Abstract
PAPf39 is a 39 residue peptide fragment from human prostatic acidic phosphatase that forms amyloid fibrils in semen. These fibrils have been implicated in facilitating HIV transmission. To enable structural studies of PAPf39 by NMR spectroscopy, efficient methods allowing the production of milligram quantities of isotopically labeled peptide are essential. Here, we report the high-yield expression, as a fusion to ubiquitin at the N-terminus and an intein at the C-terminus, and purification of uniformly labeled 13C- and 15N-labeled PAPf39 peptide. This allows the study of the PAPf39 monomer conformational ensemble by NMR spectroscopy. To this end, we performed the NMR chemical shift assignment of the PAPf39 peptide in the monomeric state at low pH.
Keywords: Aggregation-prone, HIV infection, Escherichia coli expression, Atomic Force Microscopy, Nuclear Magnetic Resonance Spectroscopy, Isotopic Labeling
Introduction
Amyloid fibril formation is a hallmark of a number of human diseases. PAPf39, or Prostatic Acidic Phosphatase fragment of 39 residues (corresponding to residues 248 to 286 in human prostatic acidic phosphatase), is a cationic peptide that forms amyloid fibrils in semen (SEVI) which increase HIV infectivity by up to five orders of magnitude [1]. Understanding the mechanism of PAPf39 fibril formation may provide insights into HIV transmission via semen and lay a foundation for the development of therapeutics against this effect. This approach requires structural information for both the monomeric and fibrillar states of PAPf39 [2, 3].
Nuclear Magnetic Resonance (NMR) spectroscopy is a powerful method for determining the structure and dynamics of biomacromolecules [4-6]. However, modern protein NMR relies on the properties of stable isotopes of carbon (13C) and nitrogen (15N). Current methods of peptide synthesis allow incorporation of isotope enriched amino acids, but synthesis is rather expensive, especially for larger peptides and certain amino acid types. In contrast, isotopic enrichment by bacterial expression in defined media using isotopically labeled metabolites such as 13C glucose and 15N ammonium chloride as the sole source of carbon and nitrogen has been proven as a cost effective strategy to generate milligram quantities of isotopically labeled proteins [4-8]. However, peptides are notoriously difficult to express in bacterial expression systems due to their rapid degradation. Peptides that have an intrinsic propensity to aggregate and, in particular, form amyloid fibrils create additional problems for recombinant expression. To overcome this challenge, various fusion constructs with larger proteins that act as solubility tags and/or protect the peptides from degradation have been used. We have previously expressed soluble peptides as ubiquitin fusion proteins [9-11]. However, when this expression system was used for the amyloidogenic peptide, PAPf39, we observed a significant level of degradation of the peptide from the C-terminus. To alleviate these substantial losses, we attached a self-cleaving intein as a C-terminal fusion. This allowed the high yield expression and easy purification of the PAPf39 peptide free of non-native sequence insertions. The fibril formation of recombinant PAPf39 was characterized by Thioflavin T (ThT) fluorescence assays and Atomic Force Microscopy (AFM) imaging, and NMR chemical shift assignment was performed for the PAPf39 monomer in solution.
Materials and Methods
Chemicals and Plasmids
All reagents were purchased from Sigma-Aldrich and Fisher Scientific unless stated otherwise. The plasmid pTXB1, restriction enzymes, and E. coli expression host BL21(DE3) were purchased from New England Biolabs (Ipswich, MA). DNA purification kits and Ni-NTA resin were obtained from Qiagen (Germantown, MD). DNA oligonucleotides were purchased from Integrated DNA technologies (Coralville, IA). 15N ammonium chloride and 13C glucose were purchased from Cambridge Isotope Laboratories (Andover, MA). SDS-PAGE analysis was done using precast Thermo Scientific Precise 4-20% gel in a Tris-HEPES buffer system, which allows all fragments to be resolved on the same gel.
Design of expression construct
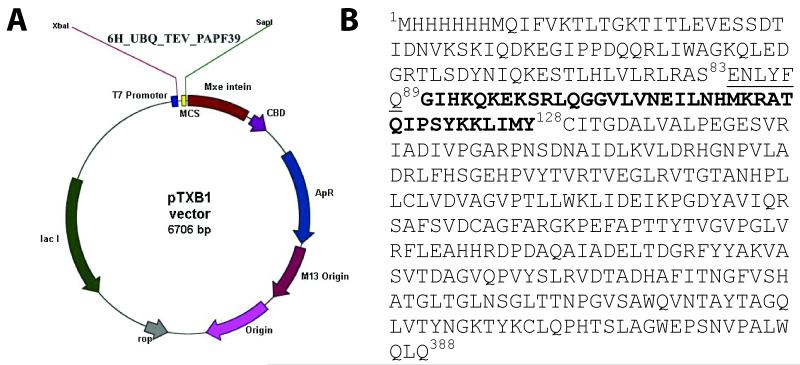
Expressing small peptides in E. coli is commonly a challenging process because the peptides are susceptible to E. coli proteases and/or are toxic to the expression host system. The use of fusion proteins may limit these problems. Our first attempt to express the PAPf39 peptide with ubiquitin attached to the N-terminus of the peptide resulted in C-terminal truncated peptides. Similar effects were observed for other fusion protein constructs such as GFP and MBP. To prevent C-terminal degradation, we created a construct with self-cleavable tag (intein) attached to the C-terminus of the peptide using a pTXB1vector [12]. The vector diagram of the protein construct is shown in Figure 1.
Figure 1.
Expression construct for PAf39 peptide. Panel A. The hexahistidine tagged-ubiquitin gene with a TEV protease cleavage site followed by the PAPf39 coding sequence was inserted into a pTXB1 construct to yield the pTXB1::6H_UBQ_TEV_PAPf39_INTEIN expression plasmid. Panel B. The sequence of the expressed construct: residues 1-83 correspond to the N-terminal 6H-tagged ubiquitin, residues 84-90 comprise the TEV protease recognition motif (cleavage between Q89 and G90), residues 90 to 128 correspond to the PAPf39 peptide, and residues 129 through 388 correspond to the intein sequence.
The protein construct was designed to have a N-terminal hexahistidine (6H) tag for purification, followed by the ubiquitin (UBQ) sequence for solubility, a TEV protease cleavage site to allow removal of the 6H-UBQ tag [9-11], the PAPf39 sequence, and an intein tag to stop the C-terminal degradation of PAPf39. The construct was inserted in a pTXB1 vector using the Xbal and Sap1 restriction enzyme sites. The gene sequence of 6H_UBQ_TEV_PAPf39 (6H-UBQ attached to PAPf39 with a TEV cleavage site in between) was amplified from the pGia vector by PCR with the following primers: forward GGTTTCCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGG and reverse GGTGGTTGCTCTTCCGCAGTACATAATTAATTTTTTATAGGATGG. The amplified PCR product, and pTXB1vector were digested with Xba1 and Sap1, and gel purified.
The digested vector was treated with SAP (Shrimp Alkaline Phosphatase) overnight, which was then inactivated by heating at 65°C. The SAP treated vector and PCR product were ligated using T4 ligase overnight at 4°C. The ligated products were transformed into DH5α competent cells using electroporation. The transformed cells were plated on LB agar plates with 100 μg/ml ampicillin and incubated overnight at 37°C. Individual colonies were picked and grown overnight in LB medium with ampicillin. Plasmid was isolated from the cells followed by sequence verification. The sequence of the verified plasmid used for expression is shown in Figure 1B, where residues 1-83 correspond to the N-terminal 6H-tagged ubiquitin, residues 84-90 comprise the TEV protease recognition motif (cleavage between Q89 and G90), residues 90 to 128 correspond to the PAPf39 peptide, and residues 129 through 388 correspond to the intein sequence followed by a chitin binding domain as encoded by the pTXB1 vector.
Recombinant expression of uniformly 15N- and 13C-enriched PAPf39
The pTXB1-6H_UBQ_TEV_PAPf39_INTEIN expression construct was transformed into E. coli BL21(DE3) and plated on LB agar plates containing 100 μg/ml of ampicillin. A single colony was picked and grown in 2 ml of LB medium to make a starter culture. From the starter culture, a 100 ml overnight culture (Neidhardt medium [13]) was inoculated. For uniform labeling of 15N and/or 13C, the bacteria were grown at 37°C in Neidhardt medium containing 0.5 g/liter of 15NH4Cl and 2 g/liter of 13C glucose as the sole nitrogen and carbon sources [13]. When the cell density reached ~0.8 O.D. at 600 nm, expression of the 6H_UBQ_TEV_PAPf39_INTEIN fusion protein was induced by adding 1 mM isopropyl-D-thiogalactopyranoside and incubating at 25°C for 6 hrs. Cells were harvested and lysed using a French pressure cell. Soluble 6H_UBQ_TEV_PAPf39_INTEIN fusion protein in the cell extract was purified using Ni-NTA affinity resin under native conditions and eluted in 250 mM imidazole buffer following the manufacturer’s protocol.
To remove the intein tag, the pH of the protein solution was adjusted to pH 8.5 using 1 M Tris (pH 8.5) followed by the addition of 125 mM dithiothreitol (DTT). The reaction was incubated at 4°C for 12 to 24 hours and the degree of intein cleavage was assessed by SDS-PAGE. Once cleavage was completed, the DTT was removed by overnight dialysis against 4 liters of modified TEV buffer (50 mM TRIS, 5% Glycerol, 300 mM sodium chloride, pH 8 with 0.5 mM EDTA). To remove the 6H-UBQ tag, 1 mg of TEV was added for every 40 mg of substrate and the reaction mixture was incubated at 4°C for up to 24 hours. Cleavage of the 6H-UBQ tag was assessed by SDS-PAGE. After TEV cleavage, the pool containing PAPf39, intein, and ubiquitin was titrated with concentrated HCl to pH ~3.5-4. This causes precipitation of the intein tag, which can be easily separated from the soluble fraction via centrifugation (~12,000 g for 20 min at 4°C). The supernatant was loaded onto a Sephadex G-75 size exclusion chromatography (SEC) column (2.5 × 150 cm) equilibrated with 5% acetic acid. Since 6H-UBQ and PAPf39 elute very closely together, the fractions containing PAPf39 and a mixture of PAPf39 and 6H-UBQ were pooled and lyophilized together. The lyophilized powder was dissolved in 0.05% TFA in water and injected onto a C18 RP-HPLC (reverse-phase high performance liquid chromatography) column (Discovery Bio Wide pore C18 10 μm, Supelco Sigma-Aldrich, Bellefonte, PA). The column was washed with 0.05% TFA in water at a flow rate of 2 ml/min and PAPf39 was eluted in 5 column volumes during a water/methanol gradient at ~80-85% methanol. Sample purity was verified by SDS-PAGE. The peptide concentration was determined using a molar extinction coefficient of 2,980 M-1 cm-1 at 280 nm. The molecular mass of the purified peptide and the degree of isotope incorporation was determined by MALDI mass spectrometry (Bruker Ultraflex III MALDI TOF/TOF).
Fibril formation and Thioflavin T Fluorescence Assay
Fibril formation assays were conducted in order to test if recombinant PAPf39 forms amyloid fibrils similar to synthetically produced peptides. As described in Ye et al. [2], 2 mg/ml of fresh recombinant un-labeled PAPf39 was agitated at 37°C for two days in 1.5 ml microfuge tubes in phosphate buffered saline pH 7.4 (PBS; 137 mM NaCl, 2.7 mM KCl, 10.14 mM sodium phosphate dibasic, 1.76 mM potassium phosphate monobasic, and 0.2% w/v sodium azide pH 7.4). After 48 hours, 50 μL of incubated sample were withdrawn and mixed with 60 μL of 100 μM Thioflavin T (ThT) in 20 mM sodium phosphate buffer pH 7.4 and 440 μL of PBS pH 7.4 to a total volume of 550 μL in a quartz cuvette. The samples were excited at 440 nm and the emission intensity was monitored between 450 and 550 nm. Appropriate blank measurements were performed without PAPf39 and subtracted.
Atomic Force Microscopy (AFM)
Samples for atomic force microscopy (AFM) were prepared and recorded as described by us previously [2]. Briefly, AFM plates were prepared by spotting 20 μL of sample on freshly cleaved mica and incubating at room temperature for twenty minutes followed by a gentle wash with 4 ml of Milli-Q water to remove extra sample layers and buffer salts. The plates were allowed to dry overnight prior to imaging. AFM amplitude and height images were acquired using AC tapping mode in air at room temperature and humidity on a MFP-3D AFM (Asylum Research, Santa Barbara, CA), using a silicon, Al reflex coated cantilever (AC240TS, Olympus America Inc., Center Valley, PA). AFM images were analyzed in the Igor Pro MFP3D software (Wavemetrics Inc., Portland, OR).
NMR experiments
NMR spectra were acquired at 5°C on Bruker 800-MHz and 600-MHz Avance II spectrometers equipped with cryoprobes with z-axis gradients (Billerica, MA). Complete resonance assignments of the backbone amides for all residues, with the exception of Gly1 and Pro31 that do not have amide protons, were obtained. This was done through the analysis of atomic correlations detected in two-dimensional 1H-15N and 1H-13C single quantum coherence (HSQC), and three-dimensional 15N-separated TOCSY, 15N-separated NOESY, HNCO, HNCACB, HNCOCACB, (H)CC(CO)NH, and H(CCCO)NH spectra using the auto-assignment program MONTE [14]. NMR samples were prepared at a final concentration of approximately 1mM PAPf39 by dissolving lyophilized protein in buffer containing 5% acetic acid, pH 2.0, and 10% D2O [9, 12]. These buffer conditions were chosen because, as shown previously [2, 3] using analytical centrifugation, PAPf39 remains monomeric in broad range of concentrations for extended periods of time under these conditions. The NMR data were processed using TopSpin 2.1 and analyzed using the software package Sparky [15].
Results and discussion
Expression and purification of isotopically labeled PAPf39
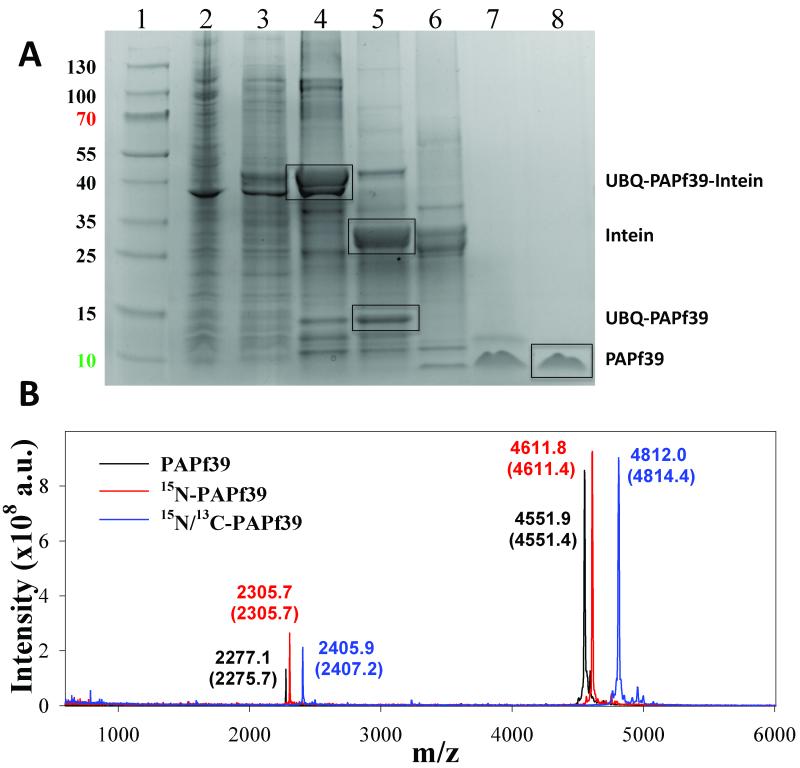
The expression system for the 6H_UBQ_TEV_PAPf39_INTEIN fusion protein produced protein product at high levels that remained in the soluble fraction of the cell extract (see Figure 2). The 6H_UBQ_TEV_PAPf39_INTEIN fusion protein was efficiently isolated from the cell lysate using Ni-NTA affinity resin under native conditions. Removal of the C-terminal INTEIN and N-terminal 6H_UBQ tags was achieved by promoting intein self-cleavage followed by digestion using TEV protease, respectively. As shown in Figure 2, the fusion protein was found to readily and specifically undergo the intein self-cleavage and proteolytic digestion to near completion. Therefore, the TEV protease was able to access and effectively cleave the engineered restriction site and the intein was able to properly fold and undergo self-cleavage without requiring alteration of the native sequence of the PAPf39 peptide. The final yield was ~7 mg (1.4 ml of 1 mM 15N/13C peptide sample for NMR) of pure PAPf39 peptide per 3 L of Neidhardt minimal media (0.5 g/liter of 15NH4Cl and 2 g/liter of 13C glucose). The measured molecular masses for unlabeled, 15N-labeled, and 15N/13C-labeled PAPf39 were determined by MALDI mass spectrometry (Figure 2). The mass spectrometry data contain peak envelopes with m/z distributions consistent with near complete degrees of isotope incorporation.
Figure 2.
Purification of the recombinant PAPf39 peptide. Panel A. Coomassie blue stained SDS-PAGE analysis of recombinant soluble lysates in E. coli strain BL21(DE3): (Lane1: Thermo Scientific Page Rules Prestained Protein Ladder 10-130 kDa) (lane 2: whole cell lysate), (lane 3: soluble protein fraction), (lane 4: fractions after Ni-NTA), (lane 5: cleavage of the intein), (lane 6: TEV-protease digest), (lane 7: PAPf39 containing fraction after SEC), (lane 8: pure PAPf39 fraction after RP-HPLC). The position of migration of all fragments, including PAPf39, is shown. Panel B. Mass spectrometry analysis of purified PAPf39: unlabeled PAPf39 (black line), 15N-labeled PAPf39 (red line), and 15N/13C-labeled PAPf39 (blue line). Determined masses are shown above each peak, with expected masses given in parenthesis.
Fibril formation in recombinant PAPf39
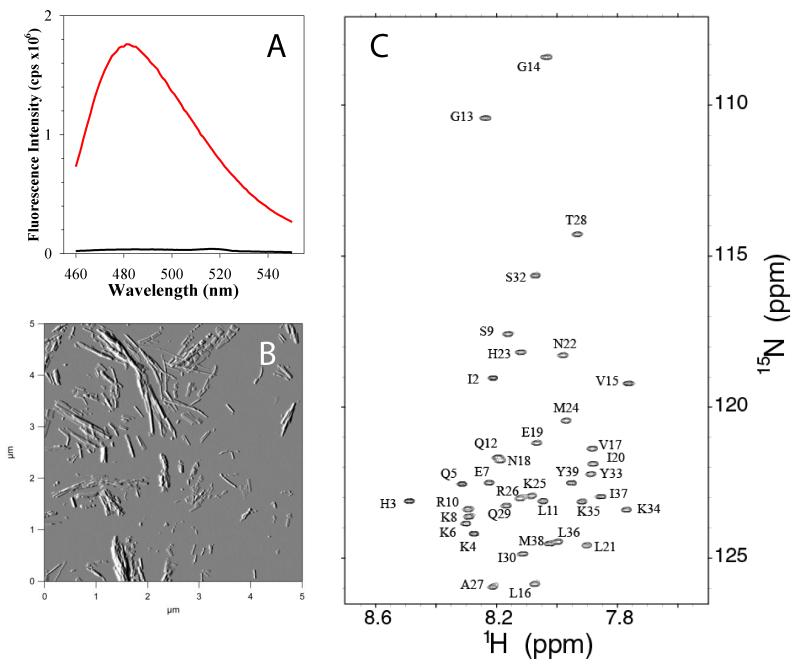
Fibril formation of recombinant PAPf39 was assayed using a previously developed protocol based on the ThT fluorescence and AFM imaging [2]. Figure 3A shows the change in ThT fluorescence after 48 hours of incubation of PAPf39 peptide at 37°C in PBS pH 7.4. The dramatic increase in fluorescence intensity is indicative of fibril formation. This is supported by AFM amplitude images that show an abundance of fibrils at 48 hours (Figure 3B). These results suggest that recombinantly expressed PAPf39 shows the same properties as chemically synthesized peptide [2, 3].
Figure 3.
Biophysical characterization of the recombinant PAPf39 peptide. Panel A. ThT fluorescence spectra before (black line) and after (red line) binding to PAPf39 fibrils. Panel B. Morphology of the PAPf39 fibrils determined by AFM. Panel C. 1H-15N chemical shift correlation spectrum (HSQC) of the PAPf39 peptide with the complete assignments of the backbone 15N amide protons.
NMR spectroscopy and assignment of chemical shifts
The high quantities of isotopically enriched PAPf39 enabled the rapid and sensitive acquisition of high-quality multidimensional heteronuclear NMR spectra. The lineshapes of resonances detected in one dimensional 1H and two-dimensional 1H-15N HSQC spectra of PAPf39 are consistent with PAPf39 behaving as a monomer in solution and free of significant aggregation. As shown in Figure 3C, the two-dimensional 1H-15N spectrum is largely free of peak overlap and overcomes the extensive chemical shift degeneracy for amide protons. Chemical shift assignments of PAPf39 resonances were completed through the analysis of correlations detected by standard triple resonance techniques. Resonances for backbone atoms were assigned using the auto-assignment program MONTE [4, 14] with the completion of the resonance assignments for side-chain atoms being manually performed.
Conclusions
A method for the recombinant expression of PAPf39, a 39-residue peptide that forms amyloid fibrils, was developed. Recombinant PAPf39 forms amyloid fibrils as shown by both ThT fluorescence assays and AFM imaging. High yields of PAPf39 from expression in minimal medium allowed isotopic labeling of the peptide for NMR experiments. Furthermore, NMR chemical shift assignment was completed. We suggest that our method will be useful to generate milligram quantities of other peptides that exhibit aggregation, toxicity toward, and/or rapid degradation within the host bacterium.
Highlights.
A protocol was developed to prepare pure isotopically labeled PAPf39 peptide for NMR
PAPf39 peptide was purified by intein mediated self-cleavage and TEV protease cleavage
Recombinant PAPf39 showed amyloid fibril formation
Chemical shift for backbone atoms were assigned
Acknowledgments
We thank Dr. Dmitri Zagorevski for assistance with mass spectrometry. We acknowledge the use of the Core Facilities in the Center for Biotechnology and Interdisciplinary Studies at Rensselaer Polytechnic Institute. This work was supported by the National Institute of General Medical Science of the National Institutes of Health under award number 1R21GM101134 (to G.I.M.). K.C.F. is supported by a National Science Foundation Graduate Research Fellowship.
Abbreviations used
- PAPf39
a 39residue peptide fragment from human prostatic acidic phosphatase
- GFP
green fluorescent protein
- MBP
maltose binding protein
- UBQ
ubiquitin
- TEV
tobacco etch virus
- PAGE
polyacrylamide gel electrophoresis
- SEC
size exclusion chromatography
- PBS
phosphate buffered saline
- Ni–NTA
nickel–nitrilotriacetic acid
- TFA
Trifluoroacetic acid
- 6H
hexahistidine tag
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Munch J, Rucker E, Standker L, Adermann K, Goffinet C, Schindler M, Wildum S, Chinnadurai R, Rajan D, Specht A, Gimenez-Gallego G, Sanchez PC, Fowler DM, Koulov A, Kelly JW, Mothes W, Grivel JC, Margolis L, Keppler OT, Forssmann WG, Kirchhoff F. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell. 2007;131:1059–1071. doi: 10.1016/j.cell.2007.10.014. [DOI] [PubMed] [Google Scholar]
- [2].Ye Z, French KC, Popova LA, Lednev IK, Lopez MM, Makhatadze GI. Mechanism of fibril formation by a 39-residue peptide (PAPf39) from human prostatic acidic phosphatase. Biochemistry. 2009;48:11582–11591. doi: 10.1021/bi901709j. [DOI] [PubMed] [Google Scholar]
- [3].French KC, Makhatadze GI. Core Sequence of PAPf39 Amyloid Fibrils and Mechanism of pH-Dependent Fibril Formation: The Role of Monomer Conformation. Biochemistry. 2012 doi: 10.1021/bi301406d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rule GS, Hitchens TK. Fundamentals of Protein NMR Spectroscopy. Springer; 2005. [Google Scholar]
- [5].Palmer AG, 3rd, Massi F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem Rev. 2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- [6].Wuthrich K. NMR studies of structure and function of biological macromolecules (Nobel lecture) Angewandte Chemie (International ed. 2003;42:3340–3363. doi: 10.1002/anie.200300595. [DOI] [PubMed] [Google Scholar]
- [7].Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- [8].Sgourakis NG, Yan Y, McCallum SA, Wang C, Garcia AE. The Alzheimer’s peptides Abeta40 and 42 adopt distinct conformations in water: a combined MD/NMR study. Journal of molecular biology. 2007;368:1448–1457. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Patel MM, Sgourakis NG, Garcia AE, Makhatadze GI. Experimental test of the thermodynamic model of protein cooperativity using temperature-induced unfolding of a Ubq-UIM fusion protein. Biochemistry. 2010;49:8455–8467. doi: 10.1021/bi101163u. [DOI] [PubMed] [Google Scholar]
- [10].Richardson JM, Makhatadze GI. Temperature dependence of the thermodynamics of helix-coil transition. Journal of molecular biology. 2004;335:1029–1037. doi: 10.1016/j.jmb.2003.11.027. [DOI] [PubMed] [Google Scholar]
- [11].Sgourakis NG, Patel MM, Garcia AE, Makhatadze GI, McCallum SA. Conformational dynamics and structural plasticity play critical roles in the ubiquitin recognition of a UIM domain. Journal of molecular biology. 2010;396:1128–1144. doi: 10.1016/j.jmb.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Evans TC, Jr., Benner J, Xu MQ. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. Journal of bacteriology. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hitchens TK, Lukin JA, Zhan Y, McCallum SA, Rule GS. MONTE: An automated Monte Carlo based approach to nuclear magnetic resonance assignment of proteins. Journal of biomolecular NMR. 2003;25:1–9. doi: 10.1023/a:1021975923026. [DOI] [PubMed] [Google Scholar]
- [15].Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2012. http://www.cgl.ucsf.edu/home/sparky/ [Google Scholar]