Cell. 2003;114:431–43. doi: 10.1016/S0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 12.Gudas JM, Payton M, Thukral S, Chen E, Bass M, Robinson MO, et al. Cyclin E2, a novel G1 cyclin that binds Cdk2 and is aberrantly expressed in human cancers. Mol Cell Biol. 1999;19:612–22. doi: 10.1128/mcb.19.1.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauper N, Beck AR, Cariou S, Richman L, Hofmann K, Reith W, et al. Cyclin E2: a novel CDK2 partner in the late G1 and S phases of the mammalian cell cycle. Oncogene. 1998;17:2637–43. doi: 10.1038/sj.onc.1202477. [DOI] [PubMed] [Google Scholar]
- 14.Zariwala M, Liu J, Xiong Y. Cyclin E2, a novel human G1 cyclin and activating partner of CDK2 and CDK3, is induced by viral oncoproteins. Oncogene. 1998;17:2787–98. doi: 10.1038/sj.onc.1202505. [DOI] [PubMed] [Google Scholar]
- 15.Caldon CE, Musgrove EA. Distinct and redundant functions of cyclin E1 and cyclin E2 in development and cancer. Cell Div. 2010;5:2. doi: 10.1186/1747-1028-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–6. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 17.Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–5. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–45. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klotz K, Cepeda D, Tan Y, Sun D, Sangfelt O, Spruck C. SCF(Fbxw7/hCdc4) targets cyclin E2 for ubiquitin-dependent proteolysis. Exp Cell Res. 2009;315:1832–9. doi: 10.1016/j.yexcr.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 20.Caldon CE, Sergio CM, Kang J, Muthukaruppan A, Boersma MN, Stone A, et al. Cyclin E2 overexpression is associated with endocrine resistance but not insensitivity to CDK2 inhibition in human breast cancer cells. Mol Cancer Ther. 2012;11:1488–99. doi: 10.1158/1535-7163.MCT-11-0963. [DOI] [PubMed] [Google Scholar]
- 21.Gu Z, Inomata K, Ishizawa K, Horii A. The FBXW7 β-form is suppressed in human glioma cells. Biochem Biophys Res Commun. 2007;354:992–8. doi: 10.1016/j.bbrc.2007.01.080. [DOI] [PubMed] [Google Scholar]
- 22.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–43. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 23.Grim JE, Gustafson MP, Hirata RK, Hagar AC, Swanger J, Welcker M, et al. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J Cell Biol. 2008;181:913–20. doi: 10.1083/jcb.200802076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minella AC, Welcker M, Clurman BE. Ras activity regulates cyclin E degradation by the Fbw7 pathway. Proc Natl Acad Sci U S A. 2005;102:9649–54. doi: 10.1073/pnas.0503677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minella AC, Grim JE, Welcker M, Clurman BE. p53 and SCFFbw7 cooperatively restrain cyclin E-associated genome instability. Oncogene. 2007;26:6948–53. doi: 10.1038/sj.onc.1210518. [DOI] [PubMed] [Google Scholar]
- 26.van Drogen F, Sangfelt O, Malyukova A, Matskova L, Yeh E, Means AR, et al. Ubiquitylation of cyclin E requires the sequential function of SCF complexes containing distinct hCdc4 isoforms. Mol Cell. 2006;23:37–48. doi: 10.1016/j.molcel.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Zhang X, Geng L, Teng L, Legerski RJ. Artemis regulates cell cycle recovery from the S phase checkpoint by promoting degradation of cyclin E. J Biol Chem. 2009;284:18236–43. doi: 10.1074/jbc.M109.002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, Jonkers J, et al. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39:759–69. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 29.Sieuwerts AM, Look MP, Meijer-van Gelder ME, Timmermans M, Trapman AM, Garcia RR, et al. Which cyclin E prevails as prognostic marker for breast cancer? Results from a retrospective study involving 635 lymph node-negative breast cancer patients. Clin Cancer Res. 2006;12:3319–28. doi: 10.1158/1078-0432.CCR-06-0225. [DOI] [PubMed] [Google Scholar]
- 30.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 31.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Shao Z-M. Cyclin e expression and prognosis in breast cancer patients: a meta-analysis of published studies. Cancer Invest. 2006;24:581–7. doi: 10.1080/07357900600894799. [DOI] [PubMed] [Google Scholar]
- 34.Liang Y, Gao H, Lin S-Y, Goss JA, Brunicardi FC, Li K. siRNA-based targeting of cyclin E overexpression inhibits breast cancer cell growth and suppresses tumor development in breast cancer mouse model. PLoS One. 2010;5:e12860. doi: 10.1371/journal.pone.0012860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–66. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 36.Galimberti F, Thompson SL, Liu X, Li H, Memoli V, Green SR, et al. Targeting the cyclin E-Cdk-2 complex represses lung cancer growth by triggering anaphase catastrophe. Clin Cancer Res. 2010;16:109–20. doi: 10.1158/1078-0432.CCR-09-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caldon CE, Lee CSL, Sutherland RL, Musgrove EA. Wilms’ tumor protein 1: an early target of progestin regulation in T-47D breast cancer cells that modulates proliferation and differentiation. Oncogene. 2008;27:126–38. doi: 10.1038/sj.onc.1210622. [DOI] [PubMed] [Google Scholar]
- 38.Musgrove EA, Hunter L-JK, Lee CSL, Swarbrick A, Hui R, Sutherland RL. Cyclin D1 overexpression induces progestin resistance in T-47D breast cancer cells despite p27(Kip1) association with cyclin E-Cdk2. J Biol Chem. 2001;276:47675–83. doi: 10.1074/jbc.M106371200. [DOI] [PubMed] [Google Scholar]
- 39.Caldon CE, Swarbrick A, Lee CS, Sutherland RL, Musgrove EA. The helix-loop-helix protein Id1 requires cyclin D1 to promote the proliferation of mammary epithelial cell acini. Cancer Res. 2008;68:3026–36. doi: 10.1158/0008-5472.CAN-07-3079. [DOI] [PubMed] [Google Scholar]
- 40.Prall OWJ, Sarcevic B, Musgrove EA, Watts CKW, Sutherland RL. Estrogen-induced activation of Cdk4 and Cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin-dependent kinase inhibitor association with cyclin E-Cdk2. J Biol Chem. 1997;272:10882–94. doi: 10.1074/jbc.272.16.10882. [DOI] [PubMed] [Google Scholar]
- 41.Prall OWJ, Rogan EM, Musgrove EA, Watts CKW, Sutherland RL. c-Myc or cyclin D1 mimics estrogen effects on cyclin E-Cdk2 activation and cell cycle reentry. Mol Cell Biol. 1998;18:4499–508. doi: 10.1128/mcb.18.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caldon CE, Sergio CM, Schütte J, Boersma MN, Sutherland RL, Carroll JS, et al. Estrogen regulation of cyclin E2 requires cyclin D1 but not c-Myc. Mol Cell Biol. 2009;29:4623–39. doi: 10.1128/MCB.00269-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juan G, Darzynkiewicz Z. Bivariate analysis of DNA content and expression of cyclin proteins. Current protocols in cytometry. Hoboken, NJ, USA: John Wiley & Sons, 2007. [DOI] [PubMed] [Google Scholar]
- 44.FlowJo. Ashland, OR, USA: Tree Star Inc., 2008. [Google Scholar]
- 45.Flow Cytometry. Principles and Applications. Totowa, New Jersey: Humana Press, 2007. [Google Scholar]
- 46.Ormerod MG. Flow Cytometry - A Basic Introduction. Los Angeles, CA: De Novo Software, 2008. [Google Scholar]
- 47.Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A. 2010;107:139–44. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam N, Sandberg ML, Sugden B. High physiological levels of LMP1 result in phosphorylation of eIF2 α in Epstein-Barr virus-infected cells. J Virol. 2004;78:1657–64. doi: 10.1128/JVI.78.4.1657-1664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]