Abstract
Previous work has established that heterogeneous nuclear ribonucleoprotein K (hnRNP K) is stabilized in an ATM-dependent manner in response to DNA damage and acts as a cofactor for p53-mediated transcription. Here, we show that in response to DNA damage caused by ionizing radiation, hnRNP K is phosphorylated in an ATM-dependent manner. Furthermore, our data indicate that ATM-dependent hnRNP K phosphorylation is required for its stabilization and its function as a p53 transcriptional cofactor in response to DNA damage. These findings thereby establish hnRNP K as an ATM target and help define how ATM orchestrates p53-dependent transcriptional responses in response to genotoxic stress.
Keywords: DNA damage, ATM, phosphorylation, transcription, p53
Introduction
Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is an evolutionarily conserved factor, found in the nucleus and cytoplasm, that was initially discovered as a component of hnRNP complexes in mammalian cells.1 hnRNP K is a member of the family of heterogeneous nuclear ribonucleoproteins and is predominantly present in the nucleus but shuttles between the nucleus and the cytoplasm via its K nuclear shuttling (KNS) domain.1 hnRNP K has been implicated in several cellular processes, such as chromatin remodeling, transcription and pre-mRNA splicing.2 The involvement of hnRNP K in these events appears to reflect its ability to interact with a range of molecular partners, including DNA, RNA, protein kinases and proteins involved in chromatin remodelling.2,3 Although the available data suggest that these effects reflect the ability of hnRNP K to bind with high affinity to single-stranded DNA (ssDNA),4,5 the precise mechanisms by which hnRNP K regulates transcription and the ways in which hnRNP K is itself regulated remain largely obscure. hnRNP K is phosphorylated on multiple residues, and the extent of its phosphorylation is modulated by a multitude of stimuli, including interleukin-1 and insulin exposure as well as by oxidative stress.6,7 Phospho-amino acid and western immunoblotting analyses have revealed that hnRNP K is phosphorylated on serine, threonine and tyrosine residues,6,8 and biochemical studies have indicated that hnRNP K phosphorylation regulates its interactions with RNA and DNA targets as well as with protein partners.6,9,10
DNA damage triggers a response pathway that, among other things, leads to arrest or slowing of cell cycle progression and the transcriptional induction of various genes that facilitate DNA repair.11,12 Defects in this DNA damage response (DDR) pathway can result in genomic instability, with ensuing mutagenesis frequently contributing to cancer predisposition.13 Key regulators of DDR events in mammalian cells are the PIKK (phosphoinositol 3-kinase like) family protein kinases, ATM (ataxia telangiectasia mutated) and ATR (ATM- and Rad3-related), the protein kinases CHK1 and CHK2, and the tumor suppressor protein p53.14,15 In response to DNA damage, ATM is activated and phosphorylates many checkpoint-determining and regulatory proteins, such as p53, CHK2 and BRCA1, highlighting its central position in DNA damage-activated signaling pathways.14,15
Through conducting differential proteomics screens, we previously identified hnRNP K as a factor whose levels increase in response to DNA damage in an ATM-dependent manner.16 Furthermore, we established that hnRNP K is a target for the p53 regulator HDM2 and acts as a p53 transcriptional cofactor in response to DNA damage.16,17 In this present study, we show that hnRNP K is phosphorylated in an ATM-dependent manner in response to ionizing radiation (IR), and that ATM-dependent phosphorylation of hnRNP K is required for this and for its role as a p53 transcriptional co-activator.
Results
hnRNP K is phosphorylated in an ATM-dependent manner
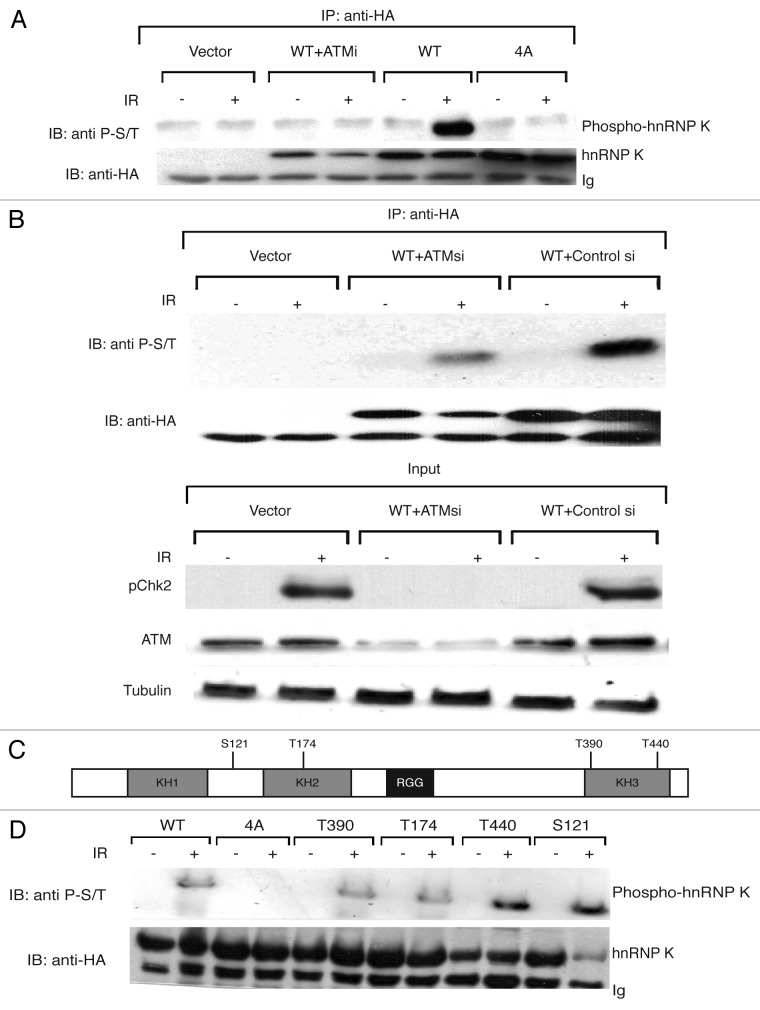
As we had already established that hnRNP K is stabilized in response to DNA damage in an ATM-dependent manner,16 we sought to determine whether hnRNP K is phosphorylated in response to DNA damage. As an approach to do this, we used an antibody that recognizes certain substrates phosphorylated on the ATM/ATR Ser/Thr-Gln (S/TQ) consensus target motif (see “Materials and Methods”). Significantly, this antibody recognized HA-epitope tagged wild-type hnRNP K derived from human U2OS cells that had been IR treated but did not detect this protein when it was derived from non-irradiated cells (Fig. 1A, lanes 6 and 5, respectively). Furthermore, this IR-induced hnRNP K phosphorylation required ATM kinase activity, as it was prevented by the specific ATM inhibitor KU-55933 (ATMi)18 (Fig. 1A, lanes 3 and 4). In line with this, short-interfering RNA (siRNA)-mediated depletion of ATM markedly reduced the detection of IR-induced hnRNP K phosphorylation (Fig. 1B, top panel; see lower panels for demonstrations of ATM depletion and effects on IR-induced, ATM-mediated CHK2 Thr-68 phosphorylation). Collectively, these findings thereby established that hnRNP K is phosphorylated in response to IR in an ATM-dependent manner.
Figure 1. ATM-dependent phosphorylation of hnRNP K. (A) ATM-mediated phosphorylation of hnRNP K was assessed in U2OS cells with a phospho-ATM/ATR substrate antibody (see “Materials and Methods”). U2OS cells were transfected with an empty vector (Vector), a vector expressing wild-type HA-hnRNP K (WT), a vector expressing the Ser/Thr→Ala mutant HA-hnRNP K (4A) or vector expressing wild-type HA-hnRNP K in the presence of ATM inhibitor (WT +ATMi). Cells were untreated or treated with IR (15 Gy) for 1 h, and immunoprecipitation was performed with HA antibody followed by western blotting with phospho-ATM/ATR substrate antibody. Western blotting with the HA antibody was performed to assess transfection efficiency. (B) ATM-mediated phosphorylation of hnRNP K was assessed as in (A) in U2OS cells transfected with the WT HA-hnRNP K and treated with either GFP siRNA (WT+Control si) or ATM siRNA (WT+ATMsi). A parental plasmid vector (Vector) was used as a control. Chk2 phosphorylation was assessed with a Chk2 phospho Thr-68 specific antibody to monitor ATM function, while an ATM antibody was used to check efficiency of siRNA depletion. Anti-tubulin antibody (Tubulin) was used as a loading control. (C) Schematic of the domain organization of hnRNP K with the locations of the four SQ/TQ motifs described in the text (see Fig. S1 for these in the context of the hnRNP K sequence). (D) As in (A), except that additional hnRNP K constructs were also employed in which Thr-390, Thr-174, Thr-440 or Ser-121 were individually regenerated in the 4A mutant background.
Analysis of the hnRNP K primary amino acid sequence identified four consensus S/TQ motifs as potential ATM target sites (Fig. 1C; Fig. S1). To investigate whether these sites were required for DNA damage-dependent hnRNP K phosphorylation, we generated an hnRNP K derivative in which all four Ser/Thr residues were mutated to non-phosphorylatable Ala residues (4A). Notably, in contrast to wild-type hnRNP K, the phospho-specific antibody no longer detected the mutant hnRNP K (Fig. 1A, compare lane 6 with lane 8). To assess which of the sites contribute to hnRNP K phosphorylation, we generated four 4S/T→A derivatives in which each potential phospho-site was individually mutated back to the wild-type hnRNP K sequence (i.e., one Ser/Thr-Gln was restored to wild-type sequence, and the other three sites were as Ala-Gln). As shown in Figure 1D, IR treatment induced phosphorylation of each of the four proteins, thus implying that all four sites can serve as ATM targets.
ATM-mediated phosphorylation promotes IR-induced hnRNP K stabilization
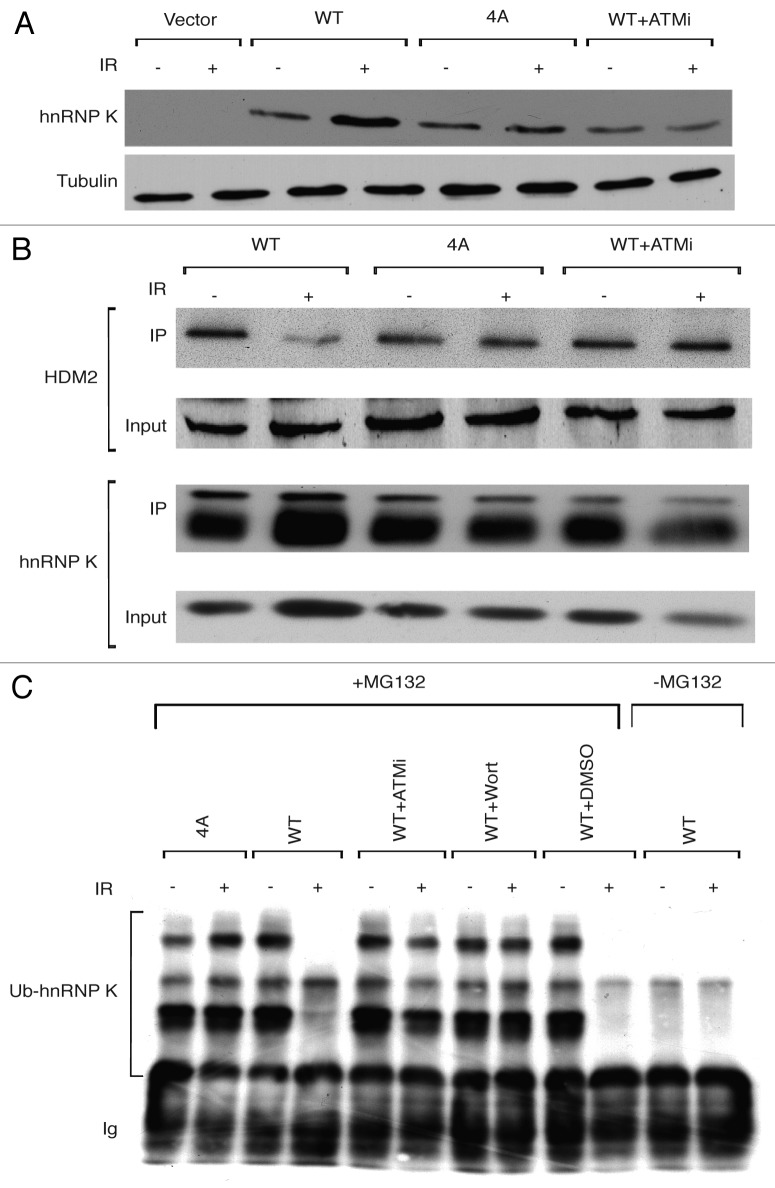
To investigate whether ATM-dependent hnRNP K phosphorylation is required for hnRNP K stabilization in response to DNA damage, we tested, by co-transfection assays, whether the IR-induced stabilization of hnRNP K is prevented by the mutations described above. Thus, we found that, while wild-type hnRNP K was stabilized following DNA damage induction, no stabilization was observed for the 4A mutant (Fig. 2A, note that stabilization of wild-type hnRNP K was prevented when cells were incubated in the presence of ATM inhibitor). We had previously shown that hnRNP K binds to and is negatively regulated by the HDM2 E3 ubiquitin ligase, which targets hnRNP K for ubiquitylation and proteosomal degradation. Moreover, we established that DNA damage causes dissociation of the HDM2-hnRNP K complex, thus stabilizing hnRNP K.16 To determine whether IR-induced dissociation of hnRNP K from HDM2 is regulated by hnRNP K phosphorylation, we assessed the binding of wild-type and 4A mutant hnRNP K proteins to HDM2 in co-immunoprecipitation assays. This revealed that, while the interaction between wild-type hnRNP K and HDM2 was reduced in response to DNA damage, this was not the case for the 4A mutant hnRNP K protein, which bound HDM2 equally well in the presence or absence of DNA damage (Fig. 2B). In line with these findings, dissociation of wild-type hnRNP K from HDM2 was prevented when cells were cultured in the presence of an ATM inhibitor (Fig. 2B).
Figure 2. ATM-dependent phosphorylation of hnRNP K controls its stabilization, its HDM2-mediated ubiquitylation and its interaction with HDM2. (A) Stabilization of hnRNP K was assessed by western blotting (with monoclonal anti-HA antibody) in U2OS cells transfected with an empty plasmid vector (Vector), a vector expressing wild type HA-hnRNP K (WT), a vector expressing the quadruple Ser/Thr→Ala mutant HA-hnRNP K (4A) or the vector expressing the wild-type HA-hnRNP K in the presence of 10 μM ATM inhibitor KU-55933 (WT +ATMi). In all cases, cells were untreated (-) or treated (+) with IR (15 Gy) for 1 h. An anti-tubulin antibody was used as loading control. (B) hnRNP K-HDM2 interaction was analyzed by IP in U2OS cells transfected as above and left untreated or treated with IR (15 Gy) for 1 h. Immunoprecipitation was with anti-HA antibody followed by western blotting with HDM2 antibody or HA antibody (hnRNP K). Inputs for both proteins are shown and represent 5% of the material used. (C) Ubiquitylation of hnRNP K was analyzed in U2OS cells transfected as above, treated or not with IR (15 Gy). Where indicated, cells were treated with the proteasome inhibitor MG132 (30 mM) for 3 h prior to IR treatment. Where indicated, U2OS cells transfected with vector expressing WT HA-hnRNP K were treated with 10 μM of ATM inhibitor (ATMi) or wortmannin (10 μM) or DMSO. Cell extracts were analyzed by immunoprecipitation with anti-HA antibody followed by western blotting with the same antibody. Bands representing ubiquitylated forms of hnRNP K (Ub-hnRNP K) and the immunoglobulin light-chain (Ig) are indicated.
To assess whether the differential behaviors of the wild-type and 4A hnRNP K proteins reflected effects on HDM2-mediated hnRNP K ubiquitylation, we mock-treated or IR-treated cells in the presence of the proteasome inhibitor MG-132 and then tested for the presence of ubiquitylated forms of hnRNP by immunoprecipitation and western blotting. Strikingly, in contrast to wild-type hnRNP K, whose ubiquitylation was abrogated upon IR treatment, the ubiquitylation status of the hnRNP K 4A mutant persisted after DNA damage induction (Fig. 2C, note that the effect of DNA damage on ubiquitylation of wild-type hnRNP K was prevented when cells were cultured in the presence of the ATM inhibitors KU-55933 or wortmannin). Together, these results support a model in which DNA damage triggers ATM-dependent hnRNP K phosphorylation and dissociation of hnRNP K from HDM2, thereby impairing HDM2-mediated hnRNP ubiquitylation and resulting in hnRNP K stabilization.
ATM-mediated hnRNP K phosphorylation promotes p53-directed transcription
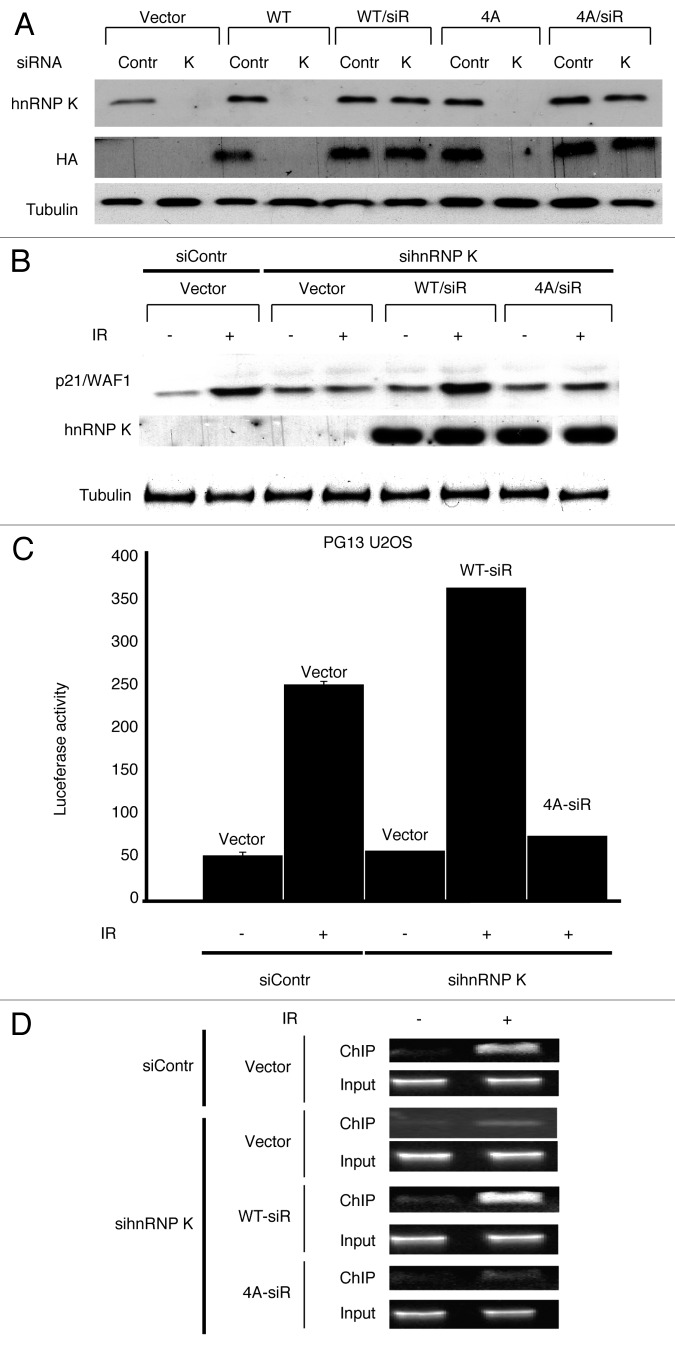
As we had previously shown that hnRNP K stabilization promotes p53-dependent transcription,16 the above findings suggested that mutating ATM target sites in hnRNP K might impair DNA damage-induced, p53-dependent transcription. To test this idea, we engineered vectors expressing HA-hnRNP K (WT or 4A) that were resistant to the hnRNP K-targeting siRNA and then expressed these in cells. Consequently, ensuing siRNA treatment led to depletion of endogenous hnRNP K (and of non siRNA-resistant 4A hnRNP K), but not the siRNA-resistant constructs (Fig. 3A). By using these vectors, we assessed the effect of the 4A mutant hnRNP K on p53-dependent expression of the p21/WAF1 protein following DNA damage induction. Strikingly, while p21 expression was normally induced in hnRNP K siRNA-treated cells transfected with the siRNA-resistant WT hnRNP K (WT/siR), siRNA-treated cells transfected with parental vector or the siRNA-resistant 4A mutant hnRNP K construct (4A/siR) failed to exhibit DNA damage-induced p21 expression (Fig. 3B).
Figure 3. ATM-dependent phosphorylation of hnRNP K promotes p53 transcriptional activity following IR. (A) Expression from hnRNP K constructs was assessed by western blotting with the indicated antibodies in cells treated or not with control siRNA (Contr) or hnRNP K siRNA (K). As indicated, these cells also contained control parental vector (Vector), or constructs expressing wild-type hnRNP (WT), siRNA resistant wild-type hnRNP K (WT/siR), 4A mutant hnRN K (4A) or siRNA resistant 4A hnRNP K (4A/siR). Anti-tubulin antibody was used as loading control. (B) p53-dependent induction of p21/WAF1 protein was assessed 6 h following IR by western blotting in cells treated with the indicated siRNAs and containing the indicated constructs. (C) U2OS cells stably transfected with a luciferase reporter gene under the control of multiple p53 binding sites (PG13 U2OS) were transfected with the indicated constructs and then IR (15 Gy) treated or mock IR treated, as indicated. Luciferase activity was measured 18 h afterwards. (D) U2OS cells were transfected with the indicated constructs, treated with control of hnRNP K siRNAs and 48 h later, they were mock treated or exposed to IR (15 Gy) and incubated for 2 h. Samples were then subjected to ChIP with anti-p53 antibody, and precipitated DNA was subjected to PCR with primers covering the p53-response elements of the p21/WAF1 promoter.
To determine whether the above findings reflected effects on p21/WAF1 transcription, we performed experiments with a human U2OS cell line containing a stably integrated luciferase construct under the control of the synthetic p53-responsive PG13 promoter.19 Thus, we found that luciferase expression was induced by DNA damage in control siRNA-treated cells (siContr) and hnRNP K siRNA-treated cells complemented with siRNA-resistant WT hnRNP K (Fig. 3C). By contrast, almost no induction was apparent in hnRNP K siRNA-treated cells containing the siRNA-resistant 4A mutant hnRNP K construct (Fig. 3C). Furthermore, since we had previously established that hnRNP K is required for efficient p53 recruitment to p53-responsive promoters,16 we also performed chromatin immunoprecipitation (ChIP) with a p53 antibody to assess whether mutation of the ATM target sites in hnRNP K affected recruitment of p53 to the p21/WAF1 promoter. As shown in Figure 3D, efficient p53 recruitment to the p21 promoter was observed in both control siRNA-treated cells and hnRNP K siRNA-treated cells complemented with siRNA-resistant wild-type hnRNP K. By contrast, promoter binding by p53 was severely compromised in hnRNP K siRNA-treated cells containing parental vector or expressing the siRNA-resistant 4A mutant hnRNP K. Taken together, these results thereby demonstrated that ATM-dependent hnRNP K phosphorylation is needed for efficient, regulated binding of p53 to the p21 promoter and ensuing transcriptional induction of this gene in response to DNA damage.
Discussion
Previous work has shown that hnRNP K is stabilized in an ATM-dependent manner and is required for effective p53 transcription following DNA damage16,17 and has indicated that such functions are controlled by hnRNP K, being arginine methylated and sumoylated.20,21 In this study, we have established that hnRNP K is phosphorylated on ATM consensus SQ/TQ target motifs in response to DNA damage in an ATM-dependent manner. Moreover, we have found that mutation of these four sites to prevent SQ/TQ phosphorylation has profound effects on hnRNP-mediated responses to DNA damage. Thus, we have shown that this 4 Ser/Thr→Ala (4A) mutation largely prevents hnRNP K stabilization in response to DNA damage and prevents DNA damage-induced dissociation of hnRNP K from the ubiquitin E3 ligase HDM2. In accord with these findings, we found that the normal reduction of hnRNP K ubiquitylation in response to DNA damage is prevented in the context of the 4A mutant. Most importantly, we have connected these observations to hnRNP K function by showing that the 4A mutant hnRNP K protein no longer promotes p53- and DNA damage-dependent induction of transcription from the p21/WAF1 gene promoter and also does not foster DNA damage-induced binding of p53 to the p21/WAF1 promoter as assessed by chromatin immunoprecipitation assays.
These findings therefore support a model in which the relatively abundant protein hnRNP K is present in various pools, with one pool possessing p53 transcriptional co-activator function. Moreover, the data indicated that this pool of hnRNP K, like p53 itself, is normally subject to rapid turnover, with its synthesis being counteracted by HDM2-mediated ubiquitylation and ensuing proteasomal degradation. Strikingly, we have found that, also like p53 itself, hnRNP K is phosphorylated by ATM in a manner that leads to hnRNP K dissociation from HDM2, thereby stabilizing the protein and allowing it to potentiate p53-mediated transcription of p21/WAF1 and likely also certain other p53 target genes. We speculate that, by targeting multiple proteins that impact on p53-dependent transcription—p53 itself, HDM2/Mdm2, hnRNP K and other p53 regulators such as HDMX22—ATM is able to achieve higher and more robust levels of regulatory control than would be possible if it were to target fewer components, or just p53 itself.
In future studies, it will clearly be interesting to explore precisely how hnRNP K interacts with HDM2, and whether its ATM-mediated phosphorylations directly induce dissociation of hnRNP K from HDM2, or whether, as is the case for p53, ATM-mediated control of hnRNP K is also associated with phosphorylation events effected by additional kinases and/or with other post-translational modifications. In this regard, we note that arginine methylation of hnRNP K has been found to potentiate p53 transcriptional activity,20 and that hnRNP K is also subject to other phosphorylations that alter hnRNP K functionality.8-10 Indeed, the cell cycle-regulated kinase Aurora A has been shown to phosphorylate hnRNP K Ser-379 in a manner that disrupts its interactions with p53.23 Moreover, it was recently established that hnRNP K Lys-422 is subject to DNA damage-induced sumoylation by the SUMO E3 ligase CBX4, and that this stimulates p53-dependent transcriptional induction of p21/WAF1.21 It is therefore tempting to speculate that, by being targeted by multiple kinases and other protein-modifying enzymes, as well as through it acting in various pathways of RNA metabolism, hnRNP K may serve to integrate DNA damage induced, p53-dependent transcriptional programs with other aspects of cell function and physiology. Finally, we note that hnRNP K was recently found to bind to the p53-induced large intergenic noncoding RNA, lincRNA-p21 and to participate with lincRNA-p21 in p53-mediated transcriptional repression programs.24 It will be interesting to ascertain whether this function for hnRNP K, like its transcriptional co-activator functions, is affected by ATM-mediated phosphorylation events on the hnRNP K protein that we have described.
Materials and Methods
Cell culture, expression vectors and transfection
Standard conditions and procedures were used for culturing mammalian cells.25 Transfections were done with calcium phosphate and cells were harvested 48 h afterwards. U2OS cells were from Cancer Research UK. HA-hnRNP K constructs were obtained from Z.A. Ronai (Sanford-Burnham Medical Research Institute).
Site-directed mutagenesis
Site-directed mutagenesis was performed by the Dpn I method, based on Stratagene's QuickChange specifications. Primers used to mutate Ser-121 to Ala were as follows: 5′-catcacccactgcaaccGCccagctcccgctcg -3′ and 5′-cgagcgggagctggGCggttgcagtgggtgatg -3′. Primers to mutate Thr-174 to Ala were: 5′-gaacttcgagagaacGctcaaaccaccatc -3′ and 5′-gatggtggtttgagCgttctctcgaagttc-3′. Primers used to mutate Thr-390 to Ala were: 5′-ggacctattattactGcacaagtaactattc-3′ and 5′-gaatagttacttgtgCagtaataataggtcc-3′.
Primers used to mutate Thr-440 to Ala were: 5′-cattaccattacaggaGcacaggaccagatac-3′ and 5′-gtatctggtcctgtgCtcctgtaatggtaatg-3′. Primers used to generate siRNA resistant hnRNP K were: 5′-aaCatCaaAgcCctAcgCacCgactacaatgccagtgtttc-3′ and 5′-GgtGcgTagGgcTttGatGttcttgcctccttttccaatc-3′. Mutations were verified by sequencing the whole hnRNP K cDNA for each mutation.
Irradiation and cell extracts
Cells were treated with γ-irradiation at 50–70% confluency in 10 ml of medium in a 10-cm Petri dish. After recovery, cells were lysed in SDS-sample buffer without bromophenol blue and equal amounts of protein analyzed by western blotting.
Antibodies, western immunoblotting and immunoprecipitation
Procedures were based on those used previously.16 Anti-p53, Chk2 (phospho-Thr68) antibodies and phospho-ATM/ATR substrate antibody (Number 6966) were from Cell Signaling Technology, Inc.; HA antibody was from Covance; β-tubulin and ATM antibodies were from Abcam; and p21 and hnRNP K antibodies were from Santa Cruz Biotechnology. Cell extracts were resolved by 10% SDS-PAGE, transferred onto nitrocellulose and blotted by standard procedures. For immunoprecipitation, cells were washed with PBS and lysed on ice in RIPA buffer supplemented with protease inhibitors. Lysates were sheared by pipetting repeatedly through a needle and cleared by centrifugation. Extracts were pre-cleared with 50 ml of Protein-A Sepharose (ThermoFisher Scientific) for 2 h at 4°C, incubated for 2 h with antibody and then overnight with 50 ml of Protein-A Sepharose. Beads were washed with RIPA buffer three times and bound proteins recovered by boiling in SDS-sample buffer. For immunoprecipitations to detect phospho-hnRNP K with the phospho-ATM/ATR substrate antibody, extracts were prepared by resuspending washed cell pellets in lysis buffer (50 mM TRIS-HCl pH 7.4, 150 mM NaCl, 1% Tween-20 detergent, 1 mM dithiothreitol) plus protease inhibitors and phosphatase inhibitors (20 mM β-glycerol phosphate and 50 nM microcystin). Samples were incubated on ice for 15 min and then clarified by centrifugation. HA-tagged recombinant proteins were immunoprecipitated from cell extracts with 4 μg of HA antibody.
siRNA design and transfection
RNA duplexes of 21 nucleotides targeting the human hnRNP K or ATM mRNAs were designed, chemically synthesized and supplied in the 2'-deprotected and desalted form by Dharmacon. Oligonucleotide sequences for hnRNP K and ATM siRNA have been previously described.16,26 In each case, the sequence was subjected to a BLAST search to ensure that the siRNA was specific to the targeted gene. U2OS cells were grown to 20–50% confluency and Oligofectamine mediated transient transfection of siRNA was done in 60 mm plates. siRNA (75 mM) and 7 ml of Oligofectamine were mixed and incubated for 20 min at room temperature and added to each plate in DMEM containing 5% serum. After 24 h the medium was changed to DMEM supplemented with 10% FBS, and cells were left in culture for an additional 24 h to bring about downregulation. GFP siRNA was used as a control.
Luciferase expression analyses
U2OS cells stably transfected with an artificial p53 binding site repeat (PG13; 13 copies of GGACGGACCTGACCGGACC)19; cloned upstream of the luciferase coding sequence in a pGL3-basic vector (Promega) were mock treated or treated with IR and allowed to recover. Cells were then harvested and luciferase activity measured according to the manufacturer’s protocol (Promega).
Chromatin immunoprecipitation (ChIP)
ChIP was done with U2OS cells as previously described.27 Primers used for PCR of p21 and GAPDH were as described previously.28,29
Supplementary Material
Acknowledgments
We thank all members of the Jackson laboratory for help and support. Research in the Jackson laboratory is funded by Cancer Research UK program grant C6/A11224, the European Research Council and the European Community Seventh Framework Programme grant agreement no. HEALTH-F2-2010-259893 (DDResponse). Core infrastructure funding was provided by CRUK (C6946/A14492) and the Wellcome Trust (WT092096). S.P.J. receives his salary from the University of Cambridge, supplemented by CRUK.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/23592
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23592
References
- 1.Matunis MJ, Michael WM, Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol. 1992;12:164–71. doi: 10.1128/mcb.12.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–38. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 3.Bomsztyk K, Van Seuningen I, Suzuki H, Denisenko O, Ostrowski J. Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 1997;403:113–5. doi: 10.1016/S0014-5793(97)00041-0. [DOI] [PubMed] [Google Scholar]
- 4.Tomonaga T, Levens D. Activating transcription from single stranded DNA. Proc Natl Acad Sci U S A. 1996;93:5830–5. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomonaga T, Levens D. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J Biol Chem. 1995;270:4875–81. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- 6.Ostrowski J, Kawata Y, Schullery DS, Denisenko ON, Higaki Y, Abrass CK, et al. Insulin alters heterogeneous nuclear ribonucleoprotein K protein binding to DNA and RNA. Proc Natl Acad Sci U S A. 2001;98:9044–9. doi: 10.1073/pnas.161284098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Seuningen I, Ostrowski J, Bustelo XR, Sleath PR, Bomsztyk K. The K protein domain that recruits the interleukin 1-responsive K protein kinase lies adjacent to a cluster of c-Src and Vav SH3-binding sites. Implications that K protein acts as a docking platform. J Biol Chem. 1995;270:26976–85. doi: 10.1074/jbc.270.45.26976. [DOI] [PubMed] [Google Scholar]
- 8.Mikula M, Karczmarski J, Dzwonek A, Rubel T, Hennig E, Dadlez M, et al. Casein kinases phosphorylate multiple residues spanning the entire hnRNP K length. Biochim Biophys Acta. 2006;1764:299–306. doi: 10.1016/j.bbapap.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, et al. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol Cell Biol. 2002;22:4535–43. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schullery DS, Ostrowski J, Denisenko ON, Stempka L, Shnyreva M, Suzuki H, et al. Regulated interaction of protein kinase Cdelta with the heterogeneous nuclear ribonucleoprotein K protein. J Biol Chem. 1999;274:15101–9. doi: 10.1074/jbc.274.21.15101. [DOI] [PubMed] [Google Scholar]
- 11.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–51. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 13.Jackson SP, Bartek J. The DNA damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–45. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 16.Moumen A, Masterson P, O’Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–78. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 17.Enge M, Bao W, Hedström E, Jackson SP, Moumen A, Selivanova G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell. 2009;15:171–83. doi: 10.1016/j.ccr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 18.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 19.Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–30. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Zhou X, Liu N, Wang C, Zhang L, Mo W, et al. Arginine methylation of hnRNP K enhances p53 transcriptional activity. FEBS Lett. 2008;582:1761–5. doi: 10.1016/j.febslet.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 21.Pelisch F, Pozzi B, Risso G, Muñoz MJ, Srebrow A. DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation. J Biol Chem. 2012;287:30789–99. doi: 10.1074/jbc.M112.390120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A. 2005;102:5056–61. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsueh KW, Fu SL, Huang CY, Lin CH. Aurora A phosphorylates hnRNPK and disrupts its interaction with p53. FEBS Lett. 2011;585:2671–5. doi: 10.1016/j.febslet.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Second Edition. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1989 [Google Scholar]
- 26.Biton S, Dar I, Mittelman L, Pereg Y, Barzilai A, Shiloh Y. Nuclear ataxia-telangiectasia mutated (ATM) mediates the cellular response to DNA double strand breaks in human neuron-like cells. J Biol Chem. 2006;281:17482–91. doi: 10.1074/jbc.M601895200. [DOI] [PubMed] [Google Scholar]
- 27.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–27. doi: 10.1016/S1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 28.Zeng SX, Dai MS, Keller DM, Lu H. SSRP1 functions as a co-activator of the transcriptional activator p63. EMBO J. 2002;21:5487–97. doi: 10.1093/emboj/cdf540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinney K, Mattia M, Gottifredi V, Prives C. p53 linear diffusion along DNA requires its C terminus. Mol Cell. 2004;16:413–24. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.