Abstract
The CCR4-NOT complex plays a crucial role in post-transcriptional mRNA regulation in eukaryotes. This complex catalyzes the removal of mRNA poly(A) tails, thereby repressing translation and committing an mRNA to degradation. The conserved core of the complex is assembled by the interaction of at least two modules: the NOT module, which minimally consists of NOT1, NOT2 and NOT3, and a catalytic module comprising two deadenylases, CCR4 and POP2/CAF1. Additional complex subunits include CAF40 and two newly identified human subunits, NOT10 and C2orf29. The role of the NOT10 and C2orf29 subunits and how they are integrated into the complex are unknown. Here, we show that the Drosophila melanogaster NOT10 and C2orf29 orthologs form a complex that interacts with the N-terminal domain of NOT1 through C2orf29. These interactions are conserved in human cells, indicating that NOT10 and C2orf29 define a conserved module of the CCR4-NOT complex. We further investigated the assembly of the D. melanogaster CCR4-NOT complex, and demonstrate that the conserved armadillo repeat domain of CAF40 interacts with a region of NOT1, comprising a domain of unknown function, DUF3819. Using tethering assays, we show that each subunit of the CCR4-NOT complex causes translational repression of an unadenylated mRNA reporter and deadenylation and degradation of a polyadenylated reporter. Therefore, the recruitment of a single subunit of the complex to an mRNA target induces the assembly of the complete CCR4-NOT complex, resulting in a similar regulatory outcome.
Keywords: deadenylation, mRNA decay, CCR4-NOT
Introduction
In eukaryotes, the removal of mRNA poly(A) tails by deadenylases, a process known as deadenylation, represses translation and generally commits the mRNA to degradation. Therefore, deadenylation provides a major mechanism for the post-transcriptional regulation of mRNA expression.1 Eukaryotic mRNAs are deadenylated by the consecutive action of two cytoplasmic deadenylase complexes.2,3 The PAN2-PAN3 complex is involved in the early phase of deadenylation and shortens mRNA poly(A) tails in a distributive manner.4,5 The second, more rapid phase of deadenylation is catalyzed by the CCR4-NOT complex.5,6 The CCR4-NOT complex is sufficient for mRNA deadenylation in the absence of the PAN2-PAN3 complex.5-7
In addition to its general role in bulk mRNA deadenylation, the CCR4-NOT complex and associated proteins have been implicated in a broad range of biological processes, including transcription initiation and elongation, ubiquitination and protein modification.3,8 Furthermore, this complex plays a key role in the post-transcriptional regulation of specific mRNAs, to which it is recruited via interactions with sequence-specific RNA-binding proteins. These RNA-binding proteins include Pumilio, Nanos, Bicaudal-C and Smaug, which regulate the temporal and spatial expression of mRNA targets during Drosophila melanogaster (Dm) oogenesis and embryogenesis.1,8 Recently, it was shown that GW182 family proteins, which are required for miRNA-mediated silencing in animal cells, directly interact with NOT1 and recruit the CCR4-NOT complex to miRNA targets.9-11
The conserved core of the CCR4-NOT complex consists of at least five subunits: NOT1, NOT2, NOT3 and two catalytically active subunits, CCR4a (or its paralog CCR4b) and POP2 (or its paralog CAF1).3,8 The catalytic subunits interact to form a catalytic module, which is recruited to the complex through interactions between POP2/CAF1 and a central MIF4G (middle domain of eukaryotic initiation factor 4G) domain in NOT1.12-18 Additional complex subunits include CAF40 (also known as NOT9, Rcd1 or RQCD1), CAF130, NOT4, NOT5, NOT10, C2orf29 and TAB182.3,8 CAF130 is a yeast-specific subunit with no metazoan counterpart.8 Although NOT4 is conserved and is an integral yeast CCR4-NOT complex subunit, it is not stably associated with the D. melanogaster (Dm) and human complexes.17,18 NOT5 is a yeast NOT3 paralog; however, in contrast with yeast, there is only one gene that encodes a NOT3/NOT5 ortholog in metazoans, termed NOT3 (or NOT3/5).8 Finally, NOT10, C2orf29 and TAB182 were identified as subunits of the human CCR4-NOT complex, and these factors lack orthologs in yeast.18-20 Unlike TAB182, NOT10 and C2orf29 are conserved in metazoans, and NOT10 copurify with the Dm CCR4-NOT complex.17 These observations point to differences in the composition of the CCR4-NOT complexes across species.
Studies of the interaction between the subunits of the CCR4-NOT complex have indicated that NOT1 acts as a scaffold for the assembly of the complex, providing binding sites for both the catalytic module and the CAF40, CAF130 and NOT2-NOT3/5 subunits.18,21-28 However, how the NOT10 and C2orf29 subunits are incorporated into the CCR4-NOT complex and the precise function of these subunits are unknown.
The differences in the composition of the CCR4-NOT complexes in yeast and metazoans and the role of the metazoan CCR4-NOT complex in post-transcriptional regulatory mechanisms such as the miRNA pathway, which has no counterpart in fungi, highlight the importance of studying the assembly and function of this complex in multicellular eukaryotes. In this study, we characterized the assembly of the CCR4-NOT complex in D. melanogaster cells. We confirmed that the C-terminal regions of NOT2 and NOT3, which contain a highly conserved NOT-box domain, interact and dock onto the NOT1 C-terminal domain. We also defined the CAF40-binding site on NOT1 and demonstrated that it overlaps with a domain of unknown function (DUF3819) that is located between the binding sites for the catalytic module and the NOT2-NOT3 module. We further show that the Dm CNOT10 and C2orf29 orthologs CG18616 and CG13567, respectively, interact thereby defining a new module of the CCR4–NOT complex. This module is recruited to the CCR4-NOT complex via an interaction with the NOT1 N-terminal domain and C2orf29. Similar results were obtained for the human CNOT10 and C2orf29 proteins in human cells. Finally, our functional studies demonstrate that all subunits in the CCR4-NOT complex trigger the degradation of a polyadenylated reporter in tethering assays and repress translation when tethered to a reporter lacking a poly(A) tail. These observations indicate that each subunit has the ability to recruit the remaining subunits of the complex to an RNA target, repressing its expression through a common mechanism.
Results
Assembly of the catalytic module of the Dm CCR4-NOT complex
To elucidate the assembly of the CCR4-NOT complex in metazoans, we investigated the interactions between the subunits of the CCR4-NOT complex in D. melanogaster Schneider cells (S2 cells). CCR4 consists of an N-terminal leucine-rich repeat (LRR) domain and a C-terminal catalytic domain (CCR4-C), and belongs to the endonuclease-exonuclease-phosphatase (EEP) enzyme family (Fig. 1A).29,30 Yeast CCR4 has been shown to interact with POP2 through its LRR domain.12-15,21,23 POP2 is a one-domain protein that adopts an RNase D-like fold and belongs to the DEDD nuclease family (Fig. 1A).15.16,31,32 POP2 interacts with NOT1 and the CCR4 LRR domain, thereby bridging the interaction between CCR4 and NOT1.12-16,18,21-23,26,27,33
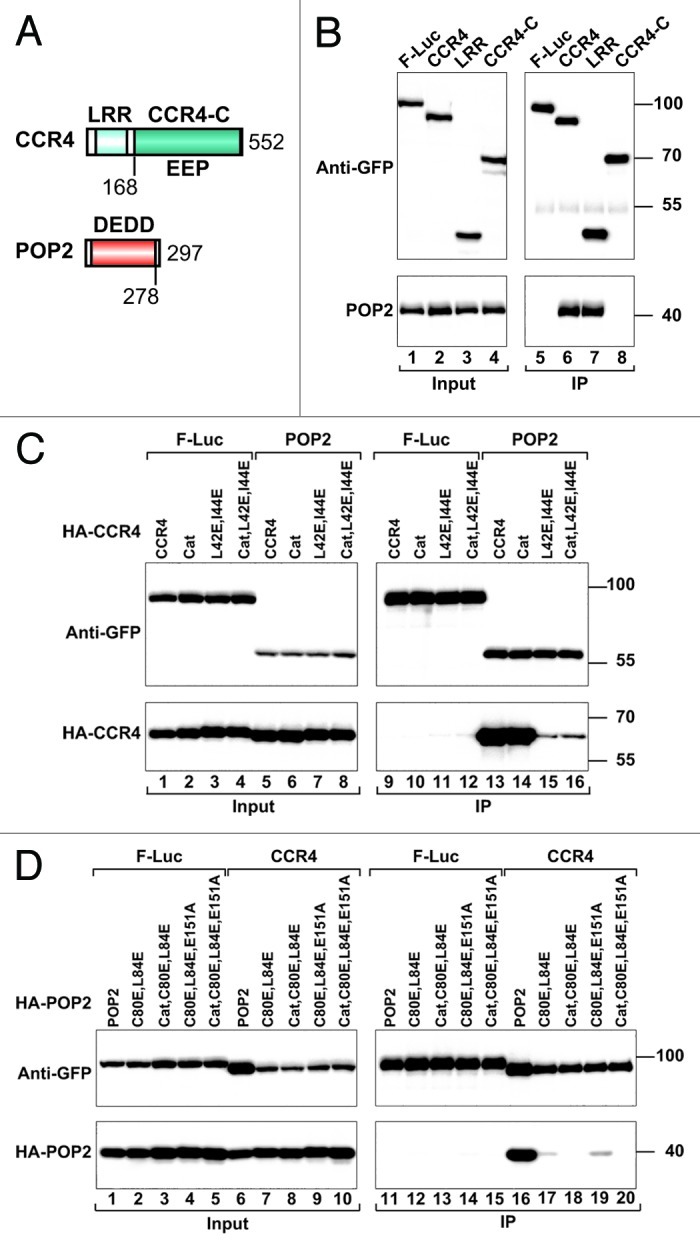
Figure 1.Dm CCR4 interacts with POP2. (A) Domain organization of Dm CCR4 and POP2. CCR4 contains a LRR domain and a catalytic EEP-nuclease domain (CCR4-C). POP2 consists of a single RNase D-like DEDD family catalytic domain. The numbers beneath the protein outline represent the amino acid position at the fragment/domain boundaries. (B) S2 cells were co-transfected with plasmids expressing GFP-tagged CCR4 (full-length or fragments) and HA-tagged POP2. GFP-tagged firefly luciferase (F-Luc) served as a negative control. Cell lysates were immunoprecipitated with polyclonal anti-GFP antibodies. Inputs (1%) and immunoprecipitates (5% GFP tagged proteins or 30% HA-tagged proteins) were subjected to western blotting using anti-GFP and anti-HA antibodies. (C) Interaction between GFP-tagged POP2 and the indicated CCR4 mutants. (D) Interaction between GFP-tagged CCR4 and the indicated POP2 mutants.
In immunoprecipitation assays using S2 cell lysates, we confirmed that Dm CCR4 and POP2 interact and that this interaction is mediated by the CCR4 LRR domain. Indeed, GFP-tagged Dm CCR4 and the isolated LRR domain coimmunoprecipitated HA-tagged POP2 (Fig. 1B, lanes 6 and 7). The CCR4 catalytic domain had no detectable interaction with POP2 (Fig. 1B, lane 8).
The crystal structure of the S. cerevisiae POP2 protein (also known as CAF1) in complex with CCR4 identified critical interface residues that mediate the interaction between the two proteins.15 Together with conservation of the protein folds, the conservation of these residues15,16,29-32 suggests that POP2 and CCR4 interact in a similar manner in all eukaryotes. Therefore, based on the structure of the S. cerevisiae POP2-CCR4 complex,15 we designed mutations in Dm POP2 and CCR4 to disrupt their interaction. A double L42E,I44E mutation on the CCR4 LRR domain strongly reduced its interaction with POP2 (Fig. 1C, lane 15), as has been described for S. cerevisiae CCR4.15 In contrast, a catalytically inactive CCR4 mutant (Cat: D412A,N414A) interacted with POP2 as efficiently as wild-type (Fig. 1C, lane 14). Accordingly, mutation of catalytic residues did not exacerbate the effect of the double L42E,I44E mutation (Fig. 1C, lane 16). Conversely, substitution of POP2 residues C80 and L84 (corresponding to S. cerevisiae POP2 residues A215 and F219, respectively) with glutamic acid strongly impaired binding to CCR4 (Fig. 1D, lane 17). These results indicate that the POP2-CCR4 interface is conserved.
NOT2 and NOT3 proteins interact through their C-terminal regions comprising the NOT-boxes
NOT2 and NOT3 are related proteins that share a conserved C-terminal motif termed the NOT-box (Fig. 2A).34 The sequence identity between the Dm NOT2 and NOT3 NOT-boxes is 28%.34 In addition to the NOT-box, NOT2 contains a less conserved N-terminal extension, which is rich in glycine (22.6%) and serine (12.4%) residues and is predicted to be unstructured (NOT2-N; Fig. 2A). NOT3 contains a highly conserved N-terminal domain (NOT3-N) that is predicted to be primarily α-helical, which is connected to the NOT-box by a linker region (NOT3-L) that is rich in serine (16.4%) and glutamine (11.8%) residues (Fig. 2A).
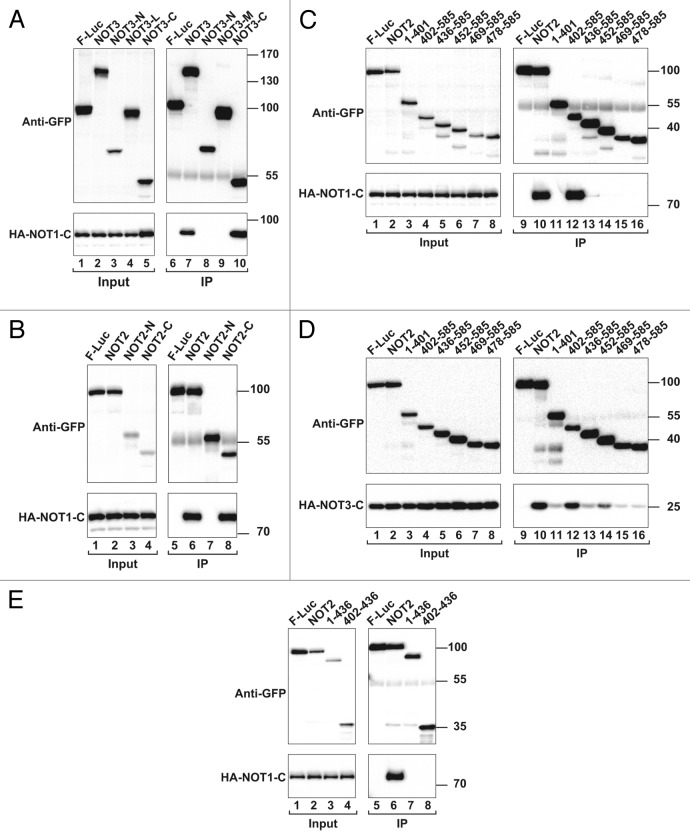
Figure 2. NOT2 and NOT3 interact via their C-terminal regions, which contain NOT-boxes. (A) NOT2 and NOT3 share a conserved C-terminal motif termed the NOT-box. NOT2 contains a less conserved N-terminal extension that is predicted to be unstructured (NOT2-N). NOT3 contains a highly conserved N-terminal domain (NOT3-N) that is connected to the NOT-box by a linker region (NOT3-L). (B) S2 cells coexpressing GFP-tagged NOT3 (full-length or fragments) and HA-tagged NOT2 were lysed 3 d after transfection. Cell lysates were immunoprecipitated using polyclonal anti-GFP antibodies and analyzed as described in Figure 1. (C and D) The interaction between GFP-tagged NOT2 (full-length or fragments) and HA-tagged NOT3 or NOT3-C was analyzed as described in Figure 1. (E) Interaction between recombinant NusA-tagged NOT3-C and GST-tagged NOT2-C. GST was used as a negative control.
To define the NOT2 and NOT3 regions that are required for their interaction, we performed coimmunoprecipitation assays using S2 cells. We observed that GFP-tagged NOT3 coimmunoprecipitated HA-tagged NOT2 and that this interaction was mediated by a NOT3 C-terminal fragment comprising the NOT-box (Fig. 2B, lanes 7 and 10). Conversely, GFP-tagged NOT2 coimmunoprecipitated full-length NOT3 and NOT3-C, and these interactions were also mediated by the NOT2 C-terminal region containing the NOT-box (Fig. 2C and D, lanes 8), which is in agreement with previous studies.18,21,27,28 The NOT2-NOT3 interaction is direct because a GST (glutathione S-transferase) tagged NOT2 C-terminal fragment (GST-NOT2-C) pulled down the C-terminal NOT3 fragment, which was expressed in Escherichia coli with a NusA tag (Fig. 2E, lane 4). We conclude that the NOT2 and NOT3 C-terminal regions containing the NOT-boxes mediate the assembly of the NOT2-NOT3 complex.
Binding of POP2-CCR4 and NOT2-NOT3 complexes to the NOT1 scaffold
Studies of the interaction of the CCR4-NOT complex subunits indicate that NOT1 serves as a scaffold protein, providing binding sites for the catalytic module and the additional subunits of the complex (Fig. 3A).18,21-28 Therefore, we investigated the interaction of NOT1 with the conserved core components of the CCR4-NOT complex. We observed that GFP-tagged NOT1 coimmunoprecipitated HA-tagged NOT2, NOT3, POP2 and CCR4 (Fig. 3B), which is in agreement with the hypothesis that NOT1 serves as a scaffold protein.
Figure 3. NOT1 interacts with the catalytic module and with the NOT2-NOT3 complex. (A) NOT1 domain organization. NOT1 consists of an N-terminal (NOT1-N), middle (NOT1-M) and C-terminal (NOT1-C) region. The NOT1-N region contains a metazoan-specific α-helical domain (residues 1–412) that interacts with NOT10 and NOT11 (NOT10/11-binding domain (NOT10/11-BD), this study). The NOT1-M region comprises a MIF4G domain that interacts with POP215,16 and a domain of unknown function (DUF3819) that interacts with CAF40 (this study). The NOT1-C region harbors a conserved NOT1 domain. (B) S2 cells were cotransfected with plasmids expressing GFP-tagged NOT1 and HA-tagged deadenylase subunits as indicated. GFP-tagged firefly luciferase served as a negative control. Cell lysates were immunoprecipitated using polyclonal anti-GFP antibodies. Inputs and immunoprecipitates were analyzed as described in Figure 1. (C) Interaction of POP2 with the NOT1-M region and the MIF4G domain. (D and E) The interaction of GFP-NOT1 (full-length or the indicated fragments) with HA-tagged NOT2 and NOT3 was analyzed as described in Figure 1.
To define the NOT1 domains involved in binding to the core complex subunits, we performed co-immunoprecipitation assays using a series of NOT1 deletion mutants. Sequence comparison, secondary structure predictions and available structural information indicate that NOT1 consists of three main regions: N-terminal (NOT1-N), Middle (NOT1-M) and C-terminal (NOT1-C) (Fig. 3A). The NOT1-N is predicted to be entirely α-helical and structural information is available for a S. cerevisiae N-terminal region corresponding to Dm NOT1 residues 415–1373.15 The NOT1-M region contains an N-terminal MIF4G domain that directly interacts with POP215,16 and a DUF3819 domain of unknown function (Fig. 3A). The NOT1-C region contains a conserved NOT1 homology domain (Fig. 3A).
First, we confirmed that the NOT1-M region interacts with POP2 via the MIF4G domain as was recently shown for human and yeast POP2 (Fig. 3C, lanes 8 and 10).15,16 We also investigated the NOT1 interaction with NOT2 and NOT3 and observed that these interactions were mediated by the NOT1-C domain (Fig. 3D and E, lanes 12), which is in agreement with previous studies.18,22-24
NOT2 and NOT3 interact with NOT1 via their C-terminal regions, which contain the NOT-boxes
We next defined the NOT2 and NOT3 regions required for NOT1-C binding. We observed that the regions mediating NOT2-NOT3 interaction were also required for NOT1-C binding. Indeed, NOT2 and NOT3 coimmunoprecipitated NOT1-C through their C-terminal regions (Fig. 4A and B). Previous studies have indicated that the interaction between NOT1 and NOT3 is mediated by NOT2.18,21,22,24,28 To more precisely define the NOT2 sequences that interact with NOT1 and NOT3, we generated a series of C-terminal fragments containing the NOT-box (residues 444–566) and increasing N-terminal extensions. However, only the NOT2 fragment 402–585 interacted with NOT1-C and NOT3-C as efficiently as full-length NOT2 (Fig. 4C and D, lane 12 vs. 10), whereas the 436–585 fragment failed to interact with NOT1-C and exhibited residual NOT3 binding (Fig. 4C and D, lane 13). These results indicate that the NOT2 NOT-box is not sufficient for NOT1 and NOT3 binding and that residues 402–436 (upstream of the NOT-box), are required for the interaction with NOT1 and NOT3. To determine whether these residues were sufficient for NOT1 binding, we performed coimmunoprecipitation assays using NOT2 fragments comprising residues 1–436 and 402–436 (fused to GFP). We observed that these fragments did not interact with NOT1 (Fig. 4E, lanes 7 and 8). Therefore, both the NOT2 NOT-box and the upstream N-terminal sequences (residues 402–436) are required for NOT1 binding.
Figure 4. The NOT2 NOT-box is not sufficient for binding to NOT1-C and NOT3-C. (A and B) Interaction between GFP-tagged NOT3 and NOT2 (full-length or fragments) and HA-tagged NOT1-C. (C–E) Interaction between GFP-tagged NOT2 (full-length or fragments) and HA-tagged NOT1-C and NOT3-C.
CAF40 interacts with a middle region of NOT1
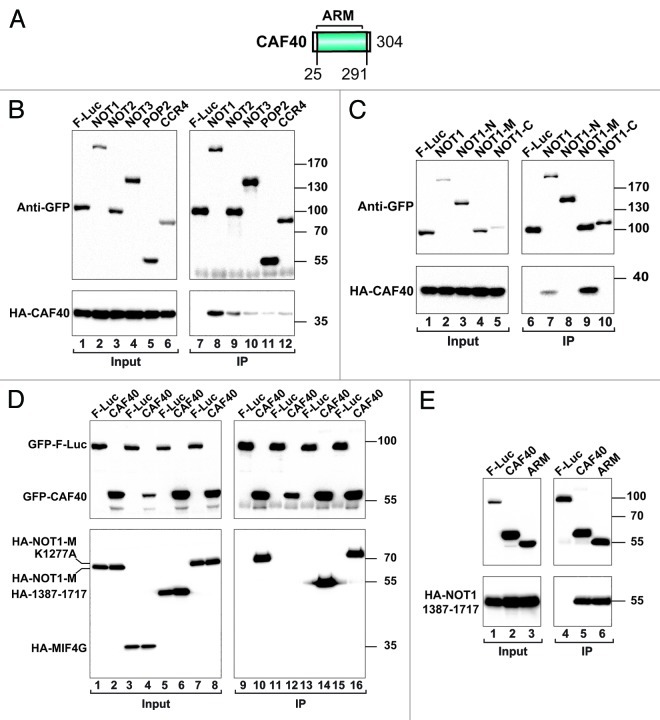
CAF40 contains a highly conserved domain comprising six armadillo repeats (Fig. 5A).35 In yeast, CAF40 interacts with NOT1; however, the domains mediating the NOT1 and CAF40 interaction have not been precisely defined.25 In immunoprecipitation assays, we confirmed that CAF40 preferentially associates with NOT1 (Fig. 5B, lane 8), specifically, the NOT1-M region (Fig. 5C, lane 9). These results indicate that CAF40 binds NOT1 independently of NOT2 and NOT3, which interact with the NOT1-C fragment. A more detailed analysis indicates that CAF40 interacts with the C-terminal portion of the NOT1-M fragment (residues 1387–1717) comprising the DUF3819 domain and not with the MIF4G domain, which interacts with POP215,16 (Fig. 5D, lane 14). Moreover, CAF40 interacts with a NOT1-M fragment harboring a mutation that abolishes its interaction with POP2 (K1277A)16 (Fig. 5D, lane 16). These results indicate that CAF40 binds NOT1 independently of POP2. Additionally, CAF40 did not detectably interact with NOT10 and C2orf29 (see below). The interaction between CAF40 and NOT1 fragment 1387–1717 was mediated by the highly conserved armadillo repeat domain in CAF40 (Fig. 5E, lane 6).
Figure 5. CAF40 interacts with a NOT1 fragment comprising the DUF3819 domain. (A) CAF40 contains a highly conserved domain comprising six armadillo repeats (ARM). (B) S2 cells were transiently transfected with expression vectors encoding GFP-tagged subunits of the CCR4-NOT complex and HA-tagged CAF40. Cell lysates were immunoprecipitated using anti-GFP antibodies and analyzed as described in Figure 1. GFP-F-Luc served as a negative control. (C) Interaction between GFP-tagged NOT1 (full-length or fragments) and HA-tagged CAF40. (D) Interaction between GFP-tagged CAF40 and the indicated HA-tagged NOT1 fragments. (E) Interaction between GFP-tagged CAF40 (full-length or the armadillo repeat domain) and a HA-tagged NOT1 fragment comprising the DUF3819 domain.
NOT10 and C2orf29 (NOT11) form a conserved module of the CCR4-NOT complex
Human CNOT10 and C2orf29 were originally identified as subunits of the human CCR4-NOT complex.18-20 These proteins are conserved in D. melanogaster (NOT10 and CG13567, respectively) and NOT10 copurify with the Dm CCR4-NOT complex.17 Both proteins are predicted to be primarily α-helical. CG13567 contains a conserved domain of unknown function named DUF2363 (Fig. 6A). In the accompanying manuscript by Mauxion et al.,36 human C2orf29 is shown to be a bona fide subunit of the human CCR4-NOT complex and is termed CNOT11. In accordance with the accompanying manuscript,36 we will refer to Dm CG13657 as Dm NOT11 hereafter.
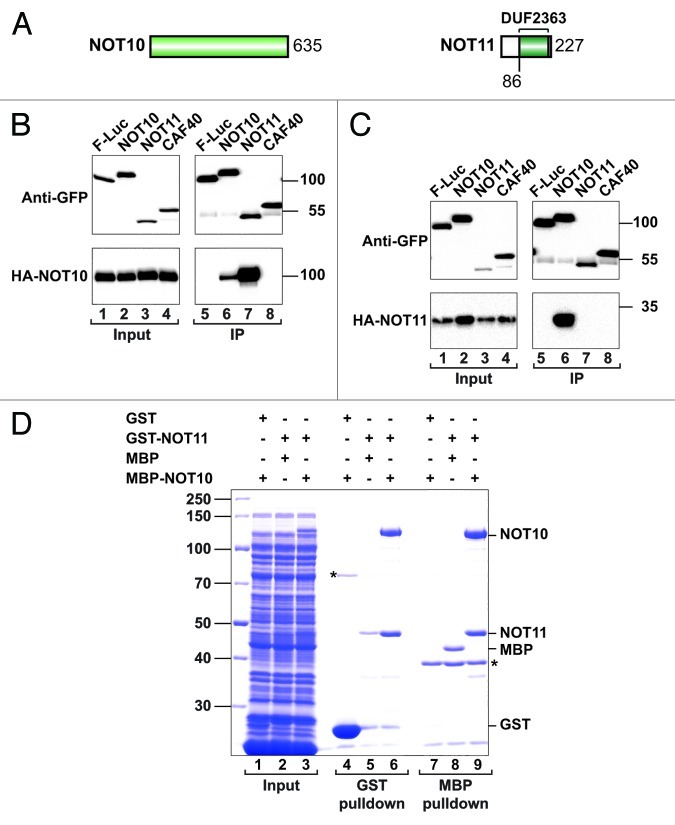
Figure 6. NOT10 directly interacts with NOT11. (A) NOT10 and NOT11 (also known as CG13567 or C2orf29) are predicted to be primarily α-helical. NOT11 contains a conserved domain of unknown function named DUF2363. (B and C) S2 cells were transiently transfected with expression vectors encoding GFP-tagged NOT10, NOT11 or CAF40 and HA-tagged NOT10 (B) or NOT11 (C). Cell lysates were immunoprecipitated with anti-GFP antibodies and analyzed as described in Figure 1. (D) The interaction between recombinant GST-tagged NOT11 and an MBP fusion of NOT10 was analyzed by SDS-PAGE followed by Coomassie blue staining. The pull-downs were performed using glutathione agarose beads (lanes 4–6) or amylose resin (lanes 7–9). Input samples (1%) and bound fractions (50%) were analyzed on SDS PAGE. The asterisk indicates a contaminant protein that copurified with GST (lane 4) or bound to the amylose resin (lanes 7–9).
In immunoprecipitation assays, we observed that NOT10 strongly interacted with NOT11, suggesting that they form a complex (Fig. 6B, lane 7). NOT10 also interacted with itself (Fig. 6B, lane 6). Conversely, NOT11 strongly interacted with NOT10 (Fig. 6C, lane 6).
To investigate whether the NOT10-NOT11 interaction was direct, we coexpressed the proteins in E. coli. We observed that MBP- (maltose binding protein) tagged NOT10 copurified with GST-tagged NOT11, but not with GST, on glutathione agarose beads (Fig. 6D, lane 6 vs. 4). Conversely, GST-NOT11 copurified with MBP-NOT10 but not with MBP on amylose resin (Fig. 6D, lane 9 vs 8). Furthermore, the expression levels of MBP-NOT10 increased by coexpression with GST-NOT11. We conclude that NOT10 and NOT11 directly interact and form a new module of the CCR4-NOT complex.
The NOT10-NOT11 complex interacts with the N-terminal NOT1 domain
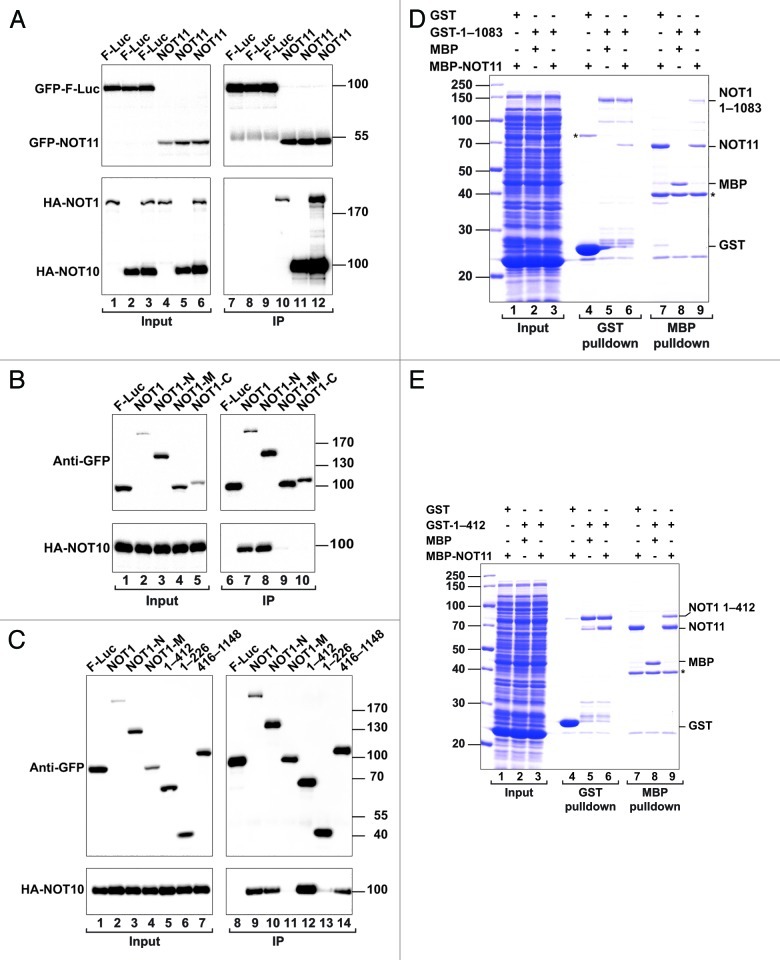
To investigate how the NOT10-NOT11 complex interacts with the CCR4-NOT complex, we performed immunoprecipitation assays in S2 cells. We observed that NOT1 and NOT10 interact with NOT11 (Fig. 7A, lanes 10 and 11, respectively). The NOT1-NOT11 interaction was detectable only when NOT11 was used as bait (Fig. 7A and data not shown) and was enhanced in S2 cells in which NOT10 was also coexpressed (Fig. 7A, lane 12 vs. 10), suggesting that NOT10 and NOT11 interact with NOT1 as a complex. Further analysis revealed that NOT10 interacts with the NOT1-N region (Fig. 7B, lane 8 and Fig. 7C, lane 10) but not with the NOT1-M or NOT1-C fragments (Fig. 7B, lanes 9 and 10 and Fig. 7C, lane 11). Moreover, the 412 N-terminal-most residues of NOT1 were sufficient for binding to NOT10 (Fig. 7C, lane 12), although the NOT1 fragment 416–1148 retained residual binding (Fig. 7C, lane 14).
Figure 7. The NOT10-NOT11 complex interacts with the NOT1 N-terminal domain. (A) S2 cells were cotransfected with a mixture of two plasmids: one expressing GFP-NOT11 and one expressing HA-NOT1. In addition, where indicated, the transfection mixtures contained a third plasmid expressing HA-NOT10 (lanes 2, 3, 5 and 6). Cell lysates were immunoprecipitated using polyclonal anti-GFP antibodies. The inputs and immunoprecipitates were analyzed by western blotting as described in Figure 1. (B and C) Interaction between GFP-tagged NOT1 (full-length or fragments) and HA-tagged NOT10. (D and E) The interaction between recombinant GST-tagged NOT1 (1–1083 or 1–412) and an MBP fusion of NOT11 was analyzed by SDS-PAGE followed by Coomassie blue staining, as described in Figure 6D.
To investigate which protein in the NOT10-NOT11 complex directly interacts with NOT1 we coexpressed GST-tagged NOT1 N-terminal fragments (1–412 or 1–1083) with MBP-tagged NOT10 or NOT11 in E. coli and performed pull-down assays. We found that NOT11, but not NOT10, interacted with the NOT1 N-terminal fragments (Figs. 7D and E, lanes 6 and 9; and data not shown), indicating that NOT11 is the subunit that docks the NOT10-NOT11 complex onto the NOT1-N domain. This result was unexpected because in immunoprecipitation assays, NOT10 interacted with NOT1 more efficiently and enhanced NOT11 binding (Fig. 7A). One possible explanation for this observation is that NOT11 is present in excess in S2 cells. Therefore, overexpressed NOT10 efficiently binds endogenous NOT11 and NOT1, whereas overexpressed NOT11 does not efficiently compete with endogenous NOT11 for NOT1 binding unless NOT10 is also overexpressed. This possibility also suggests that NOT10 facilitates the interaction between NOT11 and NOT1 either by contacting NOT1 directly or by stabilizing the NOT11 fold. Together, our results indicate that NOT10 and NOT11 form a complex that docks onto the NOT1 scaffold via interactions with NOT11 and the N-terminal NOT1 domain.
Human CNOT10 and CNOT11 form a complex that interacts with the CNOT1 N-terminal domain
To investigate whether the interactions between NOT1, NOT10 and NOT11 are conserved in humans, we performed immunoprecipitation assays of the human orthologs in human HEK293 cells (Fig. 8A). As observed for the D. melanogaster proteins, human CNOT10 interacted with CNOT1 and CNOT11 (Fig. 8B, lanes 9 and 14, respectively). More specifically, CNOT10 interacted with the N-terminal domain of CNOT1 (CNOT1-N) but not with the CNOT1-M or CNOT1-C domains (Fig. 8B, lanes 10–12). The interaction between CNOT11 and CNOT1 was weak and was enhanced when CNOT10 was coexpressed as observed in S2 cells (Fig. 8C, lane 6 vs. 8). In contrast to the D. melanogaster proteins, the 302 N-terminal-most residues of CNOT1 were not sufficient for CNOT10 binding (Fig. 8B, lane 13).

Figure 8. The NOT10-NOT11 complex is conserved in human cells. (A) Human CNOT1 consists of an N-terminal (NOT1-N), middle (NOT1-M), and C-terminal (NOT1-C) region. The POP2-binding domain adopts an MIF4G fold and is termed the CNOT1 MIF4G domain.15,16 The CNOT1-C region harbors a conserved NOT1 homology domain. (B) GFP-tagged CNOT1 (full-length or fragments) or GFP-CNOT11 were coexpressed with HA-tagged CNOT10 in HEK293 cells. The GFP-tagged proteins were immunoprecipitated from RNase A-treated cell lysates using anti-GFP antibodies. GFP-tagged MBP served as a negative control. Inputs (1.25% for the GFP-tagged proteins or 0.5% for HA-tagged proteins) and immunoprecipitates (10% for the GFP-tagged proteins or 25% for HA-tagged proteins) were analyzed by western blotting. (C) HEK293 cells were cotransfected with a mixture of two plasmids: one expressing GFP-CNOT1 and one expressing HA-CNOT11. In addition, where indicated, the transfection mixtures contained a third plasmid expressing HA-CNOT10 (lanes 3, 4, 7 and 8). Cell lysates were immunoprecipitated using polyclonal anti-GFP antibodies. The inputs and immunoprecipitates were analyzed by western blotting as described in panel B. (D) Interaction of GFP-tagged CNOT11 (full-length or DUF2363 domain) with HA-tagged CNOT10 in HEK293 cells.
Human CNOT11 consists of a highly conserved C-terminal DUF2363 domain (residues 260–496; Fig. 8A) and a less conserved N-terminal region that is absent in Dm NOT11. Because the interaction of CNOT11 with CNOT10 is conserved, we speculated that this interaction is most likely mediated by the most conserved region of CNOT11, the DUF2363 domain. Accordingly, we observed that a CNOT11 fragment comprising the DUF2363 domain interacted with CNOT10 as efficiently as full-length CNOT11 did (Fig. 8D, lanes 6 vs. 5). We conclude that CNOT10 and CNOT11 form a complex that interacts with CNOT1 in both human and D. melanogaster cells.
NOT10 and NOT11 promote the degradation of bound mRNAs
Tethering of the CCR4-NOT complex subunits promotes the degradation of polyadenylated mRNA targets.10,37 These effects have been shown for NOT1, NOT2, NOT3, CCR4 and POP2.10,37 These data suggest that tethered complex subunits recruit the catalytic module to promote the deadenylation and degradation of mRNAs. Additionally, the CCR4-NOT complex subunits repress the translation of mRNA reporters lacking poly(A) tails in tethering assays, indicating that the CCR4-NOT complex has the ability to repress translation in the absence of deadenylation.10,37 The identity of the CCR4-NOT subunits mediating translational repression and the mechanism of this repression are unknown.
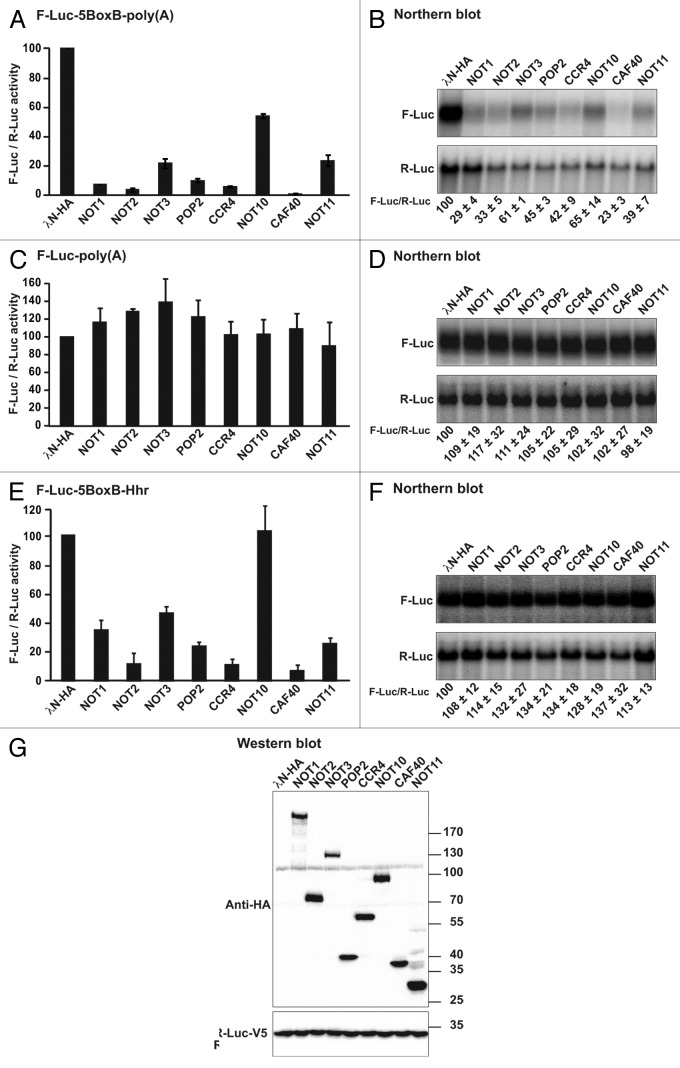
To gain insight into the role of NOT10 and NOT11, we compared the activity of these proteins with that of additional CCR4-NOT subunits in a tethering assay. For this assay, CCR4-NOT complex subunits were expressed with an N-terminal tag derived from the N protein of the bacteriophage λ (λN tag) to enable tethering to a firefly luciferase (F-Luc) reporter.38 The F-Luc reporter contained five Box B hairpins (5BoxB) inserted in the 3′ UTR;38 which bind the λN tag with high affinity and recruit CCR4-NOT complex subunits to the F-Luc-5BoxB mRNA. We observed that all of the CCR4-NOT subunits repressed the expression of the F-Luc-5BoxB mRNA reporter (Fig. 9A). Northern blot analyses revealed that all of the subunits also reduced the abundance of the F-Luc-5BoxB mRNA to different extents (Fig. 9B). CAF40 was a potent trigger of mRNA degradation, whereas although active, NOT10 and NOT11, promoted F-Luc mRNA degradation less efficiently (Fig. 9B). All subunits were expressed at comparable levels and had no effect on an F-Luc reporter lacking the BoxB hairpins (Fig. 9C, D and G).
Figure 9. All subunits of the CCR4-NOT complex elicit translational repression and mRNA degradation in tethering assays. (A and B) S2 cells were transfected with a mixture of three plasmids: one expressing the F-Luc-5BoxB reporter, one expressing Renilla luciferase (R-Luc) as a transfection control and a plasmid expressing the λN-HA or λN-HA-tagged subunits of the CCR4-NOT complex. Firefly luciferase activity was normalized to that of Renilla luciferase and set to 100 in cells expressing λN-HA. The mean values ± standard deviation of three independent experiments are shown in panel A. (B) northern blot of representative RNA samples. The numbers below the panel indicate the F-Luc-5BoxB reporter levels normalized to that of the R-Luc mRNA and set to 100 in cells expressing the λN-HA peptide. Mean values ± standard deviation of three independent experiments are shown. (C and D) An experiment similar to that described in panels A and B was performed using an F-Luc reporter lacking the Box B hairpins. (E and F) An experiment similar to that described in panels A and B was performed using an F-Luc-5BoxB reporter in which the cleavage and polyadenylation signal was substituted with a self-cleaving hammerhead ribozyme (F-Luc-5BoxB-HhR). (G) western blot showing the expression levels of the λN-HA tagged CCR4-NOT complex subunits.
To identify the CCR4-NOT complex subunits that can promote translational repression, we used an F-Luc-5BoxB reporter whose 3′ end is generated by a self-cleaving hammerhead ribozyme (F-Luc-5BoxB-HhR) and lacks a poly(A) tail.39 In agreement with previous studies,10,37 tethering of the λN-tagged NOT1, NOT2, NOT3, CCR4 and POP2 subunits reduced luciferase activity without affecting mRNA abundance (Fig. 9E and F).10,37 CAF40 was a potent translational repressor when expressed at comparable levels, whereas NOT10 was inactive at the concentration tested (Fig. 9E). These results indicate that the tethering of any subunit of the CCR4-NOT complex leads to the recruitment of additional subunits via direct and indirect protein-protein interactions (except NOT10 at this concentration), resulting in the translational repression of unadenylated reporters or the deadenylation and degradation of polyadenylated mRNAs.
Multiple NOT1 domains promote target degradation
Given that all of the CCR4-NOT complex subunits have the ability to recruit the complete complex, tethering assays using full-length proteins do not reveal the contribution of the individual subunits to translational repression and mRNA degradation. Therefore, we examined the activity of isolated protein domains. We observed that the NOT1-N fragment, which interacts with NOT10 and NOT11, was inactive in tethering assays, irrespective of the poly(A) tail (Fig. 10A–F). Accordingly, deletion of the NOT1-N region did not significantly affect the activity of the NOT1 protein in tethering assays (Fig. 10A–F). NOT1 fragments were expressed at comparable levels (Fig. 10G). This result suggests that the NOT1-N region and, thus, the NOT10-NOT11 complex may not participate in post-transcriptional mRNA regulation. In contrast, the NOT1-M and NOT1-C fragments elicited translational repression and mRNA degradation (Fig. 10A–F). The activity of the NOT1-M fragment was reduced by a single amino acid substitution (K1277A), which disrupts POP2 binding16 (Fig. 10H), indicating that the activity of the NOT1-M fragment is primarily mediated by the catalytic module but not by CAF40. However, the K1277A mutation had only a minor effect in the context of full-length NOT1 (Fig. 10I), indicating that the interaction with POP2 contributes but is not strictly required for NOT1 to promote translational repression and mRNA degradation in tethering assays.
Figure 10. Activity of NOT1 protein domains in tethering assays. (A and B) A tethering assay was performed as described in Figure 9A with NOT1 (full-length or fragments). Firefly luciferase activity was normalized to that of Renilla luciferase and set to 100 in the cells expressing λN-HA. The mean values ± standard deviation from three independent experiments are shown in panel A. (B) northern blot of representative RNA samples analyzed as described in Figure 8B. (C and D) Tethering assay using the F-Luc reporter lacking the Box B hairpins. (E and F) Tethering assay using the unadenylated F-Luc-BoxB-HhR reporter. (G) western blot showing the expression level of the protein fragments tested. (H and I) The effect of the K1277A mutation (which disrupts POP2 binding)16 on the activity of NOT1-M and full-length NOT1 was tested in tethering assays using the F-Luc-5BoxB reporter as described above.
The catalytic module requires interaction with the NOT module for full activity
We next tested the activity of POP2 and CCR4 mutants in tethering assays. A catalytically inactive POP2 mutant (Cat, D53A+E55A) promoted mRNA degradation as reported previously.10,37 In contrast, a POP2 mutant (E151A)16 that does not interact with NOT1 was impaired in tethering assays (Fig. 11A). Similarly, mutations that disrupt the interaction with CCR4 (C80E,L84E) impaired POP2 activity in tethering assays (Fig. 11A). POP2 activity was abolished when the mutations that disrupt NOT1 and CCR4 binding were combined (Fig. 11A). The effect of these mutations was independent of whether POP2 was catalytically active or inert (Fig. 11A). These results indicate that wild-type POP2 requires interaction with NOT1 and CCR4 for full activity and that the catalytic activity of POP2 is not sufficient to trigger degradation of the reporter in tethering assays. All POP2 mutants were expressed at comparable levels (Fig. 11B).

Figure 11. The catalytic module requires interaction with the NOT module for full activity in tethering assays. (A and C) Tethering assays were performed as described in Figure 9A with POP2 or CCR4 (wild-type or mutants). Firefly luciferase activity was normalized to that of Renilla luciferase and set to 100 in the cells expressing the λN-HA peptide. (B and D) western blot showing the expression level of the protein fragments and mutants tested.
For CCR4, we observed that the isolated LRR but not the catalytic domain (CCR4-C) was active in tethering assays at the concentrations tested (Fig. 11C). Accordingly, a catalytically inactive CCR4 mutant also promoted target degradation (Fig. 11C, Cat: D412A,N414A). This mutant still interacted with POP2 (Fig. 1C, lane 14). The activity of CCR4 or the isolated LRR was abolished by mutations that disrupt POP2 binding (L42E,I44E) despite the fact that these mutants were expressed at levels comparable to the wild-type (Fig. 11D). Therefore, as shown for POP2, the catalytic activity of CCR4 is not sufficient to trigger degradation of the reporter in tethering assays at the concentration tested. However, it is important to note that at higher concentrations the catalytic domain was active.
Importantly, the POP2 and CCR4 mutants that promoted degradation of the polyadenylated reporter also promoted translational repression of the unadenylated reporter (Fig. S1), indicating that these activities are interconnected. Furthermore, at the concentration tested, the proteins had no effect on a reporter lacking the BoxB hairpins, indicating that the effects are specific (Fig. S1).
The NOT2 N-terminal domain promotes the translational repression and degradation of bound mRNAs
We also analyzed the activity of NOT2 and NOT3 fragments. For NOT2, we observed that the N-terminal extension and the C-terminal region were active in tethering assays (Fig. 12A and B). In contrast, for NOT3, only the C-terminal region, which interacts with NOT2, was active (Fig. 12C and D), suggesting that NOT3 promotes mRNA degradation through its interaction with NOT2, which, in turn, interacts with the remainder of the CCR4–NOT complex. As shown for POP2 and CCR4, the NOT2 and NOT3 fragments that promoted degradation of the polyadenylated reporter also promoted translational repression of the unadenylated reporter but had no effect on a reporter lacking the BoxB hairpins (Fig. S1).
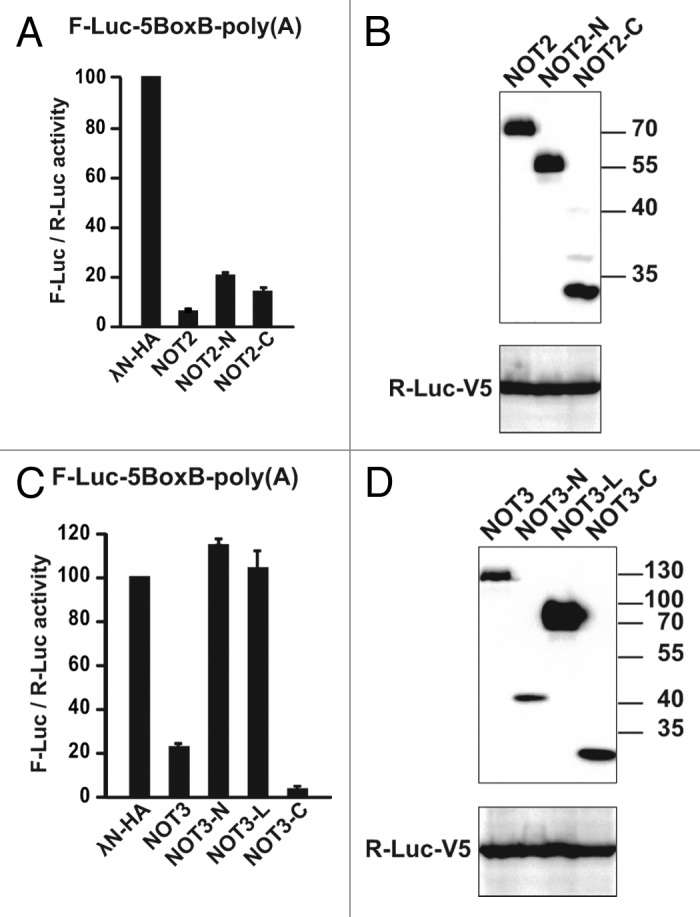
Figure 12. The NOT2-N-terminal domain is active in tethering assays. (A and C) Tethering assays were performed as described in Figure 9A with the indicated NOT2 and NOT3 fragments. Firefly luciferase activity was normalized to that of Renilla luciferase and set to 100 in the cells expressing the λN-HA peptide. (B and D) western blot showing the expression level of the protein fragments tested.
The observation that the isolated NOT2 N-terminal region promotes target degradation was unexpected because this region exhibited no detectable interaction with the core complex subunits, suggesting that this region interacts with unidentified protein partners. Our observations indicate that in contrast to the full-length proteins, only a subset of protein domains can cause translational repression and mRNA degradation independently (e.g., NOT2-N, NOT2-C, NOT1-M and NOT1-C). Further studies are required to determine whether these domains have intrinsic activity or interact indirectly with additional complex subunits or with unknown partners to regulate mRNA expression. In particular, structural studies of the interaction of subunits of the complex are needed to provide information on how to specifically disrupt these interactions, which is of critical importance to evaluate the contribution of the individual subunits to mRNA degradation and translational repression.
Discussion
The CCR4-NOT complex is a master regulator of mRNA expression. It promotes translational repression, which can occur even in the absence of deadenylation, and can direct the irreversible degradation of mRNA targets.3,8,10,37 In addition to its role in mRNA regulation, the CCR4-NOT complex has been implicated in a wide range of cellular processes including transcription, ubiquitination, DNA repair and protein modification.8 Although most of the complex subunits are conserved among eukaryotes, yeast-specific and metazoan-specific subunits have been described, indicating that the complex composition differs across species. Therefore, the study of this multifunctional complex in diverse organisms is relevant and promises to further our understanding of its diverse molecular functions. In this study, we characterized the Dm CCR4-NOT complex. We confirmed and extended the interactions that have been described in other species and defined the domains mediating the NOT1-CAF40 interaction (Fig. 13). We further demonstrated that NOT10 and NOT11 interact and dock onto the N-terminal NOT1 domain through NOT11 (Fig. 13) in both D. melanogaster and human cells. Similar results are presented in the accompanying manuscript describing interactions between human CNOT10, CNOT11 and CNOT1.36 We conclude that NOT10 and NOT11 form a conserved module of the CCR4-NOT complex. Finally, our analysis of the protein domains that mediate the interactions between the subunits and play a role in mRNA degradation provides a foundation for future studies aimed at understanding how the complex assembles and regulates the expression of target mRNAs.
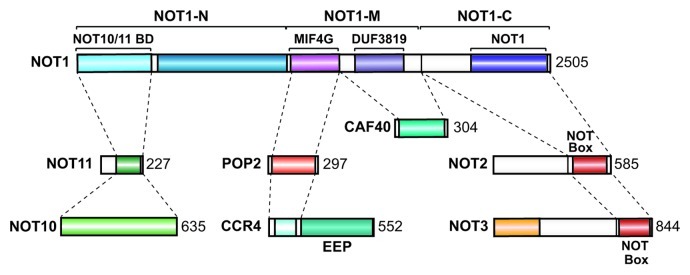
Figure 13. Diagram summarizing the interactions described in this study.
Materials and Methods
Coimmunoprecipitation assays in D. melanogaster and human cells
The plasmids encoding the deadenylase subunits for expression in D. melanogaster S2 cells are described in Table S1. Plasmids encoding Dm NOT10 (CG18616), Dm C2orf29 (CG13567) and Dm CAF40 (CG14213) were generated by inserting the corresponding cDNAs into the pAc5.1-EGFP and pAc5.1-λNHA vectors using the following restriction sites: EcoRV-XbaI (NOT10) and HindIII-XbaI (CG13567 and CAF40). Coimmunoprecipitation assays using S2 cells were performed as previously described.9 S2 cells were grown in 6-well dishes, transfected using Effectene (Qiagen) transfection reagent and harvested 3 d after transfection. The transfection mixtures contained a total of 2–5 μg of plasmid, including both HA-tagged and GFP-tagged proteins. A plasmid expressing GFP-F-Luc served as a negative control. HA and GFP-tagged proteins were detected using HRP-conjugated monoclonal anti-HA (Roche 3F10; 1:5,000) and anti-GFP antibodies (Roche, catalog number 11814460001; 1:2,000), respectively. All western blots were developed using the ECL western blotting detection system (GE Healthcare) as recommended by the manufacturer. Coimmunoprecipitation assays in human HEK293 cells were performed as described previously.16 Plasmids expressing deadenylase subunits in human cells were described previously.9 Plasmids encoding GFP or HA-tagged human CNOT10 were generated by inserting the CNOT10 cDNA (clone on15275; Kazusa DNA Research Institute) into the pEGFP-C1 and pλN-HA-C1 vectors, respectively, using the BamHI and XhoI restriction sites. Plasmids encoding GFP or HA-tagged human CNOT11 (full-length or fragment 260–496) were generated by inserting the CNOT11 cDNA into the pT7-EGFP-C1 and pλN-HA-C1 vectors, respectively, using the BamHI and KpnI (CNOT11 full-length) or the BamHI and XhoI (CNOT11 260–496) restriction sites.
In vitro pull-down assays
To express the Dm NOT2-C fragment in E. coli, the corresponding cDNA was cloned into the pnEA-NvG vector,40 resulting in an N-terminal TEV protease-cleavable GST-tagged NOT2-C protein (Table S1). Dm NOT3-C was cloned into the pETM-60 plasmid and resulted in a vector encoding N-terminal NusA-tagged proteins. To express the Dm NOT1 fragments 1–412 and 1–1083 in E. coli, the corresponding cDNAs were cloned into the pnEA-NvG and pnEA-NpG vectors, respectively,40 resulting in N-terminal GST fusion proteins. For coexpression, the full-length Dm NOT10 cDNA was cloned into pnYC-NpHM, and the full-length Dm NOT11 cDNA was cloned into both pnYC-NpHM and pnEA-NpG vectors,40 resulting in N-terminal MBP or GST fusion proteins, respectively. GST-NOT11 or GST-NOT1 was coexpressed with MBP-NOT10 or MBP-NOT11 in E. coli BL21 cells at 20°C overnight. Cells were resuspended in lysis buffer [50 mM Tris-Cl (pH 7.5), 200 mM NaCl, 2 mM MgCl2, 2 mM ATP, 1 mM DTT] supplemented with lysozyme (1 mg/ml), DNase I (5 µg/ml) and protease inhibitors. Cell lysates were incubated on ice for 10 min, lysed by sonication and cleared by centrifugation. Cleared lysates were incubated with 50 µl (50% slurry) of Protino Glutathione Agarose 4B beads (Macherey Nagel) or 50 µl (50% slurry) of amylose resin (New England BioLabs) for 1 h at 4°C with gentle rotation. Beads were washed three times with lysis buffer. The bound proteins were eluted with 40 μl of sample buffer [50 mM TRIS-HCl (pH 6.8), 2% SDS, 10% (v/v) glycerol, 100 mM DTT and 0.05% bromophenol blue] and separated on an 11% SDS-PAGE.
Tethering assays in S2 cells
For the λN-tethering assay, S2 cells were grown in 6-well dishes and transfected using Effectene transfection reagent (Qiagen). The transfection mixtures contained the following plasmids: 0.1 μg of reporter plasmid (F-Luc-5BoxB, F-Luc or F-Luc-5BoxB-Hhr), 0.4 μg of pAc5.1-R-Luc as a transfection control and various quantities of pAc5.1λN-HA constructs encoding the CCR4-NOT subunits that were adjusted to obtain comparable protein expression levels as follows: 1,000 ng for the λN-HA control, NOT1 and NOT11; 100 ng for NOT10 and CAF40; 70 ng for CCR4; 30 ng for NOT2 and 5 ng for NOT3 and POP2. When necessary, the total amount of transfected DNA was adjusted to 1.5 µg using the pAc5.1A plasmid lacking an insert. Firefly and Renilla luciferase activities were measured three days after transfection using the Dual-Luciferase Reporter Assay System (Promega). Total RNA was isolated using TriFast (Peqlab Biotechnologies) and analyzed as previously described.38
Supplementary Material
Acknowledgments
This work was supported by the Max Planck Society and by grants from the Deutsche Forschungsgemeinschaft (DFG, FOR855 and the Gottfried Wilhelm Leibniz Program awarded to E.I.). We thank N. Bercovich for providing plasmids for expression of D. melanogaster NOT1, NOT10 and NOT11 in bacteria. We are grateful to E. Conti, F. Mauxion and B. Séraphin for sharing results prior to publication.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23018
References
- 1.Weill L, Belloc E, Bava FA, Méndez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19:577–85. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- 2.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–76. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Bartlam M, Yamamoto T. The structural basis for deadenylation by the CCR4-NOT complex. Protein Cell. 2010;1:443–52. doi: 10.1007/s13238-010-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–59. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–63. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 6.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–86. doi: 10.1016/S0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 7.Brown CE, Tarun SZ, Jr., Boeck R, Sachs AB. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5744–53. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collart MA, Panasenko OO. The Ccr4--not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 9.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120–33. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18:1218–26. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18:1211–7. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 12.Draper MP, Liu HY, Nelsbach AH, Mosley SP, Denis CL. CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol Cell Biol. 1994;14:4522–31. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark LB, Viswanathan P, Quigley G, Chiang YC, McMahon JS, Yao G, et al. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J Biol Chem. 2004;279:13616–23. doi: 10.1074/jbc.M313202200. [DOI] [PubMed] [Google Scholar]
- 15.Basquin J, Roudko VV, Rode M, Basquin C, Séraphin B, Conti E. Architecture of the nuclease module of the yeast ccr4-not complex: the not1-caf1-ccr4 interaction. Mol Cell. 2012;48:207–18. doi: 10.1016/j.molcel.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Petit AP, Wohlbold L, Bawankar P, Huntzinger E, Schmidt S, Izaurralde E, et al. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex. Nucleic Acids Res. 2012;40:11058–72. doi: 10.1093/nar/gks883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, et al. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–70. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, et al. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J. 2009;422:443–53. doi: 10.1042/BJ20090500. [DOI] [PubMed] [Google Scholar]
- 19.Morita M, Suzuki T, Nakamura T, Yokoyama K, Miyasaka T, Yamamoto T. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol Cell Biol. 2007;27:4980–90. doi: 10.1128/MCB.02304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyasaka T, Morita M, Ito K, Suzuki T, Fukuda H, Takeda S, et al. Interaction of antiproliferative protein Tob with the CCR4-NOT deadenylase complex. Cancer Sci. 2008;99:755–61. doi: 10.1111/j.1349-7006.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol Cell Biol. 1999;19:6642–51. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HT. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res. 2000;28:809–17. doi: 10.1093/nar/28.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maillet L, Tu C, Hong YK, Shuster EO, Collart MA. The essential function of Not1 lies within the Ccr4-Not complex. J Mol Biol. 2000;303:131–43. doi: 10.1006/jmbi.2000.4131. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol. 2001;314:683–94. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 26.Russell P, Benson JD, Denis CL. Characterization of mutations in NOT2 indicates that it plays an important role in maintaining the integrity of the CCR4-NOT complex. J Mol Biol. 2002;322:27–39. doi: 10.1016/S0022-2836(02)00707-6. [DOI] [PubMed] [Google Scholar]
- 27.Winkler GS, Mulder KW, Bardwell VJ, Kalkhoven E, Timmers HT. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006;25:3089–99. doi: 10.1038/sj.emboj.7601194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito K, Takahashi A, Morita M, Suzuki T, Yamamoto T. The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell. 2011;2:755–63. doi: 10.1007/s13238-011-1092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dlakić M. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem Sci. 2000;25:272–3. doi: 10.1016/S0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Morita M, Yang X, Suzuki T, Yang W, Wang J, et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 2010;29:2566–76. doi: 10.1038/emboj.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daugeron MC, Mauxion F, Séraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–55. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thore S, Mauxion F, Séraphin B, Suck D. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 2003;4:1150–5. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–86. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwartjes CG, Jayne S, van den Berg DL, Timmers HT. Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4-not complex. J Biol Chem. 2004;279:10848–54. doi: 10.1074/jbc.M311747200. [DOI] [PubMed] [Google Scholar]
- 35.Garces RG, Gillon W, Pai EF. Atomic model of human Rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci. 2007;16:176–88. doi: 10.1110/ps.062600507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mauxion F, Preve B, Séraphin B. C2ORF29/CNOT11 and CNOT10 form a new module of the CCR4-NOT complex. RNA Biol. 2012 doi: 10.4161/rna.23065. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem. 2010;285:28506–13. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Gregorio E, Preiss T, Hentze MW. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–74. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–53. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 40.Diebold ML, Fribourg S, Koch M, Metzger T, Romier C. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J Struct Biol. 2011;175:178–88. doi: 10.1016/j.jsb.2011.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.