Abstract
Recombinant adenoviral vectors (rAds) are the most potent recombinant vaccines for eliciting CD8+ T cell-mediated immunity in humans; however, prior exposure from natural adenoviral infection can decrease such responses. Here we show low seroreactivity in humans against simian- (sAd11, sAd16), or chimpanzee-derived (chAd3, chAd63) compared to human-derived (rAd5, rAd28, rAd35) vectors across multiple geographic regions. We then compared the magnitude, quality, phenotype and protective capacity of CD8+ T cell responses in mice vaccinated with rAds encoding SIV Gag. Using a dose range (1 × 107 to 109 PU), we defined a hierarchy among rAd vectors based on the magnitude and protective capacity of CD8+ T cell responses, from most to least as: rAd5 and chAd3, rAd28 and sAd11, chAd63, sAd16, and rAd35. Selection of rAd vector or dose could modulate the proportion and/or frequency of IFNγ+TNFα+IL-2+ and KLRG1+CD127- CD8+ T cells, but strikingly ~30–80% of memory CD8+ T cells co-expressed CD127 and KLRG1. To further optimise CD8+ T cell responses, we assessed rAds as part of prime-boost regimens. Mice primed with rAds and boosted with NYVAC generated Gag-specific responses that approached ~60% of total CD8+ T cells at peak. Alternatively, priming with DNA or rAd28 and boosting with rAd5 or chAd3 induced robust and equivalent CD8+ T cell responses compared to prime or boost alone. Collectively, these data provide the immunologic basis for using specific rAd vectors alone or as part of prime-boost regimens to induce CD8+ T cells for rapid effector function or robust long-term memory, respectively.
Introduction
The majority of approved vaccines against viral and bacterial infections mediate protection through antibody production. By contrast, there are no highly effective vaccines for infections in which Th1 CD4+ T cells, CD8+ T cells or both play critical roles in pathogen control or elimination, such as Mycobacterium tuberculosis infection (Tb), Malaria or HIV [1–3]. The development of vaccines capable of generating potent and durable T cell immunity has been limited by the availability of suitable vectors and adjuvants. Accordingly, replication deficient recombinant adenoviral vectors (rAds) have held great promise based on their ability to generate strong T cell immunity in mice, non-human primates (NHP) and humans [4–8]. As a reflection of their potential importance, rAds have been and are being tested in a number of clinical vaccine studies against HIV, Tb and Malaria [6, 7, 9–13].
The vaccine vector based on adenovirus serotype 5 (rAd5) has been the most comprehensively studied rAd in humans and was the first to be assessed in clinical efficacy trials against HIV [6, 7]. However, the clinical utility of rAd5 may be limited in populations that are key targets for HIV, Malaria and Tb vaccines, such as sub-Saharan Africa, due to high prevalence of pre-existing immunity from prior natural infection [4, 14]. Prior immunity to rAd5 has been shown to decrease antigen expression presumably by inhibiting infection of target cells, leading to sub-optimal conditions for induction of immune responses [6, 13, 15–17], particularly within the CD8+ T cell compartment [17]. Moreover, prior immunity to rAd5 may transiently increase the relative risk of infection with HIV through undefined mechanisms [18–20]. To circumvent these potential limitations, a major research goal has been to develop rAd vectors from lower seroprevalence human-derived adenoviruses [4, 21, 22] or from non-human sources, such as monkeys and apes [23–26]. These non-human vectors can minimise issues of seroprevalence but potentially retain mechanisms of adenoviral immune activation and potency.
There are 65 serologically distinct adenoviruses that have been isolated from humans (HAd) and they can be organised into at least 7 subgroups, denoted by the letters A through G [27, 28]. Sequencing information of the common hexon gene can also be used to classify animal-derived adenoviruses into these same subgroups. The rAd5 vector was derived from an HAd in subgroup C [29], the rAd35 vector from a subgroup B virus [21], and the rAd26 and rAd28 vectors from subgroup D viruses [4, 22]. HAdB-35 exhibits much lower seroprevalence than HAdC-5 globally [4, 14, 21], while exposure rates to HAdD-26 and HAdD-28 are low in the United States but marginally higher in target populations for Tb, Malaria and HIV vaccines [14, 22]. The rAd5 vector has been evaluated in numerous pre-clinical studies, as have rAd35, rAd26 and rAd28 to a lesser extent, and a hierarchy has emerged according to which rAd5 induces the most robust CD8+ T cell responses, followed by rAd26/rAd28 and then rAd35 [4, 5, 22]. More recently, a number of simian- and chimpanzee (chimp)-derived rAds have also been developed. The simian-derived vectors, sAd11 and sAd16, were developed from monkey adenovirus strains, but their phylogenetic classification based on the human sub-grouping system has not yet been defined and their seroprevalence in human populations is unknown. Ertl and Wilson were the first to report on the potency of chimp-derived vectors [23, 24, 30, 31] and, more recently, chAd3 and chAd63 have been developed and used in clinical studies [25, 26, 32]. Hexon sequencing suggests chAd3 and chAd63 classify into subgroups C and E respectively [26]. The chAd3 vector is of particular interest as it clusters by phylogeny in the same subgroup as rAd5. It has also been used in clinical trials and shown to prime robust T cell responses against hepatitis C virus to levels consistent with protective immunity [32]. Preliminary assessment of both of these chimp-derived rAds has demonstrated low seroprevalence in a European human population [26], although seroreactivity to chimp-derived rAds can be higher in sub-Saharan Africa where chimpanzees are endemic [33]. The in vitro activity or phylogenetic similarities of novel vectors compared to established rAd vectors has been used to make predictions about how these vectors will behave in vivo [21, 22, 26]. However, there has not been a comprehensive comparative analysis in a single study using the same antigen insert to characterise the magnitude, quality, phenotype and protective capacity of CD8+ T cells using human-, simian- and chimp-derived rAd vectors in a prime and/or boost setting.
Here, we first evaluate rates of seroreactivity against the seven vectors, rAd5, rAd28, rAd35, sAd11, sAd16, chAd3 and chAd63, across different vaccine target populations, demonstrating that simian- and chimp-derived vectors have very low seroprevalence. Using a mouse model, we then directly compare rAds over a broad dose range with respect to the magnitude, quality, phenotype and protective capacity of CD8+ T cells elicited using SIV Gag as the target antigen. This approach illustrates that titration of vectors is critical to correlate results obtained for immunogenicity and protection in mouse models with response hierarchies observed in NHP and human clinical studies. Importantly, we show that rAd5 and chAd3 vectors are similarly protective in a CD8+ T cell-dependent listerial infection model, consistent with their phylogenetic similarities. We also demonstrate qualitative and phenotypic differences in CD8+ T cells induced across rAd vectors and doses, and show that rAd vaccination induces a substantial population of cells at memory that co-express CD127 and killer cell lectin-like receptor subfamily G member 1 (KLRG1). Finally, we assessed rAds both as prime vaccines with a heterologous pox-derived vector boost and as boost vaccines after priming with DNA or a heterologous rAd vector. The data demonstrate the versatility of rAds, which were effective as primes or boosts, in comparison to the pox vector, which was an ineffective prime but a robust boost for CD8+ T cell responses. These insights should inform a rational approach for using rAd vectors in prime-boost regimens to optimise robust and durable CD8+ T cell immunity, which is critical for the development of preventive and therapeutic vaccines against a variety of infections.
Materials and Methods
Adenovirus serum neutralisation assay
Volunteer sera were collected in accordance with local Institutional Review Board approvals and evaluated to determine the relative concentration of Ad neutralising antibodies using the method described by Sprangers et al. [34]. Briefly, sera were heat inactivated for 60 min at 56°C and serially diluted (covering a final sample dilution range from 1:12 to 1:8748) in a final sample volume of 50 μL D10 (Dulbecco’s modified Eagle’s medium supplemented with 10% heat inactivated fetal bovine serum, penicillin 100U/mL, and streptomycin 100μg/mL). An optimised dilution of rAd vector, each encoding a luciferase reporter gene, was added to each well in a volume of 50 μL. The rAd and sera were co-incubated for 30 min at room temperature followed by addition of 1 × 104 A549 cells (human lung carcinoma), or 293T/17 cells for chAd63, per well in 100 μL of D10. The samples were incubated at 37°C in 10 % CO2 for 24 hours. To evaluate luciferase activity, cells were pelleted and resuspended in 100μL of Glo Lysis Buffer after removal of the culture medium. The cell suspension was transferred to a Black and White isoplate (Perkin Elmer) and 100 μL of Steady-Glo Luciferase Assay System Reagent (Promega) was added per well. After incubation for 15 min at room temperature, luminescence was measured on a luminometer. The 90% inhibition serum titre was determined to be the serum dilution that could be interpolated to have 10% of the maximum luciferase activity, as determined by the assay run without the presence of a serum sample.
Mice
C57BL/6 mice, for use with vectors encoding SIV Gag, or Balb/c mice, for use with vectors encoding HIV Env, were obtained from Jackson Laboratory (Bar Harbor, ME) and housed at the Vaccine Research Center Biomedical Research Unit (Bethesda, MD). Mice were 6 to 12 weeks old at the time of vaccination. All experimental animal protocols were approved by the Vaccine Research Center Animal Care and Use Committee.
Vectors and vaccinations
The vector stocks were grown and purified with a two-step cesium chloride purification protocol and stored at ≤−70°C. Virus stocks were titrated to determine the particle units (PU) per mL via high-pressure liquid chromatography. rAd5, rAd28, rAd35, sAd11 and sAd16 expressing SIV Gag were obtained from GenVec Inc. (Gaithersburg, MD). sAd11 and sAd16 vectors were derived from the wild type viruses Simian adenovirus 11 (ATCC #VR-196) and Simian adenovirus 16 (ATCC #VR-944). Vectors were derived, built, and produced as described previously for rAd28 and rAd35 [22, 35]. chAd3 and chAd63 backbones were obtained from Okairos (Italy) and SIV Gag was cloned in to these vectors before purification of viral particles as previously described [25, 26]. All rAd vectors were rendered replication deficient through targeted deletion of the E1 adenoviral gene, although the E3 gene was additionally deleted in chAd3 and chAd63 and E3 and E4 genes in rAd5. The SIV Gag gene was inserted into the E1 locus for all constructs and is under the control of the CMV promoter. NYVAC expressing SIV Gag, DNA encoding SIV Gag (kindly provided by Zhi-Yong Yang (VRC, NIAID, NIH, MD, USA)), rAd28 encoding SIV Gag and modified vaccinia virus Ankara (MVA) encoding HIV gp140 Envelope protein (Env) were used for prime-boost experiments. All vectors, except DNA, were prepared in PBS and the indicated doses were administered subcutaneously in the rear footpads, given as a 100 μL dose split into 50 μL per footpad. DNA was prepared in PBS and given intramuscularly as a 100 μL dose split into 50 μL per gluteal muscle; two doses were administered 3 weeks apart. All vectors contained a codon-optimised version of Gag/Pol from SIV strain mac239, except for the rAd vectors and MVA used in Figure 7 E–G and Supplemental Figure 1 C and D, which contain Env from the HIV hybrid strain IIIB/BaL.
Figure 7.
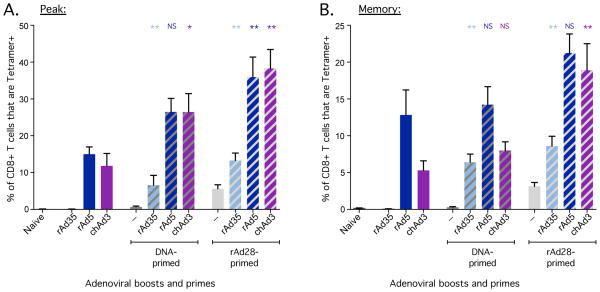
Assessment of priming by rAd vectors for a common boost. C57BL/6 mice (n=5) were primed with 1 × 107 PU of rAd vectors and boosted 8 weeks later with 1 × 107 PFU of NYVAC. At the indicated time points, SIV Gag-derived AL11-specific CD8+ T cells were quantified in peripheral blood by tetramer staining. (A) Frequency of CD8+ T cells that were tetramer+ at the time of boost, 8 weeks after priming. (B) Frequency of CD8+ T cells that were tetramer+ at the peak of the response to the boost, 2 weeks after boosting. (C) Frequency of CD8+ T cells that were tetramer+ at memory, 10 weeks after boosting. (D) Mice were primed with 1 × 109, 1 × 108 or 1 × 107 PU of rAd35 and boosted with 1 × 107 PFU of NYVAC. Gag-specific CD8+ T cells were quantified by tetramer staining over time after boosting. Alternatively, Balb/c mice (n=4–5) were primed with 1 × 107 PU of the indicated vectors and boosted 4 weeks later with 1 × 107 PFU of MVA. A group vaccinated with chAd3 expressing SIV Gag was used as a negative control for Env priming. At the indicated time points, HIV Env-derived PA9-specific CD8+ T cells were quantified in peripheral blood by tetramer staining. (E) Frequency of CD8+ T cells that were tetramer+ at the time of boost. (F) Frequency of CD8+ T cells that were tetramer+ at the peak of the response to the boost, 2 weeks after boosting. (G) Frequency of CD8+ T cells that were tetramer+ at memory, 10 weeks after boosting. Significant differences in frequency were assessed compared to rAd5 primed animals, where * = p ≤ 0.05. Bars or points on the line graph and error bars represent mean ± SEM. Each group is representative of at least two independent experiments.
Tetramer staining
Peripheral blood was harvested and red blood cells were lysed using ACK lysis buffer (Lonza, Switzerland). Splenocytes were harvested, homogenised to single cell suspensions and red blood cells were lysed using ACK lysis buffer (Lonza). Cells were washed in PBS and stained with LIVE/DEAD® Fixable Red viability dye (Life Technologies, NY, USA). Subsequently, cells were stained with PE-labeled H-2Db tetramer loaded with the immuno-dominant SIV Gag peptide AL11 (AAVKNWMTQTL; [16]) or PE-labeled H-2Dd tetramer loaded with the immuno-dominant HIV BaL Env peptide PA9 (IGPGRAFYA; [36]). After blocking with anti-FcγRIII antibody (clone 2.4G2; 5 μg/mL; BD Pharmingen, CA, USA), cells were surface stained with anti-CD8-APC-Cy7 (clone 53-6.7; Biolegend, CA, USA), anti-CD62L-PE-Cy7 (clone MEL-14; Abcam, MA, USA), anti-KLRG1-FITC (clone 2F1; Southern Biotech, AL, USA) and anti-CD127-AF647 (clone A7R34; eBiosciences, CA, USA). Cells were then fixed and permeabilised using the Fix/Perm and Perm Wash buffer system (BD Biosciences, CA, USA), before intracellular staining with anti-CD3-PerCP-Cy5.5 5 (clone 145-2C11; BD Pharmingen).
Intracellular cytokine staining
For assessment of antigen-specific cytokine production, spleens were harvested at the indicated times, homogenised to single cell suspensions and red blood cells were lysed using ACK lysis buffer (Lonza). Splenocytes were then used for in vitro restimulation, where 1.5 × 106 cells were incubated for 5 hours with anti-CD28 (1 μg/mL; BD Pharmingen), brefeldin A (BFA; 10 μg/mL; Sigma-Aldrich, MO, USA) and the following antigens as indicated: (i) the immuno-dominant MHC class I- and II-restricted SIV-Gag peptides AL11 and DD13 (DRFYKSLRAEQTD; [37]) (each at 2 μg/mL); (ii) full-length SIV Gag protein (20 μg/mL); or (iii) a peptide pool comprising 15-mers spanning HIV strain IIIB/BaL Env (each at 2 μg/mL) [36]. Samples were also incubated with anti-CD28 and BFA alone to establish background cytokine production. BFA was withheld from samples undergoing protein stimulation for 2 hours to permit processing of the protein. For staining of samples after stimulation, cells were washed in PBS and stained with LIVE/DEAD® Fixable Violet viability dye (Life Technologies). Cells were blocked with anti-FcγRIII antibody (clone 2.4G2; 5 μg/mL; BD Pharmingen) before surface staining with anti-CD8-APC-Cy7 (clone 53-6.7; Biolegend) and anti-CD4-AF700 (clone RM4-5; BD Pharmingen). Cells were fixed and permeabilised using the Fix/Perm and Perm Wash buffer system (BD Biosciences), before intracellular staining with anti-CD3-PerCP-Cy5.5 (clone 145-2C11; BD Pharmingen), anti- IFNγ-APC (clone XMG1.2; BD Pharmingen), anti-IL-2-PE (clone JES6-5H4; BD Pharmingen), anti-TNFα-APC-Cy7 (clone MP6-XT22; BD Pharmingen) and anti-IL-10-AF488 (clone JES5-16E3; eBiosciences).
Flow cytometry
Samples were resuspended in 0.5% paraformaldehyde before acquisition using a modified LSR II flow cytometer (BD Biosciences, CA, USA). Results were analysed using FlowJo version 9.3, Pestle version 1.6.2 and SPICE version 5.22 software (Mario Roederer, VRC, NIAID, NIH, MD, USA). Background cytokine staining was subtracted, as defined by staining in samples incubated without peptide or protein.
Total Gag-specific IgG enzyme-linked immunosorbent assays (ELISAs)
Nunc-Immuno 96-well plates (Nunc A/S, Roskilde, Denmark) were incubated overnight at 4°C with coating buffer (0.1M sodium carbonate/bicarbonate) containing 1 μg/mL of SIV Gag protein, blocked with 10% fetal bovine serum/PBS, then incubated for 2 hours with serum prepared from a 1:10 dilution in a six-fold dilution series for each individual sample. Wells were subsequently incubated with a 1:20000 dilution of AffiniPure Goat Anti-Mouse IgG (subclasses 1+2a+2b+3) Fc Fragment Specific (Jackson Immuno Research, PA, USA) followed by a 1:1000 dilution of avidin-Horse Radish Peroxidase (BD Pharmingen). Finally 100 μL of TMB One-Step Substrate System (Dako, CA, USA) was added followed by 100 μL of 2N H2SO4. Absorbance was measured at 450 nm and the endpoint titre for each dilution series was calculated as 3 standard deviations above the mean of the PBS vaccinated control group.
Infections and antibody-mediated depletions
For infectious challenge, we used either attenuated Listeria monocytogenes (ΔactA, ΔintB) or vaccinia virus (thymidine kinase-deficient Western Reserve strain), each expressing Gag from SIV strain mac239. Listeria:Gag was kindly provided by ANZA Therapeutics (CA, USA) and rVACV:Gag was kindly provided by Glennys Reynoso (NIAID, NIH, MD, USA). A 2 × 107 colony forming unit (CFU) dose of Listeria:Gag was intravenously administered in a 300 μL volume. Spleens were harvested 42 hours later, mechanically homogenised in 1 mL of PBS using a TissueRuptor (Qiagen Inc., CA, USA) and plated in duplicate as a tenfold dilution series on brain-heart infusion agar (Difco, MI, USA). Plates were incubated at 37°C for 24–32 hours before colonies were counted and back-calculated to yield values for total CFU per spleen. Alternatively, a 6.5 × 106 plaque forming unit (PFU) dose of rVACV:Gag was intranasally administered to anaethetised mice in a 50 μL volume. Mice were subsequently followed individually and weighed daily to assess infection-induced weight loss until 6 days after infection.
For antibody-mediated depletion of T cells, 1 mg of control (clone J1.2; rat IgG2b, anti-influenza NP), anti-CD4 (clone GK1.1; rat IgG2b) or anti-CD8 (clone 2.43; rat IgG2b) antibody was administered intraperitoneally 3 days before infection with Listeria:Gag. Mice were bled the day before infection and stained with non-competing antibodies targeting CD4 (clone RM4-4) and CD8 (clone 53-6.7) to confirm depletion of target T cell populations. Depleting antibodies were kindly provided by Fred Finkelman (University of Cincinnati, OH, USA).
Statistics
Statistical significance was calculated using a two-tailed Student’s t test for qualitative and phenotypic data using SPICE software or a two-tailed Mann-Whitney test for all other data using PRISM software.
Results
Low Seroreactivity Against Simian- and Chimp-Derived rAd Vectors in Vaccine Target Populations
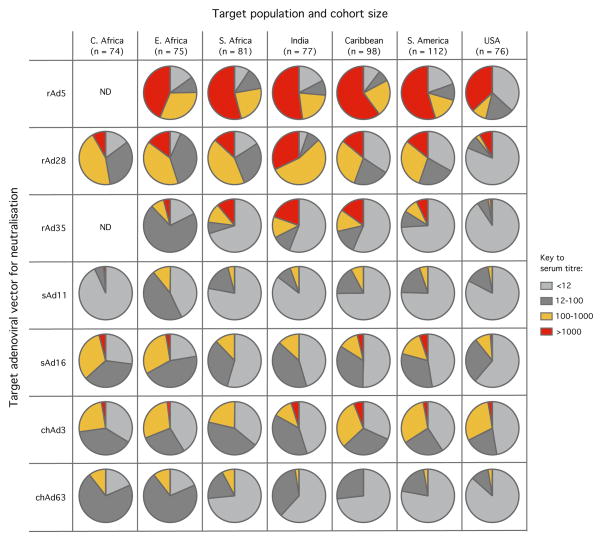
Previous studies have established that there is high and modest pre-existing immunity to rAd5 and rAd26 respectively in geographic areas where vaccines against HIV, Malaria and Tb are required, such as sub-Saharan Africa [4, 14]. In contrast, while there is limited prior exposure to rAd35 globally, it is the least immunogenic human-derived rAd, particularly within the CD8+ T cell compartment [4, 14]. To assess the potential utility of novel simian- and newly described chimp-derived vectors as vaccine candidates for the induction of CD8+ T cell immunity, we examined seroreactivity in cohorts of adults from several geographic regions where such vectors might be used: the USA, South America, the Caribbean, India, Southern Africa, Eastern Africa and Central Africa. This was evaluated using a serum neutralisation assay with a panel of representative rAd vectors expressing the luciferase reporter gene: including rAd5, rAd28, rAd35, sAd11, sAd16, chAd3, and chAd63 (Fig. 1). Neutralisation of the rAd26 vector was also evaluated and was similar to rAd28 (data not shown). Seroreactivity directed against rAd5 was common, with titres >1000 (represented by red sections in the pie graphs) observed in ~50% of individuals; rAd28 exhibited somewhat lower titres and rAd35 was the least seroreactive, consistent with prior studies. Titres were markedly lower across populations for all simian- and chimp-derived vectors compared to rAd5, rAd28 and even rAd35, with sAd11 and chAd63 displaying the lowest titres. Collectively, these data suggest that multiple simian- or chimp-derived rAds could be useful in human vaccine target populations, depending on their potency.
Figure 1.
Assessment of seroreactivity against human-, simian- and chimp-derived adenoviruses in human cohorts from diverse geographical regions. Serum samples from adults in the indicated cohorts were assessed for neutralising antibodies against rAd5, rAd28, rAd35, sAd11, sAd16, chAd3 or chAd63 vectors encoding luciferase. The titres represent the serum dilution from individuals at which 90% infection inhibition was observed based on luciferase activity relative to the maximal control, after division into four groups (<12, 12–100, 100–1000, >1000) indicated in the key. Cohort size is indicated as n.
Development of SIV Gag-Specific CD8+ T Cell Immunity After rAd Vaccination
To determine the relative potency of human-, simian- and chimp-derived rAds for induction of CD8+ T cell responses, mice were vaccinated subcutaneously with 1 × 109, 1 × 108 or 1 × 107 particle units (PU) of each vector. The vectors each expressed full-length Gag antigen from SIV strain mac239, which contains the immuno-dominant MHC class I epitope denoted AL11 [16]. Gag-specific CD8+ T cell responses in peripheral blood were followed over time using an MHC class I tetramer loaded with the AL11 peptide. As shown in Figure 2A, rAd5 induced robust and comparable CD8+ T cell responses at all doses. In contrast, responses fell below the limit of detection in some individual samples for mice vaccinated either with rAd35 at the two lower doses or with rAd28 at 1 × 107 PU (Fig. 2A). The simian-and chimp-derived vectors induced substantial CD8+ T cell responses at all doses (Fig. 2A), with some decrease in magnitude for sAd11, sAd16 and chAd63 at the 1 × 107 PU dose. For these vectors, the effect of dose titration manifested primarily as a delay in response kinetics with a lower peak magnitude. A comparison of CD8+ T cell responses induced by all vectors at the 1 × 107 PU dose (Fig. 2B) revealed a clear hierarchy between vectors as follows, from most to least potent: rAd5; chAd3, sAd11 and chAd63; sAd16; rAd28; and lastly rAd35.
Figure 2.
Tetramer+ CD8+ T cell responses in peripheral blood after vaccination with dose titrations of rAd vectors. C57BL/6 mice (n=4–5) were vaccinated with 1 × 109, 1 × 108 or 1 × 107 PU of the indicated rAd vector expressing SIV Gag. Peripheral blood samples were analysed sequentially at days 14, 21, 28, 35 and 70 using Tetramer staining to identify SIV Gag-derived AL11-specific cells. (A) Longitudinal comparison of frequencies of CD8+ T cells that were tetramer+ after vaccination with the indicated dose of each vector. (B) Longitudinal comparison for each vector at the lowest dose (1 × 107 PU), where rAd5 was compared to rAd28, rAd35, sAd11 and sAd16 (left panel) or compared to chAd3 and chAd63 (right panel) in independent experiments. The p values for each vector at 1 × 107 PU compared to rAd5 are displayed in the table, where NS= not significant, * = p ≤ 0.05 and ** = p ≤ 0.01. Data points and error bars represent mean ± SEM. Each time course is representative of at least two independent experiments.
SIV Gag-Specific Cytokine Production by CD8+ T Cells
To extend the immunologic analysis, intracellular cytokine staining and multi-parameter flow cytometry was used to assess the magnitude of Gag-specific CD8+ T cell cytokine responses following rAd vaccination at peak and memory time points. In our analysis of peripheral blood, peak responses varied from day 14 to 28 for different rAds and different doses (Fig. 2A). Therefore, the time point used to approximate the “peak” for all vectors and doses was 23 days after vaccination, but this may antecede or precede maximal responses, particularly in mice that received the lowest dose of 1 × 107 PU. Additionally, the “memory” time point used was 70 days after vaccination but later time points (100+ days) have been performed with 1 × 108 PU and similar results to the following were observed (data not shown).
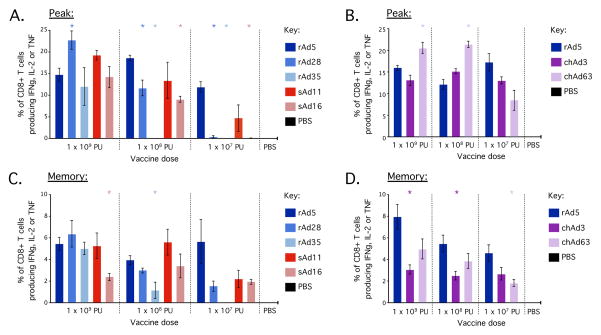
Analysis of total Gag-specific cytokine (IFNγ, IL-2 or TNFα) production by splenocytes after in vitro stimulation with the AL11 peptide revealed a pattern similar to the hierarchy observed in the peripheral blood with tetramer staining. At 1 × 109 PU, all vectors induced high frequency CD8+ T cell responses at both peak (Fig. 3A, B) and memory (Fig. 3C, D) that were generally comparable to rAd5. At 1 × 108 PU, all vectors again induced potent CD8+ T cell responses, with the exception of rAd35; in this case, responses could not be detected at peak and were significantly lower at memory. At 1 × 107 PU, rAd5 still induced robust CD8+ T cell responses at peak and memory. In comparison, rAd28 and sAd16 were significantly lower at peak, chAd63 was significantly lower at memory and rAd35 did not exhibit detectable responses at either peak or memory. Although chimp-derived vectors induced responses comparable to or even greater than rAd5 at peak at all doses, these responses contracted substantially by memory, which mirrors the contraction of Gag-specific CD8+ T cell responses observed in the peripheral blood with tetramer staining (Fig. 2). We also assessed tetramer staining and cytokine responses in the lung for all vectors at 1 × 108 PU at peak and memory time points and a similar hierarchy was observed (data not shown). Thus, a consistent hierarchy of immunogenicity across rAd vectors emerges from both antigen-specific cytokine production and tetramer staining across various tissues.
Figure 3.
Antigen-specific cytokine production by CD8+ T cells in the spleen 23 days (at peak) (A/B) or 70 days (memory) (C/D) after rAd vaccination. C57BL/6 mice (n=4–5) were vaccinated with 1 × 109, 1 × 108 or 1 × 107 PU of rAd5, rAd28, rAd35, sAd11, sAd16 (A/C), or rAd5, chAd3 or chAd63 (B/D) in independent experiments. At day 23 after vaccination, splenocytes were processed for in vitro stimulation with peptides encoding the immunodominant MHC class I and II epitopes from SIV Gag in C57BL/6 mice. The frequencies of CD8+ T cells that produced cytokine in response to antigen (i.e. stained positive for IFNγ, TNFα or IL-2) were determined. Significant differences in frequency were assessed for each vector compared to rAd5 at the equivalent dose, where * = p ≤ 0.05. Bars and error bars represent mean ± SEM. Each group is representative of at least two independent experiments.
Multiple mechanisms could account for differences in the potency and durability of CD8+ T cell responses after rAd vaccination. CD4+ T cells can influence CD8+ T cell maintenance and expansion [38, 39] and have been shown to augment CD8+ T cell responses after rAd vaccination [40]. CD4+ T cell responses primed by rAd vectors encoding SIV Gag antigen in C57BL/6 mice were detectable but low at peak (Supplemental Fig. 1A, B) and undetectable by memory time points (data not shown). Generally, responses were low and variable for rAd5, rAd28, rAd35, sAd16 and sAd11 but chimp-derived vectors induced modest responses that were significantly higher than rAd5 at 1 × 108 or 1 × 107 PU (Supplemental Fig. 1B). Similar results were obtained with rAds encoding HIV gp140 Envelope protein (Env) as a target antigen in Balb/c mice (Supplemental Fig. 1C, D). We also assessed SIV Gag-specific antibody responses and observed a hierarchy between rAds similar to that observed for CD8+ T cells at both peak and memory (Supplemental Fig. 1E–H). Overall, we cannot attribute differences in the potency of CD8+ T cell responses after vaccination with rAds to differences in the corresponding CD4+ T cell responses because, in this model, rAd vaccination strongly biases toward induction of CD8+ as opposed to CD4+ T cell immunity.
Qualitative Profiles of SIV Gag-Specific Memory CD8+ T cells
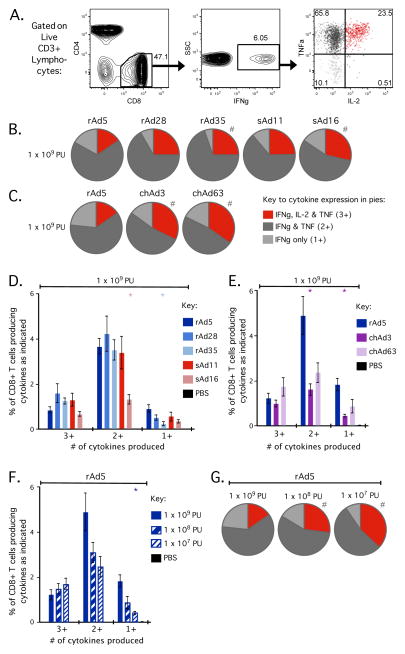
After evaluating CD8+ T cell responses induced by rAd vectors in terms of total magnitude, we then evaluated the quality of the response in terms of IFNγ, TNFα and IL-2 co-expression. The term “quality” refers to functional markers (cytokine expression, phenotypic or cytolytic markers) possessed by individual T cells [41–43]. A T cell with multiple markers is designated “multifunctional”, can secrete higher amounts of cytokine per cell and their presence correlates with protection in vaccine models of CD4+ T cell immunity and disease non-progression in HIV infection [42, 43]; thus responses that induce a higher proportion of multifunctional T cells are considered to have a better quality. As shown in Figure 4A, Gag-specific CD8+ T cells induced by rAd vaccination were either IFNγ+ TNFα+ IL-2+ (red; 23.5%), IFNγ+ TNFα+ IL-2− (dark grey; 65.8%) or IFNγ+ TNFα-IL-2− (light grey; 10.1%), but very few cells were IFNγ+ TNFα-IL-2+ (black; 0.51%). Additionally, very few (if any) Gag-specific CD8+ T cells produced TNFα and/or IL-2 without IFNγ (data not shown). Thus, quality is represented in this study by the proportion of all responding Gag-specific CD8+ T cells producing IFNγ, TNFα and IL-2 (3+, multifunctional cells), IFNγ and TNFα (2+) or IFNγ alone (1+).
Figure 4.
Qualitative profiles of CD8+ T cells in the spleen 70 days (at memory) after rAd vaccination. C57BL/6 mice (n=4–5) were vaccinated with 1 × 109, 1 × 108 or 1 × 107 PU of the indicated rAd vector expressing SIV Gag. Splenocytes were processed as in Figure 3 to determine Gag-specific production of IFNγ, TNFα and IL-2. Boolean gating was used to define subsets of CD8+ T cells expressing any possible combination of IFNγ, IL-2 and TNFα. (A) Representative flow cytometry plots after rAd5 vaccination to illustrate the populations of CD8+ T cells used for qualitative analysis. CD3+ lymphocytes were gated on CD8+ events (left plot), then IFNγ+ events (middle plot) prior to analysis of TNFα and IL-2 production (right plot). IFNγ-producing Gag-specific CD8+ T cells expressed IFNγ, TNFα and IL-2 (red), IFNγ and TNFα (dark grey) or IFNγ-alone (light grey), thereafter simplified as 3+, 2+ and 1+ cells, respectively. (B/C) The proportion of Gag-specific CD8+ T cells that are 3+, 2+ or 1+ after vaccination with 1 × 109 PU of rAd5, rAd28, rAd35, sAd11 or sAd16 (B) or rAd5, chAd3 or chAd63 (C). (D/E) The frequency of total CD8+ T cells that are 3+, 2+ or 1+ after vaccination with 1 × 109 PU of rAd5, rAd28, rAd35, sAd11 or sAd16 (D) or rAd5, chAd3 or chAd63 (E). (F) The frequency of total CD8+ T cells that are 3+, 2+ or 1+ after vaccination with each dose of rAd5. (G) The proportion of Gag-specific CD8+ T cells that are 3+, 2+ or 1+ after vaccination with each dose of rAd5. For bar graphs, significant differences in frequency were assessed compared to rAd5 at 1 × 109 PU, where * = p ≤ 0.05. Bars and error bars represent mean ± SEM. For pie graphs, significant differences in distribution were assessed compared to rAd5 at 1 × 109 PU, where # = p ≤ 0.05. Each group is representative of at least two independent experiments.
SIV Gag-specific CD8+ T cells induced by any of the rAd vectors at the 1 × 109 PU dose were predominantly 2+ cells (Fig. 4B–E). However, there were differences in the relative proportions of 3+, 2+ and 1+ cells between rAd vectors. The rAd5 vector displayed a significantly different qualitative profile compared to rAd35, sAd16, chAd3 and chAd63 (Fig. 4B, C). Underlying these differences in proportion, rAd5 generally induced higher frequencies of 2+ and/or 1+ cells compared to rAd35, sAd16, chAd3 and chAd63 (Fig. 4D, E). This resulted in a less multifunctional response after rAd5 vaccination, due to the lower proportion of 3+ cells (Fig. 4D, E). At the lower dose of 1 × 107 PU, qualitative profiles between the rAd vectors were similar (Supplemental Fig. 2A, B), although population frequencies differed significantly between vectors (Supplemental Fig. 2C, D), due to differences in total response magnitude as previously described. Thus, despite inducing similar total magnitudes at 1 × 109 PU (Fig. 3), rAds can induce CD8+ T cell responses with subtly distinct qualitative profiles. Such distinctions may have implications for the durability of CD8+ T cell immunity primed by different rAd vectors.
Importantly, we observed that decreasing the dose of rAds can alter the quality of the CD8+ T cell response. This was most evident for rAd5, where decreasing the dose resulted in lower frequencies of 2+ and 1+ cells but the frequency of 3+ cells was stable (Fig. 4F). Consequently, decreasing the dose of rAd5 lead to a significant increase in the proportion of 3+ cells (Fig. 4G). Collectively, these data show that vector selection and vector dose can influence the quality of CD8+ T cell responses. However, increasing the proportion of multifunctional cells may compromise the overall magnitude of the response.
Phenotypic Analysis of SIV Gag-Specific CD8+ T Cells
Individual CD8+ T cells also possess distinct phenotypic profiles that may be predictive of cellular fate. A well-established model for classification of effector and memory CD8+ T cells in mice is based on expression of CD127, the IL-7 receptor α chain, and KLRG1 [44, 45]. In the lymphocytic choriomeningitis virus (LCMV) infection mouse model, CD8+ T cells that expressed KLRG1 without co-expression of CD127 were shown to be terminally differentiated effector cells and designated as short-lived effector cells (SLECs) [45]. Conversely, CD8+ T cells that expressed CD127 without co-expression of KLRG1 were longer-lived and more likely to contribute to the subsequent memory population, and so were termed memory precursor effector cells (MPECs) [44].
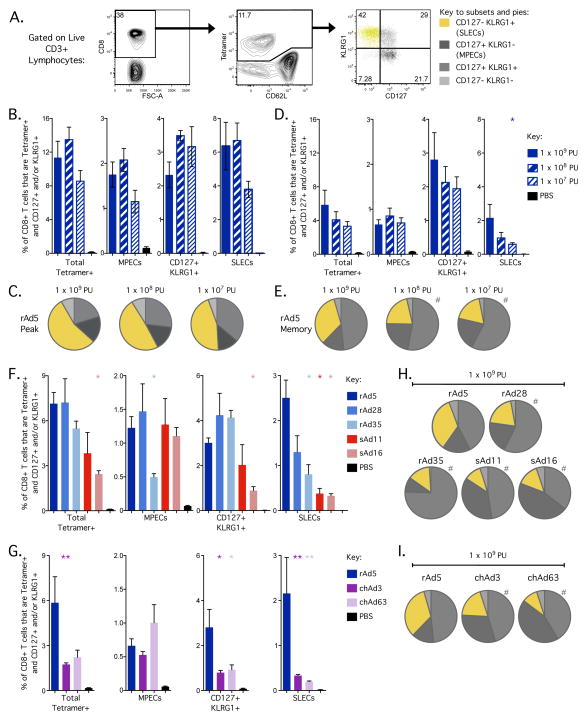
By applying these markers to SIV Gag-specific CD8+ T cells, we observed substantial populations of both CD127-KLRG1+ SLECs (yellow; 42%) and CD127+KLRG1-MPECs (dark grey; 21.7%) 23 days after rAd5 vaccination (Fig. 5A). Strikingly, we also observed a substantial population of Gag-specific CD8+ T cells co-expressing CD127 and KLRG1 (intermediate grey; 29%) (Fig. 5A). Unlike the SLEC and MPEC populations, the potential contribution of the CD127+KLRG1+ population to either immediate effector function or memory development after primary vaccination is not well defined.
Figure 5.
Phenotypic assessment of CD8+ T cells in the spleen 23 days (peak) and 70 days (memory) after rAd vaccination. C57BL/6 mice (n=4–5) were vaccinated with 1 × 109, 1 × 108 or 1 × 107 PU of the indicated rAd vector expressing SIV Gag. At days 23 and 70, splenocytes were tetramer stained to identify SIV Gag-derived AL11-specific cells. Boolean gating was used to define subsets of Gag-specific CD8+ T cells expressing any of the four possible combinations of CD127 and KLRG1. (A) Representative plots after rAd5 vaccination to illustrate gating of CD8+ T cells for phenotypic analysis. CD3+ lymphocytes were gated on CD8+ events (left plot), then tetramer+ events (middle plot) and finally assessed for KLRG1 and CD127 expression (right plot). Gag-specific CD8+ T cells expressing KLRG1 but not CD127 are termed short-lived effectors (SLECs) and cells expressing CD127 but not KLRG1 are termed memory precursor effectors (MPECs). (B/C) The frequency of total CD8+ T cells (B) and proportion of Gag-specific CD8+ T cells (C) that express each combination of CD127/KLRG1 at day 23 after rAd5 vaccination at each dose. (D/E) The frequency of total CD8+ T cells (D) and proportion of Gag-specific CD8+ T cells (E) that express each combination of CD127/KLRG1 at day 70 after rAd5 vaccination at each dose. (F/G) The frequency of total CD8+ T cells that express each combination of CD127/KLRG1 at day 70 after vaccination with rAd5, rAd28, rAd35, sAd11 and sAd16 (F) or rAd5, chAd3 and chAd63 (G) at 1 × 109 PU. (H/I) The proportion of Gag-specific CD8+ T cells that express each combination of CD127/KLRG1 at day 70 after vaccination with rAd5, rAd28, rAd35, sAd11 and sAd16 (H) or rAd5, chAd3 and chAd63 (I) at 1 × 109 PU. For bar graphs, significant differences in frequency were assessed compared to rAd5 at 1 × 109 PU, where * = p ≤ 0.05 and ** = p ≤ 0.01. Bars and error bars represent mean ± SEM. For pie graphs, significant differences in distribution were assessed compared to rAd5 at 1 × 109 PU, where # = p ≤ 0.05. Each group is representative of at least two independent experiments.
The frequency and relative proportions of these populations at peak and memory after rAd5 vaccination were then determined. At peak, SLECs were the predominant population induced at all doses (Fig. 5B, C). By memory, the frequency of total Gag-specific CD8+ T cells had contracted markedly, due to a striking reduction in the frequency of SLECs and a modest reduction in MPECs (Fig. 5D). This is most clearly seen using the relative proportions of each cell population, where there is a marked decrease in SLECs from peak to memory (Fig. 5C, E). In contrast, the frequency of CD127+KRLG1+ CD8+ T cells was well maintained (Fig. 5B, D), and thus the overall proportion of this population increased over time (Fig. 5C, E). These data show that rAd vaccination induces a large CD8+ T cell population that co-expresses CD127 and KLRG1, which is at least as stable as the MPEC population.
We also observed that the dose of vector could alter the phenotypic profile of the induced CD8+ T cell population. This effect was most evident for rAd5 at memory, which induced a substantial population of SLECs at the 1 × 109 PU dose (Fig. 5D, 5E). Decreasing the dose of rAd5 significantly decreased the frequency of SLECs (Fig. 5D) and therefore reduced the proportion of SLECs while increasing the proportion of total CD127+ cells (Fig. 5E).
Lastly, we compared phenotypic differences across the rAd vectors. At the 1 × 109 PU dose, rAd5 vaccination induced significantly more SLECs compared to other vectors (Fig. 5F–I). The simian- and chimp-derived vectors appeared to be similar, in that they induced low frequencies and proportions of SLECs with high proportions of MPECs compared to rAd5 (Fig. 5F–I). Although differences in relative proportions between rAds were most pronounced at 1 × 109 PU, they were still evident and frequently significant at 1 × 107 PU (Supplemental Fig. 3). We also analysed Gag-specific CD8+ T cells in peripheral blood and similar patterns of CD127 and KLRG1 co-expression were observed with regard to time, dose and specific rAd vector (data not shown). Overall, these data show that vector selection and dose reduction can be used to alter the phenotypic profile of CD8+ T cells by minimising induction of the SLEC phenotype, although potentially at the expense of total response magnitude. Furthermore, these results show that expression of CD127 with or without KLRG1 correlates with the stability of CD8+ T cell populations after rAd vaccination.
SIV Gag-Specific Immunity Mediates Protection Following Listeria:Gag or rVACV:Gag Challenge
There is no infectious challenge available in mice to directly model HIV infection in humans. Nonetheless, we wanted to evaluate whether Gag-specific adaptive responses at memory after rAd vaccination, and CD8+ T cells in particular, were functional in vivo. To test this, we used two infectious challenges, Listeria monocytogenes and vaccinia virus.
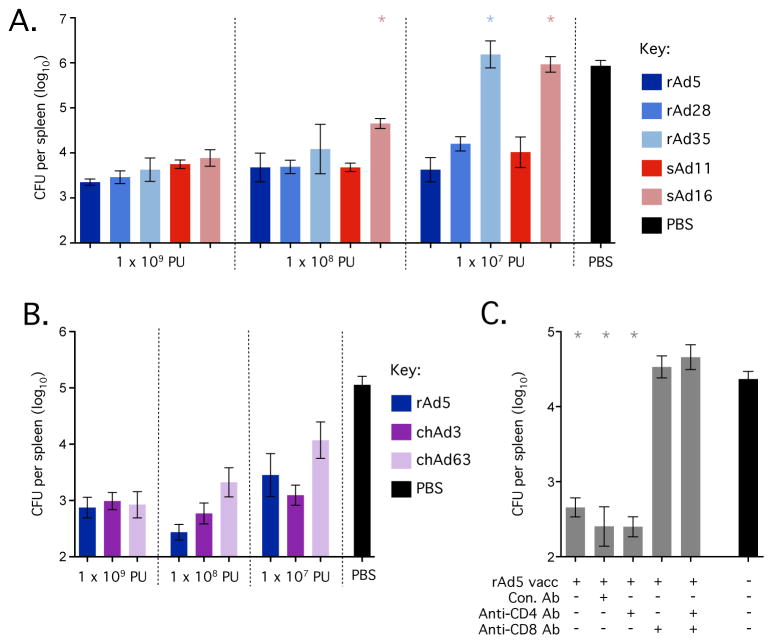
Firstly, mice were administered an attenuated strain of Listeria monocytogenes that expressed SIV Gag (Listeria:Gag) 70 days after vaccination with rAd vectors. Spleen (Fig. 6) and liver (data not shown) tissue was harvested ~42 hours later to determine bacterial load. When compared to the control group, rAd5 vaccinated mice significantly reduced bacterial load (~2.5 logs) at all doses (Fig. 6A, 6B). Mice vaccinated with rAd28 or sAd11 also significantly reduced bacterial loads at all doses compared to controls and did not differ significantly from vaccination with rAd5 at the equivalent dose, although some mice vaccinated with rAd28 or sAd11 at 1 × 107 PU exhibited higher loads, suggesting a slight loss of protection (Fig. 6A). Mice vaccinated with 1 × 109 PU of rAd35 or sAd16 were protected, but protection was diminished at the 1 × 108 PU dose and bacterial loads were equivalent to controls at the 1 × 107 PU dose. Mice vaccinated with chAd3 significantly reduced bacterial load at all doses and did not differ from rAd5 at the equivalent dose (Fig. 6B). Groups vaccinated with chAd63 were protected compared to controls but the group that received 1 × 107 PU trended towards higher bacterial loads, suggesting incomplete protection at the lowest dose (Fig. 6B). Thus, rAd5 and chAd3 consistently conferred the highest protective efficacy, followed by rAd28 and sAd11, chAd63 and finally sAd16 and rAd35.
Figure 6.
Protection afforded by vaccination with rAd vectors against intravenous challenge with Listeria:Gag. (A) Bacterial load in the spleen (colony forming units, CFU) after challenge of mice vaccinated with the indicated doses of rAd5, rAd28, rAd35, sAd11 or sAd16. (B) Bacterial load in the spleen (CFU) after challenge of mice vaccinated with the indicated doses of rAd5, chAd3 or chAd63. (C) Bacterial load in the spleen (CFU) after challenge of mice vaccinated with 1 × 109 PU of rAd5 and either left untreated or treated with a control antibody (Con. Ab), a CD4-depleting antibody (Anti-CD4 Ab), a CD8-depleting Ab (Anti-CD8 Ab) or both of the latter. Each group contained 3–6 C57BL/6 mice and all challenges used a 2 × 107 CFU dose of Listeria:Gag administered intravenously. Significant differences in bacterial load were assessed for each vector compared to rAd5 at the equivalent dose (A/B) or compared to the naïve control (C), where * = p ≤ 0.05. Bars and error bars represent geometric mean ± GEM. Each group is representative of at least two independent experiments.
To determine the relative contribution of CD4+ and CD8+ T cells to protection, mice that had been vaccinated previously with rAd5 were treated with anti-CD4 depleting antibody, anti-CD8 depleting antibody or both prior to challenge with Listeria:Gag. rAd5-mediated protection was completely abrogated in mice treated with anti-CD8 or combined anti-CD4 and CD8 depleting antibodies prior to infection (Fig. 6C), illustrating that an antigen-specific CD8+ T cell response is essential for rapid vaccine-mediated control of listerial infection.
Vaccinia virus expressing SIV Gag (rVACV:Gag) has also been used as a model to assess the protective efficacy of vaccines that elicit T cell immunity [46–48]. Mice immunized with rAds were challenged intranasally with rVACV:Gag and the loss in body weight over the next 6 days was monitored to indicate disease severity (Supplemental Fig. 4). At 6 days after infection, mice vaccinated with rAd5 had maintained their original body weight at all doses. The groups that received 1 × 109 or 1 × 108 PU of sAd16, chAd3 or chAd63, or 1 × 108 PU of sAd11, also maintained their weight. For rAd28, the group that received 1 × 109 PU maintained their original weight but those that received lower doses began to succumb. The rAd35 vector provided only partial protection at 1 × 109 PU, with all other doses succumbing. These data substantiate the potency of rAd5 and illustrate that this potency can extend to simian- and chimp-derived rAds.
rAd Priming Followed by NYVAC Boost Induces Potent CD8+ T Cell Responses
Heterologous prime-boost regimens can be used to generate high magnitude vaccine-induced CD8+ T cell populations [49]. Various regimens combining rAd vectors with other modalities, such as poxvirus-derived vectors, DNA vaccines or heterologous rAd vectors, have been tested in NHP and humans for the prevention of HIV and Malaria [5, 11, 50–54]. We therefore administered the rAd vectors as primes for a common boost or as boosts following a common prime to compare their potency in prime-boost vaccine regimens.
Priming with rAd vectors and boosting with pox vectors can induce high magnitude CD8+ T cell responses in mice, NHPs and humans [11, 50–54]. We therefore used the pox vector, NYVAC expressing SIV Gag, as a common boost for the human- and chimp-derived rAds. Mice were vaccinated with 1 × 107 PU of each rAd to best model the hierarchy of rAd-primed responses in NHP and humans and then boosted with 1 × 107 PFU of NYVAC. Mice primed with rAd5 had a modestly higher frequency of CD8+ T cells at the time of boost compared to the other rAd vectors (Fig. 7A). NYVAC did not induce detectable responses in unprimed mice but it robustly boosted all rAd-primed mice that had detectable responses at the time of boost, with the frequency of Gag-specific CD8+ T cells increasing 6–12-fold to ~60% of total CD8+ T cells at peak for most rAds (Fig. 7B). The frequency of Gag-specific CD8+ T cells after NYVAC boosting did not differ significantly between groups primed with rAd5, chAd3 and chAd63 at peak (Fig. 7B) or memory (Fig. 7C). By contrast, the rAd28-primed group was boosted by NYVAC but exhibited frequencies that were significantly lower than the rAd5-primed group at peak (Fig. 7B) and trended lower at memory (Fig. 7C). Lastly, priming with rAd35 at 1 × 107 PU did not prime detectable responses at the time of boost (Fig. 7A) and did not enable boosting in response to NYVAC (Fig. 7B, C). However, priming with higher doses of rAd35 induced responses that were potently boosted by NYVAC (Fig. 7D). In addition, we performed experiments using rAd vectors encoding HIV Env as a target antigen and another pox vector, modified vaccinia virus Ankara (MVA), expressing Env as a common boost. Consistent with the results above, there was robust boosting of Env-specific CD8+ T cell responses with all vectors (rAd5, chAd3, chAd63) that primed detectable responses at the time of boost (Fig. 7E–G). Taken together, these data show that pox vectors potently boost CD8+ T cell responses after priming with rAds, even when priming is sub-optimal.
rAd5 and chAd3 Potently Boost DNA or rAd28 Primed Responses
rAd vectors are also being evaluated in NHP and clinical trials as boosts for priming vaccines such as DNA [55–57] or heterologous rAds [5, 54]. Accordingly, we compared rAd vectors as boosts after DNA or rAd28 priming. Mice were primed with either 100 μg of DNA (two doses given 3 weeks apart) or 1 × 108 PU of rAd28 and then boosted 7 weeks later with 1 × 108 PU of rAd35, rAd5 or chAd3. After DNA priming, both rAd5 and chAd3 boosted to similar magnitudes (Fig. 8A). In contrast, rAd35-boosted responses were significantly lower than groups boosted with rAd5 or chAd3, although they were significantly higher than the DNA prime or rAd35 boost alone (Fig. 8A). As demonstrated above, the 1 × 108 PU dose of rAd35 primes CD8+ T cell responses that are very low or undetectable by tetramer staining (Fig. 2A, 7D), so this dose may be sub-optimal for boosting with rAd35. Nevertheless, in the setting of a sufficiently primed CD8+ T cell response, rAd35 is an effective boost. By day 70 after vaccination, CD8+ T cell responses contracted for all groups, but mice boosted with rAd5 or chAd3 contracted to frequencies that did not differ significantly from groups that received the vectors as boosts alone (Fig. 8B). In contrast, mice that were primed with DNA and boosted with rAd35 maintained their responses relative to peak at significantly higher levels compared to the rAd35 boost alone (Fig. 8B).
Figure 8.
Assessment of boosting by rAd vectors after a common prime. C57BL/6 mice (n=5) were primed with either 2 × 100 μg of DNA, 3 weeks apart, or 1 × 108 PU of rAd28 and boosted 7 weeks after initial priming with 1 × 108 PU of either rAd5, rAd35 or chAd3. At the indicated time points, SIV Gag-derived AL11-specific CD8+ T cells were quantified in peripheral blood by tetramer staining. (A) Frequency of CD8+ T cells that were tetramer+ at the peak of the response, 2 weeks after boosting. (B) Frequency of CD8+ T cells that were tetramer+ at memory, 10 weeks after boosting. Significant differences in frequency were assessed in primed/rAd-boosted animals compared to mice that received the equivalent rAd boost alone, where NS= not significantly different, * = p<0.05 and ** = p ≤ 0.01. Bars and error bars represent mean ± SEM. Each group is representative of at least two independent experiments.
A similar pattern was evident for rAd28 priming, where rAd5 and chAd3 strongly boosted CD8+ T cell responses at peak compared to rAd35 (Fig. 8A). Subsequently, rAd5 and chAd3 boosted responses had contracted significantly by day 70, while rAd35 responses were well maintained (Fig. 8B). Notably, rAd28-primed responses achieved a higher magnitude at peak and memory after boosting with any rAd vector compared to DNA-primed responses. This may reflect the higher frequencies of Gag-specific CD8+ T cells present at the time of boosting after rAd28 compared to DNA priming. Overall, rAd5 and chAd3 similarly boosted robust CD8+ T cell responses that then contracted, while rAd35 boosted to lower frequencies but effectively sustained these responses into memory.
Discussion
In this broad comparative analysis of seven different human-, simian- and chimp-derived rAds, a dose titration approach was used to delineate differences between vectors based on the magnitude, quality, phenotype and protective capacity of SIV Gag-specific CD8+ T cell responses. For each vector, the magnitude of CD8+ T cell responses at memory directly correlated with the protective capacity against L. monocytogenes infection. The rAd5 and chAd3 vectors induced the most robust and comparable CD8+ T cell-mediated protective immunity, followed by sAd11 and rAd28, chAd63, sAd16 and finally rAd35. Our conclusion that rAd5 is more potent than rAd28 and rAd35 is consistent with prior studies in mice and NHP using rAd26 and rAd35 with SIV Gag [4, 5] and confirms the relative potency of chAd3 and chAd63 recently described in mice, NHP and humans [25, 26, 32]. The dose titration approach also illustrated that, at the highest vector dose of 1 × 109 PU, there was little ability to discriminate between the different rAd vectors in the mouse model. This is likely due to there being sufficient antigen expression at the highest dose to prime comparable responses. Indeed, the most common dose of rAd vectors used in humans is 1 × 1010 PU, which is only 10-fold higher than the highest and most protective dose for all rAds used in this study. Additionally, viral vectors target specific receptors that mediate uptake but receptor distribution may differ from mice to humans and data generated using mouse models should be interpreted with this in mind. For example, rAd35 utilises membrane co-factor protein (CD46) [58], which is broadly expressed across nucleated cells in humans but is limited in mice to the testes. This could apply to other vectors, since receptors and soluble mediators of rAd5 uptake are incompletely characterised [59] and the primary receptors for rAd28 [22, 60] and for the simian- and chimp-derived adenoviruses have not been defined. Nevertheless, this study was able to replicate the hierarchy between rAd5, rAd28 and rAd35 seen in NHPs and humans and thus illustrates that pre-clinical mouse studies at lower doses provide predictive value for CD8+ T cell immunity in humans.
Differences between rAds in terms of the magnitude, quality, phenotype and protective capacity of CD8+ T cell responses can be used to select the optimal vectors for use as stand-alone vaccines or in prime-boost regimens. There is clear evidence that vaccine-mediated protection against malarial or SIV infection correlates with the induction of higher magnitude CD8+ T cell responses [61–63]. A stand-alone vaccine must achieve this magnitude in one vaccination, while prime-boost regimens can attain a similar or much higher magnitude in a stepwise manner over a prolonged period of time [49]. rAd5 has been the most widely studied and remains the most potent rAd vector for inducing high magnitude CD8+ T cell responses with a single immunization. Recent evidence suggests that T cell responses induced by rAd5 display a more differentiated profile based on surface markers [64] or qualitative assessments of cytokine production compared to other vaccine platforms [5, 43]. Additionally, CD8+ T cells induced by rAd5 vaccination are less proliferative on a per cell basis compared to CD8+ T cells induced by L. monocytogenes, vaccinia virus or LCMV infection and display an “exhausted” gene expression profile compared to LCMV (Sarkar and Kalia et al. Personal communication). Models for the differentiation state of T cells propose that cells sequentially lose functions, such as the production of IL-2 and TNFα, after successive or prolonged antigen encounters, becoming terminally differentiated cells that produce IFNγ only [43] or potentially exhausted cells [65, 66]. Exhausted antigen-specific CD8+ T cells are identified through high expression of markers, such as Programmed Death-1 [67] and KLRG1 [68, 69] or functional assessments, such as diminished proliferative capacity [65, 66]. Our study shows that vaccination with rAd5 at 1 × 109 PU induced a higher proportion Gag-specific CD8+ T cells that were IFNγ (1+) or IFNγ/TNFα (2+) rather than multifunctional IFNγ/IL-2/TNFα (3+) cells and a higher proportion of cells that expressed KLRG1 without CD127 co-expression. Together, these data suggest a more differentiated functional and memory phenotype, which is likely due to low-level antigen that can persist after rAd5 vaccination [17, 64, 70, 71]. Nevertheless, the high magnitude CD8+ T cell response induced by rAd5 in this study was sufficient to rapidly control pathogen load after infection, regardless of qualitative or phenotypic profiles of the CD8+ T cell population. Similarly, vaccination of NHPs with a 1 × 1010 PU dose of rAd5 but not rAd26 or rAd35 confers protection against Ebola, a rapidly progressing viral infection, by inducing a high magnitude CD8+ T cell response [72, 73]. Overall, if a stand-alone vector is required, high doses of rAd vectors such as rAd5 or chAd3 are capable of inducing high magnitude CD8+ T cell responses that may mediate rapid control of infections. Alternatively, lower doses of rAd vectors can improve the qualitative and phenotypic profiles of CD8+ T cell responses, albeit at reduced magnitudes. Further work is needed to establish whether the favourable phenotypic and qualitative profiles induced with lower doses of rAds are beneficial for boosting of primed CD8+ T cells. However, lowering the dose of rAds for priming purposes runs the risk of inducing CD8+ T cell responses below a threshold necessary for efficient heterologous boosting, particularly in a diverse human population.
A striking finding in this study is that rAd vaccination induces a substantial population of CD8+ T cells expressing both CD127 and KLRG1 at memory. The original study by Joshi et al., which assessed expression of both CD127 and KLRG1 on CD8+ T cells, defined CD127-KLRG1+ and CD127+KLRG1− CD8+ T cells as SLEC and MPEC populations respectively, but there was limited analysis of CD127+KLRG1+CD8+ T cells [45]. This population has been observed at low levels in various vaccine and infection models [74, 75]. In one study, a high frequency of CD127+KLRG1+ CD8+ T cells was detected after prime-boosting and these cells were shown to have a modestly lower proliferative potential compared to MPECs [76]. Consistent with prior reports, we observed that the frequencies of SLECs were highest at peak and decreased over time, while frequencies of MPECs and CD127+KLRG1+ cells were relatively stable (Fig. 5B–E). These data substantiate the paradigm that SLECs are short-lived while MPECs are more stable, but also suggest that CD127+KLRG1+ cells represent a relatively stable CD8+ T cell memory population that can be induced after primary vaccination. The degree to which different vaccines and infection models induce CD127+KLRG1+ CD8+ T cells is likely related to antigen persistence and/or repetitive antigenic stimulation. Thus, while acute LCMV infection is efficiently cleared [66, 77], prime-boosting re-exposes the vaccine recipient to antigen and low-level antigen can persist after rAd5 vaccination [17, 64, 70, 71]. As antigen persistence has been shown to maintain KLRG1 expression [78, 79], we speculate that CD127+KLRG1+ CD8+ T cells are a stable memory cell population associated with the repeated or protracted but not overwhelming presence of stimulating antigen.
Heterologous prime-boost immunisation can be used to dramatically expand antigen-specific CD8+ T cell responses compared to either vaccine modality alone [49]. As the magnitude and functionality of CD8+ T cell responses is critical for control of HIV viral load [63] and elimination of malarial infection during the liver stage [61, 62], rAd vectors were evaluated as part of prime-boost regimens with other vaccines. We first compared priming with human- and chimp-derived rAd vectors followed by a common boost. A recombinant pox vector was used as a boost, because poxviruses and adenoviruses do not share any homology that could lead to cross-reactive antibody or T cell responses and are therefore truly heterologous. In addition, pox vectors have been shown to act as potent boosts for CD8+ T cell immunity in NHP and human clinical trials for vaccines against HIV and Malaria [11, 51, 52, 54]. The data reported here show that, although pox vectors did not robustly prime Gag-specific CD8+ T cell responses, they potently boosted rAd-primed responses. This was true for all rAd vectors that primed detectable responses, as even sub-optimal CD8+ T cell priming with rAd35 could be boosted with pox vectors. Thus, a minimal threshold of priming is necessary for CD8+ T cells, which underscores the need to choose an rAd vector and dose that efficiently and consistently primes responses across vaccinees for clinical application of rAd prime-pox boost regimens. Additionally, pox vectors have been used as primes to induce robust antibody-mediated immunity, such as in a recent study that induced protective responses against HIV in humans [80]. Thus, pox vectors are clearly effective primes for CD4+ T cell and antibody responses combined with another vaccine modality as a boost. However, our study would suggest that if CD8+ T cell immunity is critical for protection, pox vectors should be reserved for use as a boost in prime-boost regimens.
Human- and chimp-derived rAds were also compared following common primes. We focused on rAd5, chAd3 and rAd35 as boosts, as rAd5 is currently being tested in an HIV vaccine efficacy trial following DNA priming (HIV Vaccine Trials Network (HVTN) 505), the chAd3 vector is a potential replacement for rAd5 due to its phylogenetic similarity and potency [26, 32], and rAd35 is currently the focus of a human clinical trial, comparing its efficacy as a boost to rAd5 after DNA priming (HVTN 077). We chose DNA or rAd28 as primes for several reasons. Firstly, DNA or rAd26, which is closely related to rAd28, have been already been used in NHP and/or human clinical trials as primes prior to rAd boosting for HIV, SIV, and Malaria vaccines [5, 15, 54–56]. Secondly, a viral vector such as rAd28 could induce CD8+ T cell responses in humans more efficiently than DNA, as a DNA vaccine typically requires multiple immunisations to achieve T cell response magnitudes comparable to a single immunisation with a viral vector [81]. Indeed, priming with a single shot of rAd28 induced higher magnitude CD8+ T cell responses than two shots of DNA at the time of boost, at peak after boosting and at memory, illustrating that the magnitude of the primed response was critical to the magnitude of the subsequent boost. Boosting mice primed with DNA or rAd28 with either rAd5 or chAd3 resulted in robust and comparable peak CD8+ T cell responses, which provides strong evidence that chAd3 is an appropriate alternative for rAd5. Interestingly, while rAd35 boosting did not achieve frequencies seen with rAd5 or chAd3 at peak, the response magnitude was relatively stable out to memory time points. Thus, although not an effective prime compared to other rAd vectors unless used at high doses (Fig. 7D), rAd35 may be valuable as a boost [82], especially given its low rates of seroreactivity in human populations and relative amenability to large-scale production for clinical use.
As a practical concern, the development of a large array of well-characterised rAd vectors allows versatility in their clinical application. Clinical trials have been performed using rAd vectors for multiple infections, including Malaria, Tb and HIV ([9, 12, 13] HVTN 077). Until recently, a limited number of rAd vectors (rAd5, rAd26 and rAd35) were available for clinical use, which complicated the application of a restricted pool of vectors to target multiple infections since prior immunity limits the potency of subsequent vaccination. The development of simian- and chimp-derived rAds has provided a broader array of vectors to choose from, with low seroprevalence and considerable potency [23, 30]. Accordingly, despite the constraint that prior immunity could place on repeated rAd administration, having a wide range of vectors will mitigate this issue.
In conclusion, these data can be used to both refine clinical selection of currently available rAd vectors as well as extend our understanding of mechanisms that control the potency of rAd vectors. Our data indicate that low potency vectors such as rAd35 would not be sufficient as stand-alone vaccines for prophylactic vaccination against infections requiring CD8+ T cell immunity but multiple rAd vectors such as rAd5 and chAd3 can efficiently induce such responses. By contrast, after priming with DNA or a heterologous rAd, all rAd vectors including rAd35 are capable of boosting CD8+ T cell immunity. Importantly, pox vectors such as NYVAC and MVA provide truly heterologous and very robust boosting to any rAd prime and this may be the optimal combination for induction of high magnitude CD8+ T cell responses. Finally, these data permit us to dissect requirements for induction of robust CD8+ T cell immunity. We characterised quantitative, qualitative and phenotypic differences in CD8+ T cell immunity induced by rAd vectors but early mechanisms after rAd vaccination responsible for subsequent induction of potent CD8+ T cell immunity remain unknown. A number of mechanisms have been suggested, including increased uptake or targeting of specialised APC subsets [83], expression of large amounts of antigen for a relatively prolonged time [17, 64, 70, 71] and robust activation of innate immune mechanisms [21, 22]. In ongoing work, we are examining the mechanistic bases for differences observed in this study in potency of CD8+ T cell responses between rAd vectors by exploring differences in innate immunity, the amount and duration of antigen expression and antigen presentation (manuscript in preparation). Taken together, our direct comparison of rAd efficacy and the definition of mechanisms leading to potent CD8+ T cell responses will facilitate rational development of rAd-based vaccination strategies that address the unique requirements of different infections.
Supplementary Material
Acknowledgments
Grant Support:
This research has been supported in part by a grant from the Foundation for the National Institutes of Health with support from the Collaboration for AIDS Vaccine Discovery (CAVD) award OPP1039775 from the Bill & Melinda Gates Foundation.
We would like to acknowledge the U.S. Military HIV Research Program, the International AIDS Vaccine Initiative and the HIV Vaccine Trials Network for their kind provision of sera samples used to establish rAd neutralising titres, in particular the following site investigators: Eftyhia Vardas (Perinatal HIV Research Unit, University of the Witwatersrand, Soweto, South Africa), Heather Jaspan (Desmond Tutu HIV Centre, University of Cape Town, Cape Town, South Africa), Gavin Churchyard (Aurum Institute for Health Research, Klerksdorp, South Africa), Ramesh Paranjape (National AIDS Research Institute, Pune, India), Jean Pape (Cornell University, New York NY, USA and Centres GHESKIO, Port-au-Prince, Haiti), J Peter Figueroa (Epidemiology Research and Training Unit, Ministry of Health, Kingston, Jamaica), Noreen Jack (Caribbean Care, Prevention, and Research Institute, Port of Spain, Trinidad and Tobago), Yeycy Donastorg (Unidad de Vacunas IDCP-COIN-DIGECITSS, Santo Domingo, Dominican Republic), Paulo Barroso (Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil), Artur Kalichman (Centro de Referencia e Treinamento em DST/AIDS, São Paulo, Brazil), Javier Lama (IMPACTA, Asociacion Civil Impacta Salud y Educacion, Lima, Perú). We additionally thank Zhi-Yong Yang (VRC, NIH, MD, USA) for provision of DNA encoding SIV Gag, Glennys V. Reynoso (NIAID, NIH, MD, USA) for provision of rVACV:Gag, Fred D. Finkelman (University of Cincinnati, OH, USA) for provision of antibodies used for depletion studies, Teresa R. Johnson (GenVec Inc, Gaithersburg, MD, USA) for assistance developing vaccinia virus infection protocols and Daniel E. Zak (Seattle BioMed, WA, USA) for helpful discussion.
Abbreviations used in this article
- rAd
recombinant adenoviral vector
- Tb
Mycobacterium tuberculosis infection
- NHP
non-human primate
- KLRG1
killer cell lectin-like receptor subfamily G member 1
- PU
particle units
- PFU
plaque forming units
- MVA
modified vaccinia virus Ankara
- Env
gp140 Envelope protein
- BFA
brefeldin A
- Listeria
Gag, recombinant Listeria monocytogenes expressing SIV Gag
- rVACV
Gag, recombinant vaccinia virus expressing SIV Gag
- CFU
colony forming units
- LCMV
lymphocytic choriomeningitis virus
- SLECs
short-lived effector cells
- MPECs
memory precursor effector cells
- HVTN
HIV Vaccine Trials Network
Footnotes
Disclosures:
J.G.D.G is an employee of GenVec Inc. A.N., A.F., S.C. and R.C. are employees of Okairos AG. All other authors have no financial conflicts of interest.
References
- 1.Thaiss CA, Kaufmann SHE. Toward novel vaccines against tuberculosis: current hopes and obstacles. Yale J Biol Med. 2010;83:209–215. [PMC free article] [PubMed] [Google Scholar]
- 2.Spence PJ, Langhorne J. T cell control of malaria pathogenesis. Curr Opin Immunol. 2012;24:444–448. doi: 10.1016/j.coi.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbink P, Lemckert AAC, Ewald BA, Lynch DM, Denholtz M, Smits S, Holterman L, Damen I, Vogels R, Thorner AR, et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J Virol. 2007;81:4654–4663. doi: 10.1128/JVI.02696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, O’Brien KL, Lynch DM, Simmons NL, La Porte A, Riggs AM, Abbink P, Coffey RT, Grandpre LE, Seaman MS, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457:87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catanzaro AT, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, Gu L, Martin JE, Novik L, Chakrabarti BK, et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J Infect Dis. 2006;194:1638–1649. doi: 10.1086/509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Priddy FH, Brown D, Kublin J, Monahan K, Wright DP, Lalezari J, Santiago S, Marmor M, Lally M, Novak RM, et al. Safety and immunogenicity of a replication-incompetent adenovirus type 5 HIV-1 clade B gag/pol/nef vaccine in healthy adults. Clin Infect Dis. 2008;46:1769–1781. doi: 10.1086/587993. [DOI] [PubMed] [Google Scholar]
- 8.Bassett JD, Swift SL, Bramson JL. Optimizing vaccine-induced CD8(+) T-cell immunity: focus on recombinant adenovirus vectors. Expert Rev Vaccines. 2011;10:1307–1319. doi: 10.1586/erv.11.88. [DOI] [PubMed] [Google Scholar]
- 9.Abel B, Tameris M, Mansoor N, Gelderbloem S, Hughes J, Abrahams D, Makhethe L, Erasmus M, de Kock M, van der Merwe L, et al. The novel tuberculosis vaccine, AERAS-402, induces robust and polyfunctional CD4+ and CD8+ T cells in adults. Am J Respir Crit Care Med. 2010;181:1407–1417. doi: 10.1164/rccm.200910-1484OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheehy SH, Duncan CJ, Elias SC, Collins KA, Ewer KJ, Spencer AJ, Williams AR, Halstead FD, Moretz SE, Miura K, et al. Phase Ia Clinical Evaluation of the Plasmodium falciparum Blood-stage Antigen MSP1 in ChAd63 and MVA Vaccine Vectors. Mol Ther. 2011;19:2269–2276. doi: 10.1038/mt.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheehy SH, Duncan CJA, Elias SC, Biswas S, Collins KA, O’Hara GA, Halstead FD, Ewer KJ, Mahungu T, Spencer AJ, et al. Phase Ia clinical evaluation of the safety and immunogenicity of the Plasmodium falciparum blood-stage antigen AMA1 in ChAd63 and MVA vaccine vectors. PLoS ONE. 2012;7:e31208. doi: 10.1371/journal.pone.0031208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sedegah M, Tamminga C, McGrath S, House B, Ganeshan H, Lejano J, Abot E, Banania GJ, Sayo R, Farooq F, et al. Adenovirus 5-vectored P. falciparum vaccine expressing CSP and AMA1. Part A: safety and immunogenicity in seronegative adults. PLoS ONE. 2011;6:e24586. doi: 10.1371/journal.pone.0024586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamminga C, Sedegah M, Regis D, Chuang I, Epstein JE, Spring M, Mendoza-Silveiras J, McGrath S, Maiolatesi S, Reyes S, et al. Adenovirus-5-vectored P. falciparum vaccine expressing CSP and AMA1. Part B: safety, immunogenicity and protective efficacy of the CSP component. PLoS ONE. 2011;6:e25868. doi: 10.1371/journal.pone.0025868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barouch DH, Kik SV, Weverling GJ, Dilan R, King SL, Maxfield LF, Clark S, Ng’ang’a D, Brandariz KL, Abbink P, et al. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine. 2011;29:5203–5209. doi: 10.1016/j.vaccine.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casimiro DR, Chen L, Fu TM, Evans RK, Caulfield MJ, Davies ME, Tang A, Chen M, Huang L, Harris V, et al. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J Virol. 2003;77:6305–6313. doi: 10.1128/JVI.77.11.6305-6313.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barouch DH, Pau MG, Custers JHHV, Koudstaal W, Kostense S, Havenga MJE, Truitt DM, Sumida SM, Kishko MG, Arthur JC, et al. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J Immunol. 2004;172:6290–6297. doi: 10.4049/jimmunol.172.10.6290. [DOI] [PubMed] [Google Scholar]
- 17.Lindsay RWB, Darrah PA, Quinn KM, Wille-Reece U, Mattei LM, Iwasaki A, Kasturi SP, Pulendran B, Gall JGD, Spies AG, et al. CD8+ T cell responses following replication-defective adenovirus serotype 5 immunization are dependent on CD11c+ dendritic cells but show redundancy in their requirement of TLR and nucleotide-binding oligomerization domain-like receptor signaling. J Immunol. 2010;185:1513–1521. doi: 10.4049/jimmunol.1000338. [DOI] [PubMed] [Google Scholar]
- 18.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duerr A, Huang Y, Buchbinder S, Coombs RW, Sanchez J, Del Rio C, Casapia M, Santiago S, Gilbert P, Corey L, et al. Extended follow-up confirms early vaccine-enhanced risk of HIV acquisition and demonstrates waning effect over time among participants in a randomized trial of recombinant adenovirus HIV vaccine (Step study) J Infect Dis. 2012;206:258–66. doi: 10.1093/infdis/jis342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng C, Wang L, Gall JGD, Nason M, Schwartz RM, McElrath MJ, DeRosa SC, Hural J, Corey L, Buchbinder SP, et al. Decreased pre-existing Ad5 capsid and Ad35 neutralizing antibodies increase HIV-1 infection risk in the Step trial independent of vaccination. PLoS ONE. 2012;7:e33969. doi: 10.1371/journal.pone.0033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogels R, Zuijdgeest D, van Rijnsoever R, Hartkoorn E, Damen I, de Béthune MP, Kostense S, Penders G, Helmus N, Koudstaal W, et al. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J Virol. 2003;77:8263–8271. doi: 10.1128/JVI.77.15.8263-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahl CA, Bonnell J, Hiriyanna S, Fultz M, Nyberg-Hoffman C, Chen P, King CR, Gall JGD. Potent immune responses and in vitro pro-inflammatory cytokine suppression by a novel adenovirus vaccine vector based on rare human serotype 28. Vaccine. 2010;28:5691–5702. doi: 10.1016/j.vaccine.2010.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farina SF, Gao GP, Xiang ZQ, Rux JJ, Burnett RM, Alvira MR, Marsh J, Ertl HC, Wilson JM. Replication-defective vector based on a chimpanzee adenovirus. J Virol. 2001;75:11603–11613. doi: 10.1128/JVI.75.23.11603-11613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Z, Gao G, Reyes-Sandoval A, Cohen CJ, Li Y, Bergelson JM, Wilson JM, Ertl HCJ. Novel, chimpanzee serotype 68-based adenoviral vaccine carrier for induction of antibodies to a transgene product. J Virol. 2002;76:2667–2675. doi: 10.1128/JVI.76.6.2667-2675.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peruzzi D, Dharmapuri S, Cirillo A, Bruni BE, Nicosia A, Cortese R, Colloca S, Ciliberto G, La Monica N, Aurisicchio L. A novel chimpanzee serotype-based adenoviral vector as delivery tool for cancer vaccines. Vaccine. 2009;27:1293–1300. doi: 10.1016/j.vaccine.2008.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colloca S, Barnes E, Folgori A, Ammendola V, Capone S, Cirillo A, Siani L, Naddeo M, Grazioli F, Esposito ML, et al. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci Transl Med. 2012;4:115ra2. doi: 10.1126/scitranslmed.3002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seto D, Chodosh J, Brister JR, Jones MS Community MotAR. Using the whole-genome sequence to characterize and name human adenoviruses. J Virol. 2011;85:5701–5702. doi: 10.1128/JVI.00354-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsushima Y, Shimizu H, Kano A, Nakajima E, Ishimaru Y, Dey SK, Watanabe Y, Adachi F, Suzuki K, Mitani K, et al. Novel human adenovirus strain, Bangladesh. Emerging Infect Dis. 2012;18:846–848. doi: 10.3201/eid1805.111584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang ZQ, Yang Y, Wilson JM, Ertl HC. A replication-defective human adenovirus recombinant serves as a highly efficacious vaccine carrier. Virology. 1996;219:220–227. doi: 10.1006/viro.1996.0239. [DOI] [PubMed] [Google Scholar]
- 30.Pinto AR, Fitzgerald JC, Giles-Davis W, Gao GP, Wilson JM, Ertl HCJ. Induction of CD8+ T cells to an HIV-1 antigen through a prime boost regimen with heterologous E1-deleted adenoviral vaccine carriers. J Immunol. 2003;171:6774–6779. doi: 10.4049/jimmunol.171.12.6774. [DOI] [PubMed] [Google Scholar]
- 31.Fitzgerald JC, Gao GP, Reyes-Sandoval A, Pavlakis GN, Xiang ZQ, Wlazlo AP, Giles-Davis W, Wilson JM, Ertl HCJ. A simian replication-defective adenoviral recombinant vaccine to HIV-1 gag. J Immunol. 2003;170:1416–1422. doi: 10.4049/jimmunol.170.3.1416. [DOI] [PubMed] [Google Scholar]
- 32.Barnes E, Folgori A, Capone S, Swadling L, Aston S, Kurioka A, Meyer J, Huddart R, Smith K, Townsend R, et al. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci Transl Med. 2012;4:115ra1. doi: 10.1126/scitranslmed.3003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang Z, Li Y, Cun A, Yang W, Ellenberg S, Switzer WM, Kalish ML, Ertl HCJ. Chimpanzee adenovirus antibodies in humans, sub-Saharan Africa. Emerging Infect Dis. 2006;12:1596–1599. doi: 10.3201/eid1210.060078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sprangers MC, Lakhai W, Koudstaal W, Verhoeven M, Koel BF, Vogels R, Goudsmit J, Havenga MJE, Kostense S. Quantifying adenovirus-neutralizing antibodies by luciferase transgene detection: addressing preexisting immunity to vaccine and gene therapy vectors. J Clin Microbiol. 2003;41:5046–5052. doi: 10.1128/JCM.41.11.5046-5052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McVey D, Zuber M, Ettyreddy D, Reiter CD, Brough DE, Nabel GJ, King CR, Gall JGD. Characterization of human adenovirus 35 and derivation of complex vectors. Virol J. 2010;7:276. doi: 10.1186/1743-422X-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honda M, Wang R, Kong WP, Kanekiyo M, Akahata W, Xu L, Matsuo K, Natarajan K, Robinson H, Asher TE, et al. Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity. J Immunol. 2009;183:2425–2434. doi: 10.4049/jimmunol.0900581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Ewald BA, Lynch DM, Nanda A, Sumida SM, Barouch DH. Modulation of DNA vaccine-elicited CD8+ T-lymphocyte epitope immunodominance hierarchies. J Virol. 2006;80:11991–11997. doi: 10.1128/JVI.01348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiesel M, Oxenius A. From crucial to negligible: functional CD83 T-cell responses and their dependence on CD43 T-cell help. Eur J Immunol. 2012;42:1080–1088. doi: 10.1002/eji.201142205. [DOI] [PubMed] [Google Scholar]
- 39.Williams MA, Holmes BJ, Sun JC, Bevan MJ. Developing and maintaining protective CD8+ memory T cells. Immunol Rev. 2006;211:146–153. doi: 10.1111/j.0105-2896.2006.00389.x. [DOI] [PubMed] [Google Scholar]
- 40.Yang TC, Millar J, Groves T, Zhou W, Grinshtein N, Parsons R, Evelegh C, Xing Z, Wan Y, Bramson J. On the role of CD4+ T cells in the CD8+ T-cell response elicited by recombinant adenovirus vaccines. Mol Ther. 2007;15:997–1006. doi: 10.1038/sj.mt.6300130. [DOI] [PubMed] [Google Scholar]
- 41.La Gruta NL, Turner SJ, Doherty PC. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J Immunol. 2004;172:5553–5560. doi: 10.4049/jimmunol.172.9.5553. [DOI] [PubMed] [Google Scholar]
- 42.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darrah PA, Patel DT, De Luca PM, Lindsay RWB, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 44.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 45.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belyakov IM, Earl P, Dzutsev A, Kuznetsov VA, Lemon M, Wyatt LS, Snyder JT, Ahlers JD, Franchini G, Moss B, et al. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. PNAS. 2003;100:9458–9463. doi: 10.1073/pnas.1233578100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyatt LS, Earl PL, Eller LA, Moss B. Highly attenuated smallpox vaccine protects mice with and without immune deficiencies against pathogenic vaccinia virus challenge. PNAS. 2004;101:4590–4595. doi: 10.1073/pnas.0401165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trumpfheller C, Finke JS, López CB, Moran TM, Moltedo B, Soares H, Huang Y, Schlesinger SJ, Park CG, Nussenzweig MC, et al. Intensified and protective CD4+ T cell immunity in mice with anti-dendritic cell HIV gag fusion antibody vaccine. J Exp Med. 2006;203:607–617. doi: 10.1084/jem.20052005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vezys V, Yates A, Casey KA, Lanier G, Ahmed R, Antia R, Masopust D. Memory CD8 T-cell compartment grows in size with immunological experience. Nature. 2009;457:196–199. doi: 10.1038/nature07486. [DOI] [PubMed] [Google Scholar]
- 50.Tatsis N, Lin SW, Harris-McCoy K, Garber DA, Feinberg MB, Ertl HCJ. Multiple immunizations with adenovirus and MVA vectors improve CD8+ T cell functionality and mucosal homing. Virology. 2007;367:156–167. doi: 10.1016/j.virol.2007.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casimiro DR, Bett AJ, Fu TM, Davies ME, Tang A, Wilson KA, Chen M, Long R, McKelvey T, Chastain M, et al. Heterologous human immunodeficiency virus type 1 priming-boosting immunization strategies involving replication-defective adenovirus and poxvirus vaccine vectors. J Virol. 2004;78:11434–11438. doi: 10.1128/JVI.78.20.11434-11438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang G, Shi M, Conteh S, Richie N, Banania G, Geneshan H, Valencia A, Singh P, Aguiar J, Limbach K, et al. Sterile protection against Plasmodium knowlesi in rhesus monkeys from a malaria vaccine: comparison of heterologous prime boost strategies. PLoS ONE. 2009;4:e6559. doi: 10.1371/journal.pone.0006559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reyes-Sandoval A, Berthoud T, Alder N, Siani L, Gilbert SC, Nicosia A, Colloca S, Cortese R, Hill AVS. Prime-boost immunization with adenoviral and modified vaccinia virus Ankara vectors enhances the durability and polyfunctionality of protective malaria CD8+ T-cell responses. Infect Immun. 2010;78:145–153. doi: 10.1128/IAI.00740-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, SanMiguel A, Seaman MS, Ferrari G, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koblin BA, Casapia M, Morgan C, Qin L, Wang ZM, Defawe OD, Baden L, Goepfert P, Tomaras GD, Montefiori DC, et al. Safety and immunogenicity of an HIV adenoviral vector boost after DNA plasmid vaccine prime by route of administration: a randomized clinical trial. PLoS ONE. 2011;6:e24517. doi: 10.1371/journal.pone.0024517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koup RA, Roederer M, Lamoreaux L, Fischer J, Novik L, Nason MC, Larkin BD, Enama ME, Ledgerwood JE, Bailer RT, et al. Priming immunization with DNA augments immunogenicity of recombinant adenoviral vectors for both HIV-1 specific antibody and T-cell responses. PLoS ONE. 2010;5:e9015. doi: 10.1371/journal.pone.0009015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Churchyard GJ, Morgan C, Adams E, Hural J, Graham BS, Moodie Z, Grove D, Gray G, Bekker LG, McElrath MJ, et al. A phase IIA randomized clinical trial of a multiclade HIV-1 DNA prime followed by a multiclade rAd5 HIV-1 vaccine boost in healthy adults (HVTN204) PLoS ONE. 2011;6:e21225. doi: 10.1371/journal.pone.0021225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaggar A, Shayakhmetov DM, Lieber A. CD46 is a cellular receptor for group B adenoviruses. Nat Med. 2003;9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Bergelson JM. Adenovirus receptors. J Virol. 2005;79:12125–12131. doi: 10.1128/JVI.79.19.12125-12131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson MJ, Petrovas C, Yamamoto T, Lindsay RWB, Loré K, Gall JGD, Gostick E, Lefebvre F, Cameron MJ, Price DA, et al. Type I IFN Induced by Adenovirus Serotypes 28 and 35 Has Multiple Effects on T Cell Immunogenicity. J Immunol. 2012;188:6109–6118. doi: 10.4049/jimmunol.1103717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt NW, Butler NS, Badovinac VP, Harty JT. Extreme CD8 T cell requirements for anti-malarial liver-stage immunity following immunization with radiation attenuated sporozoites. PLoS Pathog. 2010;6:e1000998. doi: 10.1371/journal.ppat.1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt NW, Podyminogin RL, Butler NS, Badovinac VP, Tucker BJ, Bahjat KS, Lauer P, Reyes-Sandoval A, Hutchings CL, Moore AC, et al. Memory CD8 T cell responses exceeding a large but definable threshold provide long-term immunity to malaria. PNAS. 2008;105:14017–14022. doi: 10.1073/pnas.0805452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang TC, Millar J, Groves T, Grinshtein N, Parsons R, Takenaka S, Wan Y, Bramson JL. The CD8+ T cell population elicited by recombinant adenovirus displays a novel partially exhausted phenotype associated with prolonged antigen presentation that nonetheless provides long-term immunity. J Immunol. 2006;176:200–210. doi: 10.4049/jimmunol.176.1.200. [DOI] [PubMed] [Google Scholar]
- 65.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 68.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DAA, Wherry EJ. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bengsch B, Seigel B, Ruhl M, Timm J, Kuntz M, Blum HE, Pircher H, Thimme R. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog. 2010;6:e1000947. doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tatsis N, Fitzgerald JC, Reyes-Sandoval A, Harris-McCoy KC, Hensley SE, Zhou D, Lin SW, Bian A, Xiang ZQ, Iparraguirre A, et al. Adenoviral vectors persist in vivo and maintain activated CD8+ T cells: implications for their use as vaccines. Blood. 2007;110:1916–1923. doi: 10.1182/blood-2007-02-062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finn JD, Bassett J, Millar JB, Grinshtein N, Yang TC, Parsons R, Evelegh C, Wan Y, Parks RJ, Bramson JL. Persistence of transgene expression influences CD8+ T-cell expansion and maintenance following immunization with recombinant adenovirus. J Virol. 2009;83:12027–12036. doi: 10.1128/JVI.00593-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sullivan NJ, Hensley L, Asiedu C, Geisbert TW, Stanley D, Johnson J, Honko A, Olinger G, Bailey M, Geisbert JB, et al. CD8+ cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat Med. 2011;17:1128–1131. doi: 10.1038/nm.2447. [DOI] [PubMed] [Google Scholar]
- 73.Geisbert TW, Bailey M, Hensley L, Asiedu C, Geisbert J, Stanley D, Honko A, Johnson J, Mulangu S, Pau MG, et al. Recombinant adenovirus serotype 26 (Ad26) and Ad35 vaccine vectors bypass immunity to Ad5 and protect nonhuman primates against ebolavirus challenge. J Virol. 2011;85:4222–4233. doi: 10.1128/JVI.02407-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prlic M, Sacks JA, Bevan MJ. Dissociating markers of senescence and protective ability in memory T cells. PLoS ONE. 2012;7:e32576. doi: 10.1371/journal.pone.0032576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiesel M, Crouse J, Bedenikovic G, Sutherland A, Joller N, Oxenius A. Type-I IFN drives the differentiation of short-lived effector CD8+ T cells in vivo. Eur J Immunol. 2012;42:320–329. doi: 10.1002/eji.201142091. [DOI] [PubMed] [Google Scholar]
- 76.Obar JJ, Jellison ER, Sheridan BS, Blair DA, Pham QM, Zickovich JM, Lefrançois L. Pathogen-induced inflammatory environment controls effector and memory CD8+ T cell differentiation. J Immunol. 2011;187:4967–4978. doi: 10.4049/jimmunol.1102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thimme R, Appay V, Koschella M, Panther E, Roth E, Hislop AD, Rickinson AB, Rowland-Jones SL, Blum HE, Pircher H. Increased expression of the NK cell receptor KLRG1 by virus-specific CD8 T cells during persistent antigen stimulation. J Virol. 2005;79:12112–12116. doi: 10.1128/JVI.79.18.12112-12116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wirth TC, Martin MD, Starbeck-Miller G, Harty JT, Badovinac VP. Secondary CD8+ T-cell responses are controlled by systemic inflammation. Eur J Immunol. 2011;41:1321–1333. doi: 10.1002/eji.201040730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 81.Jaoko W, Karita E, Kayitenkore K, Omosa-Manyonyi G, Allen S, Than S, Adams EM, Graham BS, Koup RA, Bailer RT, et al. Safety and immunogenicity study of Multiclade HIV-1 adenoviral vector vaccine alone or as boost following a multiclade HIV-1 DNA vaccine in Africa. PLoS ONE. 2010;5:e12873. doi: 10.1371/journal.pone.0012873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barratt-Boyes SM, Soloff AC, Gao W, Nwanegbo E, Liu X, Rajakumar PA, Brown KN, Robbins PD, Murphey-Corb M, Day RD, et al. Broad cellular immunity with robust memory responses to simian immunodeficiency virus following serial vaccination with adenovirus 5- and 35-based vectors. J Gen Virol. 2006;87:139–149. doi: 10.1099/vir.0.81445-0. [DOI] [PubMed] [Google Scholar]
- 83.Loré K, Adams WC, Havenga MJE, Precopio ML, Holterman L, Goudsmit J, Koup RA. Myeloid and plasmacytoid dendritic cells are susceptible to recombinant adenovirus vectors and stimulate polyfunctional memory T cell responses. J Immunol. 2007;179:1721–1729. doi: 10.4049/jimmunol.179.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.