Abstract
The CCR4-NOT complex was originally identified and its composition and organization characterized in the yeast Saccharomyces cerevisiae. It was first suggested to participate in transcription regulation, but since then it has become clear that it plays a key role in mRNA decay in all eukaryotes, thereby contributing importantly to gene expression regulation. Hence, the mammalian CCR4-NOT complex was recently shown to participate in miRNA-mediated mRNA repression. A better characterization of the composition and organization of this complex in higher eukaryotes is thus warranted. Purifications of the CCR4-NOT complex, performed by others and us, suggest that the protein of unknown function C2ORF29 is associated with this assembly. We demonstrate here that C2ORF29 is indeed a bona fide subunit of the human CCR4-NOT complex and propose to rename it CNOT11. In addition, we show that CNOT11 interacts with the first amino acids of CNOT1 and with CNOT10 and is required for the association of CNOT10 with the CCR4-NOT complex. Thus, the human CCR4-NOT complex possesses in addition to the CCR4-CAF1 deadenylase module and to the NOT module, a module composed of CNOT10 and CNOT11 that interacts with the N-terminal part of CNOT1. Phylogenetic analyses indicate that the CNOT10/CNOT11 module is conserved in all eukaryotes except fungi.
Keywords: CCR4-NOT, deadenylation, mRNA turnover, poly(A) tail, protein complexes
Introduction
Together with pre-mRNA synthesis and processing, mRNA turnover contributes significantly to the precise control of mRNA steady-state levels.1,2 In eukaryotes, degradation of functional mRNAs is most often initiated by progressive shortening of their poly(A) tails. This step is predominantly catalyzed by two interacting deadenylases called Ccr4 and Caf1.
The CCR4 and POP2/CAF1 (hereafter CAF1) genes were first identified in the yeast S. cerevisiae in genetic screens aimed to study transcriptional regulation.3,4 A few years later, biochemical analyses revealed that the Ccr4 and Caf1 yeast proteins purified with the Not proteins in large assemblies that are now generically termed the CCR4-NOT complex.5 Further protein purifications have shown that the core CCR4-NOT complex is composed in yeast of at least 9 proteins: Ccr4, Caf1, Not1, Not2, Not3, Not4, Not5, Caf40 and Caf130.6 With the exception of Caf130, for which related proteins are found only in yeast species, homologs of each subunit of the yeast core CCR4-NOT complex can be found in most eukaryotes. Thus, in humans, there is one ortholog for the yeast Not1, Not2, Not4 and Caf40 proteins, respectively CNOT1, CNOT2, CNOT4 and CNOT9 (also known as RQCD1). A single ortholog, CNOT3, shows homology to the yeast Not3 and Not5 proteins that are highly related. Conversely there are two human homologs for the Ccr4 protein: CNOT6 and CNOT6L and for the Caf1 protein: CNOT7 and CNOT8.
In 2001, it was recognized that both Ccr4 and Caf1 proteins possess domains that display sequence similarity to known nucleases: the Ccr4 protein belonging to the endonuclease-exonuclease-phosphatase (EEP) superfamily while Caf1 is a member of the ribonuclease (RNase) D family. Furthermore, deletions of the corresponding genes in yeast affected global poly(A) tail length as well as the half-lives of reporter mRNAs, demonstrating that the Ccr4 and Caf1 proteins are the major deadenylases involved in mRNA degradation in yeast.7,8 Since then, overwhelming data have demonstrated that this function of the CCR4-NOT complex is conserved in all eukaryotes (for a few examples, see refs. 9–12). Moreover, in many cases,13 it was shown that recruitment of the CCR4-NOT complex to mRNA 3′ untranslated region (3′ UTR) via specific RNA binding proteins destabilizes them and inhibits their translation, suggesting that this represents a general mechanism to regulate mRNA decay. Also, the GW182 proteins, which are necessary for the silencing functions of miRNAs in animals, have been shown recently to interact directly with the CCR4-NOT complex.14-16
Even though it is clear that the CCR4-NOT complex plays a key role in mRNA degradation, several points concerning its molecular functions need still to be clarified. In particular, the precise functions of the non-catalytic subunits in the mechanism of mRNA deadenylation remain unknown: some are functionally totally uncharacterized while for others it is only known that they are required for the integrity of the complex and/or for the efficiency of mRNA deadenylation.17-19 Furthermore, other functions, in addition to mRNA deadenylation, have been assigned to the CCR4-NOT complex. Indeed, this complex was originally identified as a transcriptional regulator (for review see20-23). This hypothesis, initially supported by genetic and physical interactions of the CCR4-NOT subunits with the transcription machinery, was recently strengthened by in vitro biochemical assays.24 Moreover, altered transcription in deadenylase mutants was shown to arise in compensation of reduced mRNA decay.25 The mechanistic role(s) of the CCR4-NOT complex in transcription remains to be deciphered. More recently, the CCR4-NOT assembly was also proposed to function in the protein ubiquitination and degradation processes23 as well as in direct translational repression.15,26
A better knowledge of the composition and of the organization of the CCR4-NOT complex is required to elucidate its molecular and biological functions. Original studies of the organization of the CCR4-NOT complex were performed in S. cerevisiae. They suggested that the large Not1 subunit is a scaffold protein on which other subunits bind.27 This was strengthened by recent structural analysis of a Ccr4-Pop2-Not1 subcomplex.19 Electron microscopy analyses further suggest that the yeast CCR4-NOT assembly adopts an overall L shape structure.28 To better characterize the mammalian CCR4-NOT complex, we used the TAP strategy to purify the protein complexes associated with one of the human Caf1 homologs, CNOT7. Like others, we identified in addition to conserved yeast homologs, two proteins that have no equivalent in the predicted yeast proteome: CNOT10 and C2ORF29. The CNOT10 protein has been systematically identified in all previous analyses of the CCR4-NOT complex in human,29-31 thus, this protein has been admitted to be a bona fide subunit of the CCR4-NOT complex and was named CNOT10. By contrast the C2ORF29 protein was reported in only one previous mass spectrometric analysis of the human CCR4-NOT complex.30 In the absence of confirmatory data, it was not definitively considered as a true subunit of this complex and thus not renamed accordingly. We demonstrate here that C2ORF29 is a genuine subunit of the human CCR4-NOT complex, that it is required for the interaction of CNOT10 with the CCR4-NOT assembly and that it interacts directly with CNOT10 and with the N-terminal end of CNOT1.
Results
TAP purification of the protein complexes associated with the Caf1 deadenylase subunit
To identify proteins potentially involved in the control of mRNA deadenylation in mammals, we purified one of the mammalian ortholog of the yeast Caf1 deadenylase, namely CNOT7, and associated partners. For this purpose, a HEK293-derived cell line expressing stably a fusion protein consisting of murine CNOT7 fused to the original TAP tag32 was established. (Note that the mouse and human proteins differ by only one amino acid). Proteins purified from this cell line by the TAP strategy were fractionated by SDS-PAGE and detected by Coomassie staining. This revealed the presence of 11 major bands (Fig. 1) which were identified by mass spectrometry analyses (Table S1). Two of them: β-tubulin and the actin-capping protein CAPZA1, are abundant constituents of the cytoskeleton and unlikely to be specific partners of Caf1. Most of the other proteins identified correspond to orthologs of the yeast CCR4-NOT subunits, namely Not1, Not2, Not3/Not5, Ccr4 and Caf40. In fact, beside Caf130, which has no definitive counterpart in human, only the ortholog of yeast Not4 was not found in the purification. In addition, two proteins for which there are no orthologs encoded by the yeast genome, CNOT10 and C2ORF29 were identified. The protein profile observed on gel after independent purifications from cells grown on plate or in suspension, transiently or stably expressing the bait protein was highly similar, arguing that we have established a complete catalog of the partners stably associated with CNOT7. This conclusion was reinforced by independent mass spectrometry analysis of the purified fractions not fractionated by SDS-PAGE that failed to identify other abundant polypeptides (data not shown). These results were highly similar to a previously published mass spectrometric analysis of the human CCR4-NOT complex,30 which proposed that this complex is composed of CNOT1, CNOT2, CNOT3, CNOT6/CNOT6L, CNOT7/CNOT8, CNOT9, CNOT10, TAB182 and C2ORF29. As the association of the protein of unknown function C2ORF29 with the CCR4-NOT complex was not validated in the previous publication,30 we decided to investigate further whether this protein is a genuine member of the CCR4-NOT complex or not.
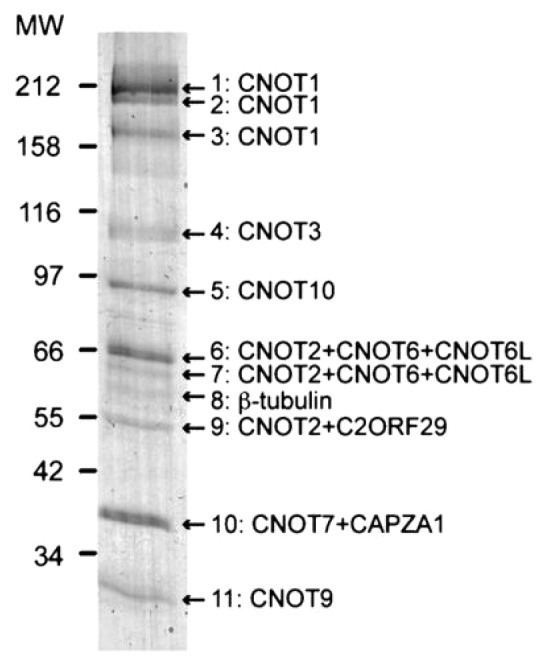
Figure 1. Purification of the proteins associated with CNOT7-TAP. The proteins associated with CNOT7-TAP were purified by the tandem affinity purification procedure from HEK293 cells expressing stably the TAP-tagged protein. After purification, proteins present in eluted fractions were fractionated by SDS-PAGE and the gel was stained with Coomassie blue. The names of the proteins identified by mass spectrometry in the 11 major bands that were analyzed are indicated.
C2ORF29 co-precipitates and co-purifies with human CCR4-NOT complex subunits
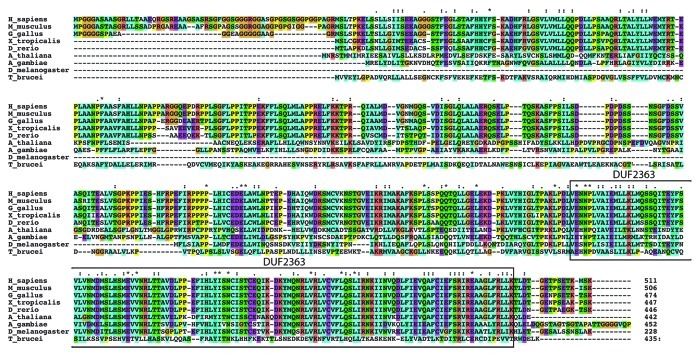
Database searches indicate that homologs for the human C2ORF29 protein can be found in all eukaryotes except fungi. A sequence alignment of C2ORF29 proteins from several species allows the distinction of three regions in C2ORF29 (Fig. 2). In amniotes, C2ORF29 possesses a Gly-rich N-terminal extension of about 60 amino acids that is absent in the protein of other organisms. The middle part of the protein is less conserved and is even absent in Drosophila melanogaster and other drosophilae. This feature is not specific for the insect kingdom as in Anopheles gambiae, the C2ORF29 protein contains this region. The C-terminal part of C2ORF29 is highly conserved but specific to this factor, thus providing little information about its function.
Figure 2. Sequence alignment for C2ORF29 proteins. Alignment of C2ORF29 homologs from various species generated with CLUSTAL X 2.1 software. Note that the Drosophila melanogaster sequence is shorter; this sequence was not included to plot the conservation of the middle part of the protein. The location of the DUF2363 domain described in Pfam database is indicated.
To verify the association of human C2ORF29 with the CCR4-NOT complex, we first performed co-precipitation experiments. The CNOT7-HA protein co-precipitated with C2ORF29-TAP (but not with the TAP tag alone) even when cell lysates were beforehand treated with micrococcal nuclease to remove detectable RNA (Fig. 3A). This demonstrated that the association of C2ORF29 in a complex containing CNOT7 resulted from specific protein-protein interaction.
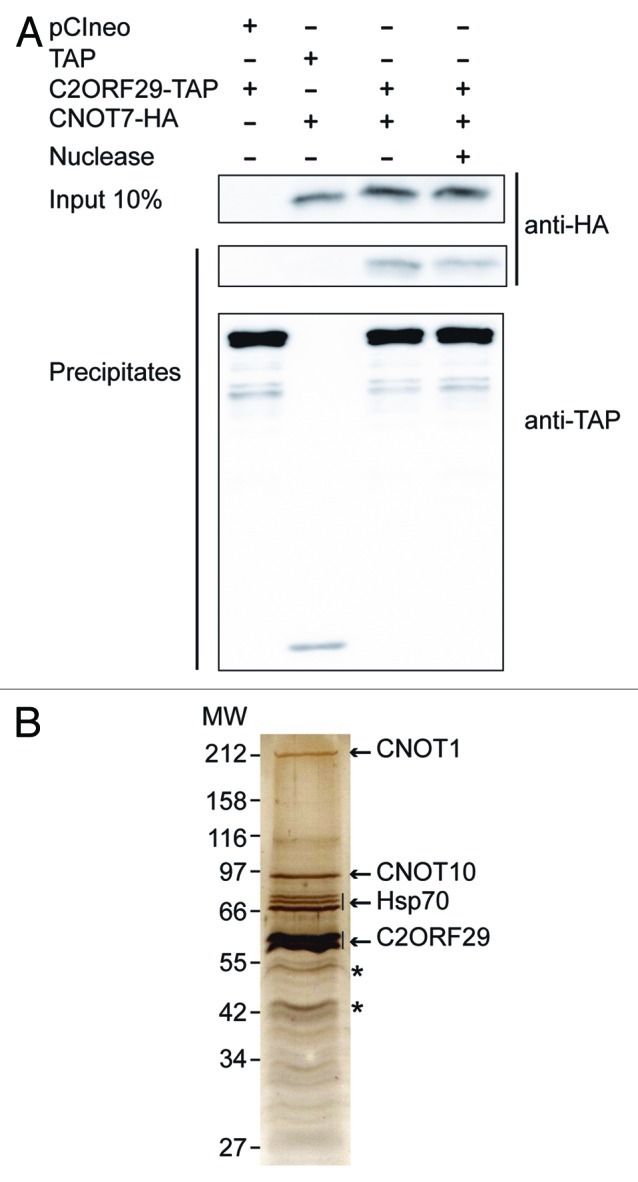
Figure 3. C2ORF29 is a bona fide subunit of the human CCR4-NOT complex. (A) HEK293 cells were co-transfected with plasmids expressing C2ORF29-TAP or the TAP tag alone and a plasmid expressing CNOT7-HA or the empty pCIneo vector. Cells were lysed and cell lysates were treated or not with micrococcal nuclease as indicated. TAP-tagged proteins were precipitated with IgG sepharose beads and the co-precipitation of the CNOT7-HA protein was analyzed by western blotting. (B) HEK293 cells were transiently transfected with a plasmid expressing C2ORF29-TAP and the proteins associated with C2ORF29 were purified by the tandem affinity purification procedure. Proteins present in eluted fractions were fractionated by SDS-PAGE and detected by silver staining. Proteins identified by mass spectrometry from the bands migrating above C2ORF29 are indicated. *: C2ORF29 breakdown products.
We then purified the proteins associated with C2ORF29 by the TAP method. Purified proteins were detected by silver staining after SDS-PAGE fractionation (Fig. 3B). Mass spectrometry analysis revealed that the major bands migrating below C2ORF29 were breakdown products of the tagged protein that is highly sensitive to proteolysis (see also Fig. 3A). The major bands migrating above C2ORF29 were identified as three isoforms of the Hsp70 protein, CNOT1 and CNOT10. The presence of the Hsp70 chaperones probably results from their association with exogenously expressed C2ORF29, which is expressed to high level in the absence of sufficient amounts of endogenous partners. Interestingly, all the other proteins co-purifying with C2ORF29 through two steps of chromatography are members of the CCR4-NOT complex. We also noticed that a single band corresponding to CNOT1 was observed in this purification (Fig. 3B) while several bands corresponding to CNOT1 were always observed when purifications were performed with CNOT7-TAP (Fig. 1). Detailed analysis of the peptides identified by mass spectrometry in the different forms of CNOT1 present in the CNOT7 purified fraction indicates that those represent staggered forms of the CNOT1 protein containing progressively shorter N-terminal regions. As C2ORF29 appears to associate with the longer form of CNOT1 protein, this suggests that C2ORF29 may associate with the N-terminal region of CNOT1.
We also attempted to determine which region of C2ORF29 was required for its incorporation in the CCR4-NOT complex. Deletion of the poorly conserved Gly-rich region found at the N-terminus of the protein did not impair its ability to co-precipitate CNOT7-HA (Fig. S1A). Further fragmentation of C2ORF29 however abolished co-precipitation of CNOT7-HA (Fig. S1A) and CNOT10 (Fig. S1B). This result indicates that the conserved C-terminal region of C2ORF29 is not sufficient to trigger association with the CCR4-NOT complex. As the above data showed that the sole proteins co-purifying with C2ORF29 are members of the CCR4-NOT complex, we concluded that C2ORF29 is a bona fide member of this assembly.
C2ORF29 and CNOT10 form a new module of the human CCR4-NOT complex
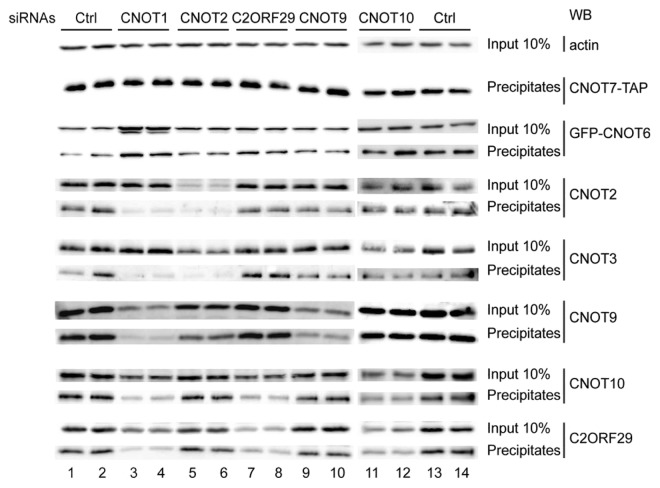
To get some information about the integration of C2ORF29 in the human CCR4-NOT complex and the overall organization of this assembly, we used the RNA interference technique to deplete individual CCR4-NOT subunits in the cells expressing stably CNOT7-TAP and tested how these depletions affected the co-precipitation of CCR4-NOT subunits with CNOT7-TAP by western blotting. In the cells treated with control siRNAs, GFP-CNOT6, CNOT2, CNOT3, CNOT9, CNOT10 and C2ORF29 were efficiently co-precipitated with CNOT7 (Fig. 4, lanes 1, 2, 13 and 14). By contrast, in the cells depleted for CNOT1, only CNOT6 was still co-precipitated (Fig. 4, lanes 3 and 4). This showed that only CNOT6 (CCR4 ortholog) interacts stably and directly with CNOT7 (Caf1 ortholog). This situation parallels the one observed in yeast where Caf1 was shown to bind directly Not1 and Ccr4 and to recruit the latter in the CCR4-NOT complex.19,27,33 This suggested also that interactions between the CNOT10 and C2ORF29 subunits and CNOT7 are, like for other NOT proteins, mediated through the CNOT1 scaffold protein. Consistent with a central role of the latter, we noticed that depletion of CNOT1 affected the steady-state levels of some of the CCR4-NOT subunits, notably CNOT9, and to a lesser extent C2ORF29 and CNOT10. Depletion of CNOT2 had no effect on the co-precipitation of CNOT6, CNOT10 and C2ORF29 with CNOT7 but affected strongly the steady-state level of CNOT3 and its co-precipitation with CNOT7 (Fig. 4, lanes 5 and 6) showing that CNOT2 is required for the stability of CNOT3 and its integration in the CCR4-NOT complex. As also observed by others,34 we noticed that depletion of CNOT2 affected the steady-state level of CNOT9 and its co-precipitation with CNOT7 but to a lesser extent than for CNOT3. As the effects observed upon CNOT2 depletion were not as strong as those observed following CNOT1 depletion, this suggested that CNOT2 is not required for CNOT9 recruitment in the CCR4-NOT complex but nevertheless impacts on CNOT9 association. In contrast, depletion of CNOT9 had no effect on the co-precipitation of the other subunits with CNOT7 (Fig. 4, lanes 9 and 10; the minor effect observed on the co-precipitation of CNOT3 is not reproducible; data not shown). Depletion of C2ORF29 did not affect the co-precipitation of CNOT6, CNOT2, CNOT3 and CNOT9 with CNOT7, but, interestingly, strongly reduced the co-precipitation of CNOT10 and the steady-state level of the protein (Fig. 4, lanes 7 and 8). The effects were quantitatively similar to those observed upon depletion of CNOT1. This demonstrated that C2ORF29 is required for the association of CNOT10 with the CCR4-NOT complex. Similarly, depletion of CNOT10 affected the steady-state level of C2ORF29 and consequently its co-precipitation with CNOT7 without affecting the co-precipitation of CNOT6, CNOT2, CNOT3 and CNOT9 (Fig. 4, lanes 11 and 12). This demonstrated that CNOT10 is reciprocally required for the association of C2ORF29 with the CCR4-NOT complex. These results suggested that C2ORF29 and CNOT10 could interact together and form a new module of the CCR4-NOT complex.
Figure 4. Analysis of proteins co-precipitating with human CNOT7 after inactivation of individual subunits of the CCR4-NOT complex by siRNA treatment. HEK293 cells expressing stably CNOT7-TAP were transfected first with the indicated siRNAs and re-transfected 24 h later with the same siRNAs and a plasmid expressing GFP-CNOT6 (because no high quality commercial antibodies against CNOT6 or CNOT6L were available). Two days later, the cells were lysed and the CNOT7-TAP protein was precipitated with IgG sepharose beads. Co-precipitation of the CNOT proteins indicated on the right side of the figure was detected by western blotting (WB). A fraction corresponding to 10% of the input fraction was also analyzed by western blotting to monitor the level of individual subunit before precipitation. To assess reproducibility, each siRNA treatment was performed in duplicate. An anti-actin western blot was also performed to verify equal loading of the input samples.
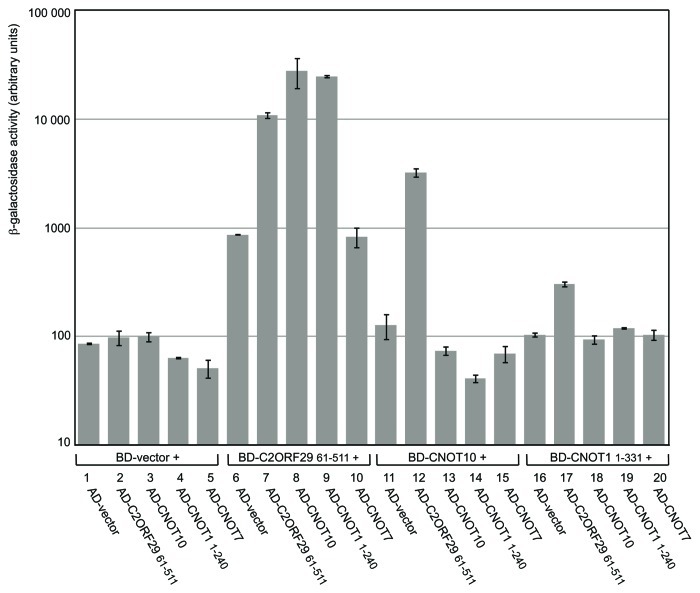
We then used the yeast two-hybrid technique to validate this hypothesis. For this purpose, cDNAs encoding full-length CNOT10, full-length CNOT7 and a C2ORF29 protein lacking its N-terminal Gly-rich region were cloned in vectors expressing fusion proteins with LexA Binding Domain (BD) or Gal4 Activating Domain (AD). Given that C2ORF29 appeared to associate with the N-terminus of CNOT1 (see above), only N-terminal regions of CNOT1 covering amino acids 1 to 331 or 1 to 240 were expressed as BD- or AD-fusion proteins respectively. Interactions were assayed by quantitating the β-galactosidase activity produced as a result of protein interaction. The assay confirmed that C2ORF29 interacts with CNOT10 (Fig. 5, lanes 8 and 12). As there are no obvious homologs of CNOT10 and C2ORF29 in yeast, it is highly improbable that the interaction could be indirectly mediated by endogenous yeast proteins. Thus this demonstrated that C2ORF29 interacts directly with CNOT10. The assay further showed that C2ORF29 could interact with itself (Fig. 5, lane 7). An interaction between C2ORF29 and the N-terminal parts of CNOT1 (Fig. 5, lanes 9 and 17) but not with CNOT7 (Fig. 5, lane 10) was also detected. By contrast, the assay did not reveal interaction between CNOT10 and the N-terminal parts of CNOT1 (Fig. 5, lanes 14 and 18). However, it is possible that CNOT10 stabilizes the interaction of C2ORF29 with CNOT1. Indeed, we observed that the β-galactosidase levels generated by the interaction of C2ORF29 and CNOT1 in the two-hybrid assay were fluctuating depending of the constructions or the substrate used (data not shown). This is reminiscent of the situation in Drosophila melanogaster where both Dm C2ORF29 and Dm Not10 are required to allow a stable interaction with Dm Not1.35 Altogether, the above data demonstrates that C2ORF29 and CNOT10 interact directly to form a new module of the CCR4-NOT complex that docks on the N-terminal part of CNOT1.
Figure 5. C2ORF29 interacts with CNOT1 and CNOT10 in two-hybrid assays. The diploid Y187/L40 yeast strain was co-transformed with plasmids expressing the indicated LexA-Binding-Domain (BD) and Gal4-Activating-Domain (AD) fusion proteins. Interaction between the different chimeric proteins was monitored by β-galactosidase assays. Activities are expressed in arbitrary units. Error bars correspond to two biological replicates.
Depletion of C2ORF29 does not affect the deadenylation rate of a stable transcript reporter
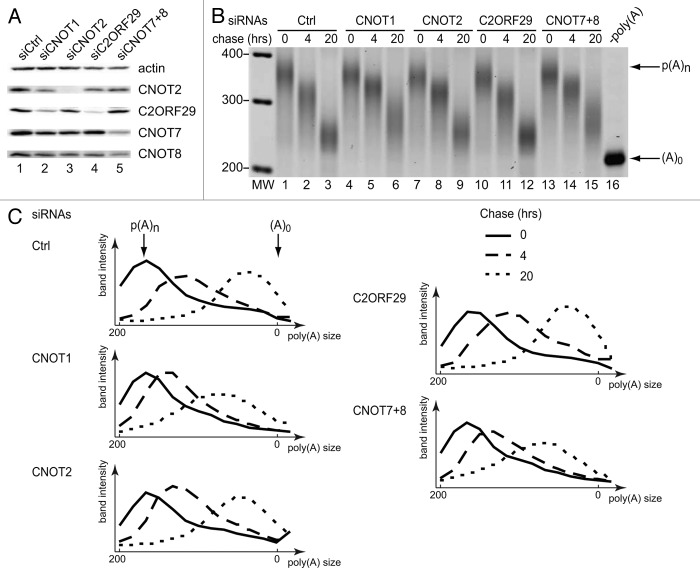
The best-characterized function of the CCR4-NOT complex is mRNA deadenylation. We thus tested whether C2ORF29 was implicated in this process. To this end, HEK293-TOF cells36 were treated with siRNAs to deplete the C2ORF29 protein (Fig. 6A, lane 4), or for comparison, CNOT1 (Fig. 6A, lane 2), CNOT2 (Fig. 6A, lane3), or the two Caf1 orthologs CNOT7 and CNOT8 (Fig. 6A, lane 5). Transcriptional pulse-chase experiments were performed in depleted cells using a plasmid reporter encoding the β-globin gene under the control of a tetracycline-regulated promoter.37 The poly(A) tail length of the β-globin mRNA was visualized using a 3′ RACE RT-PCR assay.36 This experiment revealed that, as expected, the simultaneous depletion of the deadenylases CNOT7 and CNOT8, depletion of the scaffold CNOT1 protein, or to a lesser extent CNOT2 reduced the deadenylation rate of the reporter (Fig. 6A and B). By contrast, depletion of C2ORF29, even though sufficient to partly disrupt the CCR4-NOT assembly, had no effect on the rate of deadenylation of the reporter transcript (Fig. 6A and B). This experiment suggests that, in contrast to CNOT1 or CNOT7/CNOT8, C2ORF29 is not a general deadenylation factor, even if further experiments will be required to exclude that traces of this factor could be sufficient for function.
Figure 6. Depletion of C2ORF29 does not affect the deadenylation rate of a β-globin mRNA reporter. (A) Western Blot showing the depletion levels of the indicated proteins obtained after siRNA treatment. The depletion of CNOT1 could not be monitored by western blotting because no good quality antibodies were available. (B) Poly(A) tail shortening of the β-globin mRNA reporter. HEK293-TOF cells were first transfected with the indicated siRNAs and simultaneously re-transfected 24 h later with the siRNAs and a plasmid expressing the β-globin mRNA reporter. Immediately after transfection, doxycyclin (1 ng/ml) was added to the medium to block transcription of the reporter. Two days later, cells were washed and a 4 h transcriptional pulse was performed before re-addition of doxycyclin (2 μg/ml). Chase times indicate hours after the second doxycyclin addition. The poly(A) tail length of the reporter was analyzed by a RT-PCR based assay.36 An RNA sample treated with oligo(dT) and RNase H was used as a marker for the migration of the fully deadenylated β-globin mRNA[-poly(A)]. (C) Migration profiles illustrating the extent of deadenylation of the reporter transcript. Each lane of the gel presented in Figure 6B was divided in 14 identical rectangles and the signal intensity for each rectangle quantified with the ImageQuant software (GE Healthcare). This revealed that deadenylation rates in cells depleted of CNOT1, CNOT2, or CNOT7 and CNOT8 were reduced by roughly 50% compared with control cells or C2ORF29-depleted cells. A significant fraction of the residual activity may originate from other deadenylases (e.g., PAN2).
Discussion
This study was initiated by using the TAP strategy32 to purify the protein complexes associated with one of the human Caf1 ortholog: CNOT7. In several purifications and using several strategies to identify co-purified proteins by mass spectrometry, we identified the human CNOT1, CNOT2, CNOT3, CNOT6/CNOT6L and CNOT9 proteins as stable CNOT7 partners, as well as two additional proteins with no homolog encoded in the yeast genome: CNOT10 and C2ORF29. This result is in agreement with a previously published proteomic analysis of the composition of the human CCR4-NOT complex30 that described the human CCR4-NOT complex to contain CNOT1, CNOT2, CNOT3, CNOT6/CNOT6L, CNOT7/CNOT8, CNOT9, CNOT10, TAB182 and C2ORF29, except that we did not identify significant levels of the TAB182 protein in our original purification (Fig. 1 and Table S1). This protein was however found in some subsequent purifications, but predominantly when the starting material was HEK293-derived adherent cells and not when the cells were grown in suspension (data not shown). We thus believe that TAB182 is not a stable subunit of the CCR4-NOT complex but rather interacts with it in particular conditions. By contrast, we demonstrate here through co-immunoprecipitation, co-purification and inactivation experiments that C2ORF29 is a bona fide subunit of the human CCR4-NOT complex. Thus, we propose to name it CNOT11. As we have performed an extensive analysis of the composition of the CCR4-NOT complex through independent purification and mass spectrometry analyses, we believe that, with CNOT11, a comprehensive list of the subunits present in this assembly in human cultured cells has now been established.
Our data show that CNOT10 and CNOT11 directly interact to form a module of the human CCR4-NOT complex. Interestingly, CNOT10 and CNOT11 homologs are found in all eukaryotes except fungi. The evolutionary co-occurrence of these two proteins further support that they form an optional module of the CCR4-NOT assembly. Consistently, in trypanosomes, the proteins Tb10.6k15.1820 and Tb927.8.1960, homologous to CNOT10 and CNOT11 respectively, were found to co-purify with TbCAF1-TAP.11 Similarly, the Drosophila CNOT10 homolog, protein CG18616, was found in a mass spectrometric analysis of NOT1 immunoprecipitates.17 We noticed further that the Drosophila homolog of CNOT11, protein CG13567, was also identified in the same analysis even though it was identified with 2 peptides only (see in ref. 17 and Table S2). Accordingly, the accompanying paper by Bawankar et al. presents results showing the association of a NOT10-C2ORF29 module with the N-terminal part of Dm NOT1.35
The optional presence of the CNOT10/CNOT11 module underlines the plasticity of the composition of the CCR4-NOT complex between species. This polymorphism is also supported by additional observations. Hence, if in most species, including yeast and human, the deadenylase module, which binds to a central part of NOT1 (see refs. 19, 33, 35), consists of the Ccr4 and Caf1 homologs, in trypanosomes no Ccr4-like proteins were identified in the Trypanasoma CCR4-NOT complex.11 The absence of proteins encoded in trypanosomal genomes containing simultaneously a EEP domain and leucine-rich repeats, a characteristic of Ccr4, further supports this conclusion. Interestingly, this is also the case for most plant genomes (our unpublished observation). This situation is consistent with the fact that Caf1 contacts directly Not1 while Ccr4 is indirectly associated through Caf1.19 This suggested further that in trypanosome species, Caf1 is the only CCR4-NOT subunit responsible for its deadenylating activity.11 The NOT module, that interacts with the C-terminal end of NOT1 (see refs. 33, 35), is also variable between species. In human, it is composed of CNOT2 and CNOT3, two related proteins, while it also contains Not5 in yeast. Finally, the yeast CCR4-NOT complex includes further the Not4 protein, which is however not tightly associated with the CCR4-NOT complex in other eukaryotes11,17,30 and the Caf130 protein that is found only in a restricted group of organism. In light of the inter-species variability in the CCR4-NOT complex organization, it remains to be seen whether this complex is homogenous in vivo or whether different related assemblies co-exist in a given cell.
In terms of functions, clear roles were only attributed to the deadenylase and optional Not4 modules of the CCR4-NOT complex. The former is clearly implicated in mRNA decay through poly(A) tail shortening7,8 while the later affects protein ubiquitination.39-41 Less is known about the functions of the other modules, and, even though the NOT module has been shown to influence poly(A) tail shortening (see refs. 17, 18, 34, this work), its mechanistic role in mRNA deadenylation remains unclear. This module could play a role in transcriptional regulation as well.21 For the CNOT10/CNOT11 module, the function is even more mysterious despite its conservation in a wide range of eukaryotes. Intriguingly, few phenotypes, if any, have been observed to result from depletion of these proteins in D. melanogaster or C. elegans in RNAi screens, as has been reported for the yeast Caf130 protein.20 One possibility is thus that the CNOT11/CNOT10 constitutes a functional homolog of the yeast Caf130 protein despite the absence of clear sequence similarity. Given the central role of the CCR4-NOT complex in the process of eukaryotic gene expression, there is little doubt that illuminating data will soon provide us some clues about the enigmatic functions of these CCR4-NOT subunits.
Materials and Methods
Plasmid constructions
Standard cloning strategies were used and the resulting clones were verified by sequencing when a PCR step was involved. Plasmids and oligonucleotides used for this study are reported in Tables S2 and S3.
Cell culture and Transfection
HEK293 cells and the HEK293-TOF cell line36 were maintained in DMEM medium supplemented with 10% FCS. HEK293 cells were transfected with a plasmid encoding a CNOT7-TAP fusion protein with the Effectene Transfection Reagent (Qiagen) and clones expressing stably the CNOT7-TAP fusion protein were selected with 400 μg/ml Geneticin (Invitrogen). Transient plasmid transfections were done using the Effectene Transfection Reagent (Qiagen) as recommended by the manufacturer. For siRNA and plasmid transfections, two consecutive transfections were performed as follows: cells were seeded to 80% confluency and transfected 6 h later with siRNAs (Dharmacon or Qiagen) at 25 nM final concentration with DharmaFect1 reagent (Dharmacon) as recommended by the manufacturer. Sixteen hours later, cells were trypsinized and seeded again to 80% confluency. Six hours later, cells were transfected with siRNAs at 100nM final concentration and plasmids using the DharmaFect Duo reagent (Dharmacon) as recommended by the manufacturer. Cells were harvested 48 h after the second transfection. siRNAs used in this study (Table S4) are ON-TARGETplus SMARTpools (Dharmacon) or GeneSolution siRNAs (Qiagen)
Immunoprecipitation and western blotting
Cells were lysed in IPP300 buffer (10 mM TRIS-HCl pH 8, 300 mM NaCl, 1% Igepal CA-630 and protease inhibitors) by standard procedures. When indicated, cell lysates were treated with micrococcal nuclease: CaCl2 and micrococcal nuclease (Fermentas) were added to 10 mM and 3 u/μl final concentrations, respectively, and lysates were incubated 15 min at 25°C.
For TAP co-precipitation experiments, cell lysates were incubated with IgG Sepharose 6 Fast Flow beads (GE Healthcare) for 30 min at 4°C. Beads were washed 2 times with IPP300 buffer, first eluted 5 min at room temperature in IPP150 elution buffer (10 mM TRIS-HCl pH 8, 150 mM NaCl, 1% Igepal CA-630, 1% Sodium Dodecyl Sulfate) that eluted efficiently the co-precipitated proteins but not the TAP fusion protein that was further eluted by resuspending the beads in Laemmli sample buffer (50 mM TRIS-HCl pH 6.8, 10% Glycerol, 0.05% Bromophenol Blue, 2% Sodium Dodecyl Sulfate). Western blotting was performed by standard procedures. The TAP tag was revealed with the PAP immunocomplex (DAKO) and ECL Western Blotting Detection Reagent (GE Healthcare). The HA and GFP tags were revealed with monoclonal antibody HA-11 (COVANCE) and JL8 (BD Biosciences), respectively, and the SuperSignal West Femto Maximum Sensivity Substrate kit (Pierce). Anti-CNOT2 and CNOT3 antibodies were a kind gift of Dr. Marc Timmers (University Medical Center Utrecht, Utrecht, The Netherlands), anti-CNOT9 antibodies were a kind gift of Prof. Hiroto Okayama (The University of Tokyo, Tokyo, Japan) while anti-CNOT10 antibodies were purchased from ProteinTech. Anti-CNOT7, CNOT8 and C2ORF29 antibodies were generated in the laboratory by injecting in rabbits purified recombinant His-tagged proteins produced in E. coli. For the low affinity anti-CNOT7 and CNOT8 antibodies, the SuperSignal Western Blot Enhancer kit (Pierce) was used to enhance the signal.
TAP affinity purification
The purification procedure was adapted from the procedure used in yeast with minor changes.32 Briefly, cells were lysed in IPP300 buffer (10 mM TRIS-HCl pH 8, 300 mM NaCl, 1% Igepal CA-630 and protease inhibitors) and lysates subjected to two centrifugations at 4°C: first at 20,000 g for 15 min followed by 100,000 g for 30 min. Cell supernatants were then incubated with IgG Sepharose 6 Fast Flow beads (GE Healthcare) for 2 h at 4°C and the beads were washed with 3 volumes of IPP300 buffer followed by a wash with 1 volume of TEV cleavage buffer (10 mM TRIS-HCl pH 8, 150 mM NaCl, 0.1% Igepal CA-630, 0.5mM EDTA, 1mM DTT and protease inhibitors). Beads were resuspended in 1ml of TEV cleavage buffer, 10μl AcTEV protease (Invitrogen) was added and beads were incubated 2 h at 16°C. After TEV cleavage, eluates were recovered and supplemented with 3 ml of Calmodulin Binding Buffer (10 mM TRIS-HCl pH 8, 150 mM NaCl, 1 mM MgAc, 1 mM Imidazole, 2 mM CaCl2, 0.1% Igepal CA-630, 10mM β-mercaptoethanol and protease inhibitors) and 3 μl of 1M CaCl2. These samples were then incubated with Calmodulin Affinity Resin (Agilent Technologies) for 1 h at 4°C. The beads were washed with 2 volumes of Calmodulin Binding Buffer and eluted in 5 fractions of 200μl with Calmodulin Elution Buffer (10 mM TRIS-HCl pH 8, 150 mM NaCl, 1 mM MgAc, 1 mM Imidazole, 2 mM EGTA, 0.1% Igepal CA-630, 10 mM β-mercaptoethanol). When necessary, elutions were concentrated by lyophilization before SDS-PAGE analysis. Protein identification was performed by the PAPPSO (Plateforme d'Analyze Protéomique de Paris Sud-Ouest) or IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire) mass spectrometry services using standard procedures.
Yeast two-hybrid β-galactosidase assays
A diploid yeast strain Y187/L40 resulting from the mating of Y187 and L40 haploid strains was used (MATα/MATa, ade2–101/ade2, his3–200/his3Δ200, leu2–3,-112/leu2–3,-112, lys2–801am::LYS2::(lexAop)4-HIS3/LYS2, MEL1/?, met-/MET, trp1–901/trp1–901, gal4Δ/gal4–542, gal80Δ/gal80–538, ura3–52::URA3::GAL1UAS-GAL1TATA-lacZ/URA3::(lexAop)8-lacZ).
This diploid strain was transformed simultaneously with the LexA-Binding-Domain and Gal4-Activating-Domain derived plasmids by standard LiAc procedure. Interaction between the different chimeric proteins was monitored by β-galactosidase assays performed using the kit NovaBright β-galactosidase Enzyme Reporter Gene Chemiluminescent Detection System for Yeast Cells (Invitrogen) as recommended by the manufacturer.
RNA extraction and RACE-PAT assay
Total RNA was extracted using the NucleoSpin RNA II kit (Macherey-Nagel). To monitor the length of the reporter β-globin transcript poly(A) tail, a modified PAT assay42 was used. Briefly, 50 pmoles of the R2 RNA oligonucleotide (Table S3) with a phosphate group at its 5′ end and a dideoxy C nucleotide at its 3′ end, was ligated to 1μg of total cellular RNA with T4 RNA ligase 1 (New England Biolabs) for 2 h at 37°C. The ligation reaction was then stopped 10 min at 65°C. One half of the ligation reaction was reverse-transcribed with oligonucleotide OBS4420 and RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas) as recommended by the manufacturer. PCR amplification was performed with 1μl of the reverse transcription reaction, with oligonucleotides OBS4437 and OBS4069 and with the Taq DNA Polymerase (New England Biolabs) as recommended by the manufacturer. PCR Products were resolved by migration on 3% agarose gels stained with ethidium bromide that were digitized using a Typhoon apparatus (GE Healthcare).
Supplementary Material
Acknowledgments
We thank Dr Elisa Izaurralde (Max Planck Institute for Developmental Biology, Tübingen, Germany) for sharing results prior to publication, Dr Marc Timmers (University Medical Center Utrecht, Utrecht, The Netherlands) for the gift of the anti-CNOT2 and anti-CNOT3 antibodies and Prof. Hiroto Okayama (The University of Tokyo, Tokyo, Japan) for the gift of the anti-CNOT9 antibodies, the PAPPSO (Plateforme d'Analyze Protéomique de Paris Sud-Ouest) and IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire; A. Page) mass spectrometric platforms, Benjamin Muller and Claudine Gaudon-Plesse for technical support, our lab for useful discussion and the IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire) for assistance. This work was supported by grants from the CERBM-IGBMC, the CNRS, the Ligue Contre le Cancer (Equipe Labelisée 2011) and the Agence Nationale de la Recherche (ANR-07-BLAN-0093 and ANR 11 BSV8 009 02).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23065
References
- 1.Khodursky AB, Bernstein JA. Life after transcription--revisiting the fate of messenger RNA. Trends Genet. 2003;19:113–5. doi: 10.1016/S0168-9525(02)00047-1. [DOI] [PubMed] [Google Scholar]
- 2.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 3.Denis CL. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984;108:833–44. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakai A, Chibazakura T, Shimizu Y, Hishinuma F. Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:6227–33. doi: 10.1093/nar/20.23.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol. 2001;314:683–94. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 7.Daugeron MC, Mauxion F, Séraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–55. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–86. doi: 10.1016/S0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 9.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–71. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–63. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 11.Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–88. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang W, Li C, Liu F, Jiang H, Li S, Sun J, et al. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 2009;19:307–16. doi: 10.1038/cr.2008.317. [DOI] [PubMed] [Google Scholar]
- 13.Wiederhold K, Passmore LA. Cytoplasmic deadenylation: regulation of mRNA fate. Biochem Soc Trans. 2010;38:1531–6. doi: 10.1042/BST0381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120–33. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18:1218–26. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18:1211–7. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 17.Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, et al. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–70. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–36. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basquin J, Roudko VV, Rode M, Basquin C, Séraphin B, Conti E. Architecture of the nuclease module of the yeast ccr4-not complex: the not1-caf1-ccr4 interaction. Mol Cell. 2012;48:207–18. doi: 10.1016/j.molcel.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/S0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- 21.Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 22.Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol. 2003;73:221–50. doi: 10.1016/S0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 23.Collart MA, Panasenko OO. The Ccr4--not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 24.Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 2011;25:581–93. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun M, Schwalb B, Schulz D, Pirkl N, Etzold S, Larivière L, et al. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012;22:1350–9. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem. 2010;285:28506–13. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maillet L, Tu C, Hong YK, Shuster EO, Collart MA. The essential function of Not1 lies within the Ccr4-Not complex. J Mol Biol. 2000;303:131–43. doi: 10.1006/jmbi.2000.4131. [DOI] [PubMed] [Google Scholar]
- 28.Nasertorabi F, Batisse C, Diepholz M, Suck D, Böttcher B. Insights into the structure of the CCR4-NOT complex by electron microscopy. FEBS Lett. 2011;585:2182–6. doi: 10.1016/j.febslet.2011.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 30.Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, et al. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J. 2009;422:443–53. doi: 10.1042/BJ20090500. [DOI] [PubMed] [Google Scholar]
- 31.Morita M, Suzuki T, Nakamura T, Yokoyama K, Miyasaka T, Yamamoto T. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol Cell Biol. 2007;27:4980–90. doi: 10.1128/MCB.02304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–2. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 33.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol Cell Biol. 1999;19:6642–51. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito K, Inoue T, Yokoyama K, Morita M, Suzuki T, Yamamoto T. CNOT2 depletion disrupts and inhibits the CCR4-NOT deadenylase complex and induces apoptotic cell death. Genes Cells. 2011;16:368–79. doi: 10.1111/j.1365-2443.2011.01492.x. [DOI] [PubMed] [Google Scholar]
- 35.Bawankar P, Loh B, Wohlbold L, Schmidt S, Izaurralde E. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol. 2012 doi: 10.4161/rna.23018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mauxion F, Faux C, Séraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–48. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loflin PT, Chen CY, Xu N, Shyu AB. Transcriptional pulsing approaches for analysis of mRNA turnover in mammalian cells. Methods. 1999;17:11–20. doi: 10.1006/meth.1998.0702. [DOI] [PubMed] [Google Scholar]
- 38.Seimiya H, Smith S. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182) J Biol Chem. 2002;277:14116–26. doi: 10.1074/jbc.M112266200. [DOI] [PubMed] [Google Scholar]
- 39.Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, et al. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–64. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanzawa H, de Ruwe MJ, Albert TK, van Der Vliet PC, Timmers HT, Boelens R. The structure of the C4C4 ring finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J Biol Chem. 2001;276:10185–90. doi: 10.1074/jbc.M009298200. [DOI] [PubMed] [Google Scholar]
- 41.Panasenko O, Landrieux E, Feuermann M, Finka A, Paquet N, Collart MA. The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J Biol Chem. 2006;281:31389–98. doi: 10.1074/jbc.M604986200. [DOI] [PubMed] [Google Scholar]
- 42.Sallés FJ, Strickland S. Analysis of poly(A) tail lengths by PCR: the PAT assay. Methods Mol Biol. 1999;118:441–8. doi: 10.1385/1-59259-676-2:441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.