Abstract
Regulation of gene expression in cells is mediated by protein-protein, DNA-protein and receptor-ligand interactions. PDZ (PSD-95/Discs-large/ZO-1) domains are protein–protein interaction modules. PDZ-containing proteins function in the organization of multi-protein complexes controlling spatial and temporal fidelity of intracellular signaling pathways. In general, PDZ proteins possess multiple domains facilitating distinct interactions. The human Glutaminase Interacting Protein (hGIP) is an unusual PDZ protein comprising entirely of a single PDZ domain and plays pivotal roles in many cellular processes through its interaction with the C-terminus of partner proteins. Here, we report the identification by yeast two-hybrid screening of two new hGIP-interacting partners, DTX1 and STAU1. Both proteins lack the typical C-terminal PDZ recognition motif but contain a novel internal hGIP recognition motif recently identified in a phage display library screen. Fluorescence resonance energy transfer and confocal microscopy analysis confirmed the in vivo association of hGIP with DTX1 and STAU1 in mammalian cells validating the previous discovery of S/T-X-V/L-D as a consensus internal motif for hGIP recognition. Similar to hGIP, DTX1 and STAU1 have been implicated in neuronal function. Identification of these new interacting partners furthers our understanding of GIP-regulated signaling cascades and these interactions may represent potential new drug targets in humans.
Keywords: Glutaminase Interacting Protein, PDZ domains, Yeast two-hybrid, Protein-protein interactions, Fluorescence resonance energy transfer, Confocal microscopy
1. Introduction
Protein-protein interactions are important in almost all cellular processes including cell growth, death, and intra- and intercellular communication. These interactions are mediated by distinct protein modules, of which the PDZ (PSD-95, DlgA, ZO-1) domains are a signficant class [1]. PDZ domain interactions are involved in numerous essential functions such as protein targeting, clustering of ion channels, membrane expression of receptors, cell polarity, and cell-cell communications [2]. PDZ domains typically recognize the C-terminus of their target proteins [3,4]; however, in a few cases recognition of an internal motif has also been reported [5– 7]. Because PDZ domains mediate protein-protein interactions involved in various signal transduction pathways, unravelling the complete network of interacting partners of a PDZ domain-containing protein is crucial for successful design of drugs specifically targeting its PDZ domain [8].
Human glutaminase interacting protein (hGIP) [9], also known as tax interacting protein 1 (TIP-1), is an important multifunctional protein containing a single PDZ domain. Besides the unstructured N and C-termini, the hGIP protein consists solely of the PDZ domain [10]. hGIP contains a canonical Class-I PDZ domain binding site that interacts with the C-terminus of different proteins, typical of the promiscuous target protein recognition of PDZ domains [11]. Human GIP plays an important role in cell signaling, cancer, ion transport, cell polarity and transcription through its interaction with diverse target proteins. Some of the GIP target proteins include glutaminase L [9–10], β-catenin [12], FAS [13], brain-specific angiogenesis inhibitor 2 [14], rho activator rhotekin [15], ARHGEF16 [16], potassium channel Kir2.3 [17], HPV16 E6 [18], and HTLV-1 Tax [19]. Recent findings show that GIP is involved in human invasive breast cancer [20], accelerates tumor-driven angiogenesis, and stimulates tumor formation of human glioblastoma cell lines in nude mice [21]. Since GIP is involved in various cancerous pathways and consists solely of a single PDZ domain, it has received considerable attention as a possible target for anticancer therapeutics [20–21]. We have been using various techniques including phage display library screening [6] and yeast two-hybrid (Y2H) assays [14] to find novel GIP interactors. Utilizing these techniques, we recently discovered that hGIP not only interacted via recognition of a C-terminal motif, but also recognized an internal motif with the consensus S/T-x-V/L-D sequence [6].
Here, we report the discovery through a Y2H screen of two new hGIP partner proteins containing the novel internal GIP recognition motif. Using fluorescence resonance energy transfer (FRET) and confocal microscopy, we were able to show co-localization of hGIP and these novel proteins in human cells. Similar to hGIP, the newly identified interacting proteins, DTX1 and STAU1, have been implicated in normal neuronal function. These findings further advance our understanding of GIP-regulated signaling cascades and these interactions may represent potential new targets for drugs modulating these pathways in humans.
2. Materials and Methods
2.1. Yeast strains and media
Two-hybrid reporter yeast strains used in this study were AH109 (MATα, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1) and Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, met, gal80Δ, MEL1, URA3::GAL1UAS -GAL1TATA-lacZ) (Clontech Mountain View, CA). Yeast cells were cultured at 30°C in YEPD + adenine (1% yeast extract, 2% bacto peptone, 2% dextrose, 1% adenine) or SD medium (0.67% yeast nitrogen base without amino acids, 2% dextrose, supplemented with appropriate amino acids and bases) [22]. Media were solidified with 2% agar and 40 μg/mL X-alpha-gal was added for detection of alpha-galactosidase reporter gene expression. All media reagents were obtained from DIFCO Laboratories or Sigma.
2.2. Yeast two-hybrid Screen
A yeast expression plasmid (pGBKT7-GIP) encoding hGIP fused to the carboxy terminus of the yeast Gal4 transcription factor DNA-binding domain (Gal4BD-GIP), was used in a Y2H screen as previously described [14]. Briefly, yeast strain Y187, pre-transformed with a human fetal brain cDNA library in the Gal4 activation domain (AD) vector pGADT7-Rec (Clontech, Mountain View, CA) was mated with yeast strain AH109 transformed with pGBKT7-GIP and the diploids were screened for hGIP-interacting clones [14]. A total of 1.1×107 cells were screened with mating efficiency of 10.0% in the library having 2.3×108 cells.
Clones capable of activating integrated reporter genes, namely HIS3, ADE2, MEL1 and LacZ under the control of the Gal4-responsive Upstream Activating Sequence (UAS) were identified by plating transformants on SD solid medium lacking leucine and tryptophan to select for the presence of pGBKT7-GIP and the LEU2 gene-containing library plasmid and lacking adenine and histidine and containing X-α-gal (Clontech, Mountain View, CA) (SD/-Ade/-His/-Leu/-Trp/X-α-gal). Transformants capable of growth and giving blue colonies due to conversion of X-α-gal to a blue precipitate by the alpha-galactosidase expressed by the MEL1 reporter gene, were subsequently checked for beta-galactosidase expression from the lacZ reporter gene. Clones failing to grow under more stringent conditions in the presence of 25 mM 3-aminotriazole (3-AT), a competitive inhibitor of the HIS3 gene product (SD/-Ade/-His/-Trp/-Leu/X-α-Gal/3-AT) were discarded. Plasmids were isolated from positive yeast clones and were transformed into E. coli. Plasmids from ampicillin-resistant bacterial transformants were re-tested in yeast in order to confirm their ability to activate the reporter genes when co-transformed with pGBKT7-GIP. GIP bait-dependent reporter gene expression was confirmed by transforming two-hybrid reporter yeast with plasmids expressing the putative GIP-interactors in the absence of other fusion proteins and with other unrelated fusion proteins (data not shown). For positive clones, cDNA inserts were characterized by restriction mapping, polymerase chain reaction (PCR) using MatchMaker Insert Check PCR Mix (Clontech, Mountain View, CA) and sequencing. (J. P. Robarts Research Institute, London, Ont.). A second independent cDNA library was used for confirmation experiments (Clontech, Mountain View, CA).
2.3. Plasmid construction and co-localization analyses
For confocal microscopy analyses, fluorescent protein fusion constructs of hGIP and its candidate interacting partners were created. A plasmid expressing GIP as a yellow fluorescent protein (YFP) fusion protein was created by transferring the GIP-encoding region from pGBKT7-GIP (an EcoRI-SalI fragment) into the vector pEYFP-C3 [23] that had been digested with the BglII and SalI (EcoRI and BglII ends, respectively, on each DNA were filled in by incubation with Klenow before digestion with the second restriction enzyme) to result in the final construct (YFP-hGIP). Plasmids expressing the candidate GIP-interacting partner proteins fused to cyan fluorescent protein (CFP) were similarly constructed by transferring fragments (XhoI filled-in and BglII) from the respective pGADT7-Rec AD library plasmid encoding the partner protein and ligating with the vector pECFP-C2 that had been digested with SmaI and BglII restriction enzymes to obtain the final constructs (CFP-partners). The plasmids were verified by restriction endonuclease digestions and sequencing. Plasmids expressing human Alteration/Deficiency in Activation 3 (hADA3) protein as a Gal4DB fusion protein in yeast (pGBTK7-ADA3) or as a CFP fusion protein in human cells (CFP-ADA3) were used as negative controls and have been described previously [23].
Human HeLa cells were transfected with CFP and YFP fusion protein-expression plasmids using TurboFect transfection reagent (Fermentas, Lithuania) according to the manufacturer’s instructions. Cells were incubated in 8-well coverslip-bottom chambers (Lab-Tek, Campbell, CA) in a 5% CO2 incubator at 37 °C for 24 h before being analyzed using an Olympus Fluoview FV1000 confocal laser scanning microscope (Olympus Life Science Europa GmbH, Hamburg, Germany). The microscope was configured as follows: objective lens UPLFLN 40x (water, NA 0.8) and 60x (oil, NA 1.3); sampling speed: 8μs/pixel; scanning mode: sequential unidirectional; excitation: 458 nm (CFP) and 515 nm (YFP); laser transmissivity: 30% and 10% were used for CFP and YFP, respectively; main dichroic beamsplitter: DM458/515, intermediate dichroic beamsplitter: SDM 510; CFP was detected between 470–520 nm and YFP was detected between 520–570 nm. Differential interference contrast (DIC) or standard transmission images were captured with a 515 nm laser line.
FRET analyses were performed by an acceptor photobleaching method [24] Confocal images of cotransfected cells were acquired with UPLFLN 40x Oil immersion objective. CFP and YFP images were taken before and after photobleaching of the YFP image using 514nm laser at 100% transmissivity focused onto a rectangle covering the whole cell. Gray level intensities of CFP images were pseudocolored using spec3 (heat map) lookup table of Olympus Fluoview software: Blue, green, red and white colors represent lowest, intermediate, high and highest level of gray level intensities. Using images pre- and post-bleach, the increase in average CFP fluorescence was calculated after manual tracing of each cell using the Olympus Fluoview software measurement and analysis tools. More than 10 cells were scored from each sample. Percentage increase and standard deviations of CFP fluorescence intensities were plotted using Microsoft Excel software (Microsoft Corporation, Redmond, WA).
3. Results and Discussion
3.1. Human fetal brain cDNA library screening resulted in the identification of novel GIP interacting partners
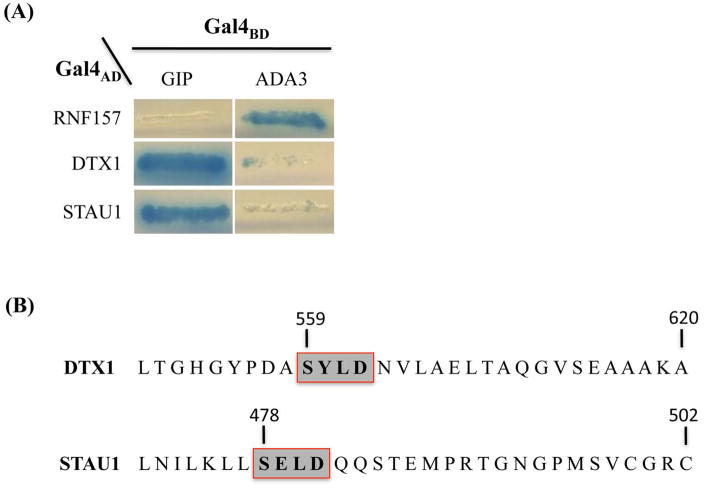
Haploid yeast transformed with a plasmid expressing human GIP fused to the yeast Gal4 DNA-binding domain (Gal4BD-GIP) were mated to 1.1×107 yeast of the opposite mating type pre-transformed with plasmids expressing human fetal brain cDNA-Gal4AD-fusion proteins [14]. Screening of the mated cells gave rise to several diploid co-transformants that were able to form colonies under conditions that required Gal4-responsive reporter gene activation. The ability of these colonies to grow in the absence of adenine and histidine suggested that they contained a library plasmid expressing a GIP-interacting protein. The specificity of GIP interactions with prospective partners was tested by a second line of Y2H experiments. For this pGBKT7-GIP and another plasmid, pGBKT7-ADA3, encoding Gal4BD fused to the hADA3 protein were introduced independently into host cells in order to compare their interaction with the newly identified putative GIP partners (Fig. 1A, left and right panels, respectively). The results of these tests indicated that the two GAL4 AD-human fetal brain cDNA plasmids were capable of activating the reporter genes when co-transformed with pGBTK7-GIP expressing Gal4BD-GIP, but not when co-transformed with pGBKT7-ADA3. The library plasmids from these yeast transformants were isolated. Sequence analyses of the cDNAs and comparisons with database sequences using BLAST searches [25] revealed that the isolated cDNAs encoded different proteins, neither of which had previously been identified as interacting with GIP.
Fig. 1.
Potential partner proteins interact specifically with GIP in a yeast two-hybrid assay and internal motifs in newly-identified hGIP partners. (A) The two-hybrid reporter yeast strain AH109 was co-transformed with plasmids expressing the Gal4AD-fusion protein indicated at left and either human GIP or ADA3 fused to Gal4BD. Representative co-transformants were streaked on selective medium (SD/-Ade/-His/-Trp/-Leu/X-α-Gal) and incubated at 30°C. Growth and expression of the alpha-galactosidase reporter gene (blue color) indicates interaction of the co-expressed fusion proteins. (B) The carboxy-terminal sequence of the two new protein partners with the potential internal GIP recognition motifs indicated by grey boxes.
The encoded proteins, Deltex homolog 1 (DTX1) and double-stranded RNA-binding protein Staufen homolog 1 (STAU1), are multi-functional proteins. DTX1 was first identified in Drosophila as a positive regulator of the Notch-signaling pathway and functions as an E3 ubiquitin ligase [26] with a possible role in negatively regulating neural-specific transcription factor activity [27]. DTX1 has also been implicated in neurogenesis, lymphogenesis and myogenesis, and may be involved in MZB (Marginal zone B) cell differentiation [28]. STAU1 is known to be expressed in brain and in a number of other tissues including pancreas, heart, skeletal muscles, liver, lung, kidney and placenta [29]. STAU1 association with double-stranded regions of RNA regulates mRNA stability [30], transport [31], and translation [32] and has been shown to be critical in maintaining neuronal function [33]. Our subsequent analyses focused on characterizing the interaction of these two novel partners with GIP and determining whether the association occurs in mammalian cells.
3.2. hGIP novel partner proteins lack a C-terminal PDZ domain-recognition motif but contain a novel internal GIP-interaction motif
The single PDZ domain in hGIP is typical of other PDZ domains, small protein-protein recognition modules that bind to well-defined C-terminal residues of a partner protein in a sequence-specific manner [7]. To determine whether the potential GIP-interacting partner proteins identified in the Y2H screen are recognized through classical C-terminal motif or via the recently identified novel internal GIP-recognition motif [6], sequence analyses were carried out and confirmed that the cDNAs for DTX1 and STAU1 inserted in the respective pGADT7-Rec AD library plasmids were full-length. Western blotting analysis of protein extracted from yeast transformed with the pGADT7-Rec AD library plasmids showed Gal4AD-fusion proteins with mobilities consistent with those expected for expression of the full-length interactor proteins indicating that the authentic C-terminus of each was likely to be present on the respective fusion proteins (data not shown).
Analysis of the predicted amino acid sequence revealed that neither of the two new partner proteins contained known PDZ domain-binding C-terminal recognition motifs [10–11]. However, the new partner proteins did contain an internal sequence (Fig. 1B) that matched the S/T-X-V/L-D consensus of the internal motif we had previously identified through phage display library screening [6].
3.3. Verification of hGIP interaction with newly identified partner proteins through co-localization studies in mammalian cells
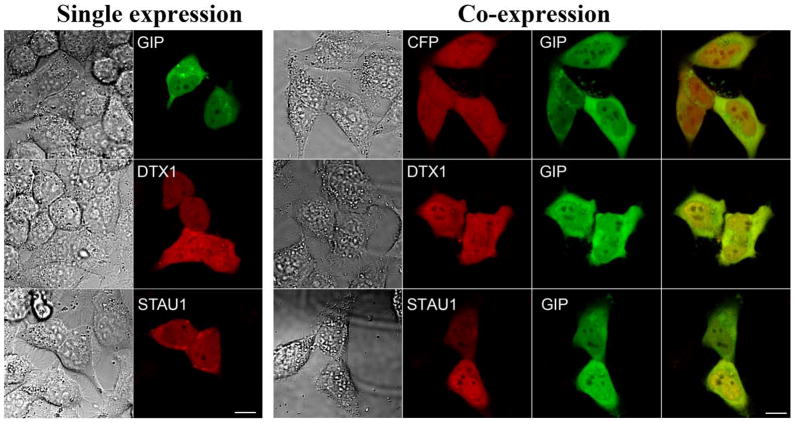
To determine whether the association of GIP with the newly identified potential partner proteins could be detected in human cells, we constructed plasmids that would express hGIP as a YFP fusion protein (YFP-hGIP) and interacting partner proteins as CFP-fusion proteins (CFP-DTX1 and CFP-STAU1). Plasmids expressing the YFP and CFP-tagged fusion proteins were first individually used to transfect HeLa cells to determine the localization of each protein (Fig. 2, left panel). Plasmids expressing only CFP or YFP without fusion to any protein were used as negative controls. HeLa cells transfected with individual plasmids expressing YFP-hGIP, CFP-DTX1 and CFP-STAU1 each displayed fluorescent signal both in the nucleus and in the cytoplasm.
Fig. 2.
Cellular localization and co-localization of hGIP and its potential partner proteins in human cells. For single expression (left panel), HeLa cells were transfected with a plasmid expressing YFP-hGIP or a potential hGIP-interacting partner, DTX1 or STAU1, as a CFP fusion protein. Images were captured by confocal microscopy. DIC images of cells are shown at left and the signal from the same cells for YFP-hGIP (GIP) (pseudo-colored green) or for the CFP fusion proteins (in red) are to the right for each. Scale bar is 10μm. For co-expression (right panel), HeLa cells were co-transfected with a plasmid expressing hGIP fused to YFP (GIP) and a second plasmid expressing CFP or CFP fused to DTX1 or to STAU1. Co-localization was analyzed by confocal microscopy. DIC images (left), CFP signals (red, middle left) and YFP-hGIP signals (green, middle right), are shown for the same cells. Yellow in the merged image at far right indicates co-localization. Scale bar is 10μm
Next, to determine whether hGIP co-localized with the potential interacting partner proteins, HeLa cells were co-transfected with two plasmids, one expressing YFP-hGIP and the other CFP or a CFP-tagged hGIP partner protein (Fig. 2, right panel). The distribution of STAU1 and DTX1 proteins between the cytoplasm and the nucleus was not significantly altered by co-expression with hGIP. The merged images of YFP-tagged hGIP with each of the CFP-tagged fusion proteins showed substantial co-localization, supporting potential interaction between hGIP and the newly identified proteins.
Taken together, the microscopy analysis demonstrates that hGIP co-localizes with both newly identified partner proteins in human cells. The co-localization along with the interactions detected in the Y2H assay suggested that these proteins might physically interact with hGIP in vivo making these interactions biologically relevant.
3.4. FRET Analysis of GIP interactions
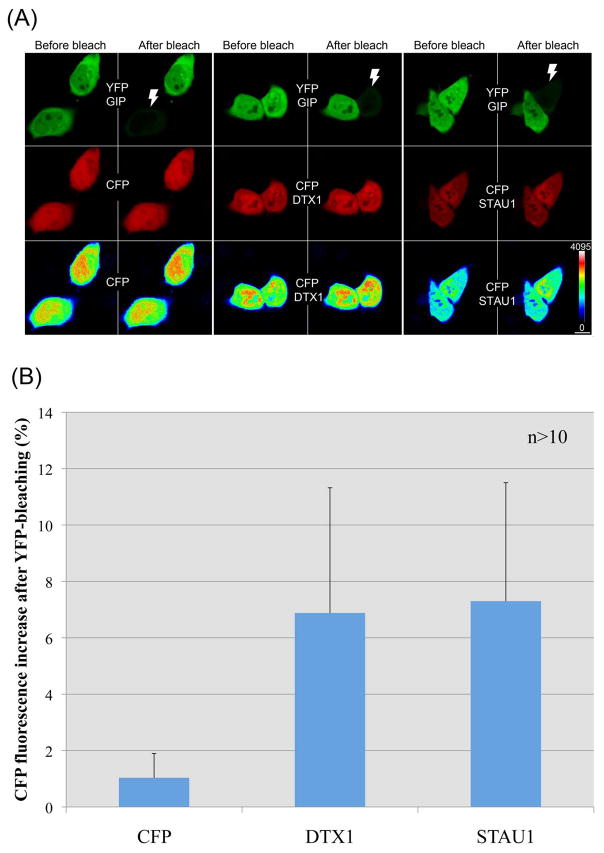
We used FRET and confocal laser scanning microscopy analysis of cells co-expressing YFP-GIP (acceptor) and CFP-conjugated proteins (donor) to examine in vivo interactions between GIP and its presumed partners at the subcellular level. Interaction between the proteins was monitored by using an acceptor photobleaching FRET approach. In this method the acceptor protein (YFP) is bleached with high laser intensity and the change in intensity of the donor (CFP) fluorescence emission is recorded. Following bleaching of the acceptor, an increase in donor fluorescence indicates that the two fluorescently-tagged fusion proteins were close enough (within 1–10 nm of each other) for direct transfer of energy from the donor to the acceptor, providing strong evidence for direct in vivo interaction of the proposed protein-binding partners. During acceptor photobleaching experiments, however, photoconversion of YFP into a CFP-like species can occur [34]. This potential non-specific increase in CFP emission was monitored by co-transfecting cells with two plasmids, one expressing CFP and the other YFP-GIP. CFP intensity images were color coded, which showed that CFP emission in the cells was not significantly affected when the acceptor protein (YFP-GIP) was bleached (Fig. 3A, CFP in the last row). To quantify the increase, average CFP intensities were scored before and after bleaching of the acceptor using cell tracing and intensity quantification tools of the Olympus Fluoview software. A mean increase in CFP fluorescence of 1.035% (±0.86) was measured in control cells (Fig. 3). Using identical detection and bleaching parameters, YFP-GIP photobleaching experiments were performed on cells cotransfected with a plasmid expressing YFP-GIP and a second plasmid expressing CFP-tagged with DTX1 or STAU1, respectively. The cells expressing CFP-DTX1 (6.88 ± 4.44) and CFP-STAU1 (7.3 ± 4.2) showed nearly a seven-fold increase in mean CFP fluorescence compared to control cells (Fig. 3B). This increase in intensity was also clearly visible on intensity color-coded images of CFP-DTX1 and CFP-STAU1 (Fig. 3A). The increase in intensity was observed both in nuclear and cytoplasmic regions suggesting that the interaction of GIP with DTX1 and with STAU1 occurs in both compartments.
Fig. 3.
FRET between YFP-GIP and DTX1 or STAU1 CFP fusion proteins in human cells. HeLa cells were co-transfected with a plasmid expressing hGIP fused to YFP (GIP) and a second plasmid expressing CFP, or CFP fused to DTX1 or STAU1. (A) Pre-bleach and post-bleach images of YFP-GIP and CFP chimeras (or CFP only) are shown in the first and second row, respectively. YFP and CFP images are pseudo-colored green and red, respectively. Last row indicates intensity color coding of the CFP images. Note the post-bleach increase in the CFP intensity of CFP-DTX1 and CFP-STAU1. The intensity change in control sample (leftmost panels for each) was almost undetectable. White thunder icons in YFP images mark the bleached cells. Color coding scale and the scale bar (10μm) are shown at the lower right corner. (B) CFP intensity from cells was measured before and after bleaching and the increase in fluorescence in the bleached cells was plotted. More than 10 cells were scored from each line and results represent the mean values with standard deviations.
The FRET analysis confirmed in vivo association of hGIP with DTX1 and STAU1. Furthermore, both DTX1 and STAU1 contain an internal motif that was previously identified as an hGIP recognition motif in a phage display library screen [6]. The demonstration of in vivo association of hGIP with partner proteins containing the same motif validates the previous discovery of S/T-X-V/L-D as a consensus internal motif for GIP recognition.
Highlights.
We identified two new human GIP partner proteins: DTX1 and STAU1.
DTX1 and STAU1 contain novel hGIP recognition motif.
DTX1 and STAU1 interact with hGIP in human mammalian cells
All the 3 proteins are involved in human neuronal function.
These interactions may represent potential new drug targets in humans.
Acknowledgments
This research was financially supported by United States Department of Agriculture PECASE award 2003-35302-12930, and AFRI award 2011-65503-20030, National Science Foundation grant IOS-0628064, and National Institutes of Health grant DK082397 to Smita Mohanty, NSERC grant 155268 to Melanie Dobson, and the grant TUBITAK 108T945 to Zeki Topcu.
Footnotes
Author contributions
S.Z. carried out cloning, yeast hybrid screening, BLAST search, verifying interactions and analyzing data, M.B. BLAST search and analysis to find the consensus peptide sequence, M.D. interpretation of data and editing the manuscript, F.A. and E.A.F. confocal analyses and mammalian cell culture, Z.T. and S.M. designed the project, analysed data and prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 2.Subbaiah VK, Kranjec C, Thomas M, Banks L. PDZ domains: the building blocks regulating tumorigenesis. Biochem J. 2011;439:195–205. doi: 10.1042/BJ20110903. [DOI] [PubMed] [Google Scholar]
- 3.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 4.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 5.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 6.Banerjee M, Zoetewey DL, Ovee M, Mazumder S, Petrenko VA, Samoylova TI, Mohanty S. Specificity and promiscuity in human glutaminase interacting protein recognition: insight from the binding of the internal and C-terminal motif. Biochemistry. 2012;51:6950–6960. doi: 10.1021/bi3008033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dev KK. Making protein interactions druggable: targeting PDZ domains. Nat Rev Drug Discov. 2004;3:1047–1056. doi: 10.1038/nrd1578. [DOI] [PubMed] [Google Scholar]
- 9.Olalla L, Aledo JC, Bannenberg G, Marquez J. The C-terminus of human glutaminase L mediates association with PDZ domain-containing proteins. FEBS Lett. 2001;488:116–122. doi: 10.1016/s0014-5793(00)02373-5. [DOI] [PubMed] [Google Scholar]
- 10.Zoetewey D, Ovee M, Banerjee M, Bhaskaran R, Mohanty S. Promiscuous binding at the crossroads of numerous cancer pathways: Insight from the binding of GIP with glutaminase L. Biochemistry. 2011;50:3528–3539. doi: 10.1021/bi102055y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 12.Kanamori M, Sandy P, Marzinotto S, Benetti R, Kai C, Hayashizaki Y, Schneider C, Suzuki H. The PDZ protein tax-interacting protein-1 inhibits beta-catenin transcriptional activity and growth of colorectal cancer cells. J Biol Chem. 2003;278:38758–38764. doi: 10.1074/jbc.M306324200. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee M, Huang C, Marquez J, Mohanty S. Probing the structure and function of human glutaminase-interacting protein: a possible target for drug design. Biochemistry. 2008;47:9208–9219. doi: 10.1021/bi800287v. [DOI] [PubMed] [Google Scholar]
- 14.Zencir S, Ovee M, Dobson MJ, Banerjee M, Topcu Z, Mohanty S. Identification of brain-specific angiogenesis inhibitor 2 as an interaction partner of glutaminase interacting protein. Biochem Biophys Res Commun. 2011;411:792–797. doi: 10.1016/j.bbrc.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynaud C, Fabre S, Jalinot P. The PDZ protein TIP-1 interacts with the Rho effector rhotekin and is involved in Rho signaling to the serum response element. J Biol Chem. 2000;275:33962–33968. doi: 10.1074/jbc.M000465200. [DOI] [PubMed] [Google Scholar]
- 16.Oliver AW, He X, Borthwick K, Donne AJ, Hampson L, Hampson IN. The HPV16 E6 binding protein Tip-1 interacts with ARHGEF16, which activates Cdc42. Br J Cancer. 2011;104:324–331. doi: 10.1038/sj.bjc.6606026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alewine C, Olsen O, Wade JB, Welling PA. TIP-1 has PDZ scaffold antagonist activity. Mol Biol Cell. 2006;17:4200–4211. doi: 10.1091/mbc.E06-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampson L, Li C, Oliver AW, Kitchener HC, Hampson IN. The PDZ protein Tip-1 is a gain of function target of the HPV16 E6 oncoprotein. Int J Oncol. 2004;25:1249–1256. [PubMed] [Google Scholar]
- 19.Rousset R, Fabre S, Desbois C, Bantignies F, Jalinot P. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene. 1998;16:643–654. doi: 10.1038/sj.onc.1201567. [DOI] [PubMed] [Google Scholar]
- 20.Han M, Wang H, Zhang HT, Han Z. The PDZ protein TIP-1 facilitates cell migration and pulmonary metastasis of human invasive breast cancer cells in athymic mice. Biochem Biophys Res Commun. 2012;422:139–145. doi: 10.1016/j.bbrc.2012.04.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han M, Wang H, Zhang HT, Han Z. Expression of Tax-interacting protein 1 (TIP-1) facilitates angiogenesis and tumor formation of human glioblastoma cells in nude mice. Cancer Lett. 2012;328:55–64. doi: 10.1016/j.canlet.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose MD, Winston FM, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press; New York: 1990. [Google Scholar]
- 23.Zencir S, Sike A, Dobson MJ, Ayaydin F, Boros I, Topcu Z. Identification of transcriptional and phosphatase regulators as interaction partners of human ADA3, a component of histone acetyltransferase complexes. Biochem J. 2012 Nov 20; doi: 10.1042/BJ20120452. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa-Ankerhold HC, Ankerhold R, Drummen GP. Advanced fluorescence microscopy techniques--FRAP, FLIP, FLAP, FRET and FLIM. Molecules. 2012;17:4047–4132. doi: 10.3390/molecules17044047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 26.Takeyama K, Aguiar RC, Gu L, He C, Freeman GJ, Kutok JL, Aster JC, Shipp MA. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J Biol Chem. 2003;278:21930–21937. doi: 10.1074/jbc.M301157200. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, Matsuno K, Nakamura K, Weinmaster G, Okano H, Nakafuku M. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J Biol Chem. 2001;276:45031–45040. doi: 10.1074/jbc.M105245200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang P, Yang Y, Nolo R, Zweidler-McKay PA, Hughes DP. Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex 1 and its role in osteosarcoma invasiveness. Oncogene. 2010;29:2916–2926. doi: 10.1038/onc.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 31.Kiebler MA, Hemraj I, Verkade P, Kohrmann M, Fortes P, Marion RM, Ortin J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebeau G, Maher-Laporte M, Topolnik L, Laurent CE, Sossin W, Desgroseillers L, Lacaille JC. Staufen1 regulation of protein synthesis-dependent long-term potentiation and synaptic function in hippocampal pyramidal cells. Mol Cell Biol. 2008;28:2896–2907. doi: 10.1128/MCB.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valentin G, Verheggen C, Piolot T, Neel H, Coppey-Moisan M, Bertrand E. Photoconversion of YFP into a CFP-like species during acceptor photobleaching FRET experiments. Nat Methods. 2005;2:801. doi: 10.1038/nmeth1105-801. [DOI] [PubMed] [Google Scholar]