Abstract
Small non-coding RNAs (ncRNAs) are important components of many regulatory pathways in bacteria and play key roles in regulating factors important for virulence. Carbon catabolite repression control is modulated by small RNAs (crcZ or crcZ and crcY) in Pseudomonas aeruginosa and Pseudomonas putida. In this study, we demonstrate that expression of crcZ and crcX (formerly designated psr1 and psr2, respectively) is dependent upon RpoN together with the two-component system CbrAB, and is influenced by the carbon source present in the medium in the model plant pathogen Pseudomonas syringae pv tomato DC3000. The distribution of the members of the Crc ncRNA family was also determined by screening available genomic sequences of the Pseudomonads. Interestingly, variable numbers of the Crc family members exist in Pseudomonas genomes. The ncRNAs are comprised of three main subfamilies, named CrcZ, CrcX and CrcY. Most importantly the CrcX subfamily appears to be unique to all P. syringae strains sequenced to date.
Keywords: psr, crcZ, crcY, crcX, Crc, ncRNA, RpoN, CbrA, CbrB, Pseudomonas syringae
Introduction
Molecular and computational analyses have revealed that bacteria contain large numbers of small, non-coding, RNA (ncRNA) molecules. Although their function in many cases is unknown, it has become widely accepted that they play critical roles in a variety of cellular processes and regulatory networks by facilitating adaptation to diverse environmental stresses and influencing the production of virulence factors.1 The majority of ncRNAs are encoded in trans and interact with their RNA targets through an antisense mechanism. In this process, complementary base-pairing with a target mRNA either activates or represses translation of the transcript, or targets the mRNA for degradation and most often requires the RNA chaperone Hfq.2 Alternatively, some ncRNAs interact directly with a protein target, sequestering the protein and preventing it from performing functions such as activating or repressing translation.2
Recently we identified and characterized two ncRNAs in the genome of Pseudomonas syringae pv tomato str. DC3000.3 One ncRNA, designated psr1 (PSPTO_5668), is located between PSPTO_0964 and PSPTO_0963. The other, psr2 (PSPTO_5669), is located between PSPTO_1621 and PSPTO_1622. These areas previously had been reported to contain a conserved RNA motif termed gamma-150.4 Both ncRNAs are significantly larger than the described motif. A third putative member of this family (psr3), located between PSPTO_2739 and PSPTO_2740, was also identified. However, psr3 is disrupted by an insertion element and therefore is not expressed in P. syringae DC3000.3 Interestingly, this RNA appears to be fully intact in P. syringae B728a and 1448A.
Homologs to DC3000 psr1 and psr3 are found in other pseudomonads. In P. aeruginosa, a psr1 homolog (crcZ) contains the gamma-150 motif4 and maps between pcnB and genes encoding the CbrA/CbrB two-component sensor-regulator system.5 The CrcZ ncRNA contains five Crc-binding sites and is thought to sequester the RNA binding protein Crc under conditions that generate low or no catabolite repression thus modulating Crc availability and the strength of catabolite repression control (CRC). CrcZ levels also vary according to the carbon source being used and are low in succinate medium (a preferred carbon source for P. aeruginosa), at an intermediate level in glucose medium and at a high level in mannitol medium (a non-preferred carbon source and a growth condition that does not generate catabolite repression).5 P. putida contains crcZ and an additional ncRNA, crcY,6 homologous to psr3 in DC3000. The crcZ and crcY ncRNAs were found to function similarly and modulate levels of Crc, controlling catabolite repression. However, in contrast to P. aeruginosa, succinate and other organic acids only have a small influence on Crc in catabolite repression in P. putida.7 Hence, the levels of crcZ and crcY are higher in P. putida when grown in succinate or citrate compared with cells grown in LB medium, where Crc-mediated catabolite repression is very strong.6 The levels of crcZ and crcY are also higher in stationary phase, where no catabolite repression is observed.6 Additionally, transcript levels of crcZ and crcY in P. putida are higher at lower temperatures.8
P. aeruginosa and P. putida do not contain psr2 homologs and a function has not yet been described for this ncRNA. In this report we analyze the role of psr1 and psr2 in modulating carbon metabolism in P. syringae DC3000 and examine their conservation in other sequenced Pseudomonas strains. Following the nomenclature of Moreno et al.,6 we refer to these ncRNAs as crcZ (psr1) and crcX (psr2).
Results
CbrA and CbrB regulate expression of crcZ and crcX in P. syringae DC3000
We previously predicted that crcZ (psr1; PSPTO_5668), crcX (psr2; PSPTO_5669) and crcY (psr3) were regulated by RpoN in P. syringae,3 based on the presence of RpoN promoter motifs upstream of each gene. Although crcY (psr3) is also flanked by a likely RpoN promoter, this gene is disrupted by an insertion sequence and is not expressed.3 To test if crcZ (PSPTO_5668) and crcX (PSPTO_5669) are regulated by RpoN in P. syringae DC3000, we compared expression of both genes in a wild-type strain vs. a mutant in which rpoN has been disrupted. Both ncRNAs had slightly reduced levels in the RpoN mutant (Fig. 1), suggesting at least partial regulation by RpoN in P. syringae DC3000, but expression was not completely eliminated. Therefore, we examined whether additional transcriptional start points for crcZ and crcX could be detected in the RpoN mutant. Our results indicate that for both crcZ and crcX, alternative transcriptional start point can be detected in the RpoN mutant (data not shown). These transcriptional start points are 2–4 bases downstream of the transcriptional start points detected in the wild-type strain. This result suggests that transcripts for crcZ and crcX arise from two promoters, one recognized by RpoN and the other by a different sigma factor. It has been shown that crcZ in P. aeruginosa and crcZ and crcY in P. putida, are regulated by RpoN5,6 but a second promoter was not reported in these species.
Figure 1. Regulation of CrcZ and CrcX in P. syringae. Relative levels of the crcZ and crcX ncRNAs, compared to WT, measured by quantitative RT-PCR, in cells grown overnight in KB medium. The levels of gene expression in each sample were calculated as fold expression ratio using gyrA as a gene for normalization. The values are averages of three independent experiments and error bars represent standard deviation.
In P. aeruginosa and P. putida, expression of crcZ is activated by the CbrA/CbrB two component sensor-regulator system.5,6 However, in P. putida activation of crcY is thought to use a different transcriptional regulator.6 To investigate the role of CbrAB in expression of crcZ and crcX in P. syringae DC3000, we used qRT-PCR to compare RNA levels in wild-type P. syringae DC3000 strain with those lacking cbrB or cbrA. In cells grown in KB medium, inactivation of the cbrB gene significantly reduced expression of both crcZ and crcX (Fig. 1). Additionally, inactivation of cbrA also reduced expression of both crcZ and crcX. The change was not as dramatic as that observed with the cbrB mutant, suggesting CbrB may interact with another histidine kinase, allowing for transcription in the absence of CbrA.
Expression of CrcZ and CrcX are influenced by the carbon source
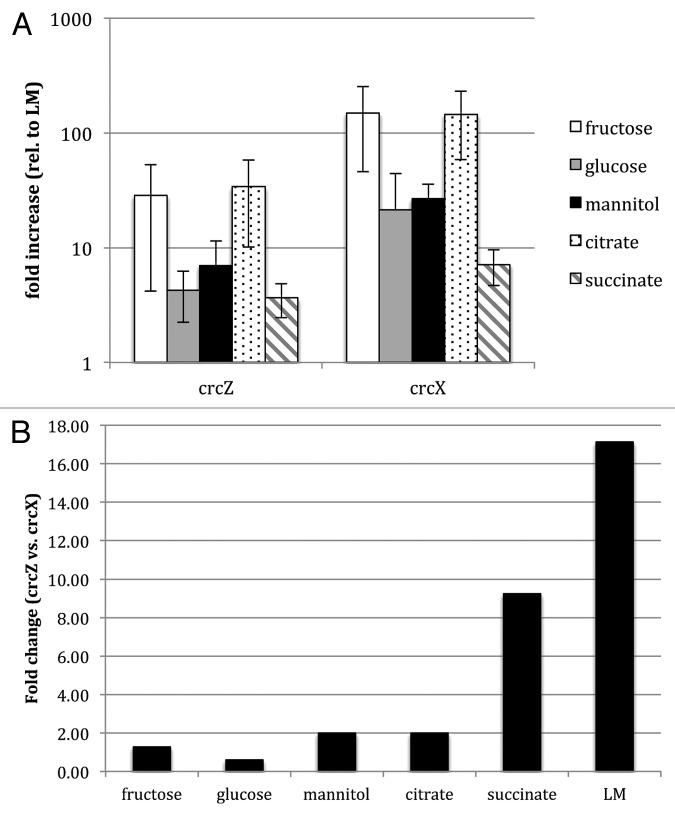
The levels of crcZ in P. aeruginosa vary according to the carbon source being used and correlate with the relief of catabolite repression.5 To analyze the influence of carbon source on mRNA levels of crcZ and crcX in P. syringae DC3000 we performed qRT-PCR to evaluate the relative levels of crcZ and crcX in cells grown in a minimal medium supplemented with various carbon sources. We found that compared with mRNA levels in LM medium at mid-exponential phase, increased levels of crcZ and crcX were observed in cells grown in fructose, glucose and mannitol, with very high levels being observed when fructose was used as the sole carbon source (Fig. 2A). mRNA levels for crcZ and crcX were also elevated when grown with the organic acid citrate as a sole carbon source. The relative abundance of crcZ vs. crcX also varies depending on the medium. In LM mid- exponential cultures and in succinate there is more crcZ than crcX, but when cells are grown in conditions that induce expression of the ncRNAs (fructose, mannitol, glucose, citrate) the ncRNAs are present in approximately the same ratio (Fig. 2B). A similar shift was reported in P. putida, where crcZ is eight times more abundant than crcY in LB-mid-log cells, but in non-repressing conditions (LB medium at stationary phase, or M9 medium supplemented with either succinate or citrate) the ratio decreases. In P. putida, levels of crcZ and crcY are high when grown in succinate (less preferred carbon source) and crcZ and crcY are lower in P. syringae when grown in succinate (a good carbon source). In contrast to what is observed in P. putida, crcZ is much more abundant than crcX in P. syringae when grown in succinate (Fig. 2B).
Figure 2. (A) Effect of carbon source on levels of crcZ and crcX. The values shown correspond to the ratios of the RNA levels observed for each condition relative to those observed in cells growing exponentially in LB medium as reference. The standard error is indicated. (B) Comparison of the abundance of crcZ relative to that of crcX under the specified growth conditions, determined by real-time RT-PCR. RNA samples were the same as those used in the assays presented in A.
CrcZ and CrcX influence growth of P. syringae
Inactivation of crcZ in P. aeruginosa, or crcZ and crcY in P. putida leads to reduced growth in non-preferred carbon sources.5,6 For P. syringae deletion of both crcZ and crcX was required to obtain a difference in growth in mannitol and fructose (Fig. 3), suggesting that crcZ and crcX are functionally redundant in DC3000. The wild-type and mutant strains grew to similar levels with succinate as the sole carbon source (Fig. 3). Decreased growth was also observed when citrate (a preferred carbon source for P. aeruginosa) was used as the sole carbon source. This might indicate that in P. syringae DC3000 the organic acid citrate is a non-preferred carbon source.
Figure 3. Effect of carbon source on growth of P. syringae crcZ and crcX mutants. Growth of wild-type DC3000 (solid black diamonds and black bar), ΔcrcZ (dark gray squares and dark gray bar), ΔcrcX (light gray triangles and light gray bar) and ΔcrcZX (white circles and white bar) in NCE supplemented with 10mM succinate, 10mM mannitol, 10 mM citrate, 12mM arabinose, or 1% myo-inositol. Samples for growth analyses were harvested at four time points (2, 4, 6 and 24 h.).
Two predicted mRNA targets for Crc in P. syringae DC3000 are the L-arabinose transporter permease protein (PSPTO_2640) and a myo-inositol 2-dehydrogenase (PSPTO_3494).9 Therefore, we tested whether deletion of crcZ and crcX influenced the growth rate of DC3000 cultured using these substrates as sole carbon sources. Reduced growth was observed for the double mutant when grown with either arabinose or myo-inositol (Fig. 3).
Sequence similarity and secondary structure of CrcX,Y,Z homologs in P. syringae
Since crcY is disrupted in DC3000, we were interested in addressing two important questions: (1) what is the prevalence of the Crc family of ncRNAs in other sequenced P. syringae pathovars and (2) are there any P. syringae strains in which crcY was disrupted by insertion sequences, as in DC3000? Since exact coordinates for the crc ncRNAs are now known and also a large number of pseudomonas genomes have been sequenced we are able to address these questions. Using the mapped transcriptional start sites, mapped 3′-ends and transcriptional activity profiles3 we determined the genomic boundaries for crcZ, crcX, and crcY in DC3000. Because crcY is disrupted by an insertion element, the two partial sequences were spliced together to form one continuous “virtual” ncRNA. This information was then used to infer the genomic boundaries for crcZ, crcX and crcY in P. syringae strains B728a and 1448A (Table 1). Putative transcript sizes varied from 342 nt to 368 nt. A consensus structure generated using LocARNA exhibits four to five conserved regions containing CA motifs (Fig. 4). The nine ncRNAs are highly conserved, but also exhibit several variable regions. For comparison, the alignments for crcZ (psr1), crcX (psr2) and crcY (psr3) from DC3000, B728a and 1448A were performed separately (see Figs. S1−S3). crcZ (psr1) and crcX (psr2) contain five conserved “CA” regions, whereas crcY (psr3) contains four “CA” regions.
Table 1. Pseudomonas syringae crcZ, crcX and crcY coordinates.
| P. syringae strain | ncRNA | 5′end | 3′end | length |
|---|---|---|---|---|
|
DC3000 |
crcZ |
1046951c |
1046608c |
343 |
| |
crcX |
1778751c |
1778383c |
368 |
| |
crcY |
3047265c |
3044933c |
361a |
|
B728a |
crcZ |
941123c |
940780c |
343 |
| |
crcX |
4476099 |
4476467 |
368 |
| |
crcY |
2865015c |
2864663c |
352 |
|
1448A |
crcZ |
1024980c |
1024637c |
343 |
| |
crcX |
1738895c |
1738527c |
368 |
| crcY | 3034385c | 3034033c | 352 |
a For P. syringae pv tomato strain DC3000 crcY, the length was determined by omitting the insertion sequence and splicing the two fragments together. CExpression occurs on the negative, complementary, strand.
Figure 4. LocARNA alignment of the nine crcZ,X,Y (psr1,2,3) genes in Pseudomonas syringae pv tomato str. DC3000, Pseudomonas syringae pv phaseolicila str. 1448A and Pseudomonas syringae pv syringae str. B728a. # = GC SS_cons represents the consensus structure. In the consensus sequence matching nested parentheses are indicated by “().” The symbol “.” indicates unpaired regions. Sequences containing “CA” motifs are highlighted with pink boxes.
Construction of a CrcZ,X,Y covariance model
Using the nine sequences from DC3000, B728a and 1448A, a co-variance model (CM) was constructed using the Infernal toolset. The model was used to search all fully sequenced and draft genomes belonging to the Pseudomonadaceae family to locate crc candidates that may have been overlooked in the earlier CMFinder search that relied on the gamma-150 motif.4 The results of the CMsearch are shown in Table 2 and Table S3. The scans retrieved all of the candidates using an earlier model based on the gamma-150 motif,4 as well as identified additional crc candidates. All pseudomonads, except one, had at least one crc candidate ncRNA (Table 2 and Table S3). The exception, P. geniculate was isolated from river water, but its lineage is currently in question10 and it is listed as an unclassified Pseudomonas in the NCBI taxonomy browser. Therefore, it is not surprising that the CMSearch did not find a crc candidate in this organism. Only one crc candidate ncRNA was found in all P. aeruginosa and P. mendocina strains. A single candidate crc ncRNA was found in P. stutzeri strain SDM, Pseudomonas 2_1_26, Pseudomonas S9 and P. psychrotolerans L19. Two crc candidates were found in other pseudomonads such as P. fluorescens, P. stutzeri and P. putida strains (Table 2 and Table S3).
Table 2. Crc ncRNA candidates identified in fully sequenced Pseudomonas genomes using CMsearch.
| Strain | Accession | Score | 5′-end | 3′-end |
|---|---|---|---|---|
|
Acinetobacter baumannii AYE |
CU459140.1 |
15.73 |
78400 |
78422 |
|
Azotobacter vinelandii DJ |
CP001157.1 |
94.02 |
4303297 |
4303638 |
|
Azotobacter vinelandii DJ |
CP001157.1 |
176.76 |
452815c |
452475c |
|
Pseudomonas aeruginosa LESB58 |
FM209186.1 |
145.66 |
5642408 |
5642759 |
|
Pseudomonas aeruginosa M18 |
CP002496.1 |
143.25 |
5361488 |
5361839 |
|
Pseudomonas aeruginosa NCGM2 S1 |
AP012280.1 |
145.03 |
888676c |
888325c |
|
Pseudomonas aeruginosa PA7 |
CP000744.1 |
136.47 |
5610663 |
5611013 |
|
Pseudomonas aeruginosa UCBPP-PA14 |
CP000438.1 |
144.16 |
5580520 |
5580871 |
|
Pseudomonas aeruginosa |
AE004091.2 |
145.4 |
5308589 |
5308940 |
|
Pseudomonas brassicacearum NFM421 |
CP002585.1 |
216.88 |
5835658 |
5836004 |
|
Pseudomonas brassicacearum NFM421 |
CP002585.1 |
244.44 |
2260512c |
2260075c |
|
Pseudomonas entomophila L48 |
CT573326.1 |
191.09 |
5029853 |
5030198 |
|
Pseudomonas entomophila L48 |
CT573326.1 |
191.06 |
3150228 |
3150563 |
|
Pseudomonas fluorescens F113 |
CP003150.1 |
243.42 |
4575452 |
4575888 |
|
Pseudomonas fluorescens F113 |
CP003150.1 |
217.08 |
5792772 |
5793163 |
|
Pseudomonas fluorescens Pf0 1 |
CP000094.2 |
237.33 |
4135995 |
4136343 |
|
Pseudomonas fluorescens Pf0 1 |
CP000094.2 |
220.71 |
5425228 |
5425569 |
|
Pseudomonas fluorescens Pf-5 |
CP000076.1 |
221.77 |
4546202 |
4546554 |
|
Pseudomonas fluorescens Pf-5 |
CP000076.1 |
211.49 |
6040962 |
6041305 |
|
Pseudomonas fluorescens SBW25 |
AM181176.4 |
217.28 |
5747060 |
5747403 |
|
Pseudomonas fluorescens SBW25 |
AM181176.4 |
207.9 |
4288010 |
4288357 |
|
Pseudomonas fulva 12 X |
CP002727.1 |
127 |
4826693 |
4826974 |
|
Pseudomonas fulva 12 X |
CP002727.1 |
90.4 |
1039803c |
1039482c |
|
Pseudomonas mendocina NK 01 |
CP002620.1 |
160.54 |
4286402 |
4286744 |
|
Pseudomonas mendocina ymp |
CP000680.1 |
158.25 |
3959213 |
3959556 |
|
Pseudomonas putida BIRD 1 |
CP002290.1 |
211.49 |
4910564 |
4910909 |
|
Pseudomonas putida BIRD 1 |
CP002290.1 |
184.83 |
2566512c |
2566177c |
|
Pseudomonas putida F1 |
CP000712.1 |
214.87 |
5102227 |
5102575 |
|
Pseudomonas putida F1 |
CP000712.1 |
190.89 |
2543815c |
2543483c |
|
Pseudomonas putida GB 1 |
CP000926.1 |
214.82 |
5246762 |
5247112 |
|
Pseudomonas putida GB 1 |
CP000926.1 |
190.63 |
2668606c |
2668277c |
|
Pseudomonas putida KT2440 |
AE015451.1 |
213.48 |
5338277 |
5338622 |
|
Pseudomonas putida KT2440 |
AE015451.1 |
198.23 |
4013244 |
4013576 |
|
Pseudomonas putida S16 |
CP002870.1 |
209.22 |
5140236 |
5140582 |
|
Pseudomonas putida S16 |
CP002870.1 |
195.87 |
3401961 |
3402289 |
|
Pseudomonas putida W619 |
CP000949.1 |
214.69 |
814356c |
814007c |
|
Pseudomonas putida W619 |
CP000949.1 |
202.25 |
2173887c |
2173555c |
|
Pseudomonas stutzeri A1501 |
CP000304.1 |
174.07 |
2072801 |
2073136 |
|
Pseudomonas stutzeri A1501 |
CP000304.1 |
133.16 |
3565446 |
3565788 |
|
Pseudomonas stutzeri ATCC 17588 LMG 11199 |
CP002881.1 |
165.37 |
1910377 |
1910712 |
|
Pseudomonas stutzeri ATCC 17588 LMG 11199 |
CP002881.1 |
132.98 |
3575287 |
3575629 |
|
Pseudomonas stutzeri DSM 4166 |
CP002622.1 |
173.57 |
2058333 |
2058668 |
|
Pseudomonas stutzeri DSM 4166 |
CP002622.1 |
134.22 |
3663966 |
3664308 |
|
Pseudomonas syringae phaseolicola 1448A |
CP000058.1 |
340.83 |
1738895c |
1738527c |
|
Pseudomonas syringae phaseolicola 1448A |
CP000058.1 |
309.67 |
3034385c |
3034033c |
|
Pseudomonas syringae phaseolicola 1448A |
CP000058.1 |
294.01 |
1024980c |
1024622c |
|
Pseudomonas syringae B728a |
CP000075.1 |
333.11 |
4476099 |
4476467 |
|
Pseudomonas syringae B728a |
CP000075.1 |
319.24 |
2865015c |
2864663c |
|
Pseudomonas syringae B728a |
CP000075.1 |
291.23 |
941123c |
940765c |
|
Pseudomonas syringae tomato DC3000 |
AE016853.1 |
337.19 |
1778751c |
1778383c |
|
Pseudomonas syringae tomato DC3000 |
AE016853.1 |
293.85 |
1046951c |
1046593c |
|
Pseudomonas syringae tomato DC3000 |
AE016853.1 |
193.29 |
3045294c |
3044933c |
| Pseudomonas syringae tomato DC3000 | AE016853.1 | 107.52 | 3047265c | 3047005c |
Using this model, multiple crc candidates were found in all P. syringae strains. The model detected at least three crc candidates in the P. syringae strains, with the exception of P. syringae pv morsprunorum M302280PT (2 candidates). P. syringae strains which potentially have more than three crc candidate ncRNAs include, P. syringae pv japonica M301072PT (5; two of which contain 7 base pair disruptions), P. syringae pv oryzae (4), and P. syringae pv pisi (5; one of which contains a 7 base pair disruption) and P. syringae pv mori 301020 (4). As for the tomato pathovars, DC3000 appears to be the only strain in which one of the crc ncRNAs (crcY) has been disrupted by an insertion.
The gamma-150 motif was also reported in Azotobacter vinelanii.4 To determine if our model yielded similar results, the new model was used to search closely related strains from the Pseudomonadaceae family (Azotobacter, Cellvibrio, Moraxella, Psychrobacter and Acinetobacter). Other than the two ncRNA candidates previously reported for Azotobacter vinelanii, we failed to detect crcZXY ncRNAs in these strains. Therefore, the Crc family of ncRNAs seems to be unique to the Pseudomonads and Azotobacter strains. The Azotobacter case is consistent with suggestions that A. vinelandii may in fact be a Pseudomonad,11 based on genome similarity. In total, our model detected 237 Crc family ncRNA candidates.
Clustering of CrcZ,X,Y candidates
The Crc ncRNA candidates identified in the closed genomes were clustered using RNAcluster, to ascertain the structural relationship between the candidates. The resulting dendogram is shown in Figure S4. The known crcZ candidates (highlighted in blue) cluster together, as well as the previously reported crcY ncRNAs (highlighted in green) and crcX ncRNAs (highlighted in red). To further investigate the relationship of the crc candidates, the Crc ncRNA candidates identified in the P. syringae genomes (closed and draft genomes) were clustered separately (Fig. S5). Overall, 105 of 107 sequences cluster into three main families.
Deciphering the relationship of the crc ncRNA candidates
crcZ
To evaluate the differences between crc ncRNA candidates we examined their genomic contexts and classified each ncRNA based on its conserved genetic linkage. For crcZ, we first identified strains containing homologs for PcnB (PSPTO_0963) and PSPTO_0964. Any crc ncRNA candidates located between the PSPTO_0963 and PSPTO_0964 homologs were considered to represent members of the crcZ subfamily. 96 CMsearch candidates shared this genomic arrangement (Table S3). Nine other CMsearch candidates were located either adjacent to a PSPTO_0964 homolog or a PSPTO_0963 homolog. In these cases complete conservation is difficult to determine since the positions of the hits within the contigs does not allow for sufficient context and therefore characterizing them as crcZ-like is less certain. Overall, with the exception of P. geniculate (which contains no instance of crcZ) all of Pseudomonas strains sequenced to date (103/104) contain at least one crc ncRNA candidate (crcZ) adjacent to pcnB. P. fluva contained two ncRNA candidates with this particular genomic arrangement, yielding a total of 105 crcZ-like candidates.
crcX
To evaluate the distribution and conservation of crcX we identified strains containing PSPTO_1621 and PSPTO_1622 homologs and compared the locations of the homologs with the coordinates from the CMSearch. Those crc ncRNA candidates located between a PSPTO_1621 and PSPTO_1622 homologs were considered to represent the crcX class. Of the 26 putative crcX ncRNAs identified all were found to be exclusive to P. syringae strains (Table S3). In addition, 15 other CMsearch candidates were located either upstream of PSPTO_1621 or PSPTO_1622 homologs. As with crcZ, some of these may be bona fide crcX instances but their status is provisional until their corresponding genome sequences can be closed. Taken together, all except one of the P. syringae strains (37 out of 38) contained a crc ncRNA candidate in proximity to PSPTO_1621 or PSPTO_1622 homologs. The single exception was P. syringae pv morsprunorum M302280PT, which does not appear to contain a crcX ncRNA. All together, the data suggest that the ncRNA crcX is exclusive to P. syringae strains.
crcY
In DC3000, PSPTO_2740 and PSPTO_2741 are transposon-related sequences that disrupt crcY, but are not found in P. syringae strains B728a or 1448A. To classify putative crcY candidates, we instead used PSPTO_2742 and PSPTO_2739 as context markers and identified strains containing their homologs. We then asked if a candidate ncRNA was located between the homologs or was located in close proximity (< 400 bps) of these homologs. Seventy-six CMsearch candidates satisfied these criteria and represent putative crcY ncRNAs. Interestingly, this ncRNA appears to be present in all Pseudomonas strains, except for P. aeruginosa and P. mendocina strains and P. syringae pv aceris M302273PT.
Several CMsearch candidates were located in genomic locations that could not be classified using the BLAST analyses described above. Some occurred in strains that have more than three crc ncRNA candidates (P. syringae japonica, P. syringae mori, P. syringae oryzae and P. syringae pisi). In these pathovars, we found that the CMsearch candidates were located on extremely short contigs (< 500 bps) and in most cases the contig is smaller than the co-variance model itself.
Regulatory features of the crc candidate ncRNAs
The upstream regions of the 96 crcZ candidates were aligned to identify potential conserved regulatory features. The analysis revealed the existence of several highly conserved motifs (Fig. S6). One motif resembles the RpoN promoter region which has a highly characteristic -24/-12-regions with the consensus sequence: 5′-YTGGCACG-N4-TTGCW-3′, (the bold G and C at positions -24 and -12 relative to the start of transcription).12 This motif is similar to the RpoN binding site for P. fluorescens and P. putida and its presence is consistent with reports that RpoN regulates the expression of crcZ.5,6 Further upstream we also identified a sequence similar to the published consensus CbrB binding site for P. aeruginosa (cTGTTACc-N3/12 -cGTAACAg)13 (Figs. S6 and S7), suggesting that all crcZ ncRNAs are regulated by the CbrA/CbrB two-component system.
Our data also suggests that crcX in DC3000 is regulated by CbrB (Fig. 1). To explore this further, regions upstream of crcX candidates were aligned (Figs. S7 and S8). As expected, a putative RpoN promoter region was present. We also found one-half of the predicted CbrB binding site at -154 to -147 (TGTTACC). A motif similar to the other half of the predicted CbrB binding site (GTAACAC) was located 29 bases away from the other half binding site. This interval is larger than the 3-bp or 12-bp spacing reported for some CbrB binding sites13 but is similar to the spacing observed upstream of crcY in P. putida.6 However, it has been reported that inactivation of CbrB has little effect on the expression of crcY in that organism.6 The same palindrome with a 29 bp spacer occurs upstream of putative crcY candidate ncRNAs (Figs. S7 and S9).
Discussion
In this study we find that the Crc ncRNA family is common throughout the genus Pseudomonas. As with the rsmX ncRNAs, P. syringae strains contain multiple copies of crc ncRNAs.14 Paralogous ncRNAs can act redundantly or additively within the bacteria cell.15-19 The growth analyses presented here suggest that CrcZ and CrcX act redundantly in at least one pathway. However, it is possible that they participate in other pathways as well, where they perform distinct functions.
crcZ is highly conserved and appears to be the only crc ncRNA present in the P. aeruginosa strains, and others that are able to cause human disease such as P. mendocina. Although some members of the Pseudomonacaeae such as Acinetobacter and Morexella contain PcnB homologs, our model did not detect candidate ncRNAs in these cases. Acinetobacter baylyi does have a Crc protein involved in catabolite repression control and aromatic compound catabolism and Crc is reported to act post-transcriptionally as in the Pseudomonads,20 but in contrast to the Pseudomonads, it displays a strong effect on transcript stability. It is possible that in Acinetobacter the levels of Crc are modulated at the transcript level and there is no requirement for ncRNAs. Alternatively, Acinetobacter strains may have ncRNAs distinct from crcX,Y and Z that regulate the levels of Crc protein. Of the 104 bacteria containing CrcY/Z/X candidates, only one (Pseudomonas aeruginosa strain 152504_uid62725) has no PSPTO_0079 homolog (data not shown). However, the absence of a Crc ortholog may be an artifact, since a “closed” genomic sequence is not yet available for this strain. Overall our results show that crcYZX ncRNAs are nearly always accompanied by a Crc ortholog.
One of the most exciting results from this study is that our analysis revealed that CrcX is only found in the P. syringae strains. This finding is important for several reasons. One is that this ncRNA or its flanking regions may be useful as a diagnostic tool to identify members of this group of plant pathogens. Second is that the presence of this region in P. syringae strains may be linked to species-specific traits found only in syringae strains. P. syringae is physiologically different from non-pathogenic Pseudomonads (using the LOPAT scheme which evaluates oxidase production, arginine dihydrolase, activity, aerobic growth and carbon source utilization). Experimental analyses of nutrient assimilation by pseudomonads indicate that plant pathogenic P. syringae strains assimilate a restricted range of nutrient sources compared with other pseudomonads21 and is physiologically specialized for growth using the most abundant amino acids in plant tissues and on the plant surface. In contrast, the P. syringae lineage lacks some metabolic reactions that are conserved in other Pseudomonads.22 CrcX may therefore be involved in the precise control of the limited pathways used for primary metabolism.
Although several reports have provided insight into the biochemistry and genetics of metabolism in pseudomonads, relatively little is known about carbon catabolism in P. syringae. Catabolite repression control (CRC) is an important global control system in Pseudomonas that fine tunes metabolism to optimize growth in a variety of different environments.9 Crc is involved in several Pseudomonas species in catabolite repression of the branched-chain keto acid dehydrogenase23 and of alkane degradation,7 as well as of a number of enzymes involved in aromatic compound degradation.24 In addition to modulating metabolism, in P. aeruginosa, Crc influences susceptibility to antibiotics, expression of Type III secretion, expression of quorum sensing-regulated virulence factors, such as pyocyanin25 and potentially other virulence functions.26 Sugars such as glucose, sucrose and fructose are known to be inducers of the P. syringae TTSS genes, whereas tricarboxylic acids (TCA) intermediates can suppress T3SS in vitro. Fructose and citrate utilization pathways used by P. syringae are upregulated when cells are exposed to apoplast extracts.21 Therefore, during plant infection we hypothesize that when T3SS is active, the expression of these ncRNAs may support utilization of these carbon sources.
In contrast to the Rsm system where there are several RNA binding proteins, the presence of only one Crc gene in the pseudomonads implies that all of the Crc ncRNAs function through a single protein. Bioinformatic analysis of predicted Crc binding sites in Pseudomonas genomes9 suggests that some targets are genus-wide and related to central metabolism, whereas others are predicted to be species-specific or unique to particular strains. Predictions for DC3000 include algP (alginate metabolism), PSPTO_3494 (myo-inositol 2-dehydrogenase), proteins involved in chemotaxis, and several transcriptional regulators. Our data implicate CrcZ and CrcX in myo-inositol utilization. Studies are underway to investigate the role of these ncRNAs in other pathways.
Materials and Methods
Bacterial growth conditions
For routine growth, Pseudomonas syringae pv tomato DC3000 was maintained at 28°C on King’s broth (KB)27 agar plates. For growth studies with various carbon sources, bacterial cells were grown in LM at 28°C with shaking overnight. The cells were sub-cultured to an OD600 of 0.1 in LM or No-carbon-E-minimal medium (NCE)28 supplemented with fructose (10 mM), glucose (10 mM), mannitol (10 mM), citrate (10 mM), succinate (10 mM), myo-inositol (1%), or arabinose (12mM) as the carbon source.
RNA isolation
Total RNA was isolated from cultured cells using the RNeasy kit (Qiagen) according to the manufacturer’s protocol using the on-column DNase treatment with the exception that lysozyme was used at a concentration of 5 mg/ml. Isolated RNA was further treated twice with 2 units of DNase (Ambion) to eliminate DNA contamination. RNA was purified from the DNase mixture using MinElute column (Qiagen).
5′ and 3′ RACE
5′ RACE assays were performed using the 5′ RACE System for the Rapid Amplification of cDNA Ends, v2.0 kit (Invitrogen) as described by Moll et al.14 Five hundred nanograms to 1 μg of isolated RNA were used in each reaction and the procedure was performed following the manufacturer’s protocol. Following amplification with a gene specific primer (GSP2), PCR products were separated on a 2% agarose gel and bands of interest were excised, gel-eluted (Zymoclean Gel DNA Recovery kit, Zymo Research) and sequenced (Genomics facility, Life Sciences Core Laboratories, Cornell University). Oligonucleotides used for reverse transcription and PCR are listed in Table S1.
3′ RACE was performed as described by Moll et al.14This protocol was adapted from Argaman et al.29 Briefly, 1 μg of RNA was mixed with 100 pmol of RNA adaptor (5′- phosphate-UUC ACU GUU CUU AGC GGC CGC AUG CUC-idT -3′), heat-denatured at 95°C for 5 min, then quick-chilled on ice. The adaptor was ligated at 17°C for 12 h in the presence of 40 units of T4 RNA ligase (New England Biolabs) and 40 units of RNase OUT (Ambion) in a buffer containing 50 mM TRIS-HCl (pH 7.9), 10 mM MgCl2, 4 mM DTT, 150 μM ATP and 10% DMSO. The ligated RNA product was purified from the reaction using the RNA Clean and Concentrator -5 kit (Zymo Research) and reverse-transcribed using 20 pmol of a single primer complementary to the RNA adaptor (A1). Reverse transcription was performed using the Thermoscript RT system (Invitrogen) according to the manufacturer’s protocol. The products of reverse transcription were amplified using a 2 μl aliquot of the RT reaction, 20 pmol of each gene-specific and adaptor-specific primer (A2) and 1X Ex-Taq Polymerase mix (TaKaRa Bio). Cycling conditions were as follows: 95°C/2 min; 35 cycles of 94°C/30 sec, 57°C/45 sec, 72°C/30 sec; 72°C/10 min. PCR separation, gel extraction, cloning and sequencing were performed as described for 5′ RACE. Primers used in 3′ RACE analyses are shown in Table S1.
Construction of the RpoN mutant
Construction of the DC3000 RpoN mutant was performed by PCR amplification of an internal sequence of rpoN, corresponding to nucleotides 100 to 700 of the coding sequence, from the DC3000 genome and cloned into pKnockout-Ω.30 The resulting plasmid was introduced into DC3000 via electroporation. Since pKnockout cannot replicate in DC3000, single-crossover integrants were selected for resistance to spectinomycin. Orientation of integration was determined by PCR.
Construction of cbrB and cbrA mutants
Flanking regions of approximately 800 bps upstream and downstream of cbrA were amplified using the oligomers ΔcbrA1fwd and ΔcbrA1rev, or ΔcbrA2fwd and ΔcbrA2rev, respectively. ΔcbrA1fwd and ΔcbrA2rev contain an external XbaI site and ΔcbrA1rev and ΔcbrA2fwd contain an NdeI site, to allow ligation of the two flanking DNA fragments. The fragments were amplified using the Expand high-fidelity PCR system (Roche). The PCR fragments were gel purified with the Zymoclean gel DNA recovery system. The two fragments, ΔcbrA1and ΔcbrA2 or ΔcbrA1 and ΔcbrA2, were digested with NdeI, joined by ligation and then amplified using the primers ΔcbrA1fwd and ΔcbrA2rev. The product was gel purified, digested with XbaI and cloned into pK18mobsacB using the XbaI restriction site. For construction of the cbrB mutant, 1.0 kb regions directly upstream and downstream of cbrB were amplified by PCR. Gel purified PCR fragments were joined by SOEing in a second PCR amplification with primers containing XbaI restriction sites. The product was gel purified using Gel DNA Recovery Kit (Zymo Research), digested XbaI and cloned into pK18mobsacB cut with the same restriction enzyme.
The plasmids were transformed into DC3000 by electroporation followed by selection for integration on LM containing 50 μg/ml kanamycin. Colonies then transferred to 10% sucrose medium to select for crossover events that resulted in the loss of the sacB gene. Sucrose resistant colonies were screened by PCR and positive clones (those containing the deletion) were confirmed by sequencing.
Construction of crcZ, crcX and crcZ/X double mutant
To make the ∆crcZ mutant strain, a deletion was created using a pK18mobsacB plasmid.31 pK18mobsacB/∆crcZ was created by PCR amplification of DNA fragments of approximately 1.0 kb upstream and downstream of the ncRNA. Gel purified PCR fragments were joined by SOEing in a second PCR amplification with primers containing HindIII and BamHI restriction sites. The product was gel purified using Gel DNA Recovery Kit (Zymo Research), digested with HindIII and BamHI and cloned into pK18mobsacB cut with the same restriction enzymes. Similarly, pK18mobsacB/∆crcX was created by PCR amplification of DNA fragments of approximately 1.0 kb that flank crcX. Gel purified PCR fragments were joined by a second PCR amplification with primers containing HindIII and BamHI restriction sites. The product was gel purified, digested with HindIII and BamHI and cloned into HindIII/BamHI digested pK18mobsacB.
The pK18mobsacB deletion constructs were confirmed by sequencing at Cornell University Life Sciences Core Laboratories Center before introducing into DC3000 via electroporation. Integration events were selected on KB medium containing 50 μg/ml kanamycin and then transferred to 10% sucrose medium to select for crossover events that resulted in the loss of the sacB gene. Sucrose resistant colonies were screened by PCR and clones containing the deletion were confirmed by sequencing. To construct the crcZX double mutant, the pK18mobsacB/∆crcX was introduced into the ∆crcZ mutant strain.
Quantitative RT-PCR
Cells were gown at 28°C in flasks in either LM or in NCE medium supplemented with the indicated carbon source. At mid-exponential phase or at stationary phase, samples were collected, harvested by centrifugation and immediately frozen at -80°C. RNA was prepared with the RNeasy Kit (Qiagen, Valencia, CA) as described above.
Total RNA (100 ng) was reverse transcribed in a thermocycler using the qScript cDNA Supermix (Quanta Biosciences) according to the manufacturer’s instructions. qPCR was performed with 10 mg of cDNA using IQ SYBR green Supermix (Bio-Rad) on a iQ5 multicolor real-time detection system (BioRad). DNA contamination and the formation of primer dimers were assessed by using controls lacking reverse transcriptase and template, respectively. The production of nonspecific products was determined by the dissociation protocol included in the software provided with the machine. The resulting cycle thresh-old (Ct) values were calculated by the software and analyzed using the 2 -∆∆Ct method. The primers are listed in Table S1. The Ct values of each gene tested were normalized to the Ct values of the housekeeping genes gyrA.
Computational analyses
A list of all genomes used in this study and their GenBank and Refseq accession numbers is provided in Table S2. The nucleotide sequences of the three crc genes from P. syringae pv tomato DC3000, P. syringae pv syringae B728a and P. syringae pv phaseolicola 1448A were manually extracted and assembled into a single FASTA file. Prediction of secondary structure, consensus model building, calibration and searching were performed as described in.14 The following software package versions were used: RNAclust v1.2.5 (www.bioinf.uni-leipzig.de/~kristin/Software/RNAclust/); RNAalifold32 ViennaRNA v1.8.5 (http://rna.tbi.univie.ac.at/cgi-bin/RNAalifold.cgi); locarna v1.6.233; and infernal v1.0.2.34 Results from the CM search were filtered to retain matches with E-values less than 1e-3. This resulted in a total of 237 CMfinder candidates.
When clustering the crc candidates, since some were substantially shorter than crc genes used to build the model, we chose to discard all sequences shorter than 280 bases. This resulted in a total of 107 sequences used in the clustering for the closed genomes and 51 sequences for clustering candidates from the P. syringae genomes (draft and closed). Nw_display from the Newick-Utils package v1.635 was used to render the clustering results as a dendogram.
Syntenic regions were identified based on homology of neighboring genes. Homology was estimated using tblastn from the NCBI Blast+ package version 2.2.26.36 All of the genomes listed in Table S2 were used to construct a single database and the following protein sequences were used as the query input: PSPTO_0963, PSPTO_0964, PSPTO_1621, PSPTO_1622, PSPTO_2739 and PSPTO_2742. An e-value cutoff of 1e-10 was used.
For each high quality CM match, a summary was generated listing the match and any high quality BLAST hit to the query protein sequences that fell within 50,000 bases of the CM match. These summaries were manually curated and given putative assignment as crcX, crcY, crcZ, or “unknown” genes.
To discover potential regulatory features upstream of crc candidates, 198 bases upstream of the putative crcZ genes was extracted and aligned using “clustalw2” from the CLUSTALW package v2.0.1237 using default parameters. This process was repeated for the putative crcX and crcY genes. Mview from the MView package v1.5238 was used to generate a custom colormap.
Supplementary Material
Acknowledgments
We would like to thank Nola Pellegrini for construction of the RpoN mutant strain, and Charlene Maciak for mapping the 3′-ends. The US. Department of Agriculture (USDA) is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely for the purposes of providing specific information and does not imply recommendation or endorsement by the USDA.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23019
References
- 1.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, et al. Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol. 2010;192:2359–72. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–19. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnleitner E, Abdou L, Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2009;106:21866–71. doi: 10.1073/pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreno R, Fonseca P, Rojo F. Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol Microbiol. 2012;83:24–40. doi: 10.1111/j.1365-2958.2011.07912.x. [DOI] [PubMed] [Google Scholar]
- 7.Yuste L, Rojo F. Role of the crc gene in catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J Bacteriol. 2001;183:6197–206. doi: 10.1128/JB.183.21.6197-6206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fonseca P, Moreno R, Rojo F. Pseudomonas putida growing at low temperature shows increased levels of CrcZ and CrcY sRNAs, leading to reduced Crc-dependent catabolite repression. Environ Microbiol. 2012 doi: 10.1111/j.1462-2920.2012.02708.x. In press. [DOI] [PubMed] [Google Scholar]
- 9.Browne P, Barret M, O’Gara F, Morrissey JP. Computational prediction of the Crc regulon identifies genus-wide and species-specific targets of catabolite repression control in Pseudomonas bacteria. BMC Microbiol. 2010;10:300. doi: 10.1186/1471-2180-10-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol. 2000;50:1563–89. doi: 10.1099/00207713-50-4-1563. [DOI] [PubMed] [Google Scholar]
- 11.Özen AI, Ussery DW. Defining the Pseudomonas genus: where do we draw the line with Azotobacter? Microb Ecol. 2012;63:239–48. doi: 10.1007/s00248-011-9914-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrios H, Valderrama B, Morett E. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–13. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdou L, Chou HT, Haas D, Lu CD. Promoter recognition and activation by the global response regulator CbrB in Pseudomonas aeruginosa. J Bacteriol. 2011;193:2784–92. doi: 10.1128/JB.00164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moll S, Schneider DJ, Stodghill P, Myers CR, Cartinhour SW, Filiatrault MJ. Construction of an rsmX co-variance model and identification of five rsmX non-coding RNAs in Pseudomonas syringae pv. tomato DC3000. RNA Biol. 2010;7:508–16. doi: 10.4161/rna.7.5.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kay E, Reimmann C, Haas D. Small RNAs in bacterial cell-cell communication. Microbe. 2006;1:63–9. [Google Scholar]
- 16.Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci U S A. 2005;102:17136–41. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–33. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–33. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmermann T, Sorg T, Siehler SY, Gerischer U. Role of Acinetobacter baylyi Crc in catabolite repression of enzymes for aromatic compound catabolism. J Bacteriol. 2009;191:2834–42. doi: 10.1128/JB.00817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rico A, Preston GM. Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol Plant Microbe Interact. 2008;21:269–82. doi: 10.1094/MPMI-21-2-0269. [DOI] [PubMed] [Google Scholar]
- 22.Mithani A, Hein J, Preston GM. Comparative analysis of metabolic networks provides insight into the evolution of plant pathogenic and nonpathogenic lifestyles in Pseudomonas. Mol Biol Evol. 2011;28:483–99. doi: 10.1093/molbev/msq213. [DOI] [PubMed] [Google Scholar]
- 23.Hester KL, Lehman J, Najar F, Song L, Roe BA, MacGregor CH, et al. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J Bacteriol. 2000;182:1144–9. doi: 10.1128/JB.182.4.1144-1149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morales G, Linares JF, Beloso A, Albar JP, Martínez JL, Rojo F. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J Bacteriol. 2004;186:1337–44. doi: 10.1128/JB.186.5.1337-1344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Sonnleitner E, Ren B, Xu Y, Haas D. Catabolite repression control of pyocyanin biosynthesis at an intersection of primary and secondary metabolism in Pseudomonas aeruginosa. Appl Environ Microbiol. 2012;78:5016–20. doi: 10.1128/AEM.00026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linares JF, Moreno R, Fajardo A, Martínez-Solano L, Escalante R, Rojo F, et al. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ Microbiol. 2010;12:3196–212. doi: 10.1111/j.1462-2920.2010.02292.x. [DOI] [PubMed] [Google Scholar]
- 27.King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–7. [PubMed] [Google Scholar]
- 28.Davis RW, Botstein D, Roth JR. Advanced bacterial genetics. Cold Spring Harbor, NY, 1980. [Google Scholar]
- 29.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–50. doi: 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 30.Windgassen M, Urban A, Jaeger KE. Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2000;193:201–5. doi: 10.1111/j.1574-6968.2000.tb09424.x. [DOI] [PubMed] [Google Scholar]
- 31.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 32.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–66. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 33.Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput Biol. 2007;3:e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Junier T, Zdobnov EM. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010;26:1669–70. doi: 10.1093/bioinformatics/btq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Brown NP, Leroy C, Sander C. MView: a web-compatible database search or multiple alignment viewer. Bioinformatics. 1998;14:380–1. doi: 10.1093/bioinformatics/14.4.380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.