Abstract
The pathogenic sequelae of BRCA1 mutation in human and mouse cells are mitigated by concomitant deletion of 53BP1, which binds histone H4 dimethylated at Lys20 (H4K20me2) to promote nonhomologous end-joining, suggesting a balance between BRCA1 and 53BP1 regulates DNA double-strand break (DSB) repair mechanism choice. Here, we document that acetylation is a key determinant of this balance. TIP60 acetyltransferase deficiency reduced BRCA1 at DSB chromatin with commensurate increases in 53BP1, while HDAC inhibition yielded the opposite effect. TIP60 -dependent H4 acetylation diminished 53BP1 binding to H4K20me2 in part through disruption of a salt bridge between H4K16 and Glu1551 in the 53BP1 Tudor domain. Moreover, TIP60 deficiency impaired HR and conferred sensitivity to PARP inhibition in a 53BP1-dependent manner. These findings demonstrate that acetylation in cis to H4K20me2 regulates relative BRCA1 and 53BP1 DSB chromatin occupancy to direct DNA repair mechanism.
Keywords: TIP60, Acetylation, BRCA1, 53BP1, Homologous recombination, PARP inhibitors
Dynamic modifications of large stretches of chromatin in cis to DNA double-strand breaks (DSBs) regulate the modular and ordered assembly of repair proteins at sites of damaged DNA. Genetic evidence suggests the balance between two such proteins, BRCA1 (Breast cancer early onset 1) and 53BP1 (p53 Binding Protein 1), influences DNA end resection and hence is a determinant of whether DSB repair ensues predominantly by either homologous recombination or nonhomologous end-joining (NHEJ)1–3. An imbalance in this competition manifests in multiple pathophysiologic processes. BRCA1 is required for embryogenesis, with complete nullizygosity in mice resulting in lethality between embryonic days E5.5-8.5. Hypomorphic BRCA1 exon 11 deleted mice survive to adulthood in the context of p53 heterozygosity, but display premature ageing, cancer susceptibility, and hypersensitivity to DNA damaging agents4. Each of these phenotypes closely correlates with homologous recombination impairment and resultant genomic instability from BRCA1 deficiency.
Conversely, mice harboring mutation in both BRCA1 and 53BP1 are viable through adulthood without displaying increased cancer susceptibility. Doubly deficient BRCA1−/−, 53BP1−/− cells have restored homologous recombination and genomic stability, and are resistant to poly(ADP)ribose polymerase inhibitors (PARPi). 53BP1 DSB localization and function in NHEJ requires its tandem Tudor repeats, which recognize mono- and di-methylated histone H4 at lysine 20 (H4K20me1 and H4K20me2) at DNA damage sites5–7. Mice expressing a 53BP1 Tudor domain mutant protein that does not bind H4K20me2 still rescued BRCA1 null phenotypes8, indicating that 53BP1 DSB chromatin association blocks an essential aspect of homologous recombination in the absence of functional BRCA1 protein. This appears to be DSB end resection, which occurs at elevated levels in 53BP1 null cells9, enabling homologous recombination to occur at sufficient levels in the context of BRCA1 mutation to maintain genomic stability1,2. These findings implicate control of end resection as the critical step that is regulated by the competition between BRCA1 and 53BP1 at DSB chromatin.
Given these observations, the identification of molecular determinants uniquely affecting BRCA1 and 53BP1 occupancy at DSBs would be a fundamentally important finding relevant to genome integrity, carcinogenesis, and potentially cancer therapy. Paradoxically, 53BP1 and BRCA1 DSB chromatin association are both dependent on a phosphorylation-ubiquitylation cascade involving γH2AX, MDC1, RNF8, and RNF16810–14. RNF8 recognizes phosphorylated MDC1 to initiate DSB ubiquitylation, which is continued by RNF168 to produce both degradative and nondegradative ubiquitin chains. BRCA1 is targeted to DSBs in part by an interaction with the ubiquitin binding protein RAP80, which recognizes RNF8- and RNF168-dependent lysine 63-linked ubiquitin (K63-Ub)15–17. BRCA1 also has additional modes of DSB recognition as it is a component of multiple protein complexes that are distinct from the RAP80 complex, most of which do not directly rely on ubiquitin recognition for DSB localization18–20.
Concurrently, 53BP1 DSB localization requires RNF8 and RNF168 E3 ubiquitin ligase activities despite the lack of an obvious ubiquitin binding domain in 53BP1 or in a 53BP1 associated protein. Elevated RNF168 activity at DSB chromatin results in increased 53BP1 and BRCA1 DSB localization, demonstrating that localization of both proteins is positively correlated with DSB ubiquitin levels21,22. 53BP1 DSB localization also requires RNF8- and RNF168-mediated degradation of chromatin proteins that compete with 53BP1 for binding to H4K20me223,24. The requirement for DSB ubiquitylation was obviated by siRNA mediated knockdown of H4K20me2 binding factors, enabling 53BP1 focus formation in RNF8 and RNF168 mutant cells24. These reports suggest that ubiquitylation modulates accessibility for H4K20me2 as a limiting component of 53BP1 DSB accumulation, and that there may be other factors that regulate this accessibility. Therefore, while DSB ubiquitin and 53BP1 levels closely parallel each other under most circumstances, they may not under scenarios that result in enhanced 53BP1 access for methylated H4K20.
Here set out to identify factors that impact the balance between BRCA1 and 53BP1 at DSBs. We document that acetylation is a critical determinant of the relative DSB chromatin occupancy and repair activities of BRCA1 and 53BP1. The underlying basis for these findings emanates from acetylation limiting 53BP1 Tudor domain recognition of methylated histone H4K20 on large stretches of DSB-associated chromatin. These findings reveal a heretofore-appreciated mechanism that sequential histone tail modifications direct DNA repair mechanism and are a key determinant of chemotherapeutic response.
Results
Acetylation differentially regulates BRCA1 and 53BP1 at DSBs
Acetylation and methylation represent chemically exclusive modifications on the ε-amino group of lysine side chains. Because 53BP1 relies on its tandem Tudor domains (53BP1-Tudor) to form DNA damage foci by recognizing dimethylated lysine 20 of histone H4 (H4K20me2)5,7,25, we postulated that acetylation would negatively impact 53BP1 chromatin association, thereby controlling the relative occupancy of BRCA1 and 53BP1 at DSBs. The acetyltransferase TIP60 could contribute to this balance, given its known involvement in early stages of the DSB response as part of the chromatin remodeling NuA4 complex26–29. To determine if TIP60 influences BRCA1 accumulation at DSBs, we transfected U2OS cells with siRNAs to either TIP60 or RVB1, each a component of the NuA4 complex and is required for TIP60 acetyltransferase activity, and examined the kinetics of BRCA1 accumulation at ionizing radiation-induced foci (IRIF). We observed strong decreases in BRCA1 foci formation at all time points following knockdown of either gene (Supplementary Fig. 1a–c), and after expression of an acetyltransferase inactive TIP60 mutant (Supplementary Fig. 1e). BRCA1 IRIF were partially restored by treatment with trichostatin A (TSA), a class I and II HDAC inhibitor (HDACi), but not with Nicotinamide (Supplementary Fig. 1d). In contrast, TSA and several other Class I and II HDACi strongly reduced 53BP1 IRIF (Supplementary Fig. 1f, g).
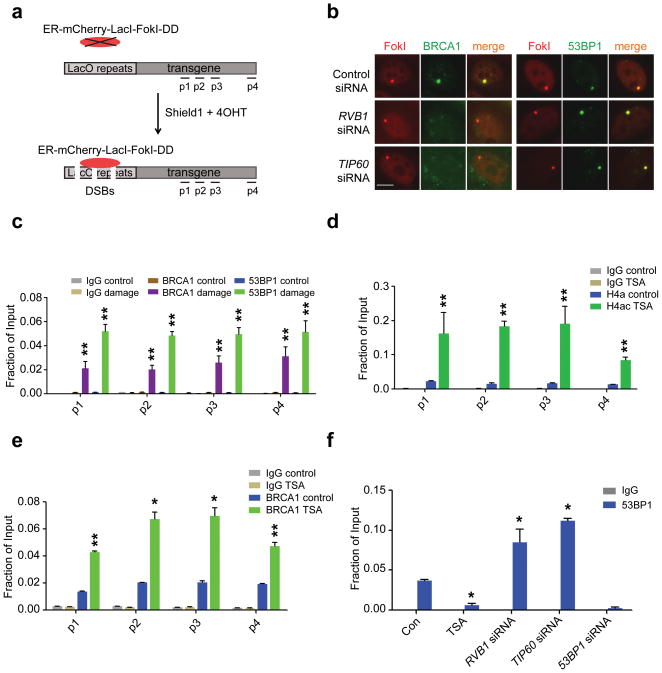
To assay how acetylation regulates the balance between BRCA1 and 53BP1 DSB localization at a single locus in a genome, we monitored their localization in a DSB reporter system that employs an mCherry-LacI-FokI nuclease fusion protein to create DSBs within a single genomic locus in U2OS cells (U2OS-DSB-reporter)30. We have equipped this cell line with stable expression of the mCherry-LacI-FokI nuclease fused to a destabilization domain and to a modified estradiol receptor. This enables inducible nuclease expression following administration of small molecules Shield1 ligand and 4-OHT. (Fig. 1a). TIP60 or RVB1 knockdown dramatically reduced BRCA1 localization to mCherry-LacI-FokI-induced DSBs, while 53BP1 exhibited a greater than 2-fold increase in DSB accumulation (Fig. 1b and Supplementary Fig. 1h), revealing concordance between chromatin responses to IR and to nuclease-induced DSBs.
Figure 1. TIP60 and HDAC inhibition differentially impact competition between BRCA1 and 53BP1 for localization to DSBs.
(a) Schematic of inducible DSB generation by mCherry-LacI-FokI at an integrated reporter transgene in U2OS cells (U2OS-DSB-reporter). ChIP-qPCR primer sets are shown as p1–p4. (b) Immunofluorescence (IF) was performed at 5 hours after induction of DSBs by mCherry-LacI-FokI in the U2OS-DSB-reporter cells transfected with the indicated siRNA. Scale bar, 10 μm. (c) ChIP-qPCR was performed with an antibody to 53BP1 or BRCA1 in the U2OS-DSB-reporter cell line with (Damage) or without (Control) induction of DSBs by mCherry-LacI-FokI. Error bars represent S.E.M. ** p<0.005. (d) ChIP-qPCR was performed with antibody to H4ac in the U2OS-DSB-reporter cell line after treatment with DMSO (Control) or 0.5 μM TSA. Induction of DSBs by mCherry-LacI-FokI was included in all conditions tested. Error bars represent S.E.M. ** p<0.005. (e) ChIP-qPCR was performed with antibody to BRCA1. Cells were treated with DMSO (Control) or 0.5 μM TSA for 5 hours and DSBs were induced by mCherry-LacI-FokI. Error bars represent S.E.M. * indicates p<0.05, ** p<0.005. (f) ChIP-qPCR was performed with antibody to 53BP1 in the U2OS-DSB-reporter cells (locus p4). DSBs were induced at 48–72 hours after siRNA transfection or 5 hours after TSA (0.5 μM) administration. Error bars represent S.E.M. * indicates p<0.05.
Chromatin IP (ChIP) for 53BP1 and BRCA1 at the U2OS-DSB-reporter locus produced results consistent with imaging studies, demonstrating 2–4 fold changes in BRCA1 and 53BP1 DSB levels depending on acetylation status. Administration of TSA resulted in elevated H4 acetylation (H4ac) and a nearly 4-fold increase in BRCA1 occupancy coupled with diminished 53BP1 association, while TIP60 or RVB1 knockdown produced the opposite effect (Fig. 1c–f). Moreover, knockdown of both HDAC1 and HDAC2 resulted in reduced 53BP1 and a corresponding increase in BRCA1 accumulation at mCherry-LacI-FokI-induced DSBs (Supplementary Fig. 1i–k).
Knockdown of 4 different members of the NuA4 histone acetyltransferase complex (TIP60, RVB1, TRRAP, and p400) increased 53BP1 DSB localization despite modestly reducing DSB associated ubiquitin foci (Supplementary Figs.2a–c, 3). Ubiquitin IRIF were substantially more reduced following RNF168 knockdown as compared to TIP60 knockdown, and this level of reduction in ubiquitin indeed abrogated 53BP1 foci formation (Supplementary Fig. 2a–c and Supplementary Fig. 3a). Notably, 53BP1 IRIF remained undetectable following either TIP60 or RVB1 knockdown in RNF168 mutated RIDDLE cells (Supplementary Fig. 2d). These findings reveal a strict requirement for RNF168 even in the context of TIP60 deficiency, indicating that RNF168-dependent ubiquitylation cannot be circumvented by decreased TIP60 acetylation. Together, these results suggest that acetylation by TIP60 regulates to the balance between BRCA1 and 53BP1 by promoting BRCA1, while limiting 53BP1, DSB localization.
BRCA1 and 53BP1 occupy distinct chromatin domains
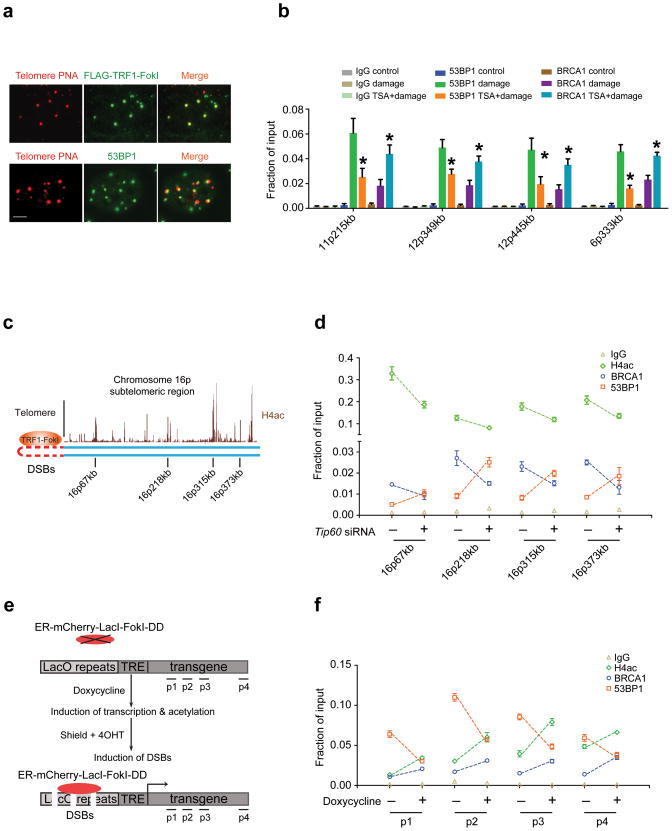
To explore the relationship between acetylation and competition between BRCA1 and 53BP1 at defined endogenous genomic sites, we developed a novel system to quantify accumulation of DSB response proteins along large stretches of subtelomeric chromatin following expression of the telomere-specific binding protein TRF1 fused to FokI. TRF1-FokI exclusively localized to telomeres and produced a focal accumulation of γH2AX, 53BP1, and BRCA1 that spreads at least several hundred kilobases into subtelomeric chromatin (Fig. 2a, b and Supplementary Fig. 4a–d). In accordance with prior results, we observed a divergent response of BRCA1 and 53BP1 at subtelomeric chromatin following TSA treatment in TRF1-FokI expressing cells (Fig. 2b). These results confirm the data from the U2OS-DSB-reporter system and from IRIF, and suggest the finding that acetylation exerts opposing regulation of BRCA1 and 53BP1 DSB chromatin association is generally relevant to DSB responses at endogenous locations.
Figure 2. DNA damage and transcription dependent acetylation regulates BRCA1 and 53BP1 DSB occupancy.
(a) TRF1-FokI colocalizes with telomeres and 53BP1 foci are observed adjacent to telomeric signals as shown by immuno-FISH using peptide nucleic acid probes against telomeres (Telomere PNA). Scale bar, 10 μm. (b) ChIP using antibodies to BRCA1 and 53BP1 at different subtelomeric regions in 293T cells with (Damage) or without (Control) expression of TRF1-FokI. Treatment with TSA is indicated. The chromosome location and distance from the telomere is indicated at each qPCR primer set. Error bars represent standard error of the mean (S.E.M). * indicates p<0.05. (c) Schematic of the subtelomeric region of 16p denoting histone H4ac pattern (UCSC genome browser) and positions of ChIP-qPCR primer sets. (d) ChIP was performed at regions of high H4ac as indicated in c in 293T cells expressing TRF1-FokI following transfection of control or TIP60 siRNA. (e) Schematic of DSB induction in transcriptionally inactive or active chromatin. Transcription is induced with doxycycline prior to DSB induction. (f) ChIP was performed in the presence or absence of doxycycline as described in e. Induction of DSBs by mCherry-LacI-FokI was included in all conditions tested following the addition of doxycycline. Error bars represent S.E.M. * indicates p<0.05.
The preceding findings demonstrate that a balance of acetyltransferase and deacetylase activities strongly influences BRCA1 and 53BP1 DSB chromatin levels. Furthermore, BRCA1 and 53BP1 ratios at DSB chromatin correlated with the degree of H4ac. High H4ac levels are present at the 5′ end of actively transcribed genes. We therefore postulated that transcription induced H4ac, in combination with DNA damage induced H4ac, would result in elevated BRCA1 and reduced 53BP1 at DSB chromatin. Moreover, this balance would be altered by TIP60 knockdown, leading to elevated 53BP1 levels in a DSB chromatin territory in which BRCA1 normally predominates.
To test this hypothesis, we monitored BRCA1 and 53BP1 association with endogenous regions on chromosome 16p that were predicted to show hyperacetylation and PolII association by the ENCODE database (Fig. 2c)31. This region demonstrated strong induction of γH2AX and increased H4K16 acetylation (H4K16ac) following TRF1-FokI expression (Supplementary Fig. 4d, e) as had originally been noted at yeast DSBs32. TIP60 knockdown consistently increased 53BP1 and decreased both H4ac and BRCA1 association at all four loci (Fig. 2d), revealing that TIP60-dependent acetylation limits 53BP1 levels at chromatin territories in cis to DSBs that are predominantly occupied by BRCA1.
To specifically address the role of transcription-dependent acetylation, we induced DSBs at either transcriptionally silent or active chromatin. mCherry-LacI-FokI DSBs can be induced in the reporter system in either transcriptionally silent or active environments through a doxycycline-regulated promoter that is downstream of tetracycline regulated elements30,33 (Fig. 2e). The presence of active transcription prior to DSB induction resulted in elevated H4ac and BRCA1 with a commensurate decline in 53BP1 at each of 4 different positions in cis to the DSB site in agreement with the hypothesis (Fig. 2f). Collectively, these data demonstrate that a combination of DNA damage- and transcription-inducible acetylation states impact the relative levels of BRCA1 and 53BP1 at DSB chromatin.
H4K16 acetylation reduces 53BP1 binding to H4K20me2
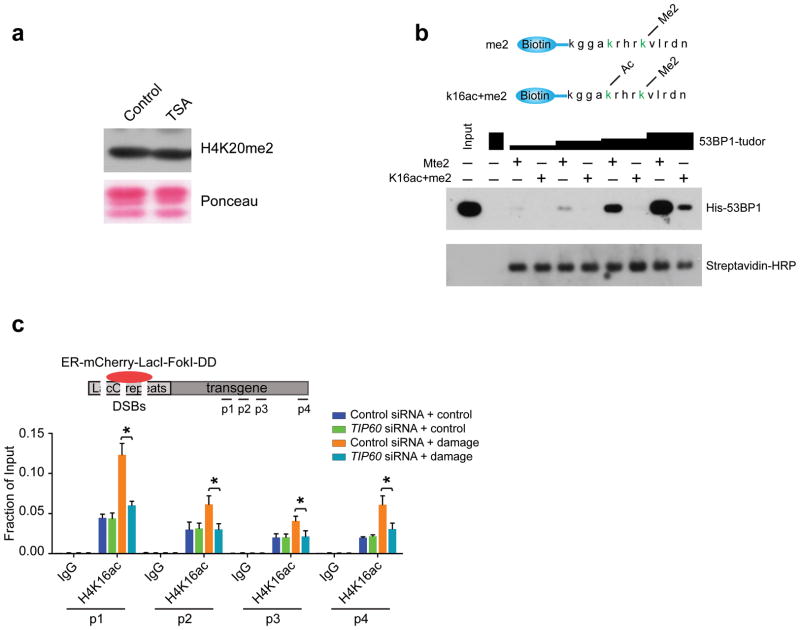
Given that dimethylation and acetylation are mutually exclusive modifications on the ε-amino group of a particular lysine residue, we initially postulated H4K20me2 levels would be decreased following TSA treatment, accounting for the absence of 53BP1 foci following HDACi. Surprisingly, 5 hours of TSA treatment did not affect total H4K20me2 as measured by Immunoblot (Fig. 3a). This result is, however, consistent with the known stability of H4K20me2, which is thought to be present at approximately 70–90% of chromatin-associated H4 molecules34.
Figure 3. Histone H4K16 acetylation impairs 53BP1 Tudor domain interaction with H4K20me2.
(a) Immunoblot (IB) was performed on acid-eluted histones from U2OS cells that had been treated for 5 hours with 0.5 μM TSA. (b) Biotinylated H4 peptide encompassing amino acids 12–25 with the relevant methylated and acetylated lysine residues highlighted in green (top panel) were used to pull down increasing amounts of His-53BP1-Tudor. IB was performed as indicated (bottom panel). (c) ChIP was performed using antibodies to H4K16ac in the absence (Control) or presence (Damage) of mCherry-LacI-FokI-induced DSBs in cells transfected with either control or TIP60 specific siRNA.
These observations suggest that acetylation at residues other than H4K20 may diminish access of 53BP1 to H4K20me2. In this regard, histone H4 is acetylated at Lys16 residues, a modification which is reported to be present in nearly 20% of the H4 amino terminal tails that include H4K20me234. To test the hypothesis that acetylation in cis to H4K20me2 could affect 53BP1 interaction, we incubated increasing amounts of the 53BP1-Tudor with biotinylated peptides encompassing amino acids 12–25 of histone H4, dimethylated at Lys20 and unmodified at every other residue, or dimethylated at Lys20 and acetylated at Lys16 (Fig. 3b top panel). 53BP1-Tudor demonstrated reduced binding to the acetylated peptide at all concentrations tested (Fig. 3b bottom panel). Meanwhile, TIP60 depletion resulted in a reduction in H4K16ac on chromatin adjacent to LacI-FokI generated DSBs at all positions tested (Fig. 3c). These findings demonstrate that TIP60-dependent acetylation of H4K16 reduces interaction between 53BP1 and its cognate recognition element H4K20me2 when present on the same H4 peptide.
H4K16 interacts with the second 53BP1 Tudor domain
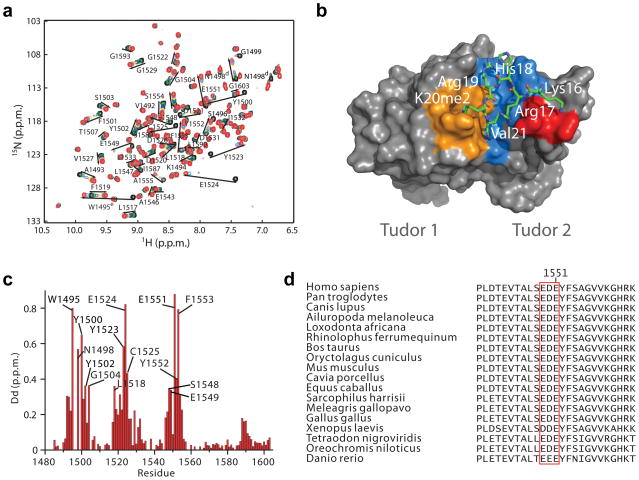
To understand the structural basis underlying the observation that H4ac reduces 53BP1 interaction with H4K20me2 peptides, we determined the three-dimensional structure of 53BP1-Tudor in complex with an H4K20me2 peptide encompassing residues 14–27 using NMR spectroscopy with 13C/15N labeling of 53BP1-Tudor and H4K20me2 containing a methyllysine analogue (H4KC20me2)35. (Fig. 4a–c, Table 1 and Supplementary Figs. 5–7). Prior crystallographic studies revealed the structure of this complex5, but electron density of the H4K20me2 peptide was only discernible for H4 amino acids Arg19 and K20me2, thus prompting an investigation of the structure using NMR. To calculate the structure of the 53BP1-Tudor–H4K20me2 complex, 100 intermolecular nuclear Overhauser enhancement signals (NOEs) involving H4 amino acids Gly14, Ala15, Lys16, Arg17, His18, Arg19, Val21, Leu22, Arg23 were used (Table 1 and Supplementary Table 1). Due to exchange broadening leading to disappearance of NMR signals of K20me2 or KC20me2 because of conformational exchange, no intermolecular NOEs involving K20me2 or KC20me2 could be detected and therefore no intermolecular distances could be determined for this amino acid. Therefore, we used 26 distance restraints derived from the crystal structure of the 53BP1-Tudor–H4K20me2 complex5 were included in the calculations to position the side chain of K20me2 in the aromatic binding cage of 53BP1. The assumption here is that one of the K20me2 conformations in exchange in the binding cage corresponds to the conformation detected in the crystal structure. While signal broadening is a clear indication of conformational exchange of K20me2, the NMR data also strongly suggest that K20me2 or KC20me2 occupy an aromatic binding cage in 53BP1-Tudor as seen in the previously determined crystal structure of 53BP1-Tudor–H4K20me25. In the 1H-15N HSQC spectrum of 53BP1-Tudor, the backbone HN resonances of Tyr1502, Phe1519, Asp1521 and Tyr1523 and side chain indole HN resonance of Trp1495 exhibit pronounced changes in chemical shift after addition of the H4K20me2 or H4KC20me2 peptides. These five 53BP1 residues form the binding cage. Notably, there is a 0.8 p.p.m. downfield shift of Trp1495 indole HN proton (Fig. 4a, c and Supplementary Fig. 6a), which is fully consistent with the establishment of a cation-π interaction between the indole ring of Trp1495 and the dimethylammonium group of K20me2. As expected, this shift is markedly reduced when 53BP1-Tudor is titrated with a non-methylated H4K20 peptide (Supplementary Fig. 6b). Also in strong agreement with a cation-π interaction between Trp1495 and K20me2 positioning K20me2 in the aromatic cage, in the 1H-13C HSQC spectrum large downfield shifts are detected for side chain resonances or Trp1495 after addition of H4K20me2 (Supplementary Fig. 6c). Further evidence for conformational exchange in the vicinity of K20me2 is the presence of multiple signals for the side chains 1H-13C resonances of Leu22 in the 13C-labeled H4K20Cme2 peptide upon titration with 53BP1-Tudor (Supplementary Fig. 6d).
Figure 4. Solution structure of 53BP1-Tudor in association with a methylated H4 peptide.
(a) Changes in the 1H-15N HSQC correlation spectrum of 53BP1-Tudor upon addition of increasing amounts of H4K20me2 peptide. The 53BP1 correlation signals affected by the interaction with the peptide are labeled. (b) NMR structure of 53BP1-Tudor in complex with a histone H4K20me2 peptide. The peptide (aa 14-27) is in stick represention with only key interacting residues shown. 53BP1-Tudor is in gray surface representation with its methyllysine binding cage highlighted in orange. Important hydrophobic residues for which intermolecular NOEs were detected are shown in blue. The red area corresponds to 53BP1 Asp1550 and 2 Glu1551. (c) Histogram of 1H-15N chemical shift changes for the backbone amide atoms and Trp1495 HN and Asn1498 HN of 53BP1-Tudor observed upon saturation with the H4K20me2 peptide. The signals of Ser1496 and Gly1499 disappeared completely after titration with H4K20me2. Backbone HN signals of Trp1495, Ser1497 and Asn1498 were not detected, even in the absence of peptide. (d) Amino acid sequence alignment of 53BP1 from various vertebrate species illustrating conservation of the 1549–1551 acidic patch in the second Tudor domain.
Table 1.
NMR and refinement statistics
| 53BP1-Tudor – H4K20me2 | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOEa | 2,766 |
| Intra-residue | 673 |
| Inter-residue | 2,093 |
| Sequential (|i − j| = 1) | 574 |
| Medium-range (|i − j| < 4) | 400 |
| Long-range (|i − j| > 5) | 1,019 |
| Intermolecular | 100 |
| Hydrogen bonds | 65 |
| Total dihedral angle restraints | 215 |
| φ | 75 |
| ψ | 76 |
| χ1 | 64 |
| Additional distance constraints | |
| Crystallography-based restraints b | 26 |
| Structure statistics | |
| Violations (mean ± s.d.) | |
| Distance constraints (Å) | 0.05 ± 0.04 |
| Dihedral angle constraints (°) | 1.9 ± 1.2 |
| Max. dihedral angle violation (°) | 4.26 ± 0.58 |
| Max. distance constraint violation (Å) | 0.22 ± 0.14 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0079 ± 0.0001 |
| Bond angles (°) | 2.11 ± 0.03 |
| Impropers (°) | 1.78 ± 0.32 |
| Average pairwise r.m.s. deviationc (Å) | |
| Heavy | 1.01 ± 0.10 |
| Backbone | 0.55 ± 0.11 |
The number of unique non-redundant NOEs is indicated.
Distance restraints involving K20me2 derived from the crystal structure of 53BP1-Tudor–H4K20me2.
Pairwise r.m.s. deviation was calculated for residues1484–1603 of 53BP1-Tudor and residues 15–23 of the H4K20me2 peptide from an ensemble of 20 structures.
In the NMR structure, the central H4K20me2 peptide region adopts a looping conformation. H4K16 is positioned in immediate vicinity to 53BP1 Glu1551, indicating a direct salt bridge between the positively charged H4K16 and negatively charged Glu1551 (Fig. 4a, b and Supplementary Fig. 7a). Acetylation of H4K16 would eliminate the positive charge from Lys16, therefore disrupting this electrostatic interaction. H4R17 occupies a pocket at the interface between the two Tudor domains and also forms a charged interaction with Glu1551 (Supplementary Fig. 7a). A stretch of acidic amino acids (Glu1549, Asp1550, Glu1551) in the second 53BP1 Tudor domain is completely conserved across all vertebrate species (Fig. 4d), suggesting the general importance of this charge-charge interaction for regulatory binding to H4K20me2. While acetylation would eliminate the salt bridge between H4K16 and the 53BP1 acidic patch, it is important to note that some interaction may be maintained between H4K16ac and the 53BP1 Tudor domain, thus accounting for the reduced, albeit not absent association in pulldown experiments (Fig. 3b). Consistent with this assertion, the side chains of H4 Lys16 and 53BP1 Tyr1552 are in contact in the NMR structure as several NOEs were detected between these two residues (Supplementary Fig. 6e, Supplementary Fig. 7a and Supplementary Table 1). The interaction between Tyr1552 and Lys16 orients Lys16 toward Glu1551. Mutations were introduced in the H4KC20me2 peptide to validate the complex structure (Supplementary Fig. 7b).
53BP1 impedes BRCA1 DSB localization in TIP60 deficient cells
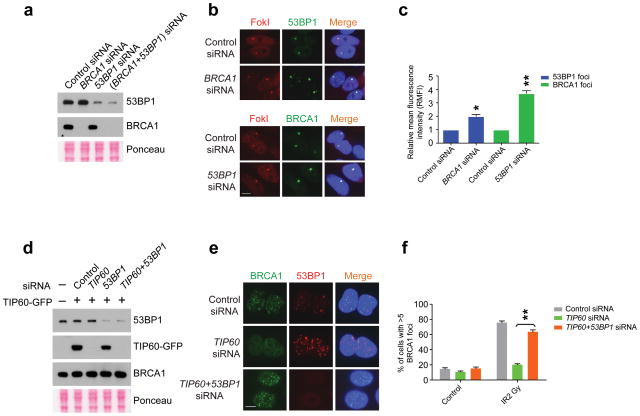
The prior results demonstrate that acetylation has opposing effects on BRCA1 and 53BP1 at DSBs, and modulates a competition between them for accumulation at sites of DNA damage. In support of this idea, we observed approximately 2-fold elevated 53BP1 levels at the DSB reporter locus following BRCA1 knockdown, and 4-fold increased BRCA1 accumulation following 53BP1 knockdown (Fig. 5a–c). Moreover, BRCA1 IRIF was largely restored in cells experiencing combined TIP60 and 53BP1 knockdown (Fig. 5d–f). These results strongly suggest that 53BP1 inhibits BRCA1 DSB access in the context of hypoacetylated chromatin.
Figure 5. 53BP1 knockdown restores BRCA1 localization to DSBs.
(a) IB with cell lysates prepared from cells treated with siRNAs as indicated. (b) Representative images of 53BP1 (upper panel) or BRCA1 (lower panel) at mCherry-LacI-FokI-induced DSBs in cells transfected with the indicated siRNA. Scale bar, 10 μm. (c), Quantification of relative mean fluorescence intensity (RMFI) from b. RMFI was calculated from 200 cells and 3 independent experiments. Error bars represent S.E.M. * indicates p<0.05, ** p<0.005. (d), IB with cell lysates prepared from TIP60-GFP expressing cells treated with siRNAs as indicated. (e) Representative images of BRCA1 and 53BP1 co-staining following fixation at 5 hours after 2 Gy IR treatment of U2OS cells that were transfected with the indicated siRNA. Scale bar, 10 μm. (f) Quantification of percentage of cells possessing greater than 5 BRCA1 IRIF per cell from e. Error bars represent S.E.M. ** indicates p<0.005.
Acetylation alleviates a 53BP1-dependent block of DSB repair
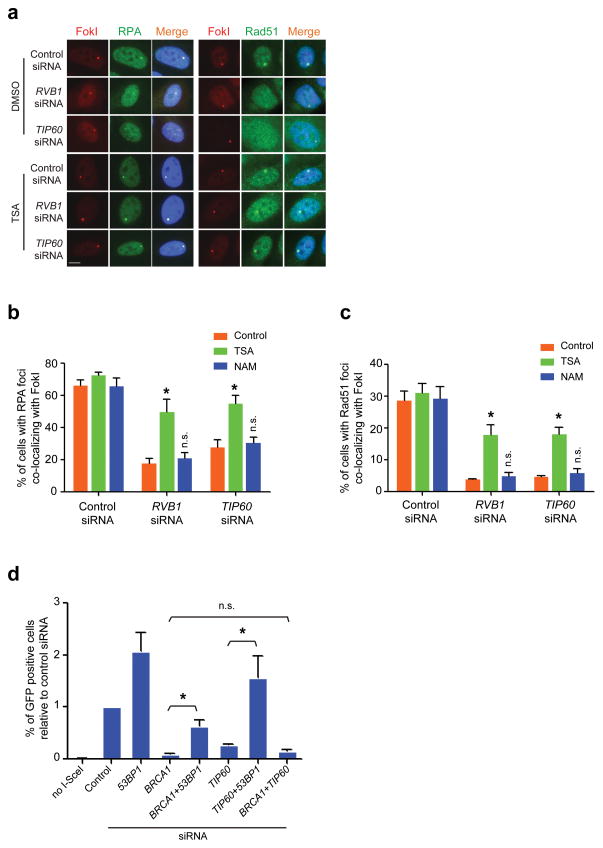
Homologous recombination requires DNA end resection and Rad51 nucleofilament formation on 3′ overhangs to initiate strand invasion into a sister chromatid. BRCA1 mutation results in diminished homologous recombination and Rad51 DSB accumulation, which has been associated with reduced end resection1,2,36–38. 53BP1 nullizygosity restores end resection, Rad51 IRIF and homologous recombination, in addition to viability through embryogenesis and resistance to PARPi in BRCA1 mutant cells1,2. These observations support a prediction that TIP60 or RVB1 deficiency would diminish end resection and Rad51 nucleofilament formation and that HDACi or co-depletion of 53BP1 would at least partially rescue these functions. Consistent with this assertion, RVB1 or TIP60 knockdown strongly reduced RPA and Rad51 accumulation at the LacI-FokI reporter, and this was restored by TSA treatment (Fig. 6a–c). Moreover, TIP60 knockdown reduced homology directed DNA repair (HDR) at an I-SceI nuclease-induced DSB to a similar extent as BRCA1 knockdown (Fig. 6d). Dual knockdown of BRCA1 and 53BP1 restored HDR as previously described. Similarly, 53BP1 knockdown rescued HDR in TIP60 deficient cells consistent with restoration of BRCA1 DSB repair function (Fig. 6d).
Figure 6. HDACi or 53BP1 deficiency restores end resection and homology directed DSB repair in TIP60 deficient cells.
(a) IF was performed with antibodies to RPA or Rad51 in siRNA transfected U2OS-DSB-reporter cells that were incubated with either DMSO control or TSA (0.5 μM). Scale bar, 10 μm. (b, c) Quantification of RPA and Rad51 DSB localization from a. Analysis was performed on 200 cells per group in at least 2 independent experiments. Error bars represent S.E.M. * p < 0.05, n.s. not significant. (d) Homology directed repair of I-SceI DSBs in siRNA transfected U2OS DR-GFP reporter cells as assessed by FACS for percent GFP positive cells. *p<0.05, n.s. = not significant.
53BP1 promotes sensitivity to PARPi during TIP60 deficiency
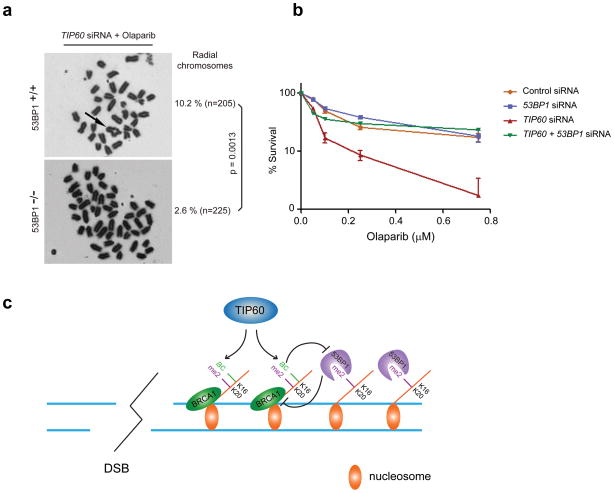
BRCA1 mutation confers hypersensitivity to the PARPi Olaparib, which is reversed by 53BP1 nullizygosity39,40. To determine if an equivalent genetic interaction exists between 53BP1 and TIP60, we transfected 53BP1 +/+ and 53BP1 −/− mouse embryonic fibroblasts with siRNA to TIP60 and subsequently exposed them to Olaparib. Nullizygosity for 53BP1 strongly mitigated the impact of TIP60 knockdown on radial chromosome formation (Fig. 7a), demonstrating similar reductions as reported for cells harboring mutations in both BRCA1 and 53BP11. Moreover, TIP60 deficiency resulted in hypersensitivity to Olaparib that was reversed by accompanying 53BP1 knockdown (Fig. 7b), demonstrating that excessive 53BP1 activity can mediate hypersensitivity to PARPi in the context of hypoacetylation. Collectively, these findings support a novel mechanism in which sequential histone modifications encompassing H4K16ac in cis to Lys20 methylation facilitates segregation of BRCA1 and 53BP1 into distinct chromatin territories to influence DSB repair mechanism (Fig. 7c).
Figure 7. 53BP1 promotes genomic instability and sensitivity to PARPi in the absence of TIP60.
(a) Representative metaphase images from 53BP1+/+ and 53BP1−/− MEFs following knockdown of TIP60 and Olaparib treatment. Significance was calculated using Fisher’s exact test. (b) Percentage survival following Olaparib administration in U2OS cells that had been transfected with the indicated siRNAs. (c) Model depicting that TIP60 -dependent acetylation limits 53BP1 binding to H4K20Me2 when present on the same histone H4 tail. TIP60 deficiency would result in hypoacetylated H4K16 and a 53BP1-dependent block of BRCA1 DSB chromatin association.
Discussion
BRCA1 and 53BP1 are chromatin-associated DNA damage response proteins with dichotomous roles in dictating DNA repair mechanism choice. BRCA1 is firmly implicated in promoting homologous recombination, whereas 53BP1 is required for certain types of NHEJ, exemplified by class switch recombination at immunoglobulin loci and fusions of deprotected telomeres41–43. Genetic evidence in mice reveals that a major component of BRCA1 function in promoting homologous recombination, genomic stability, and tumor suppression is to counterbalance 53BP1 DSB chromatin activities1,2. BRCA1 deficiency results in 53BP1-dependent illegitimate end-joining, genomic instability, and cancer susceptibility.
This study establishes that oppositional TIP60 acetylation and Class I/II HDAC-mediated deacetylation are essential determinants of a competition between BRCA1 and 53BP1 for DSB chromatin association. Hypoacetylation due to TIP60 deficiency mimicked BRCA1 mutation, resulting in reduced BRCA1 and Rad51 DSB localization, and enhanced radial chromosome formation and sensitivity to PARPi. Each of these sequelae was reverted by concomitant 53BP1 deficiency, suggesting that inappropriate accumulation of 53BP1 at DSB chromatin represents the common event responsible for genomic instability in the context of either BRCA1 mutation or hypoacetylation due to TIP60 deficiency.
The data supports a model whereby acetylation of H4K16 plays a central role in determining the balance of BRCA1 and 53BP1 at DSB chromatin by reducing, albeit not eliminating 53BP1 Tudor domain’s binding affinity for H4K20me2 when present on the same H4 tail. Thus, sequential histone modifications act in a combinatorial manner to limit 53BP1 DSB chromatin occupancy, thereby promoting BRCA1 DSB localization and HDR. The solution structure of H4K20me2 peptides in association with the 53BP1 Tudor domains reveals a direct charge-charge interaction between H4K16 and 53BP1 Tudor domain residue Glu1551, which belongs to a conserved patch of acidic residues. This information was not available from prior crystallographic studies involving the 53BP1 Tudor domains and an H4K20me2 peptide due to a lack of discernible electron density of H4 residues other than H4K20me2 and Arg195. The salt bridge interaction between H4K16 and 53BP1 Glu1551 reveals a potential common mode of regulation with respect to H4 binding proteins. Notably, H4K16 interacts with an acidic patch present at the H2A-H2B interface in nucleosomes and H4K16 also forms a salt bridge with a conserved acidic residue present in the H4K20me2 binding domain of Orc144,45. Acetylation of H4K16 would eliminate the charged interaction with 53BP1 Tudor domain residue Glu1551, as well as with each of these other interacting partners.
While biochemical and structural data indicate acetylation sequential to H4K20me2 regulates the balance between 53BP1 and BRCA1 at DSBs (Fig. 7c), other TIP60-dependent mechanisms may also contribute, such as the involvement of acetyl-lysine binding proteins and acetylation on additional TIP60 substrate besides H4K16. The solution structure of 53BP1 with an H4K20me2 peptide predicts that association of an H4K16ac binding protein would further disrupt this interaction. It will therefore be of interest to determine if a putative TIP60 -regulated H4K16ac binding protein is present at DSBs and if it exerts similar control of BRCA1 and 53BP1 DSB localization. In addition, incorporation of synthetic H4K16ac into nucleosomes prevents compaction of chromatin into 30-nm fibers in vitro46. The contribution of changes in higher order chromatin structure to BRCA1 and 53BP1 DSB chromatin occupancy due to H4K16ac may also warrant examination in future studies.
Both TIP60 and HDACs have been reported to localize to DSBs, suggesting a dynamic process of acetylation and deacetylation impacts DSB repair mechanism outcome. Consistent with our findings, HDACs 1 and 2 are required for efficient NHEJ, a 53BP1-dependent process47. Deacetylation occurred at early phases of the DNA damage response, with H4K16 acetylation accumulating over several hours after the initial wave of deacetylation. The kinetics of deacetylation and acetylation therefore closely mirrors the timing of 53BP1 and BRCA1 foci formation. 53BP1 IRIF occur within minutes of DNA damage induction, whereas BRCA1 IRIF accumulate over hours, despite each depending on RNF168 dependent ubiquitylation. In addition, BRCA1 and 53BP1 show non-overlapping localization at IRIF by super-resolution fluorescence imaging48. This difference is suggestive that additional events contribute to the later onset of BRCA1 IRIF. Our results are consistent with TIP60 -dependent acetylation as being an important determinant and help explain the divergent DSB localization properties of BRCA1 and 53BP1 despite their common reliance on γH2AX, MDC1, RNF8 and RNF168.
Hereditary breast and ovarian cancer susceptibility is a consequence of germline, heterozygous mutations in the BRCA1 and BRCA2 genes. BRCA patients are heterozygous, while their tumors generally display loss of the wild-type BRCA allele, thus creating tumor specific homologous recombination deficiency. This feature of a BRCA null tumor in the context of a heterozygous patient inspired the development of PARPi as a synthetic lethal strategy to exploit tumor specific homologous recombination deficiency39,40,49. The data presented here suggest that consideration of histone acetylation levels may be informative in predicting responses to PARPi and other DNA damaging anti-cancer therapies. Importantly, histone H4K16 acetylation is frequently reduced in cancers50. Moreover, TIP60 is a haploinsufficient tumor suppressor in mice and TIP60 mutations as well as reduced TIP60 expression has been reported in a broad range of cancer types, including breast cancer51. Our findings extend the concept that chromatin modifications play a critical role in establishing the balance between repair proteins at sites of DSBs and suggest that consideration of either TIP60 status or histone acetylation levels may be informative in predicting responses to anti-cancer therapy.
Methods
Cell culture
All cells were maintained in DMEM (Invitrogen) with 10% calf serum and pen/strep. U2OS-DSB-reporter cell line was provided by S. Janicki and D. Spector (Wistar Institute and Cold Spring Harbor Laboratories). The U2OS DR-GFP cells and 53BP1 −/− mouse embryonic fibroblasts were provided by M. Jasin (Memorial Sloan Kettering Cancer Center) and J. Chen (MD Anderson Cancer Center), respectively.
Plasmids and siRNAs
C-terminal GFP tagged TIP60 expression vector was generated by PCR amplification of TIP60 from pCR4-TOPO-TIP60 (Open Biosystems) and subsequent cloning into pDEST47 using the Gateway cloning method (Invitrogen). The TIP60 acetyltransferase inactive mutant52 (Q377E|G380E) was generated by site-directed mutagenesis. The coding sequence of TRF1 was cloned in frame to the nuclease domain of FokI to generate the TRF1-FokI fusion in pCDNA3. The following siRNA sequences were used: Control siRNA: 5 ′-ACGCGUAACGCGGGAAUUU or 5′-GCCAUUCUAUCCUCUAGAGGAUG; siTIP60: 5′-TGATCGAGTTCAGCTATGA, or smart pool siRNA against human or mouse TIP60 from Dharmacon; RVB1 siRNA: 5 ′-TAAAGGAGACCAAGGAAGT, or CTCTCTCTCTCTCTCTCTA28; 53BP1 siRNA: 5′-UAUUACCGUCUCCUCGUUC; BRCA1 siRNA: 5′-AGAUAGUUCUACCAGUAAA; TRRAP and p400 siRNA: smart pool siRNA against human TRRAP or p400 from Dharmacon; HDAC1 siRNA: 5 ′-CAGCGACUGUUUGAGAACC; HDAC2 siRNA: 5′-GCGGAUAGCUUGUGAUGAA47.
Transfections
Transient plasmid transfections were carried out with LipoD293 (Signagen), and siRNA transfections with Lipofectamine RNAiMax (Invitrogen) according to manufacturer’s instructions. Analyses were performed 16–48 hours after transfection of plasmids, and 48–72 hours after siRNA transfection.
Antibodies and chemicals
Antibodies against 53BP1 and RPA were from Novus; rabbit polyclonal anti-BRCA1 antibodies from Millipore, mouse monoclonal anti-BRCA1 antibody D9 from Santa Cruz; anti-H4ac, anti-H4K16ac, anti-γH2AX, anti-His-tag and Streptavidin-HRP from Millipore; anti-Rad51 was from Santa Cruz; anti-GFP antibody was from Roche and anti-H4K20me2 was from Active Motif; and anti-TRRAP was purchased from Cell Signaling. Trichostatin A (TSA) and nicotinamide (NAM) were purchased from Sigma. Vorinostat, MGCD0103 and Droxinostat were from SelleckChem. Olaparib was from ChemieTek.
ChIP
Chromatin immunoprecipitation (ChIP) was carried out as described previously30. Induction of DSBs at telomeric repeats was performed by transfecting 293T cells with the TRF1-FokI expression plasmid. Cells were cross-linked with 1% (v/v) formaldehyde for 10 min, 16 hours after transfection, followed by the addition of glycine to 0.125 M for 5 min to stop the cross-linking. Cells were lysed in 10 mM Tris, pH 8.0, 10 mM NaCl, 0.2% NP-40. Nuclei were isolated. resuspended in 50 mM Tris pH 8.1, 10 mM EDTA, 1% SDS and sonicated to obtain approximately 200–500 bp chromatin fragments using a Bioruptor (Diagenode). Chromatin fragments were precleared salmon sperm DNA/protein-A agarose (Millipore)and incubated with pre-bound antibody-protein A agarose overnight at 4°C. Beads were washed once in low salt buffer (20 mM Tris, pH 8.1, 2 mM EDTA, 50 mM NaCl, 1% Triton X-100, 0.1% SDS), twice in high salt buffer (20 mM Tris, pH 8.1, 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% SDS), once in LiCl buffer (10 mM Tris, pH 8.1, 1 mM EDTA, 0.25 mM LiCl, 1% NP-40, 1% deoxycholic acid) and twice in TE buffer (10 mM Tris-HCl, pH 8. 0, 1 mM EDTA). Washed beads were eluted twice with 100 μL of elution buffer (1% SDS, 0.1 M NaHCO3) and de-crosslinked (0.1 mg/ml RNase, 0.3 M NaCl and 0.3 mg/ml Proteinase K) overnight at 65°C. The DNA samples were purified with Qiaquick PCR columns (Qiagen). qPCR was carried out on an ABI 7900HT instrument using the SYBR Green detection system. For each primer set used, standard curves were created using serially diluted input samples. ChIP-qPCR primer sequences are listed in the Supplementary information. The following amplification program was used: 95°C for 10 min, 95°C for 15 sec and 60°C for 1 min, 40 cycles.
Protein purification and peptide pull-down assays
Plasmids expressing 53BP1-Tudor were transformed into E. coli BL21 and 53BP1-Tudor was purified using a Ni-NTA Fast Start Kit (Qiagen) according to the manufacturer’s instruction. Purified protein were first checked by PAGE and Coomassie staining, and further purified by size exclusion chromatography using a Superdex 75 column.
The amino acid sequence of the biotinylated peptides corresponds to histone H4 tail amino acids 12 to 25. The following 2 peptides were used:
K20me2: KGGAKRHRK(me2)VLRDN
K16acK20me2: KGGAK(ac)RHRK(me2)VLRDN
The peptides were purchased from Syd Labs, Inc. Purified protein and peptides were incubated overnight at 4°C in binding buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.05% NP-40). Varied amount of protein were used in the pull-downs while keeping the amount of peptide constant (1 μg / pull-down). Streptavidin agarose (PIERCE) slurry was added and incubated for 1 hour at 4°C. Beads were washed 3× with binding buffer and boiled in Laemmli buffer.
Immunofluorescence
Immunofluorescence was performed as described previously30. Cells grown on coverslips were fixed and permeabilized. Following incubation with primary antibody, cells were washed with PBST and then incubated with secondary antibody. After extensive washing with PBST, coverslips were mounted onto glass slides using Vectashield mounting medium containing DAPI (Vector Laboratories).
Image capture and analysis
Images were captured with a QImaging RETIGA-SRV camera connected to a Nikon Eclipse 80i miscroscope. All images for a given experiment were captured on the same day with the same exposure times in an 8-bit grayscale format. Images were analyzed using the ImageJ software (NIH). For U2OS-DSB-reporter cells, mCherry signal was used to identify the area of damage, and the presence or intensity of BRCA1, 53BP1, RPA or Rad51 foci in the area was determined. Background was subtracted for each cell. Grayscale images were pseudocolored and overlayed using Image J software. All analyses were carried out on unmodified images.
DR-GFP HDR Assay
U2OS DR-GFP cells were treated with the indicated siRNAs in 24-well plates and 24 hours later, cells were transfected with 0.4 μg of I-SceI per well. 72 hours after I-SceI transfection, cells were trypsinized and percentage of GFP positive cells was determined by flow cytometry.
Metaphase Spreads Preparation
Cells were treated with 0.5μM nocodazole for 3–4 hours, collected and washed in PBS. Subsequently, cells were lysed in 75 mM KCl, fixed on ice with fixation solution (3:1 methanol / acetic acid). Fixed cells were dropped onto slides at 55°C, allowed to dry and stained with Giemsa.
Southern blot
Southern blot was carried out with a DIG labeling and detection kit (Roche) per manufacturer’s instructions.
Statistics
Student t test was used to determine p values comparing two groups throughout the manuscript. Fisher’s exact test was used in analyzing metaphase spread results.
53BP1-Tudor protein preparation for structure determination
For NMR structure determination, unlabeled, 15N-labeled and 15N-/13C-labeled 53BP1-Tudor (aa 1484-1603) samples were prepared as reported previously5.
Preparation of H4 peptides used in NMR
An H4K20me2 peptide (aa 14-27) was chemically synthesized and purified by reversed phase HPLC. In addition, H4K20 peptides (aa 14-27 and 10-27) were produced recombinantly in E. coli as fusions to an N-terminal GB1-His6-tag cleavable by TEV protease, chemically modified to install a dimethylated lysine analogue at position 20 (KC20me2), and purified as we reported previously35,53. This procedure allows isotope-enrichment of the peptides, greatly facilitating NMR characterization35. The recombinant peptides have extra N-terminal residues (GHM) from the expression vector. Mutant peptides were obtained by site-directed mutagenesis. Amino acid sequences of the peptides are indicated below.
Wild-type H4KC20me2 (aa 14-27): GHMGAKRHRKC(me2)VLRDNIQ
A15G H4KC20me2 (aa 14-27): GHMGGKRHRKC(me2)VLRDNIQ
K16G H4KC20me2 (aa 14-27): GHMGAGRHR KC(me2)VLRDNIQ
R17G H4KC20me2 (aa 14-27): GHMGAKGHR KC(me2)VLRDNIQ
R19G H4KC20me2 (aa 14-27): GHMGAKRHGKC(me2)VLRDNIQ
L22G H4KC20me2 (aa 14-27): GHMGAKRHRKC(me2)VGRDNIQ
D24G H4KC20me2 (aa 14-27): GHMGAKRHR KC(me2)VLRGNIQ
Wild-type H4KC20me2 (aa 10-27): GHMLGKGGAKRHRKC(me2)VLRDNIQ
K12G H4KC20me2 (aa 10-27): GHMLGGGGAKRHR KC(me2)VLRDNIQ
NMR experiments and structure calculations
NMR experiments were collected at 298 K using a Bruker Avance 700 MHz spectrometer equipped with a cryoprobe. Protein and peptide samples were in a buffer solution made up of 25 mM sodium phosphate, pH 7.0, 90%H2O/10%D2O, 0.3 mM DSS, 1.5 mM NaN3. For a typical NMR titration, 1H-15N HSQC spectra were collected on a 0.3 mM 15N-labeled 53BP1-Tudor sample titrated with a nonlabeled peptide from a 15 mM stock solution. Additionally, 1H-13C HSQC spectra were collected on a 0.5 mM 13C/15N-labeled H4KC20me2 peptide (aa 14-27) titrated with nonlabeled 53BP1-Tudor from a 5 mM stock solution. For NMR structure determination, two complexes (1.7 mM 13C/15N-labeled 53BP1-Tudor and 8.5 mM H4K20me2 (aa 14-27); 4 mM 13C/15N-labeled recombinant H4KC20me2 and 5 mM nonlabeled 53BP1-Tudor) were prepared. The structure of 53BP1-Tudor–H4K20me2 was determined using simulated annealing protocols implemented in CYANA54 and AMBER55. The structural statistics are given in Table 1. The quality of the final structures was assessed by PROCHECK-NMR56. Regarding the Ramachandran plot statistics of 20 NMR models (Table 1), for the 53BP1-Tudor component of the complex (aa 1484-1603), 82.1% of the residues are in the most favored regions, 16.0% in additional allowed regions, 0.7% in generously allowed regions and 1.2% in disallowed regions. For the H4K20me2 component of the complex (aa 15-23), 55.6% of the residues are in the most favored regions, 32.8% in additional allowed regions, 7.8% in generously allowed regions and 3.9% in disallowed regions. Details of the NMR experiments and structure calculation procedure are presented in the Supplementary Methods. PyMol (http://www.pymol.org/)57 was used to create all molecular representations.
Supplementary Material
Acknowledgments
We thank J. Wu for technical support, J. Chen (MD Anderson, Houston, TX) for 53BP1−/−MEFs, M. Jasin (Memorial Sloan Kettering Cancer Center, New York, NY) for U2OS DR-GFP reporter cells, G. Stewart (University of Birmingham, Birmingham, UK) for RIDDLE cells, and S. Janicki (Wistar Institute, Philadelphia, PA) and D. Spector (Cold Spring Harbor Laboratories, Long Island, NY)for the U2OS 2-6-3 reporter cell line. We thank KM. Miller (University of Texas, Austin, TX) and A. Sfeir (New York University, New York, NY) for their critical reading of the manuscript and helpful comments. R.A.G. is supported by 1R01CA138835-01 from the NCI, a Research Scholar Grant from the American Cancer Society, a DOD Breast Cancer Idea Award, a UPENN-FCCC SPORE Pilot Grant, and funds from the Abramson Family Cancer Research Institute and Basser Research Center for BRCA. G.M. acknowledges support from NCI grant 1R01CA132878 and funds from the Mayo Clinic Breast Cancer Specialized Program of Research Excellence (SPORE) NCI grant P50CA116201.
Footnotes
ACCESSION CODES
The atomic coordinates for the NMR structure of 53BP1-Tudor–H4K20me2 have been deposited in the Protein Data Bank with accession code 2LVM. The associated NMR chemical shifts have been deposited in the Biological Magnetic Resonance Bank with accession code 18579.
AUTHOR CONTRIBUTIONS
J.T. initiated the study and performed the majority of experiments with guidance from R.A.G. N.W.C designed and validated the TRF1-FokI DSB reporter system in conjunction with N.M.S. and R.A.G. E.M.M. provided technical assistance to J.T. G.C. prepared the isotope-enriched proteins and peptides and performed the NMR spectroscopy experiments and structure calculations. M.V.B. made the protein expression constructs and helped in sample preparations for NMR studies. G.M. supervised the structural studies. The study was conceived by J.T. and R.A.G. Writing was performed by J.T. and R.A.G. with contributions from G.M., N.M.S. and N.W.C.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–95. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 4.Xu X, et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–71. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- 5.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oda H, et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–76. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–14. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–29. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripathi V, Nagarjuna T, Sengupta S. BLM helicase-dependent and -independent roles of 53BP1 during replication stress-mediated homologous recombination. J Cell Biol. 2007;178:9–14. doi: 10.1083/jcb.200610051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 11.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 12.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–34. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 13.Kolas NK, et al. Orchestration of the DNA-Damage Response by the RNF8 Ubiquitin Ligase. Science. 2007 doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huen MS, et al. RNF8 Transduces the DNA-Damage Signal via Histone Ubiquitylation and Checkpoint Protein Assembly. Cell. 2007 doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–5. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 18.Livingston DM. Cancer. Complicated supercomplexes. Science. 2009;324:602–3. doi: 10.1126/science.1174839. [DOI] [PubMed] [Google Scholar]
- 19.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–26. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li ML, Greenberg RA. Links between genome integrity and BRCA1 tumor suppression. Trends Biochem Sci. 2012;37:418–24. doi: 10.1016/j.tibs.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudjonsson T, et al. TRIP12 and UBR5 Suppress Spreading of Chromatin Ubiquitylation at Damaged Chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 22.Poulsen M, Lukas C, Lukas J, Bekker-Jensen S, Mailand N. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J Cell Biol. 2012;197:189–99. doi: 10.1083/jcb.201109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acs K, et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol. 2011;18:1345–50. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- 24.Mallette FA, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. Embo J. 2012;31:1865–78. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–11. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 26.Murr R, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 27.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–73. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 28.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Cote J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol Cell Biol. 2005;25:8179–90. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–81. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–82. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–13. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janicki SM, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–98. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–86. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui G, Botuyan MV, Mer G. Preparation of recombinant peptides with site-and degree-specific lysine (13)C-methylation. Biochemistry. 2009;48:3798–800. doi: 10.1021/bi900348z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–8. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 37.Chen L, Nievera CJ, Lee AY, Wu X. Cell Cycle-dependent Complex Formation of BRCA1{middle dot}CtIP{middle dot}MRN Is Important for DNA Double-strand Break Repair. J Biol Chem. 2008;283:7713–20. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 38.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–3. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 40.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 41.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–8. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–64. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–7. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 44.Kuo AJ, et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–9. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 46.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 47.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–51. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapman JR, Sossick AJ, Boulton SJ, Jackson SP. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci. 2012 doi: 10.1242/jcs.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 50.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 51.Gorrini C, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–7. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 52.Wolf E, et al. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell. 1998;94:439–49. doi: 10.1016/s0092-8674(00)81585-8. [DOI] [PubMed] [Google Scholar]
- 53.Simon MD, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–12. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 55.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 57.Delano WF. The PyMOL Molecular Graphics System, version 1.3r1. Schrodinger, LLC; New York: 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.