Abstract
Differential hydrogen/deuterium exchange (H/DX) coupled with mass spectrometry (H/DX-MS) offers a rapid and sensitive characterization of changes in protein following perturbations induced by changes in folding, ligand binding, oligomerization, and modification. The characterization of H/DX rates in software tools and automated data processing often relies on the centroid mass calculation, and, thereby, the deuterium distribution in the mass spectra is neglected. Here we present an example demonstrating the clear limitation of using only a centroid approach to characterize the H/DX rate, in which the change in protein is not reflected as the difference in deuterium uptake based on centroid calculation.
Introduction
Hydrogen/deuterium exchange mass spectrometry (H/DX-MS) has emerged as a powerful tool for the analysis of protein conformation and dynamics (1–4). H/DX studies focus principally on the backbone amide hydrogens (-CO-NH-) that exchange at rates that can be followed by MS. These rates are good readouts for protein dynamics at the global, peptide, and soon even at the amino-acid level (1, 5).
H/DX studies often involve a comparison of conformational dynamics, in which the H/DX rate of a region of a protein in one state is compared to that of the same region following a perturbation. The perturbation can be ligand binding, mutation, posttranslational modification, protein activation, and formation of a complex or oligomer. Changes to the H/DX rates readily reflect the changes in protein after exposure to stimuli. As a result, they can be used to identify ligand binding sites (5–7), protein-protein interfaces (8–10), and allosteric effects (11–13).
The most widely used approach for measuring H/D exchange rates at the retional or peptide level follows the isotope clusters of the peptides for various H/D exchange time points, calculates the centroid mass of the isotope cluster, and subtracts the centroid mass of undeuterated peptides from that of deuterated peptides (Supplemental Figure 1). The source of information is the mass spectrum, which originally was processed manually in a tedious and time-consuming way. To ease this burden, a number of software programs have been developed to extract automatically the deuterium isotope patterns (14–20). A possible problem associated with centroid calculation is that it neglects the deuterium distribution and outputs a single value (21). Consequently, information from the distribution is often lost. An example is evidence for EX1 behavior, which may offer an important clue about protein function.
In most cases, one is able to assess the changes because there are differences in the deuterium uptake from centroid calculation. We found, however, that simply calculating the centroid mass of target peptides can lead to a wrong conclusion. When we investigated the changes induced by Ca2+ binding to an N-terminus truncated DREAM (downstream regulatory element antagonist modulator), we found that there is no difference in the H/DX rate for one peptide from EF-hand 2 (of four EF hands) based on centroid calculation, but upon closer examination, this region shows changes upon Ca2+ binding. The purpose of this note is to illustrate this phenomenon and in so doing to urge practitioners to examine the isotopic distributions in mass spectral H/DX of proteins.
Experimental
Materials
All chemicals were from Sigma (St. Louis, MO) unless otherwise noted. Short full-length or sFL DREAM (amino acid residues 95–256, lacking the first 94 residues of the full-length protein) was expressed and purified as described previously (22).
Peptide Level H/DX
Differential, solution H/DX experiments were performed to detect the changes in full length DREAM induced by Ca2+ binding, as previously described (12, 23). DREAM (50 μM) was incubated with 5 mM Ca2+ for at least 1 h on ice before H/DX analysis. Continuous labeling was initiated by incubating 1 μL of the 50μM protein complex (with and without Ca2+) with 19 μL of D2O buffer for a predetermined time (10, 30 s, 1, 2, 15 min, 1, and 4 h) at 4 °C. The exchange reaction was quenched by mixing with 30 μL of 3 M urea, 1% TFA at 1 °C. The mixture was passed over a custom-packed pepsin column (2 mm × 2 cm) at 200 μL/min. Digested peptides were captured on a 2 mm × 1 cm C8 trap column (Agilent) and desalted with a 3 min flow. Peptides were then separated by using a 2.1 mm × 5 cm C18 column (1.9 μm Hypersil Gold, Thermo Scientific) with a 5 min linear gradient of 4%–40% CH3CN in 0.1% formic acid. Protein digestion and peptide separation were carried out in ice-water bath to reduce back exchange. Mass spectrometric analyses were with a hybrid LTQ Orbitrap with capillary temperature at 225 °C, and data were acquired with a mass resolving power of 100,000 for ions of m/z 400. Each experiment was performed in duplicate.
Peptide Identification and H/DX Data Processing
MS/MS experiments were also performed with the LTQ Orbitrap (Thermo Fisher, Walthan, MA). Production spectra were acquired in a data-dependent mode, and the six most abundant ions were selected for production analysis. The MS/MS*.raw data files were converted to *.mgf files and then submitted to Mascot (Matrix Science, London, UK) for peptide identification. Peptides included in the set used for H/DX had a MASCOT score of 20 or greater. The MS/MS MASCOT search was also performed against a decoy (reverse) sequence, and ambiguous identifications were ruled out. The production (MS/MS) spectra of all peptide ions from the MASCOT search were manually inspected, and only those verifiable were used in the coverage. The centroid masses of isotopic envelopes were calculated with HD Desktop (17), as described elsewhere: deuterium level (%) = {[m(P) − m(N)]/[m(F) − m(N)]} × 100%, where m(P), m(N) and m(F) are the centroid values of partially deuterated peptide, nondeuterated peptide, and fully deuterated peptide, respectively (3). To accommodate situations where a fully deuterated control is not available, m(F) was determined with m(F) = m(N) + ((n − p − 2)/z), where n is the number of amino acids in the peptide, p is the number of prolines, and z represents charge. Prolines, with no amide hydrogen, were not considered. The value “2” is subtracted within the equation because the first two amino acids do not retain deuterium (2). An adjustment was made for the exchange media at 95% deuterium content. No correction was made for back exchange because all values are relative and susceptible to the same back exchange (24).
Results and Discussion
DREAM is a 29.6 kDa (256 amino acids) EF-hand Ca2+-binding protein that serves as a transcriptional repressor that modulates pain through the regulation of the prodynorphin gene (25). DREAM possesses four EF hands, indicating that it can bind up to four Ca2+. Recently NMR and thermodynamic studies of N-terminal truncated mouse DREAM showed, however, that only EF hand 2, 3 and 4 are responsible for Ca2+ binding, not EF-1 (26, 27). We applied comprehensive H/DX analysis for N-terminal-deleted sFL DREAM in the absence (apo) and presence of Ca2+ (holo) to investigate the Ca2+-binding-induced changes in protein, and thereby to confirm Ca2+ binding sites.
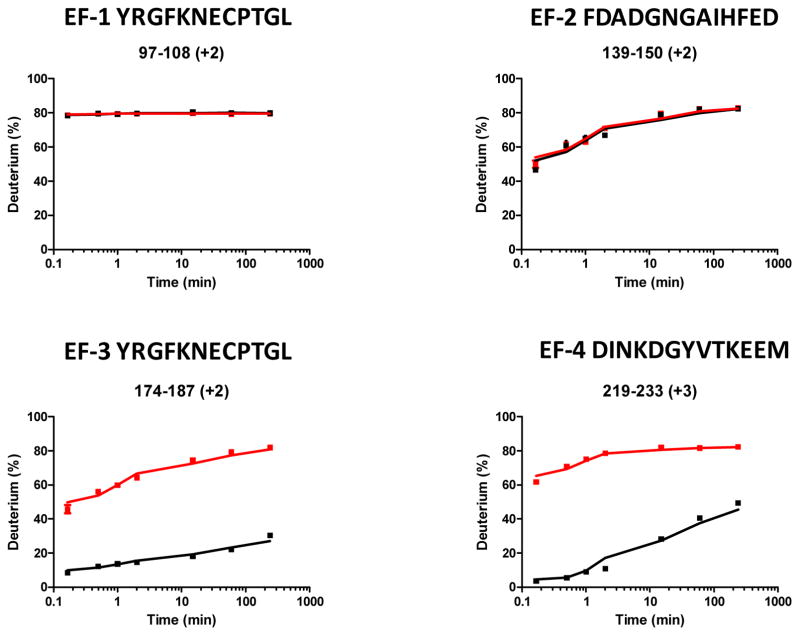
We submitted both apo and holo proteins to H/DX under identical conditions, so changes in H/DX rates would reliably reflect any Ca2+-binding-induced changes. As shown in Figure 1, EF hands 3 and 4 undergo lower exchange upon addition of Ca2+, indicating Ca2+ binding at these two regions, as was established by NMR (26). Isothermal titration calorimetry (ITC) analysis and other evidence from DREAM homolog proteins revealed that Ca2+ binds to EF hand 2 as well, but with lower affinity compared to binding at EF hand 3 and 4 (27). Unexpectedly, the deuterium uptake curves for the peptide 139–150 covering the EF hand 2 did not show any difference between apo and holo states (Figure 1). On the basis of this lack of difference, one would normally draw a conclusion that EF hand 2 is not binding Ca2+ under these conditions. Upon close inspection, however, the mass spectral isotopic distributions between apo and holo are quite different (Figure 2).
Figure 1.
Representative deuterium uptake curves for peptides from EF-1, 2, 3, and 4 of sFL DREAM. The red lines represent the deuterium incorporation of the peptides from the apo state, and the black lines represent that from Ca2+-bound (holo) state.
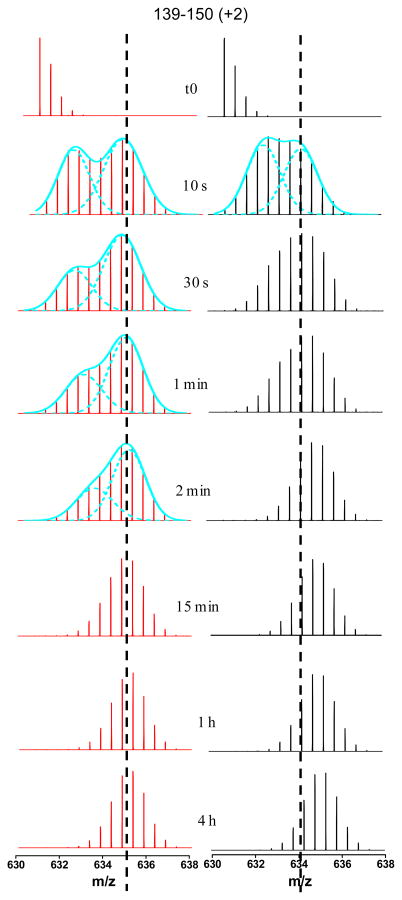
Figure 2.
Mass spectra of the peptide (AA 139–148) from EF hand 2 at different on-exchange time for both apo and holo states. The EX1 kinetics can be distinguished by the presence of two isotopic distributions. The spectra with obvious EX1 kinetics at different on-exchange time points were fitted with two Gaussian distributions shown as cyan dashed lines. The vertical black dashed lines indicate the centroid positions of the higher mass distribution in EX1 kinetics.
We know that H/DX into a protein occurs via two processes: (1) local opening and closing of the protein, and (2) chemical exchange with the solvent-accessible backbone amide hydrogens1. It follows that the kinetics for amide H/DX has two limits, EX2 and EX1. In EX1, the exchange rate is much faster than the closing rate and, therefore, the amide hydrogens in a region showing EX1 kinetics exchange with deuterium rapidly and completely during the first open event, giving rise to a bimodal deuterium distribution. By comparison, EX2 kinetics occurs when the exchange rate is much slower than the closing rate. Substantial exchange requires a few cycles of structural opening and closing. For EX2, mass spectra show a single isotopic distribution that gradually shifts to higher mass with increasing exchange time. The H/DX rate can be characterized by the shift of the deuterium distribution’s centroid mass as a function of time. Under physiological condition, most proteins follow EX2 kinetics, and the centroid approach of data processing is reliable.
For H/DX of DREAM, the average mass centroids of the entire isotopic distribution for the doubly charged peptide 139–150 from EF hand 2 are not different after Ca2+ binding. There are significant differences, however, for the mass spectra of apo and holo at the early exchange time points. The deuterium distribution of mass spectra at the early exchange times (10 s, 30 s, 1 min, and 2 min) for the apo protein exhibited a distinct EX1 signature as a broad isotopic envelope and a bimodal isotopic distribution. In contrast, this EX1 signature occurred for the holo protein only at 10 s.
To reveal the exchange mechanism’s details, we fit the mass spectra that displayed EX1 kinetics with two Gaussian distributions (28, 29). For the apo state, a high mass distribution at centroid m/z 635.14 appeared as early as 10 s, and its signal intensity increased with labeling time. The centroid mass difference between the high-mass and the undeuterated species (m/z 631.17) is ~8 Da, implicating at least 8 residues are involved in exchange through correlated local unfolding in this peptide region. Factoring in a small extent of back exchange, we conclude that nearly all 11 exchangeable amino-acid residues in this peptide must be involved in the EX1 exchange. Interestingly, the low-mass distribution continued to increase in mass, indicating EX2 kinetics while at the same time the EX1 exchange kinetics occurs, but gradually disappeared after 2 min of exchange.
The data indicate the co-existence of two major protein conformations, one of which undergoes correlated H/DX (EX1) and the other which undergoes uncorrelated H/DX (EX2). The EX1 kinetics is concurrent with EX2 because a transient, locally unfolded state arises from part of the total population undergoing EX2 kinetics. This state undergoes EX1 kinetics as seen by the appearance of the high-mass distribution (Figure 2). An example of this phenomenon is discussed in a recent review (30).
We also observed the two protein conformations for the holo state. The high-mass component, however, only displayed a ~6 (not ~8) Da mass increase from m/z 631.17 to 634.28, suggesting that two of the eight residues of apo are not involved in the EX1 exchange kinetics of the holo. The entire distribution shifted upward by the remaining 2 Da via EX2 kinetics after the EX1 kinetics was complete (from 30 s to 4 h H/DX). This is another example whereby not all residues in a given region of a protein participate in EX1 exchange kinetics, but some exchange via EX2 (28, 31, 32). The difference is due to Ca2+ binding, which shifts the exchange of those two residues from EX1 to EX2 and slows their exchange. Moreover, the low mass component in the EX1 signature had a mass increase compared to undeuterated, but diminished after 10 s of exchange, indicating that the protein population not undergoing EX1 exchange had much faster EX2 kinetics compared to that of the apo state. Overall, there is a significant difference for the EF-2 region between apo and holo protein with regard to the EX1 and EX2 kinetics, which suggest Ca2+ binding also induces changes in EF-2 region, and this is consistent with conclusions drawn from NMR and other studies mentioned earlier (26, 27).
Although the centroid approach to characterize H/DX rate offers a representation of an overall average protein conformation, it is usually reliable and, therefore, has been extensively used in H/DX software tools for automated data processing. Mass spectra may contain, however, useful information other than centroid mass values. The example shown here informs us of the occurrence of unexpected correlated unfolding (EX1 kinetics). Recently, Engen and coworkers (31) reported, using both centroid mass calculations and deuterium distribution analysis, EX1 kinetics in a comparison study of the protein conformational dynamics between E. coli β clamp homodimer and monomeric mutant. Although the centroid mass was sufficient to determine the difference between two proteins, a thorough analysis also revealed a bimodal distribution and additional features of the protein.
The example we present here demonstrates a clear limitation of using only the centroid approach to study the H/D exchange differences in a protein. Unfortunately, mass spectral isotope distributions are not considered in some software tools for H/DX. To recover the information on the multi-modal distributions, the corresponding peptides should be processed separately. Recently, two software tools were proposed to identify and extract automatically any peptides with a bimodal pattern of isotopic distribution (21, 33).
Supplementary Material
An illustration of centroid-based deuterium incorporation determination used to generate deuterium uptake kinetic curve. (A) Isotope distribution of one target peptide from a labeled protein after exposure to deuterium for a predefined period time. The cyan dashed line indicates the centroid mass of the distribution. (B) Table summarizing the centroid mass and the percentage of deuterium incorporation at each on-exchange time. *the theoretical full deuterated centroid mass. The percentage of deuterium incorporation is calculated according to the equation described in the Experimental Section. (C) The kinetic curve shows the deuterium incorporation as a function of on-exchange time.
Acknowledgments
We thank Bruce Pascal from Prof. Patrick R. Griffin’s lab at Scripps Florida for the help with HDX data analysis and helpful discussion. The work was supported by grants from the National Institute of General Medical Sciences (8 P41 GM103422-35) of the NIH.
References
- 1.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1984;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 2.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engen JR, Smith DL. Investigating protein structure and dynamics by hydrogen exchange MS. Anal Chem. 2001;73:256A–265A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 5.Hamuro Y, Coales SJ, Morrow JA, Molnar KS, Tuske SJ, Southern MR, Griffin PR. Hydrogen/deuterium-exchange (H/D-Ex) of PPARgamma LBD in the presence of various modulators. Protein Sci. 2006;15:1883–1892. doi: 10.1110/ps.062103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruning JB, Chalmers MJ, Prasad S, Busby SA, Kamenecka TM, He Y, Nettles KW, Griffin PR. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. doi: 10.1016/j.str.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 7.West GM, Chien EY, Katritch V, Gatchalian J, Chalmers MJ, Stevens RC, Griffin PR. Ligand-dependent perturbation of the conformational ensemble for the GPCR beta2 adrenergic receptor revealed by HDX. Structure. 2011;19:1424–1432. doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chik JK, Schriemer DC. Hydrogen/deuterium exchange mass spectrometry of actin in various biochemical contexts. J Mol Biol. 2003;334:373–385. doi: 10.1016/j.jmb.2003.09.044. [DOI] [PubMed] [Google Scholar]
- 9.Coales SJ, Tuske SJ, Tomasso JC, Hamuro Y. Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography and mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:639–647. doi: 10.1002/rcm.3921. [DOI] [PubMed] [Google Scholar]
- 10.Huang RY, Wen J, Blankenship RE, Gross ML. Hydrogen-deuterium exchange mass spectrometry reveals the interaction of Fenna-Matthews-Olson protein and chlorosome CsmA protein. Biochemistry. 2012;51:187–193. doi: 10.1021/bi201620y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iacob RE, Zhang J, Gray NS, Engen JR. Allosteric interactions between the myristate- and ATP-site of the Abl kinase. PLoS One. 2011;6:e15929. doi: 10.1371/journal.pone.0015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang YJ, Busby SA, Pascal BD, Garcia-Ordonez RD, Bruning JB, Istrate MA, Kojetin DJ, Dodge JA, Burris TP, Griffin PR. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nature Structural & Molecular Biology. 2011;18:556–U172. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rand KD, Jorgensen TJ, Olsen OH, Persson E, Jensen ON, Stennicke HR, Andersen MD. Allosteric activation of coagulation factor VIIa visualized by hydrogen exchange. J Biol Chem. 2006;281:23018–23024. doi: 10.1074/jbc.M602968200. [DOI] [PubMed] [Google Scholar]
- 14.Pascal BD, Chalmers MJ, Busby SA, Mader CC, Southern MR, Tsinoremas NF, Griffin PR. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinformatics. 2007;8:156. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slysz GW, Baker CA, Bozsa BM, Dang A, Percy AJ, Bennett M, Schriemer DC. Hydra: software for tailored processing of H/D exchange data from MS or tandem MS analyses. BMC Bioinformatics. 2009;10:162. doi: 10.1186/1471-2105-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 17.Pascal BD, Chalmers MJ, Busby SA, Griffin PR. HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J Am Soc Mass Spectrom. 2009;20:601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, Marciano D, Novick S, Goswami D, Chalmers MJ, Griffin PR. HDX Workbench: Software for the Analysis of H/D Exchange MS Data. J Am Soc Mass Spectrom. 2012;23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikamanon P, Pun E, Chou W, Koter MD, Gershon PD. “TOF2H”: a precision toolbox for rapid, high density/high coverage hydrogen-deuterium exchange mass spectrometry via an LC-MALDI approach, covering the data pipeline from spectral acquisition to HDX rate analysis. BMC Bioinformatics. 2008;9:387. doi: 10.1186/1471-2105-9-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hotchko M, Anand GS, Komives EA, Ten Eyck LF. Automated extraction of backbone deuteration levels from amide H/2H mass spectrometry experiments. Protein Sci. 2006;15:583–601. doi: 10.1110/ps.051774906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreshuk A, Stankiewicz M, Lou XH, Kirchner M, Hamprecht FA, Mayer MP. Automated detection and analysis of bimodal isotope peak distributions in H/D exchange mass spectrometry using HeXicon. International Journal of Mass Spectrometry. 2011;302:125–131. [Google Scholar]
- 22.Craig TA, Benson LM, Venyaminov SY, Klimtchuk ES, Bajzer Z, Prendergast FG, Naylor S, Kumar R. The metal-binding properties of DREAM - Evidence for calcium-mediated changes in DREAM structure. Journal of Biological Chemistry. 2002;277:10955–10966. doi: 10.1074/jbc.M109660200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Garcia-Ordonez RD, Pascal BD, Burris TP, Dodge JA, Griffin PR. Hydrogen/Deuterium Exchange Reveals Distinct Agonist/Partial Agonist Receptor Dynamics within Vitamin D Receptor/Retinoid X Receptor Heterodimer. Structure. 2010;18:1332–1341. doi: 10.1016/j.str.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 25.Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 26.Lusin JD, Vanarotti M, Li C, Valiveti A, Ames JB. NMR structure of DREAM: Implications for Ca(2+)-dependent DNA binding and protein dimerization. Biochemistry. 2008;47:2252–2264. doi: 10.1021/bi7017267. [DOI] [PubMed] [Google Scholar]
- 27.Osawa M, Dace A, Tong KI, Valiveti A, Ikura M, Ames JB. Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J Biol Chem. 2005;280:18008–18014. doi: 10.1074/jbc.M500338200. [DOI] [PubMed] [Google Scholar]
- 28.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J Am Soc Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 29.Chik JK, Vande Graaf JL, Schriemer DC. Quantitating the statistical distribution of deuterium incorporation to extend the utility of H/D exchange MS data. Anal Chem. 2006;78:207–214. doi: 10.1021/ac050988l. [DOI] [PubMed] [Google Scholar]
- 30.Percy AJ, Rey M, Burns KM, Schriemer DC. Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry-a review. Anal Chim Acta. 721:7–21. doi: 10.1016/j.aca.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 31.Fang J, Engen JR, Beuning PJ. Escherichia coli processivity clamp beta from DNA polymerase III is dynamic in solution. Biochemistry. 2011;50:5958–5968. doi: 10.1021/bi200580b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan CR, Hebling CM, Rand KD, Stafford DW, Jorgenson JW, Engen JR. Conformational transitions in the membrane scaffold protein of phospholipid bilayer nanodiscs. Mol Cell Proteomics. 2011;10:M111, 010876. doi: 10.1074/mcp.M111.010876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kan ZY, Mayne L, Chetty PS, Englander SW. ExMS: data analysis for HX-MS experiments. J Am Soc Mass Spectrom. 2011;22:1906–1915. doi: 10.1007/s13361-011-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An illustration of centroid-based deuterium incorporation determination used to generate deuterium uptake kinetic curve. (A) Isotope distribution of one target peptide from a labeled protein after exposure to deuterium for a predefined period time. The cyan dashed line indicates the centroid mass of the distribution. (B) Table summarizing the centroid mass and the percentage of deuterium incorporation at each on-exchange time. *the theoretical full deuterated centroid mass. The percentage of deuterium incorporation is calculated according to the equation described in the Experimental Section. (C) The kinetic curve shows the deuterium incorporation as a function of on-exchange time.