1. Introduction
Transcription of the ~200 copies of ribosomal RNA genes (rDNA), present in the mammalian haploid genome, by the dedicated RNA Polymerase I (Pol I) enzyme and subsequent processing of the ribosomal RNA (rRNA) are fundamental control steps in the synthesis of functional ribosomes (reviewed in [1–3]). If rRNA synthesis is inhibited, cells undergo cell cycle arrest associated with apoptosis, senescence or autophagy depending on the cell type. Conversely, accelerated rRNA synthesis tightly correlates with cellular proliferation rates. Not surprisingly, it is becoming increasingly clear that dysregulation of Pol I transcription and ribosome biogenesis is linked to the etiology of a broad range of human diseases.
Perhaps the best recognized diseases associated with dysregulated ribosome biogenesis are caused by loss of function mutations in the molecular constituents of the ribosome or factors intimately associated with Pol I transcription and processing collectively termed ribosomopathies (Table 1)[4]. This class of genetic diseases includes those caused by mutations in ribosomal proteins for example Diamond-Blackfan anemia and 5q-syndrome[4–7]. Alternatively they are associated with mutations in modulators or components which impact on Pol I transcription, such as is the case for Treacher Collins Syndrome[8–10], or Blooms and Werner syndrome[11–13]. Other ribosomopathies are associated with mutations that affect rRNA processing and modification such as Shwachman-Diamond Syndrome[14], Dyskeratosis Congenita[15], Cartilage Hair Hypoplasia[16, 17], North American Indian childhood cirrhosis[18, 19], Bowen-Conradi syndrome[20] and alopecia, neurological defect and endocrinopathy (ANE) Syndrome[21] (Table 1). Ribosomopathies are generally rare and treatment options are unfortunately extremely limited tending to be more palliative than curative.
Table.
| Genetic diseases associated with dysregulation of Pol I transcription | |||||||
|---|---|---|---|---|---|---|---|
| Disease | Gene | Molecular function at rDNA |
Consequences of impaired function |
Clinical manifestations | Inheritance | OMIM # | References |
| Treacher Collins Syndrome | TCOF1 | interacts with UBF and Pol I | reduced rDNA transcription and cell growth | craniofacial abnormalities, hearing loss | autosomal dominant | #154500 | [8–10] |
| POLRIC | subunit of Pol I and III | [29] | |||||
| POLRID | subunit of Pol I and III | [29] | |||||
| Blooms Syndrome | BLM | interacts with rDNA to promote Pol I transcription | reduced genomic stability, at the rDNA suppression of rDNA transcription | growth retardation, premature aging, juvenile cataracts, skin atrophy, diabetes mellitus cancer predisposition | autosomal reccessive | #210900 | [11,12] |
| Werner Syndrome | WRN | interacts with the Pol I transcriptional complex to mediate promoter clearance | suppression of rDNA transcription | growth retardation, premature aging, juvenile cataracts, skin atrophy, cancer predisposition | autosomal reccessive | #277700 | [12,13] |
| Cockayne Syndrome | ERCC6 | interacts with Pol I and SL-1 | suppression of rDNA transcription | severe growth retardation, mental retardation, hearing loss, cataracts neurological dysfunction | autosomal reccessive | #133540 | [49,50] |
| CSB interacts with TTF1 to activate rDNA chromatin | |||||||
| Siderius X-linked Mental Retardation | PHF8 | interacts with the Pol I transcriptional complex to activate rDNA transcription | reduced rRNA synthesis | mental retardation craniofacial abnormalities | X-linked recessive | #300263 | [66,67] |
| Roberts Syndrome | ESCO2 | Not defined | reduced rRNA synthesis | growth retardation, craniofacial abnormalities, limb malformations | autosomal reccessive | #268300 | [68] |
| Cordelia de Lange Syndrome | NIPBL | Not defined | reduced rRNA synthesis | growth retardation, craniofacial abnormalities, upper limb malformations, hirsutisin, mental retardation gastrointestinal abnormalities, | autosomal dominant | #268300 | [68] |
| SMC1A | |||||||
| SMC3 | |||||||
| 1. Periventricular Nodular Heteropia | FLNA | interacts with the Pol I transcriptional complex to suppress transcription | altered rRNA synthesis | developmental malformations of the brain, skeleton and heart | X-linked dominant | #300049 | [77] |
| 2. Otopalatodigital Syndrome | #311300 | ||||||
| 3. Frontometaphyseal Dysplasia | #305620 | ||||||
| 4. Melnick-Needles Syndrome | #309350 | ||||||
| 5. X-linked Cardiac Valvular Dystroph | #314400 | ||||||
| Genetic diseases associated with ribosome biogenesis | |||||||
|---|---|---|---|---|---|---|---|
| Diamond Blackfan anemia | RPS19, RPS24 | components of ribosomal subunits | impairment of 18S rRNA processing and 40S ribosomal subunit formation | growth retardation, craniofacial abnormalities, thumb abnormalities, macrocytic anemia | autosomal dominant | #105650 | [5] |
| RPS17, RPL35A | |||||||
| RPL5, RPL11 | |||||||
| RPS7, RPL36 | |||||||
| RPS15, RPS27A | |||||||
| 5q-Syndrome | PS14 | maturation of 40S ribosomal subunits | deregulated biogenesis ribosome leading to impaired protein synthesis and translation | Macrocytic anemia, hypolobulated micromegakaryocytes | autosomal dominant | #153550 | [7] |
| Shwachman-Diamond Syndrome | SBDS | rRNA processing | block in rRNA maturation and decreased levels of the 60S ribosomal subunit | growth retardation, neutropenia, panceatic insufficiency, skeletal abnormalities, cancer | autosomal recessive | #260400 | [14] |
| X-linked Dyskeratosis Congenita | DKC1 | components of H/ACA small nucleolar RNAs and telomerase RNA | H/ACA small nucleolar RNAs pseudouridiylation defects, telomerase deficiency | premature aging, cytopenias, skin hyperpigmentation, nail dystrophy, cancer predisposition | X-linked recessive | #305000 | [15] |
| Dyskeratosis Congenita | NOP10 | autosomal recessive | #224230 | ||||
| NHP2 | autosomal recessive | #224230 | |||||
| Anauxetic Dysplasia Cartilage Hair Hypoplasia | RMRP | noncoding RNA, component of Rnase MRP | maturation of 5.8S rRNA of 60S ribosomal subunit | severe growth retardation, mild mental retardation, reduced chondrocytes | autosomal recessive | #607095 | [16,17] |
| Bowen-Conradi Syndrome | EMG1 | 18S assembly factor, methyltransferase | methylates the 18S rRNA | impaired pre- and post-natal growth, psychomotor retardation | autosomal recessive | #211180 | [20] |
| North American Indian Childhood Cirrhosis | CIRH1A | component of the SSU processome | maturation of 18S and 25S rRNAs | transient neonatal jaundice that progresses to biliary cirrhosis | autosomal recessive | #604901 | [18,19] |
In addition to ribosomopathies, dysregulation of Pol I activity is common in diseases associated with profound changes in cellular growth such as cardiac hypertrophy, atrophy or cancer. Indeed abnormal nucleoli, the site of Pol I transcription, has been used as a marker of aggressive tumours for over 100 years, well before the function of the nucleolus was known. In contrast to ribosomopathies, altered Pol I transcriptional activity in these diseases largely results from dysregulated upstream signaling pathways and consequently altered expression or activity of factors directly involved in Pol I transcription. In the case of cancer, this includes hyperactivation of classic oncogenes and upstream oncogenic signaling pathways, (e.g., epidermal growth factor (EGF) receptor, c-MYC and mammalian target of rapamycin (mTOR)/PI3K/AKT), or release from repression by tumour suppressors, (e.g., p53, retinoblastoma protein (pRb)). While it has been debated for some time whether the dysregulation of Pol I is a cause or a consequence in diseases such as cancer, recent studies have gone a long way to answer this question[22, 23]. Using genetic approaches and small molecule inhibitors of Pol I activity Bywater et al.,[22] provided definitive proof that hyperactive Pol I transcription is required for the malignant phenotype of certain cancers and targeting Pol I can be used as a therapeutic approach to treat malignancy with few side effects on normal cells[22].
These examples illustrate how far the concept of dysregulated Pol I transcription and its contribution to human disease has come in the past 10 years. However, in reality we are just at the beginning of the long journey to fully understanding the etiology and development of the diverse array of pathologies and proliferative disorders associated with ribosomopathies and deranged Pol I transcription. While a number of recent publications have covered ribosomopathies associated with mutation in ribosomal proteins and processing /assembly factors (see reviews[4, 24, 25]), here we review our current knowledge of human diseases specifically associated with dysregulation of Pol I transcription and its associated regulatory apparatus, including some cases where this dysregulation is directly causative. Through out the review, for clarity, we will utilize the mammalian/human terminology for the Pol I transcription factors. We will also provide insight into and discussion of possible therapeutic approaches to treat patients with dysregulated Pol I transcription.
2. Diseases with mutations in factors directly associated with RNA Polymerase I transcription
A number of factors have been identified that co-immunoprecipitate with the Pol I transcription components and whose encoding genes when mutated result in both dysregulated Pol I transcription and a specific human disease syndromes (Table 1; Figure 1). These include the proteins treacle, blooms syndrome helicase, werner helicase, cockayne syndrome B, plant homeodomain finger protein 8 and Filamin A which underlie the diseases Treacher Collins Syndrome, Blooms and Werner syndrome, Cockayne Syndrome, Siderius X-linked mental retardation and a group of Filamin A associated diseases. Other proteins, which either regulate or are structural components of the cohesin complex, have also been shown to modulate Pol I transcription. However, their mechanism of action with respect to Pol I transcription has not been well established. These include the proteins nipped-B-like (NIPBL), structural maintenance of chromosomes (SMC) 1A, SMC3 and establishment of cohesin 1 homologue 2 (ESCO2), which when mutated are associated with the cohesinopathy diseases, Cornelia de Lange Syndrome or Roberts Syndrome.
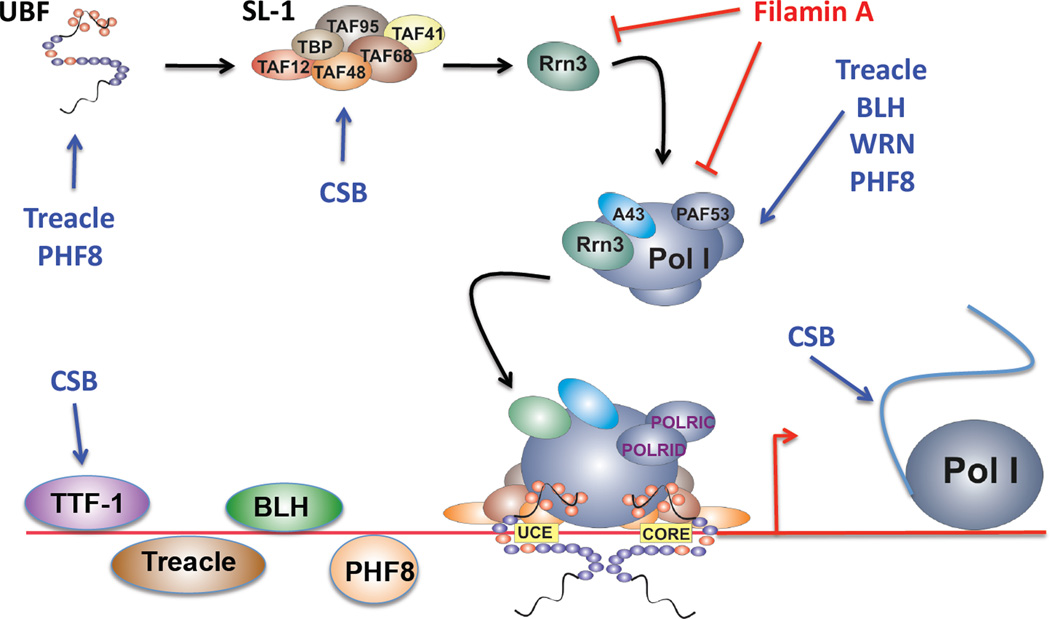
Figure 1. Regulation of RNA Polymerase I transcription by factors whose mutation is associated with genetic diseases.
A schematic illustrating the sites of action where disease associated factors, including treacle, blooms syndrome helicase (BLM), werner helicase (WRN), cockayne syndrome B (CSB), plant homeodomain finger protein 8 (PHF8) and Filamin A, regulate RNA Polymerase I transcription. UBF: upstream binding factor; SL-1: selectivity factor 1; Pol I: RNA polymerase I; TTF-1: termination transcription factor 1; UCE: upstream control element;
Interestingly, while these diseases are all unique, collectively they possess many overlapping symptoms. For instance, the majority of these patients present with symptoms of aging including hearing loss, cataracts and decreased subcutaneous fat. Two thirds have neuronal issues typically stemming from altered migration or development of the progenitor cells. Five of these diseases exhibit malformations in the skeletal or facial tissues and also limb or body growth defects. It is tempting to speculate that these overlapping symptoms provide an insight into the consequence of dysregulated Pol I transcription, where as the unique features of each disease are due to the "other" functions of the disease causing protein in question.
For ribosomopathies associated with ribosomal protein mutations (eg., Diamond Blackfan anaemia, 5q- syndrome) or those associated with rRNA processing defects (eg., North American Indian childhood cirrhosis, Bowen-Conradi syndrome) the causative lesions have been definatively linked to a ribosome biogenesis factor (Table 1). In contrast, for Pol I transcription-linked diseases, many of the proteins we will discuss were first described in the context of Pol II transcription or DNA damage/p53 mediated responses. It has only been in the last 10 years that their role in Pol I transcription has been uncovered. As such there are varying degrees of confidence for the direct impact of their role in dysregulation of Pol I activity on disease etiology. However, in a few cases the evidence for a causative role is overwhelming. For example, in the case of Treacher Collins Syndrome, mouse models have confirmed that deletion of TCOF1 is sufficient for development of this disease[26–28]. Moreover, despite some non-Pol I functions attributed to the TCOF1 encoded protein treacle, Treacher Collins Syndrome patients without TCOF1 mutations exhibited mutations in two subunits common to Pol I and III, POLRIC (AC40 in yeast) and POLRID (AC19 in yeast)[29]. This strongly suggests that the disease primarily results from defects in Pol I activity and thus can be considered a bone fide ribosomopathy. Of course the list of potential ribosomopathy causing genes is not static and with the proteomic analysis of the nucleolus coming to fruition[30, 31] and the advent of massively parallel sequencing, new gene candidates whose mutation can induce disease through defects in Pol I transcription are likely to be described in the near future.
2.1 Treacher Collins Syndrome (TCS)
TCS (Treacher Collins-Franceschetti syndrome) is classified as a mandibulofacial dysostosis and is extremely rare (1/50,000 live births)[32, 33]. TCS is an autosomal dominant disorder with 60% of the cases resulting from new or de novo mutations rather than being hereditary[32, 33].
TCS results from abnormal differentiation of the first and second pharyngeal arches during the fourth week of fetal development. Specifically the defect lies with the neuroepitheium which gives rise to the neural crest cells, a migratory cell population from which the cartilage, bone and connective tissue of the head and face are generated[9]. Thus TCS is characterized by abnormal craniofacial development in early embryogenesis and also hearing loss. Distinguishing features include cleft palate, down slanting palpebral fissures, coloboma of the lower eyelid, micrognathia, microtia, hypoplastic zygomatic arches and macrostomia[9].
Most cases of TCS are caused by mutations in one copy of the TCOF1 gene (81–93% of cases)[34]. No patients have been reported with both copies of the gene mutated. Over 120 different mutations have been described, predominantly in the coding region on the gene resulting in an aberrant, truncated protein, which mislocalizes to the cytoplasm[33, 35]. This observation was supported by the phenotype of TCOF1+/− mouse embryos, which showed similar craniofacial defects and growth retardation as the human disease[26–28]. Those patients with an absence of a TCOF1 mutation instead exhibited mutations in the subunits common to Pol I and Pol III (POLRIC and POLRID)[29]. As described above together this data suggests that TCS is a heterogeneous disease primarily resulting from defects in ribosome biogenesis and thus is a true ribosomopathy.
TCOF1 encodes the serine/alanine-rich phosphoprotein treacle which has been reported to associate with the centrosomes, kinetochores[36], the Nop65p-associated pre-ribosomal ribonucleoprotein (pre-rRNP) complex, and localize to the dense fibrillar centre (DFC) of the nucleolus[9]. Treacle’s role as a centrosome- and kinetochore-associated protein is mediated by its interaction with Polo-like kinase 1 (Plk1) and was found to be critical for spindle fidelity and mitotic progression. Thus, when absent, spindle orientation and cell cycle progression is disrupted, which perturbs maintenance, proliferation and localization of the neural progenitors during cortical neurogenesis[36]. As a component of the human Nop65p-associated pre-rRNP complex, treacle participates in 2'-O-methylation of pre-rRNA. This occurs at an early stage in processing and is important for ribosome maturation[9].
However, it is treacle's localization to the nucleolus and the nucleolar organizer region (NOR) that suggested a role in Pol I transcription. Indeed, treacle promotes Pol I transcription by interacting with the transcription factor upstream binding factor (UBF)[8, 10], rDNA chromatin[10], and the Pol I enzyme itself (Figure 1)[9]. Conversely, treacle knock down leads to inhibition of rDNA transcription and cell growth, which was associated with dispersion of Pol I and UBF from the nucleolus (Figure 1)[8]. Consistent with an essential function for treacle in rDNA transcription, mice haploinsufficient for TCOF1 exhibited reduced ribosomal production associated with decreased cell proliferation, increased neuroepithelial apoptosis and deficient formation of migrating cranial neural crest cells which are responsible for the craniofacial anomalies characteristic of TCS[27].
2.2 Blooms (BS) and Werner Syndrome (WRNS)
BS and WRNS are both rare autosomal recessive disorders with very similar characteristics (Table 1). Specifically, they both exhibit severe growth retardation (proportional dwarfism), cancer predisposition (particularly to sarcomas), juvenile cataracts, atrophy of the skin, faciocranial abnormalities, genome instability and premature aging, which characterizes them as Progeria diseases. They also develop hypogonadism, osteoporosis, diabetes mellitus, and arthrosclerosis. Blooms syndrome is characterized by a high level of sister chromatid exchange and dysregulated insulin signaling, manifested as insulin resistance in children or insulin-resistance diabetes mellitus in young adults[37]. Onset typically occurs in the third decade of life with a significant decline in health resulting in death at ~50[38]. A number of these human phenotypes have been recapitulated in knock out mouse models[39–41].
The cause of BS is a mutation in the blooms syndrome helicase (BLH) encoding gene (BLM) and in the case of WRNS a mutation in the WRN gene resulting in the production of a truncated protein (WRN). BLH and WRN are both nuclear helicases and members of the RecQ subfamily of ATP dependent 3'–5' DNA helicases that localize to the nucleolus in S phase[42] or when quiescent cells are re-activated[12]. BLH and WRN co-localized in the nucleolus, while they do not interact by immunoprecipitation[42], they both positively regulate rDNA transcription by Pol I[11–13].
WRN was shown to be required for vascular EGF (VEGF), fibroblast GF-β (FGF-β) and EGF stimulation of Pol I, but not for platelet derived GF (PDGF)-AB or insulin GF-1 (IGF-1)[13] presumably due to differences in the signaling pathways. Low dose actinomycin D treatment, which selectively inhibits rDNA transcription, caused both helicases to relocate out of the nucleolus[11, 12]. Similarly, serum starvation promotes WRN relocation from the nucleolus[12]. In quiescent cells any residual nucleolar WRN was found exclusively bound to the remaining active rDNA[13]. Consistent with direct roles in the regulation of rDNA transcription, both helicases co-immunopreciptiate with subunits of Pol I (Figure 1)[11, 12]. BLH was also shown to associate with telomeres and the rDNA repeats by chromatin immunoprecipitation, binding in the non-transcribed spacer region of the rDNA, which presumably are the sites of replication initiation[43]. Recently, it's been shown that BLH directly interacts with the rDNA and "unwinds these GC-rich rDNA-like substrates that normally inhibit transcription"[11]. WRN also appears to be associated with Pol I to mediate promoter clearance rather than elongation[38].
While both BLM and WRN appear to contribute to the regulation of rDNA transcription, critical details are still missing as to how these helicases mechanistically modulate their effects on Pol I transcription and how they themselves are regulated. One publication demonstrated that WRN activity is inhibited by the serine/threonine kinase DNA-dependent protein kinase, and stimulated by p300 acetylation[38]. Understanding this regulation and adequately assessing the effect of BLM and WRN mutations on the modulation of Pol I transcription is important if we are to establish the role of rDNA transcription in the etiology of Blooms and Werner Syndromes. Furthermore, these studies will be necessary if we are to uncover the relative contibution of inhibition of Pol I activity to the disease phenotype in comparison to other functions of the two proteins. In this vein, it should be noted that these two helicases have also been shown to be important for p53 regulation, telomere maintance, DNA repair and Pol II transcription, with an overriding influence on genomic stability[37, 38, 44, 45].
2.3 Human Cockayne Syndrome (CS)
CS is an inherited autosomal recessive disease, which is extremely rare. For example, only 2.7 cases occur per million births in Western Europe[46]. In the majority of cases, the individual is characterized by severe postnatal growth failure (cachectic dwarfism), premature aging and progressive neurological dysfunction (demyelination, brain atrophy, calcification), that results in physical and mental retardation. The symptoms vary but can include photosensitivity, microcephaly, very low body weight, gait defects, ocular and skeletal abnormalities, high pitched voice, and dental caries. A number of symptoms also phenocopy aging, such as retinal degeneration, sensorineural hearing loss, cataracts and loss of subcutaneous fat. The majority of afflicted people die in childhood (average survival 12 years) or as they age they will progressively lose skills such as walking, talking, sitting and are often prone to pneumonia, kidney and liver dysfunction. To date there is no cure and treatment is palliative[46].
CS is caused by mutations in ERCC8 or ERCC6, which encode the cockayne syndrome WD repeat protein (CSA) and cockayne syndrome B protein (CSB) respectively. The majority of cases (62–80%) are attributed to mutations in ERCC6[47]. Both CSA and CSB are required for the transcription coupled repair branch of the nucleotide excision repair pathway[48] and initially the phenotype for CS was ascribed to these roles. However, Bradsher et al.[49] demonstrated a role for CSB in regulating Pol I transcription. They identified the presence of a complex containing CSB, Pol I, selectivity factor 1 (SL-1), two transcription initiation factor II H (TFIIH) subunits (the helicases xeroderma pigmentosum D (XPD) and XPB) and XPG, which modulated Pol I transcription (Figure 1). Moreover, mutations in CSB, XPB and XPD destablized this complex and reduced rRNA synthesis[49]. Subsequently, CSB, XPD and XPB were implicated in the positive regulation of elongation by Pol I[50, 51]. It has also been reported that CSB recruits DNA repair and chromatin remodeling factors to UV-stalled Pol I in vivo[48]. Yuan et al.[52] reported data consistant with an alternative mechanism. They demonstrated that CSB binds the Pol I transcription terminator factor, TTF-1 (Figure 1), to form a complex at the active rRNA genes with Pol I and the histone methyltransferase G9a. In the absence of CSB this complex is disrupted and pre-rRNA synthesis reduced. Since CSB knockout mice are viable this would suggest that CSB may function more as a facilitator, rather than as an essential component in Pol I transcription[52]. Alternatively, another protein may compensate for CSB under these circumstances.
Like BS and WRNS, there is also evidence that CSB modulates and is modulated by p53 levels[53–55]. CSB knockdown also causes metaphase chromosome fragility of human small RNA U1, U2 and 5S rRNA genes suggesting a further role, this time in Pol II and Pol III transcription[56]. Thus CSB can affect transcription by all three Pol's and further investigation will be required to elucidate the relative contributions of dysregulation of each polymerase. Strikingly however, a number of the hallmarks characterizing CS, including defective growth and neuronal development and premature aging, are similar to those observed in TCS, consistent with the model that dysregulation of Pol I is a significant determinant of these phenotypes.
2.4 Siderius X-linked mental retardation (XLMR-CL/P)
XLMR-CL/P itself is rare, although it is a subset of the much larger group of diseases, X-linked mental retardation (XLMR). XLMR is considered a common cause of intellectual disability which affects about 1.6/1000 males. Since it is an X-linked inherited recessive trait female carriers only occasionally display symptoms and these are mild[57]. To put this in perspective, mental retardation affects 1–3% of the population, of these cases 25–35% have a genetic background and 25–30% of these are classified as XLMR[58]. To date ~90 genes have been implicated as the causative genetic abnormalities in XLMR. Typically these genes encode proteins involved in transcription regulation, either functioning as transcription factors or chromatin structure modifiers[59].
XLMR-CL/P is a syndrome characterized by mild mental retardation and facial issues such as the presence of a cleft lip and palate, broad nasal tip and large hands[57, 58, 60] very similar to those features observed in TCS. XLMR-CL/P is caused by a mutation in the PHF8 gene. Plant homeodomain finger protein 8 (PHF8) is a histone lysine demethylase belonging to the Jmjc domain-containing histone demethylase family[61, 62].
PHF8 is thought to function in chromatin remodelling thus affecting global gene transcription[61, 63, 64]. In particular, PHF8 is instrumental in the regulation of neuronal differentiation[57, 63], cell survival, brain and jaw development in Zebrafish[64], and regulation of the cell cycle[65]. Potentially, the mental retardation symptoms associated with XLMR-CL/P are accounted for by PHF8 role in early brain development; via regulation of the transcription of genes essential for neurogenesis, particularly in the neuronal populations involved in memory and learning[59]. Importantly however, with respect to Pol I, PHF8 has 4–6 nucleolar localization sequences (NLS)[57, 60] and localizes to the nucleolus [66]. In the nucleolus PHF8 binds to hypomethylated rRNA genes (transcriptionally competent), co-localizing with the euchromatin histone marker trimethylated histone H3 K4 (H3K4me3) throughout the rDNA repeat[66]. Knockdown of PHF8 reduced rRNA expression, which correlated with decreased dimethylated H3K9 (H3K9me2) levels at the rDNA promoter and was reversed by overexpression of PHF8[67]. Consistent with a role in the control of rRNA synthesis, PHF8 was shown to co-immunoprecipate with Pol I and UBF, and was required for Pol I localization at the promoter (Figure 1)[66]. It is tempting to speculate that perhaps treacle, which also regulates the association of UBF with Pol I, may cooperate with PHF8 in this function. However, as is the case for the other ribosomopathies, further study will be required to evaluate the consequence of PHF8 dysregulating Pol I transcription in these diseases.
2.5 Cohesinopathies
Cohesin is a complex that is critical for sister chromatid cohesion, chromosome segregation during S phase, chromosome condensation, DNA damage repair and gene regulation including Pol I transcription of the rRNA[68, 69]. Mutations in modulators or components of the cohesion complex have been associated with two human cohesinopathy diseases; Roberts syndrome (RS) (caused by homozygous mutations in ESCO2 which encodes an cohesin acetylase) and Cornelia de Lange syndrome (CdLS) (60% of cases are attributed to heterozygous mutations in genes coding for the cohesin subunits, SMC1A or SMC3, and the regulator NIPBL). RS is a rare autosomal recessive disorder (~1 per 160,000 births) whereas CdLS is an autosomal dominant disorder. Both CdLS and RS are multisystem developmental disorders characterized by pre- and postnatal growth retardation, cognitive impairment, severe limb growth deficiency, external and internal structural malformations, and facial dysmorphia (such as cleft lip and palate)[70].
The cohesinopathies are thought to stem not from defects in chromosome segregation but from altered gene expression. However, the precise mechanism involved is not well understood. Intriguingly, yeast strains bearing mutations (eco1-W216G and scc2-D730V) analogous to those associated with human RS and CdLS respectively, exhibit reduced rRNA levels and as a consequence protein translation is impaired[68]. The reduced protein translation was sufficient to account for the large number of mRNAs misregulated in response to these mutations. These observations were confirmed in a human RS cell line. Thus the observed decrease in rRNA levels and resultant reduced ribosome capacity and protein synthesis may be the drivers for cohesinopathies [68]. However, a publication, using Zebrafish as a model, demonstrated that mutating the ESCO2 acetylase generated a different gene expression profile than that observed when mutating a cohesin component[71]. This data suggests that ESCO2 role in RS may be independent of cohesin and thus differs to CdLS. Since both ESCO2 and cohesin affect Pol I transcription perhaps the similarities in phenotype observed between these two diseases are due to their effects on Pol I transcription[71].
2.6 Filamin A associated diseases
Mutations in the gene FLNA have been associated with a number of rare diseases, including periventricular nodular heteropia (PVNH) caused by a null mutation and otopalatodigital syndrome (OPD), frontometaphyseal dysplasia (FMD), Melnick-Needles syndrome (MNS) and X-linked cardiac valvular dystrophy (XCVD) which are the result of missense mutations generating a gain of function for Filamin A. Filamin A is one of three isoforms (Filamin A, B and C) and is the most abundant in humans. Filamin A was first reported as a F-actin-binding protein functioning as an intracellular signaling scaffold and thus has been ascribed roles in modulating three dimensional shape, cell motility and transcriptional regulation (reviewed in [72–76]).
A recent study demonstrated that Filamin A localizes to the nucleolus[77]. This observation was supported by the identification of Filamin A peptides in the human nucleolus using large-scale mass spectrometry (Nucleolar Online Proteomics Database, http://www.lamondlab.com/NOPdb3.0). Filamin A was found to interact with components of a Pol I complex, including RRN3, and RPA40 (Figure 1). In the absence of Filamin A Pol I occupancy at the rDNA promoter increases[77], thus Filamin A functions to suppress Pol I transcription.
2.7 How do mutations in Pol I components cause specific disease syndromes?
How do mutations in components of a ubiquitously required processes, such as Pol I transcription, ribosome processing and assembly, have such specific effects on certain cell lineages and tissues remains a fundamental unanswered question. A number of plausible hypotheses have been put forward. When tested in mouse models or tissue culture, some of these mutations reproduce many aspects of ribosomopathies. However, no single mechanism can at this stage account for the full spectrum of the disease phenotype.
One of the most intensively studies ribosomopathies is Diamond Blackfan Anemia (DBA), a disease associated with defects in erythropoiesis[78]. The etiology of this disease has been definitively linked, in the majority of cases, to haploinsufficient mutations in ribosomal proteins (Rps). This allowed modeling of the syndrome in cell culture systems by RNAi mediated knockdown of specific Rps and by knockouts of select Rps in mice[79–82]. One of the leading hypotheses to account for DBA centers on the recently discovered “nucleolar surveillance pathway” (also called nucleolar stress pathway)[25]. In this model, mutations or insults that disrupt ribosome biogenesis at the level of rRNA synthesis, processing or assembly, result in the sequestration of the E3 ubiquitin ligase murine double minute 2 (MDM2) by free Rps (predominantly the 60S Rps, L5 and L11) in a complex with 5S rRNA. This then leads to accumulation of p53. The elevated levels of p53 subsequently induce cell cycle arrest or apoptosis depending on the cell type[25, 83–85]. In the case of DBA, the nucleolar stress and activation of p53 results in preferential apoptosis or cell cycle arrest of the erythroid progenitors leading to anemia[86]. It has also been proposed that the reduced levels of functional ribosomes in surviving erythroid cells results in altered translation of mRNAs that encode proteins critical for erythropoiesis[6, 87]. Another proposal is that the aberrant accumulation of defective ribosomal precursors somehow contributes to the disease, for example perhaps via ribophagy[88]. On balance, it seems most plausible that the disease results from a combination of nucleolar stress and altered patterns in mRNA translation due to competition for the remaining ribosomes.
Some of the general mechanisms hypothesized to account for DBA may be relevant to almost all of the ribosomopathies associated with defects in Pol I associated factors described above. For example, as most of the mutations are predicted to be associated with defective rDNA transcription (possibly with the exception of Filamin A which normally functions to repress Pol I) they would be expected to exhibit, to varying degrees, activation of the nucleolar surveillance pathway associated with p53 accumulation and apoptosis. The findings with TCS, arguably the ribosomopathy most robustly linked to defective Pol I transcription, supports this model. For example, reduced rDNA transcription associated with haploinsufficiency of TCOF1 results in stabilization of p53, cell-cycle arrest and apoptosis of the migrating cranial neural crest cells[28]. Moreover, both pharmacological and genetic inhibition of p53 prevents apoptotic elimination of neural crest cells while rescuing the craniofacial abnormalities associated with mutations in TCOF1[9, 28].
What is more difficult to explain is why the TCS phenotype is restricted to the craniofacial tissues. Why isn’t p53 activated in all tissue haploinsufficient for TCOF1? One hypothesis is that specific cell and tissue types exhibit significant differences in the threshold for activation of p53 in response to reductions in Pol I transcription. This concept has now been experimentally confirmed in a cancer model where malignant hemapoietic cells were profoundly more sensitive to a selective small molecule inhibitor of Pol I transcription compared to normal hematologic cells of the same lineage[22]. This was shown to be due to differential activation of p53 even though Pol I transcription was inhibited to the same extent in normal and cancer cell[22]. Interestingly, loss of treacle has also been proposed to make cells more sensitive to oxidative stress, reducing their threshold for p53 activation[89].
However, this model would predict that all ribosomopathies associated with mutations that affect Pol I transcription should have the same phenotypes, which is not the case. One non–mutually exclusive possibility to explain this is that different cells and tissues exhibit varing expression levels of critical Pol I components. Thus a mutation in any given Pol I factor may only reduce its expression below a critical level to inhibit rDNA transcription, sufficient to activate nucleolar stress, in a subset of cells at a specific stage in development. Alternatively, a given Pol I associated component might be more essential for optimal ribosome biogenesis in certain tissues (i.e., tissue specific regulation of Pol I transcription), and thus deficiencies in that factor would be more likely to cause nucleolar stress in that tissue type. In this regard it is interesting to note that the high sensitivity of the rapidly proliferating neuroepithelium may be linked to the observation that the highest expression of treacle is found in the brain[90].
With respect to defective translation, a given ribosomopathy disease phenotype might also reflect specific changes in translation of mRNAs that encode proteins critical for development of that particular tissue and this may underlie the pathology of the disease. For example, TCS might reflect selective defects in the translation of mRNA encoding factors critical for the development of the neuroepithelium which gives rise to the neural crest cells from which the cartilage, bone and connective tissue of the head and face are generated[27]. In contrast to this specific model, a recent study demonstrated that the rescue of apoptosis and the normalization of craniofacial abnormalities in a mouse model of TCS, in response to p53 inhibition, occurred independently of the effects on ribosome biogenesis[28]. This suggests that p53-dependent neuroepithelial apoptosis is the primary mechanism underlying the pathogenesis of TCS. It will be interesting to see in future studies if this can be replicated in mice harboring mutations in POLR1C or POLR1D.
With respect to the other putative ribosomopathies, such as Blooms and Werner Syndrome, Siderius X-linked mental retardation and Cohesinopathy, it is highly likely that nucleolar stress and/or deficiencies in mRNA translation contribute to a component of their disease phenotype. However, as the transcription factors and chromatin modifiers implicated in these ribosomopathies have additional roles in Pol II transcription/replication/repair, it is most likely that these diseases reflect a mosaic of these different functions. As a first step in better defining the contribution of nucleolar stress to these diseases, future studies could examine the effect of modulating the activity of p53 in transgenic models that faithfully reflect the mutations and disease phenotypes observed in these complex syndromes.
3. Diseases associated with modulation of Pol I transcriptional activity through dysregulation of upstream signaling pathways
Proliferative growth requires that protein synthesis, and thus ribosome availability, can match cell cycle rates. Insufficient protein synthesis and the daughter cells will progressively get smaller, where as a surplus results in enlarged cells and is often associated with cellular transformation. Consequently, in normal cells Pol I transcription and ribosome biogenesis is tightly coordinated with the cell cycle in order to respond to changes in demand for proliferative growth. Interestingly, inhibition of cell cycle progression typically does not prevent cell growth, where as blocking growth invariable leads to cell cycle arrest[91, 92]. Thus ribosome biogenesis is upstream of and dominant to cell cycle regulation. However, it is not only proliferating cells that require tight regulation of ribosome biogenesis as it is also essential for terminally differentiated cells, particularly those with specialized functions that require a high demand for protein synthesis, such as muscle or secretory cells[93]. For example fetal and adult hearts have the same number of cardiomyocytes at birth, thus, with post-natal development the hypertrophic growth (an increase in size and mass of cell in the absence of proliferation) of the cardiomyocytes is critical to meet the increasing circulatory demand and pressure load on the heart. Hypertrophic growth of the heart is also observed in response to prolonged training exercise or pregnancy. This growth is achieved to a large extent by increasing the protein synthesis capacity of the cells through accelerated ribosome biogenesis and up regulation of rDNA transcription[93]. Similar regulation is used in reverse to promote atrophy rather than hypertrophy[94]. For example atrophy is required during normal development to cause shrinking and involution of the thymus in early childhood or the tonsils in adolescence. Atrophy of skeletal muscle is also observed naturally with aging, which is know as sarcopenia, and presumably this too, at least in part, is linked to decreased ribosome output.
Regulation of cellular growth utilizes a complicated network of signaling molecules, which can respond to environmental or cellular cues to impact on pivotal points to modulate Pol I transcription. As a consequence, the signaling network maintaining cellular growth during normal development frequently goes awry contributing to diseases, such as pathological hypertrophy observed with heart disease or hyper proliferation in cancer. Thus dysregulation of signaling pathways and their impact on modulators of Pol I transcription underlies growth dependent diseases.
3.1 Muscle Hypertrophy and Atrophy
3.1.1 Cardiac Hypertrophy
Pathological events including myocardial infarction, pressure or volume overload and congenital factors can initiate pathophysiological hypertrophic growth of the myocardium. This inappropriate and sustained cardiac hypertrophy is associated with a re-expression of fetal genes, sarcomere remodeling, thickening of the heart muscle and enlargement of the ventricles (reviewed in [95, 96]). Such growth initially compensates for the increased work demands on the heart muscle, but often deteriorates into structural and electrophysiological remodelling that results in reduced cardiac output and contractile dysfunction, increasing the likelihood of stroke and heart failure (ischemia and dilated cardiomyopathy)[97]. Clinically, cardiac hypertrophy is an independent risk factor for heart failure. Any deterioration of heart muscle function is broadly classified as cardiomyopathy, which is also a symptom often associated with RASopthies, that is disorders caused by germline mutations in the RAS pathway, one of the key regulators of rDNA transcription and ribosome biogenesis[98]. For example cardiomyopathy is common in Noonan syndrome patients (mutated RAF1), Leopard syndrome (mutated PTPN11: SHP2), and Costello syndrome (mutated HRAS)[99].
In 1985 Morgan and colleagues[100, 101] published landmark studies demonstrating that increased ribosome synthesis, as opposed to increased protein translation rate, predominantly mediated the muscle cell growth associated with cardiac hypertrophy. They went on to demonstrate that it was the rate of Pol I transcription, which dictated the number of ribosomes in this system[102]. It was a further 6 years before the Pol I specific transcription factor, UBF, was established as a key modulator of Pol I transcription in cardiac hypertrophy. Utilizing a rat cardiomyocyte model, hypertrophic stimuli such as contraction, norepinephrine and endothelin where shown to increase ribosome biogenesis via elevating the amount and activity of UBF[103–105]. Hannan and Rothblum extended this work to demonstrate that overexpression of UBF, or reducing UBF expression with antisense RNA, increased or prevented rDNA transcription and cardiac hypertrophy respectively[106–108]. While this mechanism has not been evaluated in humans it was reported that heart failure patients display morphological changes in the nucleus of their myocytes including an increase in size of the nucleolus[109].
Since these initial reports, few studies have evaluated the involvement of rDNA transcription in cardiac hypertrophy, although numerous additional components of the Pol I transcription apparatus have been cloned, including the key transcription factor RRN3[2]. Many of these factors play critical roles in growth factor mediated control of rRNA synthesis. Furthermore, our understanding of the growth factors and signaling pathways activated during pathophysiology has grown immensely but have not been examined in the context of modulating Pol I transcription. Thus ironically, although cardiac hypertrophy was one of the first diseases to be associated with deranged rDNA transcription, our knowledge of the exact mechanism of action by which Pol I transcription is modulated during hypertrophy lags behind other diseases such as cancer.
Although numerous signaling pathways have been reported to play a role in cardiac hypertrophy, these often depend on the model system being evaluated (reviewed in [110]). In addition to mechanical load[111], the RAS and PI3K/AKT/mTOR signaling pathways have been shown to modulate cardiac hypertrophy and there is increasing evidence that they may cooperate in this process. In one study PI3K and p21-activated kinase (PAK) signaling pathways were both shown to co-operate with RAS to activate RAF1 kinase[112] which promotes hypertrophy[113]. More recently it was suggested that α1A-adrenergic receptor activation of PI3K and Rac1 GTP exchange factor T-cell lymphoma invasion and metastasis factor 1 (TIAM) leads to activation of ERK in neonatal cardiomyocytes[114]. Other studies have linked cyclic AMP stimulation of G-protein coupled receptors (GPCR) and RAS signaling or GPCR activation of ERK in promoting cardiac hypertrophy[99]. In addition, a fundamental regulator of cardiac growth is the oncogene c-MYC[115, 116] (discussed in section 3.2.1.1).
While a number of the above signaling pathways have been shown to mediate phosphorylation and/or increased expression of UBF in non cardiac systems such as smooth muscle[117], to what extend they do so in myocytes and the contribution of additional Pol I components, such as RRN3 or SL-1, remains fertile ground for further investigations. It is important to note however, that many of the pathological signaling pathways thought to contribute to left ventricular hypertrophy including RAS/ERK, PI3K/AKT/mTOR and c-MYC play prominent roles in regulating Pol I transcription in tumor cells (section 3.2). It is possible that the dysregulation of signaling to Pol I transcription during pathological cardiac hypertrophy will be very similar to the regulation of Pol I transcription in cancer. Consistent with this notion, more contemporaneous studies investigating the regulation of 5S rRNA, transcribed by Pol III, demonstrated that oncogenic and tumor suppressor pathways involved in the regulation of ribosome biogenesis in tumor cells also regulate 5S synthesis in cardiac myocytes [118].
3.1.1 Renal hypertrophy
Loss of a kidney can promote compensatory hypertrophy in the remaining kidney, resulting in no net loss of kidney function. The resultant hypertrophy is associated with newly formed epithelium and growth of the glomeruli and capsules. However, in disease states induced by diabetes, renal hypertrophy is deleterious, resulting in reduced function and kidney failure. Renal hypertrophy and the accumulation of extracellular matrix proteins are hallmarks of kidney disease observed early in the development of both Type 1 and 2 diabetes[119, 120]. Both hallmarks require elevated protein synthesis rates, which was initially ascribed to reduced degradation of the rRNA[121, 122], whereas a more recent publication reported increased rDNA transcription followed by elevated protein translation[120]. In the glomerular epithelial cells, Mariappan et al.[120] demonstrated that high glucose induction of Pol I transcription was mediated via the PI3K signaling pathway activating both ERK and S6 Kinase which resulted in phosphorylation and thus activation of UBF at serine 338. Phosphorylated UBF was released from an inhibitory complex with p19ARF and could now freely bind the RPA194 subunit of Pol I thus promoting rDNA transcription. Interestingly, elevated UBF phosphorylation has previously been reported in vivo in the kidney of Type 1 or 2 diabetic rats and mice respectively[120]. However, it has not been established whether elevated UBF phosphorylation occurs in diabetic patients with renal hypertrophy.
These findings raise the tantalizing prospect that selective Pol I inhibitors being developed to treat cancer[22](see section 4) may be efficacious in treating cardiac and renal hypertrophy. This hypothesis must now be tested in animal models of these diseases. Encouragingly, mice treated with these inhibitors, during the course of cancer studies, do not exhibit any renal or cardiac dysfunction nor muscular atrophy (R. Hannan, M. Bywater and D. Drygin; unpublished data) consistent with the idea that only cells with abnormal growth are preferentially sensitive to the modulation of Pol I activity.
3.1.4 Atrophy
In contrast to the heart where hypertrophy is the major clinical issue, in skeletal muscle atrophy is an important pathophysiological feature of disease and aging. Muscular atrophy is broadly defined as a decrease in muscle mass due to the process of reabsorbing and breaking down tissues via apoptosis[123]. Consequently, the hallmarks of atrophy include reduced cell number and size of the myofibers and often a switch from type II fast to slow fibres. This leads to reduced muscle contractile function and critically, the loss of more than 40% of the body cellular mass can be fatal[123, 124].
As with hypertrophy, at a cellular level atrophy is caused by an imbalance between proteins synthesis and degradation. To date only one publication has addressed the association between skeletal muscle atrophy and reduced protein synthesis[125] and one other investigated skeletal muscle hypertrophy[126]. Using a denervated muscle model in which myostatin levels were reduced, activation of the mTOR signaling pathway was observed. The PI3K/AKT/mTOR pathway has already been established as essential for correct muscle growth, both by positively regulating protein synthesis via increased ribosome biogenesis or via AKT phosphorylation of forkhead transcription factor (FOXO) driving the transcription of ubiquitin ligases and autophagy-related genes to promote protein degradation[123, 125]. Intriguingly, a recent study demonstrated that denervation induced skeletal muscle atrophy was associated with dramatic decreases in de novo rRNA synthesis however, activation of mTOR failed to rescue muscle cell growth and rDNA transcription[125]. While the mRNA levels for UBF, and SL-1 subunits Tata binding protein (TBP) and TBP-associated factor (TAF) 1B were all elevated in response to mTOR activation, there was a significant decrease in the level of the SL-1 subunit TAF1A suggesting that SL-1 may be functionally limiting for muscle growth in this model[125]. This study supports the hypothesis that dysregulation of rRNA synthesis triggers atrophy even in the presence of positive growth signals, such as activated mTOR pathway. Thus one of the ongoing challenges for the development of therapeutic approaches to combat diseases associated with atrophy will be to define the signaling pathways that mediate the mTOR independent down regulation of rDNA transcription.
In the converse situation, skeletal muscle hypertrophy induced by mechanical loading, the increased skeletal muscle growth was shown to be associated with increased rDNA transcription and rRNA content [126]. The skeletal muscle hypertrophy was preceded by an increase in c-MYC expression and c-MYC dependent increased expression of Pol I components[126] previously termed the Pol I regulon[127]. Similarly, factors involved in rDNA chromatin remodeling, such as the Williams syndrome transcription factor, were enriched at the rDNA promoter by mechanical loading. Since the two examples above used different skeletal muscle systems it is not clear whether the factors required to activate ribosome biogenesis during skeletal muscle hypertrophy are the same pathways defective during diseases associated with skeletal muscle atrophy. Clearly further studies in this area are required to resolve these outstanding questions.
3.2 Cancer
Over 100 years ago, well before the function of the nucleolus in ribosome biogenesis was understood, it was recognized that the size of the nucleoli were increased in tumour cells. Indeed, pathologists have used abnormal nucleolar size as an indicator of particularly aggressive tumours and in some cases, such as malignant melanocarcinoma, nucleolar size is an accurate clinical marker for disease[128]. As the nucleolus is the site of Pol I transcription and its size correlates with the rate of rRNA synthesis, this data suggests that overactive Pol I transcription is a frequent occurrence during malignant transformation[3, 128]. Indeed, using comparative expressed sequence hybridization, which identifies chromosomal regions corresponding to differential gene expression, Williamson et al.[129] demonstrated consistently abnormally high levels of rRNA in all six tumour types tested and these increased with cancer stage. This was supported by a recent study which demonstrated that 45S, 28S, 18S and 5.8S rRNA were increased in human primary prostate cancers[130]. In particular, c-MYC driven cancers are almost universally associated with hyperactivated Pol I transcription[1]. These findings have led to two major questions; i) How is Pol I transcription accelerated in cancer; and ii) Is the dysregulated transcription required for the malignant phenotype?
With respect to the former, the initial clues for how Pol I transcription might be dysregulated in cancer came from the unexpected finding that the tumour suppressor protein, pRb, directly repressed rDNA transcription[131]. Subsequent evidence came from studies demonstrating that mammalian rDNA transcription is not simply a slow and indirect consequence of altered nutrient signaling, but dynamically regulated by the mitogen activated kinase ERK1/2[98]. This opened the door for a flurry of studies demonstrating that pathways typically associated with malignancy via signaling to Pol II also coregulated Pol I transcription (reviewed in [1, 132]). It is now recognized that elevated Pol I transcription during cancer progression and tumour maintance is typically mediated by either overactivation of oncogenes or an oncogenic signaling pathway, or release from inhibition by tumour suppressors and tumour suppressor signaling (Figures 2 and 3). In both cases, dysregulated signaling converges to modulate formation of the core pre-initiation complex and or alter the transcriptional activity of Pol I including elongation. With respect to the contribution of dysregulated Pol I transcription to cancer, very recent data has shown unequivocally that dysregulated Pol I transcription is required for the maintenance of the malignant phenotype of certain hematologic cancers and can be target to therapeutically treat cancer in vivo (discussed in more detail in section 4 [22]).
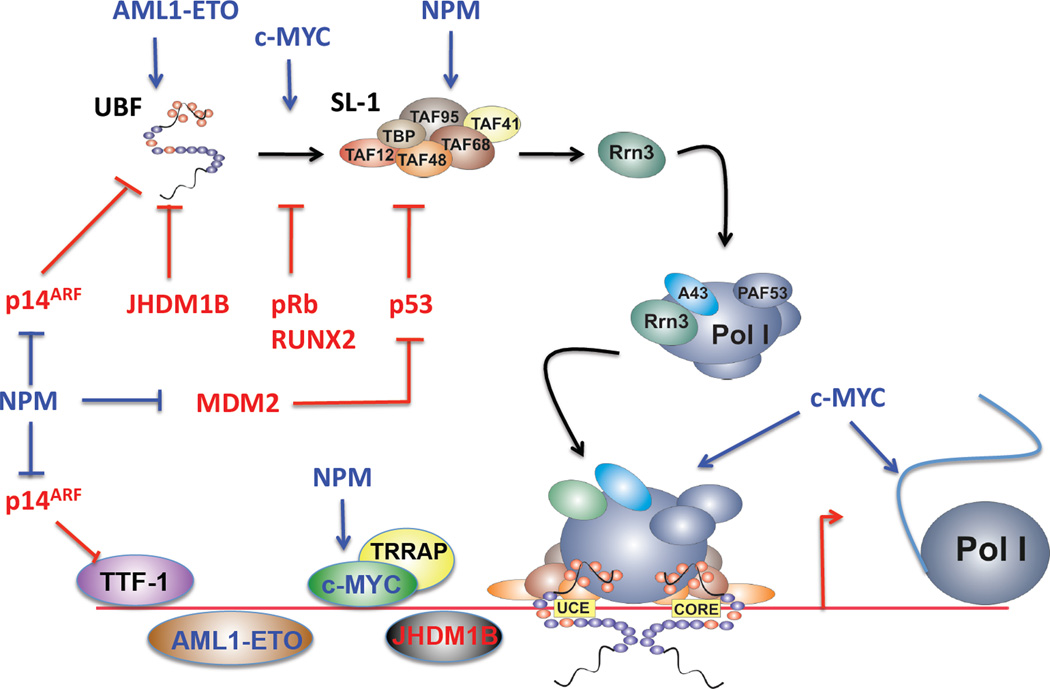
Figure 2. Modulation of RNA Polymerase I transcription by oncogenes and tumour suppressors.
A schematic illustrating the sites of action where oncogenes and tumour suppressors regulate RNA polymerase I transcription in a positive (blue) or negative (red) fashion. UBF: upstream binding factor; SL-1: selectivity factor 1; Pol I: RNA polymerase I; TTF-1: termination transcription factor 1; UCE: upstream control element. Note; in addition to directly interacting with the rDNA repeat and core transcription components, c-MYC also indirectly modulates RNA Polymerase I transcription by coordinating the increased expression of a cohort of Pol II transcribed genes termed the "Pol I regulon" which comprises over 90% of the core Pol I transcription factor complex including UBF, RRN3 and RNA Polymerase I subunits.
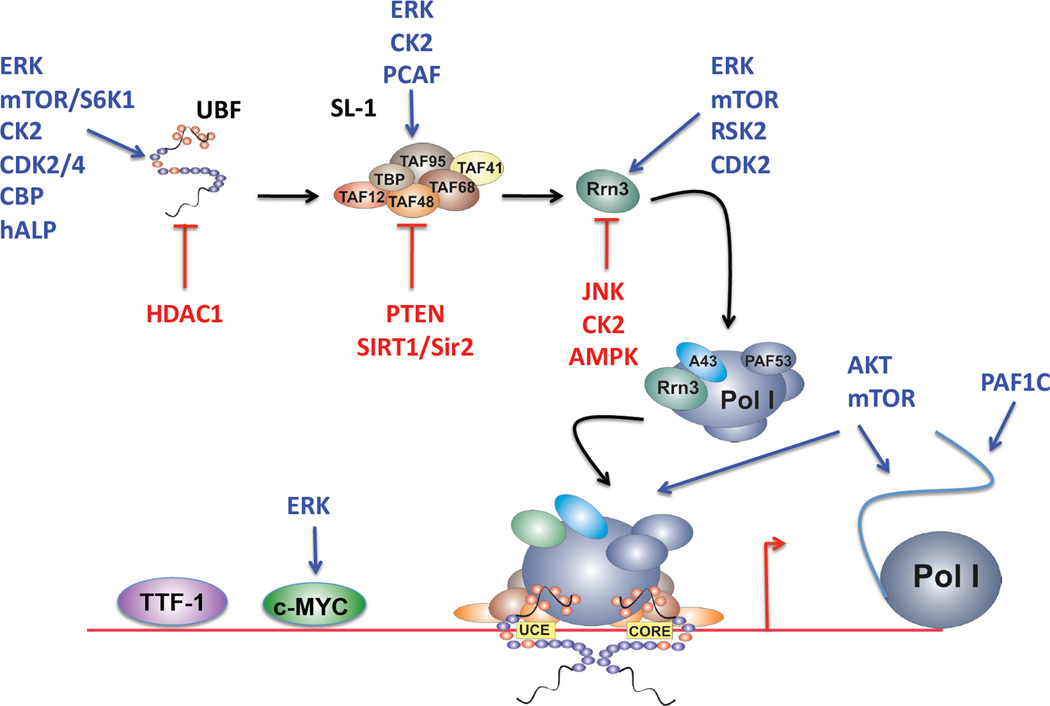
Figure 3. Modulation of RNA Polymerase I transcription by oncogenic and tumor suppressor signaling pathways.
A schematic illustrating the sites of action where signaling molecules regulate RNA polymerase I in a positive (blue) or negative (red) fashion. UBF: upstream binding factor; SL-1: selectivity factor 1; Pol I: RNA polymerase I; TTF-1: termination transcription factor 1; UCE: upstream control element;
3.2.1 Oncogenes and Tumour Suppressors
A number of oncogenes and tumour suppressors have been demonstrated as bona fide direct regulators of Pol I transcription (Figure 2): including the oncogenes c-MYC and AML1-ETO and the tumour suppressors p53, pRb, and p14ARF. In addition, an increasing list of other factors with oncogenic or tumor suppressor-like activity including nucleophosmin (NPM), RUNX2, ZNF545/ZFP82 and JHDM1B have been shown to play direct roles in modulating Pol I transcription during malignancy (reviewed in [1, 2, 92]).
3.2.1.1 c-MYC
c-MYC is a potent oncoprotein and transcription factor which is overexpressed in ~50% of all cancers[3, 133], notably leukemia, sarcoma, lymphoma and its gene is frequently translocated in multiple myeloma[134], Burkitt lymphoma, neuroblastoma and colon carcinomas. Moreover, its dysregulated expression correlates with poor prognosis[134]. As a transcription factor, c-MYC forms a heterodimer with Max to bind site-specific sequences in the genome termed Enhancer Box sequences (E-boxes). This leads to the recruitment of factors such as histone acetyltransferases, acetylation or methylation of the nucleosomal histones and the modulation of chromatin structure making it permissive for transcription by all three RNA Polymerases[133, 135, 136]. c-MYC-mediated global changes in the chromatin allows for coordinate transcriptional regulation of ~15% of all the genes in the genome[137, 138]. Predominant amongst this cohort of c-MYC-transcriptional gene targets are those essential for cell growth, including regulators of ribosome biogenesis, protein synthesis and metabolism[133, 139].
Of specific relevance for this review, c-MYC stimulates Pol I transcription via at least two mechanisms (Figure 2). Firstly, c-MYC coordinates the increased expression of a cohort of Pol II transcribed genes termed the "Pol I regulon" which comprises over 90% of the core Pol I transcription factor complex including UBF, RRN3 and Pol I subunits[127, 140]. The net result is two fold: i) an increase in the abundance of the Pol I transcription apparatus; and ii) an increase in the number of transcriptionally active rDNA repeats mediated via the increased abundance of the cytoarchitectural transcription factor UBF[127, 141, 142]. Consistent with this mechanism, components of the Pol I transcription apparatus are highly overexpressed in c-MYC driven malignancies and normalisation of their expression leads to selective apoptotic death of c-MYC driven malignancy, thus their elevated expression is necessary for the c-MYC-driven malignant phenotype[22]. Numerous other malignancies also demonstrate overexpression of Pol I components[90, 143] although it is less clear if this is related to dysregulated c-MYC expression.
Secondly, c-MYC has been reported to function in Pol I transcription by directly binding to the rDNA. This leads to recruitment of its co-factor transformation/transcription domain-associated protein (TRRAP) and eventually Pol I. It is believed that the binding of c-MYC mediates the looping of the rDNA resulting in elevated rRNA synthesis[144–146]. More recently it was also demonstrated that c-MYC binds SL-1 to stabilise the SL-1/UBF complex, thus increasing UBF recruitment to the rDNA promoter and leading to upregulated Pol I transcription[2]. Interestingly, c-MYC can also function in a non-DNA binding manner at Pol III promoters via recruitment of TRRAP and the histone acetyltransferase GCN5 to the tRNA and 5S promoters, thus enhancing TFIIIB binding and histone H3 acetylation to elevate Pol III transcription rate[147]. Overall c-MYC functions as a regulator of all three RNA polymerases, a role that may underlie its potency as an oncogenic transcription factor. Indeed as outlined below, recent studies have demonstrated at least part of the oncogenic activity of c-MYC is dependent on its ability to modulate Pol I transcription[22].
3.2.1.2 AML1-ETO
The oncoprotein AML1-ETO is the most frequent chromosomal translocation associated with acute myeloid leukemias and encodes a fusion protein between the Runt-related transcription factor 1/acute myeloid leukemia 1 (RUNX1/AML1) and Myeloid transforming gene on chromosome 8/Eight-Twenty-One (MTG8/ETO)[148]. The fusion protein AML1-ETO retains the DNA binding ability of AML1, but not its transactivation or nuclear matrix target signaling. AML1 binds DNA via its runt domain and together with core-binding factor β (CBFβ) forms part of the CBF that binds enhancers and promoters to alter Pol II transcription. Typical targets of CBF include genes involved in cell cycle, hematopoietic-specific genes and proliferation. On the other hand, ETO is an auxiliary protein that interacts with transcription factors, recruits a range of corepressors such as histone deacetylases (HDACS), and modulates Pol II transcription repression[148]. Both AML1 and AML1-ETO can bind to the rDNA repeats and associate with UBF. AML1 predominantly associates with hypermethylated rDNA whereas AMLl-ETO binds where the H3K4 methylation is highest (i.e., transcriptionally active rDNA). Thus AML-1 antagonises, where as AML1-ETO promotes Pol I transcription[149]. This suggests that AML-1 mediated down regulation of rRNA synthesis may be integral in the process of myeloid differentiation while activation of Pol I transcription may be essential for AML1-ETO oncogenicity associated with a global block in cellular differentiation (M. Bywater personal communication).
3.2.1.3 Nucleophosmin
Nucleophosmin (NPM: B23, NOR38) belongs to the nucleoplasmin family of chaperones and has been strongly implicated in the pathogenesis of numerous malignancies. Paradoxically, it has been described both as an oncogene and a tumor suppressor, depending on cell type and abundance. While NPM overexpression is associated with different types of solid tumors, mutations or translocations that impair NPM function or reduce functional levels, are observed in 30% of acute myeloid leukemias[2, 150–153]. NPM is predominantly localized to the nucleolus, but it can shuttle to the cytoplasm and nucleoplasm in response to its phosphorylation at particular sites[154]. NPM has been shown to play a role in pre-mRNA processing, the response to genotoxic stress, apoptosis, control of ploidy, DNA repair, cellular transport and maintenance of chromatin structure.
With respect to its predominant role in the nucleolus, NPM is typically described as a nucleolar endoribonuclease. However, more recently NPM was shown to associate with the rDNA chromatin[150], and promote increased recruitment of TAF148 to the rDNA promoter (Figure 2), thus stimulating rDNA transcription[1, 155]. Nucleophosmin also modulates rDNA transcription indirectly through the c-MYC-ARF-MDM2 axis[156]. For example, NPM interacts with the E3 ligase MDM2 (HDM2 in humans) to control p53 levels in response to nucleolar stress[157, 158]. NPM also targets the tumour suppressor p14ARF to the nucleoli which in turn inhibits Pol I transcription (section 3.2.1.5)[159]. Finally, overexpression of NPM has been shown to enhance c-MYC nucleolar localization (Figure 2) and promote c-MYC-driven rDNA transcription[150]. Despite the slew of studies described above linking NPM and cancer, definitive evidence demonstrating that dysregulation of the nucleolar related functions of NPM, as opposed to its other chaperone related functions, are responsible for its contribution to malignant transformation, are not yet available [151].
3.2.1.4 RUNX2
Runt-related transcription factor 2 (RUNX2) is a transcription factor, which predominantly plays a role in osteoblast proliferation and differentiation, and drives bone specific gene expression. RUNX2 was first shown to be overexpressed in c-MYC driven T cell lymphoma, with more recent publications suggesting it plays a positive role in promoting invasive breast cancer and prostate cancer (reviewed in [160]). RUNX2 acts with its cofactors to remodel chromatin and either activates or suppresses Pol II transcription depending on the cellular context. In what seems to be a recurring theme, recently RUNX2 was also shown to localize to the nucleolus. Paradoxically, RUNX2 is associated with open chromatin and when complexed with UBF and SL-1 inhibits rDNA transcription (Figure 2)[161] by complexing with HDAC1 and inducing deaceylation of both UBF and the histone proteins[162]. How inhibition of rRNA synthesis by RUNX2 contributes to cancer, or whether RUNX2 can activate rDNA transcription in other contexts, remains to be established.
3.2.1.5 pRB, p53 and p14ARF
The concept that tumour suppressors can directly repress Pol I transcription and their inactivation during cancer would facilitate upregulation of Pol I transcription, first came to fruition with the demonstration that the tumour suppressor pRb directly inhibited Pol I transcription in experiments using cultured cells and purified rDNA transcription components[131]. This observation provided a paradigm shift in our understanding of how rDNA transcription was modulated and immediately highlighted the potential importance of dysregulated Pol I activity in cancer biology. pRb functions to mediate G1-S phase arrest in response to cellular insults such as DNA damage, and this is effected primarily by binding E2F transcription factors[163–166]. The relationship between pRb and E2F is also important for the cellular processes of DNA repair, differentiation, metabolism and cancer[2, 167, 168]. pRb can also affect genomic stability by modulating chromosome condensation and E2F transcriptional targets critical for spindle assembly checkpoint (SAC) function[164]. Although pRb is deleted in all retinoblastomas, activating mutations in the gene in other cancers are rare, rather it is dysregulation of its key modulators, including E1A, CDK-cyclin complexes and the caspase dependent proteolytic pathway, which promotes elimination of pRb function that is typical in all cancers[163]. As highlighted above, pRb can directly repress Pol I transcription, as can its related pocket protein p130[131, 169]. One publication suggested pRb blocked the ability of SL-1 to bind the rDNA[170], however, other studies suggest the mechanism is due to pRb interfering with the binding of UBF to SL-1 (Figure 2)[131, 169, 171]. Subsequently it has been shown that pRb can also repress 5S synthesis by Pol III[172–174] leading to the general paradigm that some of the most potent tumour suppressors and oncogenes mediate their potent effects on cancer via controlling all three RNA polymerases[3].
p53 has been coined the guardian of the genome[175], and its loss of activity through mutation or deletion is associated with accelerated proliferation and genomic instability. Consequently, p53 is inactivated in the majority of human cancers, with over 50% of tumours characterized by either mutated or deleted p53 and it has been suggested the rest have impaired p53 signaling[176]. p53 protein expression is tightly regulated by targeted degradation via the 26S proteasome and E3 ligase MDM2 (HDM2 in humans) which itself is over expressed in 5–10% of all human cancers[176]. p53 is stabilized via two mechanisms: i) checkpoint homology 1 (CHK1), CHK2 or ataxia telangiectasia mutated (ATM) phosphorylation[176]; and ii) oncogenic activation of p14ARF which blocks p53 interaction with MDM2[177]. p53 activity is also modified by acetylation and methylation, the result of which is varied transcription of p53-dependent targets. While p53 typically mediates transcriptional activation of its targets[176], in the context of Pol I it represses transcription by binding to the TBP and TAF1110 subunits of SL-1, thus preventing SL-1 from interacting with UBF and forming the Pol I pre-initiation complex (PIC) (Figure 2)[2, 178]. Thus p53 promotes a feed forward repression loop in which the inhibition of Pol I transcription causes activation of p53 via the nucleolar stress pathway (section 2.7) which in turn further represses Pol I transcription. This concept is important as it suggests that a major function of Pol I transcription and nucleolar integrity is to titrate p53 levels, further underlining the importance of Pol I transcription in malignancy. Thus, in normal cells there is a break in this positive feedback loop, where p53 induces MDM2 expression leading to downregulation of p53 activity. Perhaps not unexpectedly, cancers with both dysregulated pRb and mutated p53 are the most aggressive and also have the highest rate of rRNA synthesis[1].
The 9p21 gene cluster, harboring the tumour suppressive genes p14ARF and p16INK4a, is a major mutation hotspot in human cancers. p14ARF is produced from the same gene that encodes p16INK4A. The two transcripts arise through the utilisation of alternative promoters and an alternate reading frame for translation[179–182]. As described above, p14ARF restrains cell growth by binding and abrogating MDM2 inhibition of p53 activity, and therefore facilitates p53 mediated cell cycle arrest and apoptosis[177]. p14ARF is also a central regulator of ribosome biogenesis; modulating rDNA transcription by both indirect and direct mechanisms. For example, p14ARF indirectly modulates rDNA transcription via interactions with NPM (section 3.2.1.3 and [183]). In addition, p14ARF directly functions as a repressor of rDNA transcription via at least two separate mechanisms: i) interfering with UBF phosphorylation inhibiting PIC formation[184]; and ii) inhibiting nucleolar import of TTF-I by binding to this nucleolar localization sequence and causing TTF-1 to accumulate in the nucleoplasm (Figure 2). p14ARF also binds MDM2 thus preventing MDM2 binding and ubiquitinylation of TTF-I. The overall result is that p14ARF inhibits both rRNA synthesis and processing[185, 186].
3.2.1.6 ZNF545/ZFP82 and JHDM1B
Over the last few years a number of other proteins implicated in the etiology of cancer have also been shown to modulate Pol I transcription. For many of these factors our knowledge on their role in cancer is insuffient to confidently ascribe them functions as bone fide tumour suppressors or oncogenes. These include the nuclear 19q13 KRAB domain-containing zinc finger protein ZNF545/ZFP82 and the nucleolar histone demethylase JHDM1B. When sequestered to the nucleoli, ZNF545/ZFP82 represses NF-κB and AP-1 pathway dependent ribosome biogenesis specifically by inhibiting Pol I transcription[187, 188]. However, the mechanism of action was not determined. ZNF545/ZFP82 is ubiquitously expressed in cells and downregulated in primary tumours such as nasopharyngeal carcinoma, esophageal squamous cell carcinoma, gastric colon carcinomas and infrequently reduced in hepatocellular, lung, breast, renal, prostate and cervical cancer cell lines[188]. This pattern of expression and action on Pol I transcription suggests it may be a tumour suppressor.
JHDM1B interacts with the rDNA genes to repress transcription, in this case by demethylating Lys4 of histone H3 causing dissociation of UBF from the rDNA (Figure 2). Although JHDM1B represses Pol I transcription its expression was increased in aggressive primary glioblastomas[3, 189]. Thus it is unclear if it acts as an oncogene or tumour suppressor. Formal experiments demonstrating a causative role for ZNF545/ZFP82 and JHDM1B in the initiation or progression of human cancer are ongoing. It may be some time until the consequences of their control of Pol I transcription for malignant transformation is entirely clear.
3.2.2 Oncogenic signaling
Overactivation of oncogenic signaling pathways are a primary and pervasive mechanism by which Pol I transcription is hyperactivated during malignant transformation (Figure 3). These pathways converge primarily on the Pol I transcription factor RRN3, UBF and to a lesser extent SL-1. Two of the major growth regulatory pathways known to modulate Pol I transcription include the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR signaling cascades[132]. However, other signaling molecules have also been implicated in cancer-associated regulation of Pol I transcription, including casein kinase 2 (CK2), Cyclin-dependent kinase 2 and 4 (CDK2, CDK4) and AMP-activated protein kinase (AMPK).
3.2.2.1 RAS/RAF/ERK pathway
The RAS/RAF/MEK/ERK pathway is hyperactivated by mutations in RAS or RAF. RAS mutations have been reported in up to 30% of all cancers (range of 10–90% depending on disease site) especially in lung, pancreatic and colon cancer. RAF mutations have been identified in 6–7% of human cancers with an increased prevalence in melanomas and thyroid cancers[190].
To date, published data suggests that the RAS/RAF/MEK/ERK pathway can regulate Pol I transcription specifically through the activities of ERK (Figure 3). ERK has been shown to phosphorylate, and thus activate both: i) UBF resulting in an increased rate of Pol I transcription elongation[98, 191]; and ii) RRN3 promoting Pol I initiation[192]. ERK also phosphorylates c-MYC promoting its stabilization[193], which may result in increased Pol I transcription. Interestingly MAPK activity was shown to induce TBP expression[194], which may lead to increased levels of functional SL-1 and thus promotes Pol I transcription. This may be functionally relevant to cancer since increased TBP expression has been observed in a subset of colon carcinomas[194].
3.2.2.2 PI3K/AKT/mTOR pathway
Common PI3K/AKT/mTOR pathway mutations or amplifications found in cancer include those of the PIK3CA catalytic subunit for PI3K, which are more prevalent in breast, colorectal, glioblastoma, ovarian and prostate cancer[195, 196]. Alternatively PTEN, which negatively affects the pathway, expression is reduced in a broad range of cancer types, including breast, prostate, renal cancer and approximately 30–50% of melanomas[195]. Furthermore, the pathway is constitutively activated in cells transformed by dysregulation of receptor tyrosine kinases. AKT, 4EBP1, eIF4E, Rheb and S6K1 have all been reported as overexpressed in a subset of cancers. Surprisingly there are few reported cases of mTOR mutations[195, 197, 198].
The PI3K/AKT/mTOR pathway, like ERK, modulates the phosphorylation and activity of critical Pol I components (Figure 3). For example S6K1, downstream of mTOR, was shown to indirectly regulate phosphorylation of the UBF acidic tail thus enhancing its ability to activate Pol I transcription[199]. mTORC1/S6K signaling may also phosphorylate, and thus activate, RRN3 on residue serine 44[200]. Increased expression of PTEN was shown to repress Pol I transcription by selectively dissociating the SL-1 complex, thus reducing SL-1 occupancy on the rDNA promoter[201]. More recently AKT was shown to potently activate Pol I transcription at multiple stages, including transcription initiation, elongation and cotranscriptional processing[202]. Moreover, AKT activity cooperates with c-MYC to promote Pol I transcription. AKT inhibition in a model of Burkitt lymphoma promoted apoptosis[202], suggesting that decreased ribosome biogenesis is likely to be a fundamental component of the therapeutic response to AKT inhibitors in cancer. The direct Pol I dependent targets of AKT are not known but intriguingly at least part of its effect on rRNA synthesis is independent of mTORC1[202].
3.2.2.3 Co-regulation by the RAS and PI3K pathways
In some cases, signaling down the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR cascades converge to regulate Pol I transcription leading to cancer. The HER2 and EGF receptors themselves are also subject to amplification and activating mutation in cancer[190, 195, 197, 203]. The overactivity of these receptors can "hyperactivate" the RAS and PI3K pathways resulting in the stimulation of rDNA transcription.
Both the ERK and PI3K signaling cascades can also signal to ribosomal S6 kinase 2 (RSK2) and c-jun N-terminal kinase (JNK), which have also been implicated in cancer development although their specific role requires further verification. RSK2 was shown to increase proliferation and anchorage independent transformation in mouse skin epidermal cells[204] and to play a role in mediating FGFR-3 dependent transformation of hemopoietic cells[205] (reviewed in [206]). JNK is downstream of MAPK (MKK4/7) and PI3K/RAS/RAC and is predominantly activated by stress. JNK plays a role in development, apoptosis, growth, inflammatory and immune responses, which are all processes regulated in cancer development and progression. JNK expression or activity has been shown to be upregulated in retinoblastoma, melanoma, breast carcinoma, invasive ovarian cancer and downregulated in colorectal cancer although the mechanism of action mediating this phenotype is not clear (reviewed in [207]). With respect to Pol I transcription, RSK2 phosphorylation and activation of RRN3 stimulated Pol I activity[192]. In contrast, JNK dependent phosphorylation at threonine 200 inhibited RRN3 activity by impairing the interaction between RRN3, Pol I and SL-1[2] (Figure 3).
3.2.2.4 Other cancer associated signaling pathways
In addition to the RAS and PI3K pathways, signaling cascades involving CK2, CDK2, CDK4, AMPK and ATM are also consistently dysregulated in cancer and regulate Pol I transcription control.
CK2 is a heterotetramer composed of two catalytic and one regulatory subunit, which mediates phosphorylation of over 300 substrates[208]. Signaling pathways modulated by CK2 include PI3K, NFκB, Wnt, PTEN[209] and more recently the Hh/Gli signaling pathways[210]. These pathways are involved in the regulation of cellular proliferation, survival and the DNA damage response[208]. The level of CK2 in normal tissues is tightly regulated[211, 212], however, its overexpression or increased activity has been reported in many cancers, including leukemia and solid tumours such as prostate and colorectal cancer[1, 208]. Moreover, CK2 expression/activity was shown to correlate positively with malignant transformation and aggressive tumour behavior. A number of CK2 inhibitors have demonstrated a therapeutic activity in a wide range of human cancer cell lines[213]. The highly selective, small-molecule inhibitor, CX-4945, has shown positive results in Phase I trials targeting multiple cancer types[214, 215]. CK2 regulates Pol I transcription via two mechanisms (Figure 3): i) CK2 copurifies with and phosphorylates Pol I subunits; and ii) CK2 phosphorylates Pol I associated proteins including UBF, RRN3, SL-1, Topoisomerase IIa, nucleolin and NPM[1, 216, 217]. CK2 phosphorylation of the transcription initiation factor RRN3 at serines 170 and 172 inactivates RRN3, and triggers the release of RRN3 from Pol I after transcription initiation, which, in mammals is thought to be essential for transcription elongation[218], although studies from yeast suggest that dissociation of RRN3 from the Pol I holoenzyme is not required for efficient rDNA transcription[219].
The CDKs, cyclins and CDK inhibitors are key elements in the regulation of the cell cycle and thus the control of cell fate and differentiation[168, 220]. For example, G1 to S transition requires CDK2 and CDK4 with their cyclin partners, E and D1 respectively. One role for these kinases is the phosphorylation and inactivation of pRb, thus permitting E2F-mediated transcription which is essential for DNA replication[220]. This process is highly regulated by the CDK inhibitors p16INK4, p21CIP1 and p27KIP1[220]. With respect to cancer, CDK2 expression is frequently upregulated in a wide array of cancers in particular melanoma, head and neck cancer and cervical cancer[90]. Moreover, overexpression of CDK4 has been reported in liposarcomas, oral squamous cell carcinoma, pancreatic, lung and nasopharyngeal cancer[221–224].
Both CDK2-cyclin E and CDK4-cyclin D phosphorylate UBF at serine 388 and 484 respectively, both phosphorylations are believed to be required to maintain UBF activity (Figure 3) during cell cycle progression[225, 226]. CDK2-Cyclin E was also reported to phosphorylate RRN3 at serine 44[2]. Despite these observations, the specific contribution of CDK2 and CDK4 regulation of Pol I to malignancy has not been defined. However, a recent study demonstrated that selective ablation of CDK2 activity disrupted the growth of nontransformed mouse embryonic fibroblast (MEFS) and human colon cancer cells. Consquently the authors proposed that CDK2 is not dispensable for proliferation in normal or malignant cells as previous suggested[227]. Although not formally demonstrated, it is possible that the control of Pol I is critical for CDK2 regulation of cell survival.
AMP-activated protein kinase (AMPK) is a master regulator of energy homeostasis, which modulates heavily energy-dependent processes such as ribosome biogenesis. Thus AMPK is activated by energy deficiency and as a result shuts down high-energy dependent pathways (reviewed in [228–230]). AMPK is overepressed in ovarian carcinomas[231] and endocrine related cancers[228]. The involvement of AMPK in cancer is controversial, some studies suggest that it acts as a tumour suppressor or as an oncogene depending on the cellular context[232, 233]. In terms of Pol I transcription, phosphorylation of RRN3 at residue serine 635 by AMPK impaired the interaction of RRN3 with SL-1 (Figure 3)[234]. Thus, AMPK adapts rRNA synthesis to nutrient availability and the cellular energy status, two processes integral to tumor development and survival.
Finally there are links emerging between the DNA damage response and Pol I transcription mediated via ATM signaling[235]. Genomic instability is a persistent feature of cancer cells, driving the accumulation of oncogenic mutations (reviewed in [236]). Conversely radiation and diverse genotoxic agents, which activate DNA damage pathways are still therapeutic mainstays of many cancer treatment regimens. The response to DNA damage is regulated predominantly by two distinct kinase signaling cascades, the ataxia telangiectasia and Rad3 related (ATR)-CHK1 and ATM-CHK2 pathways, which are activated by single-stranded DNA and DNA double-strand breaks (DSB) respectively (reviewed in [235, [237]). In response to DSBs, ATM functions to initiate DNA repair via homologous recombination by promoting formation of single-stranded DNA at sites of damage through nucleolytic resection[237]. Mutations in ATM cause Ataxia-telangiectasia, a rare autosomal recessive disorder characterized by progressive cerebellar ataxia, oculocutaneous telangiectasias, and variable degrees of immunodeficiency. Moreover somatic mutations in ATM lead to genomic instability and cancer predisposition[239]. Of the DNA damage response genes that function upstream of p53, ATM (which phosphorylates p53 in response to DNA damage) is the most frequently mutated in human cancers, particularly in lung adenocarcinomas, pancreatic cancer and certain hematological malignancies[235, 238–241]. With respect to Pol I, induction of DNA breaks leads to a transient repression in Pol I transcription mediated by ATM kinase activity and the repair factor proteins nijmegen breakage syndrome 1 (NBS1) and mediator of DNA damage checkpoint protein 1 (MDC1). Specifically, DNA lesions interfere with the assembly of the Pol I initiation complex thus leading to a premature displacement of the elongating holoenzymes from the rDNA[234]. What is less clear is the target(s) in the Pol I apparatus modulated by the ATM/NBS1/MDC1-dependent pathways. Similarly, to what extent repression of Pol I activity in cancer is associated with mutated ATM or Ataxia-telangiectasia and the extent to which this contributes to the disease etiologies[1, 242–244].
To further complicate the situation, some of the key regulators of Pol I transcription are also subjected to postranslational modifications other than phosphorylation, such as acetylation (Figure 3), mediated by factors also implicated in malignant transformation. For example, UBF is acetylated by the CREB-binding protein and p300 leading to increased rDNA transcription[245, 246]. Conversely, UBF deacetylation by HDAC1 inhibits the ability of UBF to interact with PAF53, thus preventing the efficient assembly of functional Pol I transcription complexes[162, 247]. A further study suggested histone acetyl-transferase (hALP) acetylates UBF and increased its association with PAF53 thus promoting rDNA transcription[248, 249]. The TAFI68 subunit of SL-1 is also acetylated, in this case, by PCAF and deacetlyated by SIRT1/mSir2 resulting in enhancement and repression of transcription respectively[250].
The above list of factors dysregulated in cancer and reported to modulate Pol I transcription is not exhaustive. A host of other factors have been implicated in regulating Pol I transcription and malignancy, including the proline, glutamate and leucine rich protein 1 (PELP1)[251], the histone methyltransferase G9a[52], nuclear actin, myosin (NM1)[244], PAF1C involved in Pol I and Pol II elongation[252, 253], components of the DNA repair and replication process[216]such as Topoisomerase I and IIα, Ku70/80, PCNA, TFIIH and CSB. Indeed, as our knowledge of the components and modulators of Pol I transcription increases so will the number of targets dysregulated in cancer. The major challenge will be to determine the extent to which Pol I regulation by these pathways contributes to their oncogenic signaling ability and whether any Pol I components specifically modulated by these pathways might serve as targets for novel therapeutics to treat cancer.
4. Targeting Pol I transcription as a therapy for disease
The previous sections have summarized the plethora of data demonstrating that Pol I transcription of the rRNA genes is aberrantly regulated in a broad range of diseases associated with dysregulation of cellular growth, most notable muscle hypertrophy and cancer. This dysregulation is achieved either through direct mutations in components of the Pol I transcription apparatus, as found in ribosomopathies, or more commonly through direct or indirect effects of oncogenic and tumour suppressor signaling. With respect to the ribosomopathies, it is highly likely that, at least in some cases, lesions in the Pol I apparatus may be directly causative in the disease pathology[10, 29]. Unfortunately, in terms of therapy for ribosomopathies, short of gene therapy approaches to correct these lesions, there are no current treatments for these diseases.
In contrast to ribosomopathies, to date, there is no direct evidence that the accelerated rDNA transcription associated with cancer is sufficient to initiate malignant transformation. Demonstrating this has remained elusive as in the experimental setting it is difficult, if not impossible, to selectively drive ribosome biogenesis through the over-activation of a single rDNA transcription component. This is most likely because rDNA transcription is tightly coupled to downstream processes, such as rRNA processing, ribosome assembly and transport. Thus “gain of function” at any single step simply leads to a further downstream step becoming rate limiting. The only factors that can affect a robust increase in Pol I activity and cause accelerated ribosome synthesis are broad regulators of cell growth, such as c-MYC and AKT. c-MYC and AKT activate multiple targets of the Pol I transcription system and also regulate downstream processes such as processing[133, 202]. However, these factors are also transforming due to their pleiotropic effects on many aspects of malignant transformation, in addition to their role in ribosome biogenesis.
However, this does not mean that tumour cells cannot become ”addicted" to accelerated ribosome biogenesis and therefore selectively vulnerable to therapeutics that block or inhibit rRNA synthesis. Indeed, historically there have been numerous clinically approved drugs whose therapeutic affect is, at least in part, mediated through disrupting ribosome biogenesis including actinomycin D, cisplatin, ironotican/topotican, mitomycin C, 5-Fluorouracil and temsirolimus (reviewed in [1]). Actinoymcin D intercalates GC-rich duplex DNA in the rDNA repeats. Cisplatin crosslinks DNA with a high affinity for HMG-proteins, thus hijacking UBF from its site of action and inhibiting Pol I activity. There is also a tight correlation between UBF level and cisplatin sensitivity. Irinotican and topotican function by trapping Topoisomerase I to the rDNA resulting in DNA strand breaks thus mechanically prevents Pol I transcription. Mitomycin C alkylates guanosines thus inducing crosslinking in the rDNA while 5-Fluorouracil incorporates into the 47S pre-RNA thus inhibits processing. Further examples are rapamycin/temsirolimus, which inhibit rRNA synthesis by interfering with mTORC1 activity[199]. However, none of these drugs are selective enough for Pol I transcription to allow definitive conclusions on how much their therapeutic effect is mediated via Pol I.
An important recent advance in this respect has been the emergence of the first small molecule inhibitors, which preferentially target Pol I transcription. The first of these was CX-3543 (quarfloxin), which disrupts nucleolin/rDNA G-quadruplex complexes thus inhibiting Pol I transcription and inducing apoptotic death of cancer cells[254]. The next generation of these targeted inhibitors includes the small molecule, CX-5461, which specifically prevents SL-1 binding to the rDNA promoter resulting in potent inhibition of Pol I transcription[23]. Building on this data, the same group used CX-5461 and genetic approaches to provide unequivocal evidence that accelerated rDNA transcription and nucleolar integrity are necessary for oncogenic activity in hematologic tumor cells[22]. Furthermore, they demonstrated that Pol I transcription could be selectively targeted in vivo to therapeutically treat tumors in both genetically engineered and xenograft models of lymphoma and leukemia through the activation of p53-dependent apoptosis, while sparing normal cells[22]. Intriguingly, the induction of p53 mediated apoptotic death of the tumor cells was rapid, occurring within hours of treatment as a result of nucleolar stress and was independent of changes in total ribosome levels or protein translation. This later observation is important as it demonstrates that Pol I transcription and nucleolar integrity are required for the survival of certain tumour cells, independent of their function in regulating ribosome levels, protein translation and proliferative growth. These findings lend strong support for the evolving paradigm of the nucleolus being a key regulator of the biology of the cancer cells distinct from its role in determining the abundance of ribosomes[255].
A number of important questions about the concept of targeting Pol I in disease arise from the Pol I inhibitor studies. Firstly, what predicts sensitivity to selective Pol I inhibition? The malignant B cells underwent apoptosis in response to Pol I inhibition, but normal B cells did not; Why?[22]. In the same vein, while hematologic malignancies appear to be universally sensitive to Pol I inhibition, which is dependent on a functional p53 pathway, the sensitivity of solid tumours to CX-5461, is more variable and not p53 dependent[23]. One possibility is that hematologic malignancies have a unique nucleolar biology that makes them extremely sensitive to induction of p53-mediated apoptosis following inhibition of Pol I transcription. This is consistent with the observation that mutations in genes encoding Rps mediate reduced ribosome biogenesis and activation of p53 which is a common theme among bone marrow failure syndromes such as Diamond Blackfan Anemia and the acquired 5q-myelodysplastic syndrome which also exhibit increased cancer susceptibility (Table 1).
An equally interesting question is what confers the pre-existing or acquired resistance to Pol I inhibitors. Prolonged dosing of CX-5461 in mice bearing Eμ-MYC lymphomas resulted in a period of disease remission but the mice eventually relapse due to acquired resistance[22]. Understanding the mechanism behind such resistance should allow for the prediction of which cancers might respond to Pol I inhibition and aid in the rational design of combination therapies.
Currently data from murine models and xenograft studies in cancer suggest that patients with hematologic malignancies might represent a highly sensitive population for the first trial of CX-5461 in humans. Indeed, one of the most exciting developments for the Pol I transcription field with respect to human disease is the initiation of the first clinical trial of CX-5461 in lymphoma and leukaemia patients in Melbourne, Australia by the Peter MacCallum Cancer Centre in collaboration with Cylene Pharmaceuticals (RD Hannan, personal communication). The trial consists of a dose escalation phase and an expansion cohort at the maximum tolerated dose that will more robustly examine proof-of-mechanism and predictive biomarkers. Optimistically, this may represent the dawn of a new age for targeting Pol I transcription in human malignancies.
5. Summary and Perspectives
Ribosome biogenesis is fundamental for cell growth, proliferation and survival. However, it is now becoming clear that deranged ribosome synthesis and function underlies a growing list of ribosomopathies that often result in catastrophic outcomes for patients. These syndromes are due to germline and/or somatic mutations, and include Treacher Collins and Shwachman-Diamond syndrome. While deranged rDNA transcription almost certainly drives Treacher Collins syndrome, the diverse phenotypes observed with other ribosomopathies specifically associated with defective Pol I transcription, and potentially additional extra-ribosomal effects of the mutated genes involved, means that the direct role of the altered Pol I transcription remains to be elucidated. Strikingly however, ribosomopathies share a number of common symptoms including bone marrow failure, hematological dysfunction, immune abnormalities, cranio-facial defects, premature aging and cancer predisposition[4, 24]. This commonality of symptoms is highly supportive of the importance ribosome dysfunction plays in these diseases. While the underlying mechanisms remain elusive, it seems likely that they are associated with cell type specific effects on nucleolar surveillance pathways and changes in translational profiles. Intensive efforts are being made to define these mechanisms and hopefully these will provide at least partial treatment options for these severe disease syndromes.
It is also clear that Pol I transcription is consistently dysregulated in a wide range of diseases associated with maladaptive growth including pathological muscle hypertrophy and cancer. In these diseases altered Pol I transcription is achieved through direct modulation by oncogenes, tumor suppressors (eg., c-MYC, p53 and pRb) or via altered tumour signaling pathways. While tumour signaling has been the focus of considerable attention by pharmaceutical companies, due to their broad effects on a multitude of cellular processes it has not been possible to determine the precise contribution, if any, that deranged Pol I transcription makes to the disease phenotype. However, the recent discovery of selective, small molecule inhibitors of Pol I provides an unheralded opportunity to probe the relationship between deranged Pol I transcription and disease. In particular the demonstration that inhibitors of Pol I can selectively kill cancer cells provides an exciting new class of anti-neoplastic drugs that may significantly advance cancer treatment[22]. In particular, hematologic cancers driven by oncogenes, such as c-MYC, that are addicted to dysregulated ribosome biogenesis are often associated with poor prognosis, represent therapeutic opportunities for inhibitors of Pol I transcription.
Of particular relevance to understating the role of Pol I in human disease is the accumulating evidence supporting an evolving paradigm of the nucleolus being a key regulator of cellular biology, distinct from its role in determining the abundance of the ribosomes[255]. These functions notably include titration of oncogenes and tumor suppressors (eg., pRb-MDM2-p53: the nucleolar surveillance pathway), and also the sequestration and regulation of numerous other factors with critical roles in cellular homeostasis [256–259]. Thus it follows that as the assembly of the nucleolus is dependent on ongoing rDNA transcription, any perturbations in Pol I transcription associated with disease, has the potential to directly contribute to the disease pathology through the disruption of extra-ribosomal functions of the nucleolus[128]. Defining precisely the relationship between nucleolar homeostasis in normal and diseased cells is a major challenge for the future. Importantly, recent advances in defining the nucleolar proteome in response to various stresses will facilitate the advancement of this area of research (reviewed in [260]).
It is evident from the application of deep sequencing to specific disease genomes, that the list of factors directly implicated in Pol I transcription and are dysregulated or subject to mutation in disease is going to increase. In particular, it is becoming increasingly apparent that genetic lesions in factors which mediate chromatin reading, writing or assembly, plays significant roles in the etiology and maintenance of a wide variety of hematologic and solid cancers, plus genetically inherited disease[261]. Along with chromatin modifiers that seem to preferential associate with Pol I[236, 262, 263], there is an expanding list of Pol II related chromatin modifiers that also directly interact with, and control the epigenetic status of the Pol I transcription apparatus[236, 262, 263]. Indeed, the observations in yeast and Drosophila that the level of epigenetic rDNA silencing has profound effects on genome wide heterochromatin, genomic stability and aging (reviewed in [69, 259, 264, 265]) suggests that deranged Pol I transcription may contribute in a much broader sense to disease and function at multiple levels including: rDNA transcription and proliferative capacity; extra-ribosomal functions of the nucleolus; and genome wide effects on the heterochromatin.
In summary, with a few exceptions, the last two decades of study into Pol I transcription have largely focused on purified in vitro systems and cultured cells to dissect the molecular mechanisms by which rDNA transcription is regulated. These studies have been essential to lay a basic framework for understanding Pol I transcription in eukaryotic cells. However, the challenge for the future is to translate this fundamental knowledge into cell and tissue biology based approaches to understand the role PoI I transcription plays in cellular homeostasis and the consequences when it is perturbed during diseases such as cancer. To achieve this will require increased efforts in the design and application of selective inhibitors and conditional genetic approaches that allow specific steps in Pol I transcription and ribosome biogenesis to be manipulated in model systems that faithfully recapitulate their human disease counterparts.
Highlights.
dysregulation of RNA Polymerase I transcription and ribosome biogenesis is linked to a broad range of human diseases
ribosomopathies, hypertrophy, atrophy and cancer are all associated with dysregulation of RNA polymerase I transcription
possible therapeutic approaches to treat patients with dysregulated RNA Polymerase I transcription.
Acknowledgments
Due to space limitations we have not been able to individually cite many of the original publications that have contributed substantially to the field. We sincerely apologize to the authors of these publications. This work was supported by the National Health and Medical Research Council (NHMRC) of Australia project grants; NHMRC Research Fellowship to R.D.H and R.B.P; Grants in Aid from the Prostate Cancer Foundation of Australia and the Leukemia Foundation of Australia; and NIH RO1 to L.I.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
R.D. Hannan is a member of the Scientific Advisory Board for Cylene.
References
- 1.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 2.Grummt I. Wisely chosen paths--regulation of rRNA synthesis: delivered on 30 June 2010 at the 35th FEBS Congress in Gothenburg, Sweden. FEBS J. 2010;277:4626–4639. doi: 10.1111/j.1742-4658.2010.07892.x. [DOI] [PubMed] [Google Scholar]
- 3.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Narla A, Ebert BL. Translational medicine: ribosomopathies. Blood. 2011;118:4300–4301. doi: 10.1182/blood-2011-08-372250. [DOI] [PubMed] [Google Scholar]
- 5.Chiabrando D, Tolosano E. Diamond Blackfan Anemia at the Crossroad between Ribosome Biogenesis and Heme Metabolism. Adv Hematol. 2010;2010 doi: 10.1155/2010/790632. 790632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball S. Diamond Blackfan anemia. Hematology Am Soc Hematol Educ Program. 2011;2011:487–491. doi: 10.1182/asheducation-2011.1.487. [DOI] [PubMed] [Google Scholar]
- 7.Pellagatti A, Hellstrom-Lindberg E, Giagounidis A, Perry J, Malcovati L, Della Porta MG, Jadersten M, Killick S, Fidler C, Cazzola M, Wainscoat JS, Boultwood J. Haploinsufficiency of RPS14 in 5q- syndrome is associated with deregulation of ribosomal- and translation-related genes. Br J Haematol. 2008;142:57–64. doi: 10.1111/j.1365-2141.2008.07178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin CI, Yeh NH. Treacle recruits RNA polymerase I complex to the nucleolus that is independent of UBF. Biochem Biophys Res Commun. 2009;386:396–401. doi: 10.1016/j.bbrc.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 9.Trainor PA, Dixon J, Dixon MJ. Treacher Collins syndrome: etiology, pathogenesis and prevention. Eur J Hum Genet. 2009;17:275–283. doi: 10.1038/ejhg.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci U S A. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grierson PM, Lillard K, Behbehani GK, Combs KA, Bhattacharyya S, Acharya S, Groden J. BLM helicase facilitates RNA polymerase I-mediated ribosomal RNA transcription. Hum Mol Genet. 2012;21:1172–1183. doi: 10.1093/hmg/ddr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiratori M, Suzuki T, Itoh C, Goto M, Furuichi Y, Matsumoto T. WRN helicase accelerates the transcription of ribosomal RNA as a component of an RNA polymerase I-associated complex. Oncogene. 2002;21:2447–2454. doi: 10.1038/sj.onc.1205334. [DOI] [PubMed] [Google Scholar]
- 13.Lutomska A, Lebedev A, Scharffetter-Kochanek K, Iben S. The transcriptional response to distinct growth factors is impaired in Werner syndrome cells. Exp Gerontol. 2008;43:820–826. doi: 10.1016/j.exger.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Ganapathi KA, Austin KM, Lee CS, Dias A, Malsch MM, Reed R, Shimamura A. The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood. 2007;110:1458–1465. doi: 10.1182/blood-2007-02-075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 16.Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, Makitie O, Kaitila I, de la Chapelle A. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 17.Thiel CT, Rauch A. The molecular basis of the cartilage-hair hypoplasia-anauxetic dysplasia spectrum. Best Pract Res Clin Endocrinol Metab. 2011;25:131–142. doi: 10.1016/j.beem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Freed EF, Baserga SJ. The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Res. 2010;38:4798–4806. doi: 10.1093/nar/gkq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed EF, Prieto JL, McCann KL, McStay B, Baserga SJ. NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002892. e1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer B, Wurm JP, Kotter P, Leisegang MS, Schilling V, Buchhaupt M, Held M, Bahr U, Karas M, Heckel A, Bohnsack MT, Wohnert J, Entian KD. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Psi1191 in yeast 18S rRNA. Nucleic Acids Res. 2011;39:1526–1537. doi: 10.1093/nar/gkq931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nousbeck J, Spiegel R, Ishida-Yamamoto A, Indelman M, Shani-Adir A, Adir N, Lipkin E, Bercovici S, Geiger D, van Steensel MA, Steijlen PM, Bergman R, Bindereif A, Choder M, Shalev S, Sprecher E. Alopecia, neurological defects, and endocrinopathy syndrome caused by decreased expression of RBM28, a nucleolar protein associated with ribosome biogenesis. Am J Hum Genet. 2008;82:1114–1121. doi: 10.1016/j.ajhg.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe MK, Ryckman DM, Rice WG, Schmitt C, Lowe SW, Johnstone RW, Pearson RB, McArthur GA, Hannan RD. Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, Streiner N, Quin JE, Sanij E, Bywater MJ, Hannan RD, Ryckman D, Anderes K, Rice WG. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- 24.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fumagalli S, Thomas G. The role of p53 in ribosomopathies. Semin Hematol. 2011;48:97–105. doi: 10.1053/j.seminhematol.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Dixon J, Dixon MJ. Genetic background has a major effect on the penetrance and severity of craniofacial defects in mice heterozygous for the gene encoding the nucleolar protein Treacle. Dev Dyn. 2004;229:907–914. doi: 10.1002/dvdy.20004. [DOI] [PubMed] [Google Scholar]
- 27.Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, Dixon MJ, Trainor PA. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci U S A. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, Glynn EF, Ellington L, Du C, Dixon J, Dixon MJ, Trainor PA. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dauwerse JG, Dixon J, Seland S, Ruivenkamp CA, van Haeringen A, Hoefsloot LH, Peters DJ, Boers AC, Daumer-Haas C, Maiwald R, Zweier C, Kerr B, Cobo AM, Toral JF, Hoogeboom AJ, Lohmann DR, Hehr U, Dixon MJ, Breuning MH, Wieczorek D. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- 30.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 31.Boisvert FM, Ahmad Y, Gierlinski M, Charriere F, Lamont D, Scott M, Barton G, Lamond AI. A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.011429. M111011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edery P, Manach Y, Le Merrer M, Till M, Vignal A, Lyonnet S, Munnich A. Apparent genetic homogeneity of the Treacher Collins-Franceschetti syndrome. Am J Med Genet. 1994;52:174–177. doi: 10.1002/ajmg.1320520210. [DOI] [PubMed] [Google Scholar]
- 33.Conte C, D'Apice MR, Rinaldi F, Gambardella S, Sangiuolo F, Novelli G. Novel mutations of TCOF1 gene in European patients with Treacher Collins syndrome. BMC Med Genet. 2011;12:125. doi: 10.1186/1471-2350-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon MJ, Read AP, Donnai D, Colley A, Dixon J, Williamson R. The gene for Treacher Collins syndrome maps to the long arm of chromosome 5. Am J Hum Genet. 1991;49:17–22. [PMC free article] [PubMed] [Google Scholar]
- 35.Marszalek-Kruk BA, Wojcicki P, Smigiel R, Trzeciak WH. Novel insertion in exon 5 of the TCOF1 gene in twin sisters with Treacher Collins syndrome. J Appl Genet. 2012;53:279–282. doi: 10.1007/s13353-012-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakai D, Dixon J, Dixon MJ, Trainor PA. Mammalian neurogenesis requires Treacle-Plk1 for precise control of spindle orientation, mitotic progression, and maintenance of neural progenitor cells. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002566. e1002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tikoo S, Sengupta S. Time to bloom. Genome Integr. 2010;1:14. doi: 10.1186/2041-9414-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chester N, Kuo F, Kozak C, O'Hara CD, Leder P. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom's syndrome gene. Genes Dev. 1998;12:3382–3393. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc Natl Acad Sci U S A. 1998;95:13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombard DB, Beard C, Johnson B, Marciniak RA, Dausman J, Bronson R, Buhlmann JE, Lipman R, Curry R, Sharpe A, Jaenisch R, Guarente L. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol Cell Biol. 2000;20:3286–3291. doi: 10.1128/mcb.20.9.3286-3291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yankiwski V, Marciniak RA, Guarente L, Neff NF. Nuclear structure in normal and Bloom syndrome cells. Proc Natl Acad Sci U S A. 2000;97:5214–5219. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schawalder J, Paric E, Neff NF. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell Biol. 2003;4:15. doi: 10.1186/1471-2121-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Smith K, Waldman BC, Waldman AS. Depletion of the bloom syndrome helicase stimulates homology-dependent repair at doublestrand breaks in human chromosomes. DNA Repair (Amst) 2011;10:416–426. doi: 10.1016/j.dnarep.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaneko H, Fukao T, Kasahara K, Yamada T, Kondo N. Augmented cell death with Bloom syndrome helicase deficiency. Mol Med Report. 2011;4:607–609. doi: 10.3892/mmr.2011.484. [DOI] [PubMed] [Google Scholar]
- 46.Natale V. A comprehensive description of the severity groups in Cockayne syndrome. Am J Med Genet A. 2011;155A:1081–1095. doi: 10.1002/ajmg.a.33933. [DOI] [PubMed] [Google Scholar]
- 47.Laugel V, Dalloz C, Durand M, Sauvanaud F, Kristensen U, Vincent MC, Pasquier L, Odent S, Cormier-Daire V, Gener B, Tobias ES, Tolmie JL, Martin-Coignard D, Drouin-Garraud V, Heron D, Journel H, Raffo E, Vigneron J, Lyonnet S, Murday V, Gubser-Mercati D, Funalot B, Brueton L, Sanchez Del Pozo J, Munoz E, Gennery AR, Salih M, Noruzinia M, Prescott K, Ramos L, Stark Z, Fieggen K, Chabrol B, Sarda P, Edery P, Bloch-Zupan A, Fawcett H, Pham D, Egly JM, Lehmann AR, Sarasin A, Dollfus H. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31:113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- 48.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 49.Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 50.Lebedev A, Scharffetter-Kochanek K, Iben S. Truncated Cockayne syndrome B protein represses elongation by RNA polymerase I. J Mol Biol. 2008;382:266–274. doi: 10.1016/j.jmb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 51.Assfalg R, Lebedev A, Gonzalez OG, Schelling A, Koch S, Iben S. TFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res. 2012;40:650–659. doi: 10.1093/nar/gkr746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 53.Latini P, Frontini M, Caputo M, Gregan J, Cipak L, Filippi S, Kumar V, Velez-Cruz R, Stefanini M, Proietti-De-Santis L. CSA and CSB proteins interact with p53 and regulate its Mdm2-dependent ubiquitination. Cell Cycle. 2011;10:3719–3730. doi: 10.4161/cc.10.21.17905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lake RJ, Basheer A, Fan HY. Reciprocally regulated chromatin association of Cockayne syndrome protein B and p53 protein. J Biol Chem. 2011;286:34951–34958. doi: 10.1074/jbc.M111.252643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frontini M, Proietti-De-Santis L. Interaction between the Cockayne syndrome B and p53 proteins: implications for aging. Aging (Albany NY) 2012;4:89–97. doi: 10.18632/aging.100439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu A, Fan HY, Liao D, Bailey AD, Weiner AM. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol Cell. 2000;5:801–810. doi: 10.1016/s1097-2765(00)80320-2. [DOI] [PubMed] [Google Scholar]
- 57.Laumonnier F, Holbert S, Ronce N, Faravelli F, Lenzner S, Schwartz CE, Lespinasse J, Van Esch H, Lacombe D, Goizet C, Phan-Dinh Tuy F, van Bokhoven H, Fryns JP, Chelly J, Ropers HH, Moraine C, Hamel BC, Briault S. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lisik MZ, Sieron AL. X-linked mental retardation. Med Sci Monit. 2008;14:RA221–RA229. [PubMed] [Google Scholar]
- 59.Dawson MA, Bannister AJ. Demethylases go mental. Mol Cell. 2010;38:155–157. doi: 10.1016/j.molcel.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 60.Abidi FE, Miano MG, Murray JC, Schwartz CE. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin Genet. 2007;72:19–22. doi: 10.1111/j.1399-0004.2007.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, Helin K. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CD, Klose RJ, Ratcliffe PJ, Schofield CJ. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum Mol Genet. 2010;19:217–222. doi: 10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu J, Shi G, Jia Y, Li J, Wu M, Dong S, Wong J. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 2010;20:908–918. doi: 10.1038/cr.2010.81. [DOI] [PubMed] [Google Scholar]
- 64.Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, Yaghi NK, Lim H, Garcia BA, Brizuela L, Zhao K, Roberts TM, Shi Y. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Z, Wang Y, Li X, Xu L, Wang X, Sun T, Dong X, Chen L, Mao H, Yu Y, Li J, Chen PA, Chen CD. PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res. 2010;20:794–801. doi: 10.1038/cr.2010.75. [DOI] [PubMed] [Google Scholar]
- 68.Bose T, Lee KK, Lu S, Xu B, Harris B, Slaughter B, Unruh J, Garrett A, McDowell W, Box A, Li H, Peak A, Ramachandran S, Seidel C, Gerton JL. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002749. e1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327:693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 70.Liu J, Krantz ID. Cohesin and human disease. Annu Rev Genomics Hum Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Monnich M, Kuriger Z, Print CG, Horsfield JA. A zebrafish model of Roberts syndrome reveals that Esco2 depletion interferes with development by disrupting the cell cycle. PLoS One. 2011;6:e20051. doi: 10.1371/journal.pone.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Najib S, Saint-Laurent N, Esteve JP, Schulz S, Boutet-Robinet E, Fourmy D, Lattig J, Mollereau C, Pyronnet S, Susini C, Bousquet C. A switch of G protein-coupled receptor binding preference from phosphoinositide 3-kinase (PI3K)-p85 to filamin A negatively controls the PI3K pathway. Mol Cell Biol. 2012;32:1004–1016. doi: 10.1128/MCB.06252-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou AX, Hartwig JH, Akyurek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 2010;20:113–123. doi: 10.1016/j.tcb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 74.Velkova A, Carvalho MA, Johnson JO, Tavtigian SV, Monteiro AN. Identification of Filamin A as a BRCA1-interacting protein required for efficient DNA repair. Cell Cycle. 2010;9:1421–1433. doi: 10.4161/cc.9.7.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan Y, Shen Z. Interaction with BRCA2 suggests a role for filamin-1 (hsFLNa) in DNA damage response. J Biol Chem. 2001;276:48318–48324. doi: 10.1074/jbc.M102557200. [DOI] [PubMed] [Google Scholar]
- 76.Yue J, Lu H, Liu J, Berwick M, Shen Z. Filamin-A as a marker and target for DNA damage based cancer therapy. DNA Repair (Amst) 2012;11:192–200. doi: 10.1016/j.dnarep.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deng W, Lopez-Camacho C, Tang JY, Mendoza-Villanueva D, Maya-Mendoza A, Jackson DA, Shore P. Cytoskeletal protein filamin A is a nucleolar protein that suppresses ribosomal RNA gene transcription. Proc Natl Acad Sci U S A. 2012;109:1524–1529. doi: 10.1073/pnas.1107879109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shenoy N, Kessel R, Bhagat T, Bhattacharya S, Yu Y, McMahon C, Verma A. Alterations in the ribosomal machinery in cancer and hematologic disorders. J Hematol Oncol. 2012;5:32. doi: 10.1186/1756-8722-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Flygare J, Kiefer T, Miyake K, Utsugisawa T, Hamaguchi I, Da Costa L, Richter J, Davey EJ, Matsson H, Dahl N, Wiznerowicz M, Trono D, Karlsson S. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105:4627–4634. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- 80.Matsson H, Davey EJ, Draptchinskaia N, Hamaguchi I, Ooka A, Leveen P, Forsberg E, Karlsson S, Dahl N. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol. 2004;24:4032–4037. doi: 10.1128/MCB.24.9.4032-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miyake K, Flygare J, Kiefer T, Utsugisawa T, Richter J, Ma Z, Wiznerowicz M, Trono D, Karlsson S. Development of cellular models for ribosomal protein S19 (RPS19)-deficient diamond-blackfan anemia using inducible expression of siRNA against RPS19. Mol Ther. 2005;11:627–637. doi: 10.1016/j.ymthe.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 82.Choesmel V, Fribourg S, Aguissa-Toure AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17:1253–1263. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 83.Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fumagalli S, Ivanenkov VV, Teng T, Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26:1028–1040. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, Kutok JL, Ebert BL. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moniz H, Gastou M, Leblanc T, Hurtaud C, Cretien A, Lecluse Y, Raslova H, Larghero J, Croisille L, Faubladier M, Bluteau O, Lordier L, Tchernia G, Vainchenker W, Mohandas N, Da Costa L. Primary hematopoietic cells from DBA patients with mutations in RPL11 and RPS19 genes exhibit distinct erythroid phenotype in vitro. Cell Death Dis. 2012;3:e356. doi: 10.1038/cddis.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sieff CA, Yang J, Merida-Long LB, Lodish HF. Pathogenesis of the erythroid failure in Diamond Blackfan anaemia. Br J Haematol. 2010;148:611–622. doi: 10.1111/j.1365-2141.2009.07993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 89.Duan X, Kelsen SG, Clarkson AB, Jr, Ji R, Merali S. SILAC analysis of oxidative stress-mediated proteins in human pneumocytes: new role for treacle. Proteomics. 2010;10:2165–2174. doi: 10.1002/pmic.201000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Bjorling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 91.Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 92.White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 93.Hannan RD, Jenkins A, Jenkins AK, Brandenburger Y. Cardiac hypertrophy: a matter of translation. Clin Exp Pharmacol Physiol. 2003;30:517–527. doi: 10.1046/j.1440-1681.2003.03873.x. [DOI] [PubMed] [Google Scholar]
- 94.Onyezili FN. On two paradoxical side-effects of prednisolone in rats, ribosomal RNA biosyntheses, and a mechanism of action. Biochem Pharmacol. 1986;35:2309–2312. doi: 10.1016/0006-2952(86)90456-9. [DOI] [PubMed] [Google Scholar]
- 95.Kuwahara K, Nishikimi T, Nakao K. Transcriptional regulation of the fetal cardiac gene program. J Pharmacol Sci. 2012;119:198–203. doi: 10.1254/jphs.12r04cp. [DOI] [PubMed] [Google Scholar]
- 96.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 97.Chein KR, Grace AA, Hunter JJ. Molecular basis of cardiac hypertrophy and failure. Philadelphia: WB Saunders Co; 1998. [Google Scholar]
- 98.Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell. 2001;8:1063–1073. doi: 10.1016/s1097-2765(01)00384-7. [DOI] [PubMed] [Google Scholar]
- 99.Sala V, Gallo S, Leo C, Gatti S, Gelb BD, Crepaldi T. Signaling to cardiac hypertrophy: insights from human and mouse RASopathies. Mol Med. 2012 doi: 10.2119/molmed.2011.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morgan HE, Siehl D, Chua BH, Lautensack-Belser N. Faster protein and ribosome synthesis in hypertrophying heart. Basic Res Cardiol. 1985;80(Suppl 2):115–118. [PubMed] [Google Scholar]
- 101.Siehl D, Chua BH, Lautensack-Belser N, Morgan HE. Faster protein and ribosome synthesis in thyroxine-induced hypertrophy of rat heart. Am J Physiol. 1985;248:C309–C319. doi: 10.1152/ajpcell.1985.248.3.C309. [DOI] [PubMed] [Google Scholar]
- 102.McDermott PJ, Rothblum LI, Smith SD, Morgan HE. Accelerated rates of ribosomal RNA synthesis during growth of contracting heart cells in culture. J Biol Chem. 1989;264:18220–18227. [PubMed] [Google Scholar]
- 103.Hannan RD, Luyken J, Rothblum LI. Regulation of ribosomal DNA transcription during contraction-induced hypertrophy of neonatal cardiomyocytes. J Biol Chem. 1996;271:3213–3220. doi: 10.1074/jbc.271.6.3213. [DOI] [PubMed] [Google Scholar]
- 104.Hannan RD, Luyken J, Rothblum LI. Regulation of rDNA transcription factors during cardiomyocyte hypertrophy induced by adrenergic agents. J Biol Chem. 1995;270:8290–8297. doi: 10.1074/jbc.270.14.8290. [DOI] [PubMed] [Google Scholar]
- 105.Luyken J, Hannan RD, Cheung JY, Rothblum LI. Regulation of rDNA transcription during endothelin-1-induced hypertrophy of neonatal cardiomyocytes. Hyperphosphorylation of upstream binding factor, an rDNA transcription factor. Circ Res. 1996;78:354–361. doi: 10.1161/01.res.78.3.354. [DOI] [PubMed] [Google Scholar]
- 106.Hannan RD, Stefanovsky V, Taylor L, Moss T, Rothblum LI. Overexpression of the transcription factor UBF1 is sufficient to increase ribosomal DNA transcription in neonatal cardiomyocytes: implications for cardiac hypertrophy. Proc Natl Acad Sci U S A. 1996;93:8750–8755. doi: 10.1073/pnas.93.16.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brandenburger Y, Jenkins A, Autelitano DJ, Hannan RD. Increased expression of UBF is a critical determinant for rRNA synthesis and hypertrophic growth of cardiac myocytes. FASEB J. 2001;15:2051–2053. doi: 10.1096/fj.01-0853fje. [DOI] [PubMed] [Google Scholar]
- 108.Brandenburger Y, Arthur JF, Woodcock EA, Du XJ, Gao XM, Autelitano DJ, Rothblum LI, Hannan RD. Cardiac hypertrophy in vivo is associated with increased expression of the ribosomal gene transcription factor UBF. FEBS Lett. 2003;548:79–84. doi: 10.1016/s0014-5793(03)00744-0. [DOI] [PubMed] [Google Scholar]
- 109.Rosello-Lleti E, Rivera M, Cortes R, Azorin I, Sirera R, Martinez-Dolz L, Hove L, Cinca J, Lago F, Gonzalez-Juanatey JR, Salvador A, Portoles M. Influence of heart failure on nucleolar organization and protein expression in human hearts. Biochem Biophys Res Commun. 2012;418:222–228. doi: 10.1016/j.bbrc.2011.12.151. [DOI] [PubMed] [Google Scholar]
- 110.Rohini A, Agrawal N, Koyani CN, Singh R. Molecular targets and regulators of cardiac hypertrophy. Pharmacol Res. 2010;61:269–280. doi: 10.1016/j.phrs.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 111.Ruwhof C, van der Laarse A. Mechanical stress-induced cardiac hypertrophy: mechanisms and signal transduction pathways. Cardiovasc Res. 2000;47:23–37. doi: 10.1016/s0008-6363(00)00076-6. [DOI] [PubMed] [Google Scholar]
- 112.Chaudhary A, King WG, Mattaliano MD, Frost JA, Diaz B, Morrison DK, Cobb MH, Marshall MS, Brugge JS. Phosphatidylinositol 3-kinase regulates Raf1 through Pak phosphorylation of serine 338. Curr Biol. 2000;10:551–554. doi: 10.1016/s0960-9822(00)00475-9. [DOI] [PubMed] [Google Scholar]
- 113.Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, Bos JM, Ommen SR, Esposito G, Lepri F, Faul C, Mundel P, Lopez Siguero JP, Tenconi R, Selicorni A, Rossi C, Mazzanti L, Torrente I, Marino B, Digilio MC, Zampino G, Ackerman MJ, Dallapiccola B, Tartaglia M, Gelb BD. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 114.Vettel C, Wittig K, Vogt A, Wuertz CM, El-Armouche A, Lutz S, Wieland T. A novel player in cellular hypertrophy: G(i)betagamma/PI3K-dependent activation of the RacGEF TIAM-1 is required for alpha(1)-adrenoceptor induced hypertrophy in neonatal rat cardiomyocytes. J Mol Cell Cardiol. 2012 doi: 10.1016/j.yjmcc.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 115.Zhong W, Mao S, Tobis S, Angelis E, Jordan MC, Roos KP, Fishbein MC, de Alboran IM, MacLellan WR. Hypertrophic growth in cardiac myocytes is mediated by Myc through a Cyclin D2-dependent pathway. EMBO J. 2006;25:3869–3879. doi: 10.1038/sj.emboj.7601252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xiao G, Mao S, Baumgarten G, Serrano J, Jordan MC, Roos KP, Fishbein MC, MacLellan WR. Inducible activation of c-Myc in adult myocardium in vivo provokes cardiac myocyte hypertrophy and reactivation of DNA synthesis. Circ Res. 2001;89:1122–1129. doi: 10.1161/hh2401.100742. [DOI] [PubMed] [Google Scholar]
- 117.Hershey JC, Hautmann M, Thompson MM, Rothblum LI, Haystead TA, Owens GK. Angiotensin II-induced hypertrophy of rat vascular smooth muscle is associated with increased 18 S rRNA synthesis and phosphorylation of the rRNA transcription factor, upstream binding factor. J Biol Chem. 1995;270:25096–25101. doi: 10.1074/jbc.270.42.25096. [DOI] [PubMed] [Google Scholar]
- 118.Goodfellow SJ, Innes F, Derblay LE, MacLellan WR, Scott PH, White RJ. Regulation of RNA polymerase III transcription during hypertrophic growth. EMBO J. 2006;25:1522–1533. doi: 10.1038/sj.emboj.7601040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vallon V, Thomson SC. Renal function in diabetic disease models: the tubular system in the pathophysiology of the diabetic kidney. Annu Rev Physiol. 2012;74:351–375. doi: 10.1146/annurev-physiol-020911-153333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mariappan MM, D'Silva K, Lee MJ, Sataranatarajan K, Barnes JL, Choudhury GG, Kasinath BS. Ribosomal biogenesis induction by high glucose requires activation of upstream binding factor in kidney glomerular epithelial cells. Am J Physiol Renal Physiol. 2011;300:F219–F230. doi: 10.1152/ajprenal.00207.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Melvin WT, Kumar A, Malt RA. Conservation of ribosomal RNA during compensatory renal hypertrophy. A major mechanism in RNA accretion. J Cell Biol. 1976;69:548–556. doi: 10.1083/jcb.69.3.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Melvin WT, Kumar A, Malt RA. Synthesis and conservation of ribosomal proteins during compensatory renal hypertrophy. Biochem J. 1980;188:229–235. doi: 10.1042/bj1880229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fanzani A, Conraads VM, Penna F, Martinet W. Molecular and cellular mechanisms of skeletal muscle atrophy: an update. J Cachexia Sarcopenia Muscle. 2012 doi: 10.1007/s13539-012-0074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Powers SK, Smuder AJ, Judge AR. Oxidative stress and disuse muscle atrophy: cause or consequence? Curr Opin Clin Nutr Metab Care. 2012;15:240–245. doi: 10.1097/MCO.0b013e328352b4c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Machida M, Takeda K, Yokono H, Ikemune S, Taniguchi Y, Kiyosawa H, Takemasa T. Reduction of ribosome biogenesis with activation of the mTOR pathway in denervated atrophic muscle. J Cell Physiol. 2012;227:1569–1576. doi: 10.1002/jcp.22871. [DOI] [PubMed] [Google Scholar]
- 126.von Walden F, Casagrande V, Ostlund Farrants AK, Nader GA. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. Am J Physiol Cell Physiol. 2012;302:C1523–C1530. doi: 10.1152/ajpcell.00460.2011. [DOI] [PubMed] [Google Scholar]
- 127.Poortinga G, Wall M, Sanij E, Siwicki K, Ellul J, Brown D, Holloway TP, Hannan RD, McArthur GA. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 2011;39:3267–3281. doi: 10.1093/nar/gkq1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shiue CN, Arabi A, Wright AP. Nucleolar organization, growth control and cancer. Epigenetics. 2010;5 doi: 10.4161/epi.5.3.11376. [DOI] [PubMed] [Google Scholar]
- 129.Williamson D, Lu YJ, Fang C, Pritchard-Jones K, Shipley J. Nascent pre-rRNA overexpression correlates with an adverse prognosis in alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2006;45:839–845. doi: 10.1002/gcc.20347. [DOI] [PubMed] [Google Scholar]
- 130.Uemura M, Zheng Q, Koh CM, Nelson WG, Yegnasubramanian S, De Marzo AM. Overexpression of ribosomal RNA in prostate cancer is common but not linked to rDNA promoter hypomethylation. Oncogene. 2012;31:1254–1263. doi: 10.1038/onc.2011.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 132.Hannan KM, Sanij E, Hein N, Hannan RD, Pearson RB. Signaling to the ribosome in cancer--It is more than just mTORC1. IUBMB Life. 2011;63:79–85. doi: 10.1002/iub.428. [DOI] [PubMed] [Google Scholar]
- 133.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 134.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gomez-Roman N, Felton-Edkins ZA, Kenneth NS, Goodfellow SJ, Athineos D, Zhang J, Ramsbottom BA, Innes F, Kantidakis T, Kerr ER, Brodie J, Grandori C, White RJ. Activation by c-Myc of transcription by RNA polymerases I, II and III. Biochem Soc Symp. 2006:141–154. doi: 10.1042/bss0730141. [DOI] [PubMed] [Google Scholar]
- 136.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010;20:91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Varlakhanova NV, Knoepfler PS. Acting locally and globally: Myc's ever-expanding roles on chromatin. Cancer Res. 2009;69:7487–7490. doi: 10.1158/0008-5472.CAN-08-4832. [DOI] [PubMed] [Google Scholar]
- 139.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 140.Poortinga G, Hannan KM, Snelling H, Walkley CR, Jenkins A, Sharkey K, Wall M, Brandenburger Y, Palatsides M, Pearson RB, McArthur GA, Hannan RD. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–3335. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, Moss T, Rothblum L, Hannan KM, McArthur GA, Pearson RB, Hannan RD. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics. 2009;4:374–382. doi: 10.4161/epi.4.6.9449. [DOI] [PubMed] [Google Scholar]
- 143.Huang R, Wu T, Xu L, Liu A, Ji Y, Hu G. Upstream binding factor up-regulated in hepatocellular carcinoma is related to the survival and cisplatin-sensitivity of cancer cells. FASEB J. 2002;16:293–301. doi: 10.1096/fj.01-0687com. [DOI] [PubMed] [Google Scholar]
- 144.Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG, Wright AP. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 145.Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 146.Shiue CN, Berkson RG, Wright AP. c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene. 2009;28:1833–1842. doi: 10.1038/onc.2009.21. [DOI] [PubMed] [Google Scholar]
- 147.Kenneth NS, Ramsbottom BA, Gomez-Roman N, Marshall L, Cole PA, White RJ. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc Natl Acad Sci U S A. 2007;104:14917–14922. doi: 10.1073/pnas.0702909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Reikvam H, Hatfield KJ, Kittang AO, Hovland R, Bruserud O. Acute myeloid leukemia with the t(8;21) translocation: clinical consequences and biological implications. J Biomed Biotechnol. 2011;2011:104631. doi: 10.1155/2011/104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bakshi R, Zaidi SK, Pande S, Hassan MQ, Young DW, Montecino M, Lian JB, van Wijnen AJ, Stein JL, Stein GS. The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. J Cell Sci. 2008;121:3981–3990. doi: 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li Z, Hann SR. Nucleophosmin is essential for c-Myc nucleolar localization and c-Myc-mediated rDNA transcription. Oncogene. 2012 doi: 10.1038/onc.2012.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Colombo E, Alcalay M, Pelicci PG. Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene. 2011;30:2595–2609. doi: 10.1038/onc.2010.646. [DOI] [PubMed] [Google Scholar]
- 152.Meani N, Alcalay M. Role of nucleophosmin in acute myeloid leukemia. Expert Rev Anticancer Ther. 2009;9:1283–1294. doi: 10.1586/era.09.84. [DOI] [PubMed] [Google Scholar]
- 153.Rau R, Brown P. Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: towards definition of a new leukaemia entity. Hematol Oncol. 2009;27:171–181. doi: 10.1002/hon.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ramos-Echazabal G, Chinea G, Garcia-Fernandez R, Pons T. In silico studies of potential phosphoresidues in the human nucleophosmin/B23: its kinases and related biological processes. J Cell Biochem. 2012;113:2364–2374. doi: 10.1002/jcb.24108. [DOI] [PubMed] [Google Scholar]
- 155.Bergstralh DT, Conti BJ, Moore CB, Brickey WJ, Taxman DJ, Ting JP. Global functional analysis of nucleophosmin in Taxol response, cancer, chromatin regulation, and ribosomal DNA transcription. Exp Cell Res. 2007;313:65–76. doi: 10.1016/j.yexcr.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li Z, Hann SR. The Myc-nucleophosmin-ARF network: a complex web unveiled. Cell Cycle. 2009;8:2703–2707. doi: 10.4161/cc.8.17.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 159.Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Blyth K, Vaillant F, Jenkins A, McDonald L, Pringle MA, Huser C, Stein T, Neil J, Cameron ER. Runx2 in normal tissues and cancer cells: A developing story. Blood Cells Mol Dis. 2010;45:117–123. doi: 10.1016/j.bcmd.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 161.Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 162.Ali SA, Dobson JR, Lian JB, Stein JL, van Wijnen AJ, Zaidi SK, Stein GS. A RUNX2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation. J Cell Sci. 2012;125:2732–2739. doi: 10.1242/jcs.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chinnam M, Goodrich DW. RB1, development, and cancer. Curr Top Dev Biol. 2011;94:129–169. doi: 10.1016/B978-0-12-380916-2.00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Manning AL, Dyson NJ. pRB, a tumor suppressor with a stabilizing presence. Trends Cell Biol. 2011;21:433–441. doi: 10.1016/j.tcb.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sun A, Bagella L, Tutton S, Romano G, Giordano A. From G0 to S phase: a view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J Cell Biochem. 2007;102:1400–1404. doi: 10.1002/jcb.21609. [DOI] [PubMed] [Google Scholar]
- 166.Polager S, Ginsberg D. E2F - at the crossroads of life and death. Trends Cell Biol. 2008;18:528–535. doi: 10.1016/j.tcb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 167.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Henley SA, Dick FA. The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 2012;7:10. doi: 10.1186/1747-1028-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hannan KM, Hannan RD, Smith SD, Jefferson LS, Lun M, Rothblum LI. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene. 2000;19:4988–4999. doi: 10.1038/sj.onc.1203875. [DOI] [PubMed] [Google Scholar]
- 170.Voit R, Schafer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hannan KM, Kennedy BK, Cavanaugh AH, Hannan RD, Hirschler-Laszkiewicz I, Jefferson LS, Rothblum LI. RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene. 2000;19:3487–3497. doi: 10.1038/sj.onc.1203690. [DOI] [PubMed] [Google Scholar]
- 172.White RJ, Trouche D, Martin K, Jackson SP, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 173.Chu WM, Wang Z, Roeder RG, Schmid CW. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272:14755–14761. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 174.Larminie CG, Cairns CA, Mital R, Martin K, Kouzarides T, Jackson SP, White RJ. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Efeyan A, Serrano M. p53: guardian of the genome and policeman of the oncogenes. Cell Cycle. 2007;6:1006–1010. doi: 10.4161/cc.6.9.4211. [DOI] [PubMed] [Google Scholar]
- 176.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 178.Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20:5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics. 2010;5:685–690. doi: 10.4161/epi.5.8.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Dominguez-Brauer C, Brauer PM, Chen YJ, Pimkina J, Raychaudhuri P. Tumor suppression by ARF: gatekeeper and caretaker. Cell Cycle. 2010;9:86–89. doi: 10.4161/cc.9.1.10350. [DOI] [PubMed] [Google Scholar]
- 181.Matheu A, Maraver A, Serrano M. The Arf/p53 pathway in cancer and aging. Cancer Res. 2008;68:6031–6034. doi: 10.1158/0008-5472.CAN-07-6851. [DOI] [PubMed] [Google Scholar]
- 182.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 183.Brady SN, Yu Y, Maggi LB, Jr, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ayrault O, Andrique L, Fauvin D, Eymin B, Gazzeri S, Seite P. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene. 2006;25:7577–7586. doi: 10.1038/sj.onc.1209743. [DOI] [PubMed] [Google Scholar]
- 185.Lessard F, Morin F, Ivanchuk S, Langlois F, Stefanovsky V, Rutka J, Moss T. The ARF tumor suppressor controls ribosome biogenesis by regulating the RNA polymerase I transcription factor TTF-I. Mol Cell. 2010;38:539–550. doi: 10.1016/j.molcel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 186.Lessard F, Stefanovsky V, Tremblay MG, Moss T. The cellular abundance of the essential transcription termination factor TTF-I regulates ribosome biogenesis and is determined by MDM2 ubiquitinylation. Nucleic Acids Res. 2012;40:5357–5367. doi: 10.1093/nar/gks198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cheng Y, Liang P, Geng H, Wang Z, Li L, Cheng SH, Ying J, Su X, Ng KM, Ng MH, Mok TS, Chan AT, Tao Q. A Novel 19q13 Nucleolar Zinc Finger Protein Suppresses Tumor Cell Growth through Inhibiting Ribosome Biogenesis and Inducing Apoptosis but Is Frequently Silenced in Multiple Carcinomas. Mol Cancer Res. 2012 doi: 10.1158/1541-7786.MCR-11-0594. [DOI] [PubMed] [Google Scholar]
- 188.Wang S, Cheng Y, Du W, Lu L, Zhou L, Wang H, Kang W, Li X, Tao Q, Sung JJ, Yu J. Zinc-finger protein 545 is a novel tumour suppressor that acts by inhibiting ribosomal RNA transcription in gastric cancer. Gut. 2012 doi: 10.1136/gutjnl-2011-301776. [DOI] [PubMed] [Google Scholar]
- 189.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 190.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Stefanovsky VY, Moss T. The splice variants of UBF differentially regulate RNA polymerase I transcription elongation in response to ERK phosphorylation. Nucleic Acids Res. 2008;36:5093–5101. doi: 10.1093/nar/gkn484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhao J, Yuan X, Frodin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol Cell. 2003;11:405–413. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 193.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 194.Johnson SA, Dubeau L, Kawalek M, Dervan A, Schonthal AH, Dang CV, Johnson DL. Increased expression of TATA-binding protein, the central transcription factor, can contribute to oncogenesis. Mol Cell Biol. 2003;23:3043–3051. doi: 10.1128/MCB.23.9.3043-3051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Populo H, Lopes JM, Soares P. The mTOR Signalling Pathway in Human Cancer. Int J Mol Sci. 2012;13:1886–1918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sheppard K, Kinross KM, Solomon B, Pearson RB, Phillips WA. Targeting PI3 kinase/AKT/mTOR signaling in cancer. Crit Rev Oncog. 2012;17:69–95. doi: 10.1615/critrevoncog.v17.i1.60. [DOI] [PubMed] [Google Scholar]
- 197.Solomon B, Pearson RB. Class IA phosphatidylinositol 3-kinase signaling in non-small cell lung cancer. J Thorac Oncol. 2009;4:787–791. doi: 10.1097/JTO.0b013e3181a74dba. [DOI] [PubMed] [Google Scholar]
- 198.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhang C, Comai L, Johnson DL. PTEN represses RNA Polymerase I transcription by disrupting the SL1 complex. Mol Cell Biol. 2005;25:6899–6911. doi: 10.1128/MCB.25.16.6899-6911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS, Hung S, Astle MV, Bywater M, Wall M, Poortinga G, Jastrzebski K, Sheppard KE, Hemmings BA, Hall MN, Johnstone RW, McArthur GA, Hannan RD, Pearson RB. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci Signal. 2011;4:ra56. doi: 10.1126/scisignal.2001754. [DOI] [PubMed] [Google Scholar]
- 203.McArthur GA. The coming of age of MEK. Lancet Oncol. 2012;13:744–745. doi: 10.1016/S1470-2045(12)70317-0. [DOI] [PubMed] [Google Scholar]
- 204.Cho YY, Yao K, Kim HG, Kang BS, Zheng D, Bode AM, Dong Z. Ribosomal S6 kinase 2 is a key regulator in tumor promoter induced cell transformation. Cancer Res. 2007;67:8104–8112. doi: 10.1158/0008-5472.CAN-06-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kang S, Dong S, Gu TL, Guo A, Cohen MS, Lonial S, Khoury HJ, Fabbro D, Gilliland DG, Bergsagel PL, Taunton J, Polakiewicz RD, Chen J. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–214. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Romeo Y, Zhang X, Roux PP. Regulation and function of the RSK family of protein kinases. Biochem J. 2012;441:553–569. doi: 10.1042/BJ20110289. [DOI] [PubMed] [Google Scholar]
- 207.Sabapathy K. Role of the JNK pathway in human diseases. Prog Mol Biol Transl Sci. 2012;106:145–169. doi: 10.1016/B978-0-12-396456-4.00013-4. [DOI] [PubMed] [Google Scholar]
- 208.Piazza F, Manni S, Ruzzene M, Pinna LA, Gurrieri C, Semenzato G. Protein kinase CK2 in hematologic malignancies: reliance on a pivotal cell survival regulator by oncogenic signaling pathways. Leukemia. 2012;26:1174–1179. doi: 10.1038/leu.2011.385. [DOI] [PubMed] [Google Scholar]
- 209.Silva A, Jotta PY, Silveira AB, Ribeiro D, Brandalise SR, Yunes JA, Barata JT. Regulation of PTEN by CK2 and Notch1 in primary T-cell acute lymphoblastic leukemia: rationale for combined use of CK2- and gamma-secretase inhibitors. Haematologica. 2010;95:674–678. doi: 10.3324/haematol.2009.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhang S, Wang Y, Mao JH, Hsieh D, Kim IJ, Hu LM, Xu Z, Long H, Jablons DM, You L. Inhibition of CK2alpha Down-Regulates Hedgehog/Gli Signaling Leading to a Reduction of a Stem-Like Side Population in Human Lung Cancer Cells. PLoS One. 2012;7:e38996. doi: 10.1371/journal.pone.0038996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ruzzene M, Pinna LA. Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim Biophys Acta. 2010;1804:499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 212.Tawfic S, Yu S, Wang H, Faust R, Davis A, Ahmed K. Protein kinase CK2 signal in neoplasia. Histol Histopathol. 2001;16:573–582. doi: 10.14670/HH-16.573. [DOI] [PubMed] [Google Scholar]
- 213.Hundsdorfer C, Hemmerling HJ, Hamberger J, Le Borgne M, Bednarski P, Gotz C, Totzke F, Jose J. Novel indeno[1,2-b]indoloquinones as inhibitors of the human protein kinase CK2 with antiproliferative activity towards a broad panel of cancer cell lines. Biochem Biophys Res Commun. 2012;424:71–75. doi: 10.1016/j.bbrc.2012.06.068. [DOI] [PubMed] [Google Scholar]
- 214.Pierre F, Chua PC, O'Brien SE, Siddiqui-Jain A, Bourbon P, Haddach M, Michaux J, Nagasawa J, Schwaebe MK, Stefan E, Vialettes A, Whitten JP, Chen TK, Darjania L, Stansfield R, Anderes K, Bliesath J, Drygin D, Ho C, Omori M, Proffitt C, Streiner N, Trent K, Rice WG, Ryckman DM. Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J Med Chem. 2011;54:635–654. doi: 10.1021/jm101251q. [DOI] [PubMed] [Google Scholar]
- 215.Pierre F, Chua PC, O'Brien SE, Siddiqui-Jain A, Bourbon P, Haddach M, Michaux J, Nagasawa J, Schwaebe MK, Stefan E, Vialettes A, Whitten JP, Chen TK, Darjania L, Stansfield R, Bliesath J, Drygin D, Ho C, Omori M, Proffitt C, Streiner N, Rice WG, Ryckman DM, Anderes K. Pre-clinical characterization of CX-4945, a potent and selective small molecule inhibitor of CK2 for the treatment of cancer. Mol Cell Biochem. 2011;356:37–43. doi: 10.1007/s11010-011-0956-5. [DOI] [PubMed] [Google Scholar]
- 216.Hannan RD, Hempel WM, Cavanaugh A, Arino T, Dimitrov SI, Moss T, Rothblum L. Affinity purification of mammalian RNA polymerase I. Identification of an associated kinase. J Biol Chem. 1998;273:1257–1267. doi: 10.1074/jbc.273.2.1257. [DOI] [PubMed] [Google Scholar]
- 217.Hannan RD, Cavanaugh A, Hempel WM, Moss T, Rothblum L. Identification of a mammalian RNA polymerase I holoenzyme containing components of the DNA repair/replication system. Nucleic Acids Res. 1999;27:3720–3727. doi: 10.1093/nar/27.18.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bierhoff H, Dundr M, Michels AA, Grummt I. Phosphorylation by casein kinase 2 facilitates rRNA gene transcription by promoting dissociation of TIF-IA from elongating RNA polymerase I. Mol Cell Biol. 2008;28:4988–4998. doi: 10.1128/MCB.00492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Stone A, Sutherland RL, Musgrove EA. Inhibitors of cell cycle kinases: recent advances and future prospects as cancer therapeutics. Crit Rev Oncog. 2012;17:175–198. doi: 10.1615/critrevoncog.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- 221.Thway K, Flora R, Shah C, Olmos D, Fisher C. Diagnostic utility of p16, CDK4, and MDM2 as an immunohistochemical panel in distinguishing well-differentiated and dedifferentiated liposarcomas from other adipocytic tumors. Am J Surg Pathol. 2012;36:462–469. doi: 10.1097/PAS.0b013e3182417330. [DOI] [PubMed] [Google Scholar]
- 222.Poomsawat S, Buajeeb W, Khovidhunkit SO, Punyasingh J. Alteration in the expression of cdk4 and cdk6 proteins in oral cancer and premalignant lesions. J Oral Pathol Med. 2010;39:793–799. doi: 10.1111/j.1600-0714.2010.00909.x. [DOI] [PubMed] [Google Scholar]
- 223.Wu A, Wu B, Guo J, Luo W, Wu D, Yang H, Zhen Y, Yu X, Wang H, Zhou Y, Liu Z, Fang W, Yang Z. Elevated expression of CDK4 in lung cancer. J Transl Med. 2011;9:38. doi: 10.1186/1479-5876-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Tang LH, Contractor T, Clausen R, Klimstra DS, Du YC, Allen PJ, Brennan MF, Levine AJ, Harris CR. Attenuation of the Retinoblastoma Pathway in Pancreatic Neuroendocrine Tumors Due to Increased Cdk4/Cdk6. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-11-3264. [DOI] [PubMed] [Google Scholar]
- 225.Voit R, Hoffmann M, Grummt I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Voit R, Grummt I. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc Natl Acad Sci U S A. 2001;98:13631–13636. doi: 10.1073/pnas.231071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Horiuchi D, Huskey NE, Kusdra L, Wohlbold L, Merrick KA, Zhang C, Creasman KJ, Shokat KM, Fisher RP, Goga A. Chemical-genetic analysis of cyclin dependent kinase 2 function reveals an important role in cellular transformation by multiple oncogenic pathways. Proc Natl Acad Sci U S A. 2012;109:E1019–E1027. doi: 10.1073/pnas.1111317109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Brown KA, Samarajeewa NU, Simpson ER. Endocrine-related cancers and the role of AMPK. Mol Cell Endocrinol. 2012 doi: 10.1016/j.mce.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 229.Carling D, Thornton C, Woods A, Sanders MJ. AMP-activated protein kinase: new regulation, new roles? Biochem J. 2012;445:11–27. doi: 10.1042/BJ20120546. [DOI] [PubMed] [Google Scholar]
- 230.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Li C, Liu VW, Chiu PM, Chan DW, Ngan HY. Over-expressions of AMPK subunits in ovarian carcinomas with significant clinical implications. BMC Cancer. 2012;12:357. doi: 10.1186/1471-2407-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hardie DG. AMP-activated protein kinase: a cellular energy sensor with a key role in metabolic disorders and in cancer. Biochem Soc Trans. 2011;39:1–13. doi: 10.1042/BST0390001. [DOI] [PubMed] [Google Scholar]
- 233.Wu CL, Qiang L, Han W, Ming M, Viollet B, He YY. Role of AMPK in UVB-induced DNA damage repair and growth control. Oncogene. 2012 doi: 10.1038/onc.2012.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, Voit R. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc Natl Acad Sci U S A. 2009;106:17781–17786. doi: 10.1073/pnas.0909873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 236.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 237.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: The molecular choreography. Mutat Res. 2012 doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 238.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba II, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mansouri L, Gunnarsson R, Sutton LA, Ameur A, Hooper SD, Mayrhofer M, Juliusson G, Isaksson A, Gyllensten U, Rosenquist R. Next generation RNA-sequencing in prognostic subsets of chronic lymphocytic leukemia. Am J Hematol. 2012;87:737–740. doi: 10.1002/ajh.23227. [DOI] [PubMed] [Google Scholar]
- 241.Roberts NJ, Jiao Y, Yu J, Kopelovich L, Petersen GM, Bondy ML, Gallinger S, Schwartz AG, Syngal S, Cote ML, Axilbund J, Schulick R, Ali SZ, Eshleman JR, Velculescu VE, Goggins M, Vogelstein B, Papadopoulos N, Hruban RH, Kinzler KW, Klein AP. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, Zomerdijk JC. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 2007;26:1560–1568. doi: 10.1038/sj.emboj.7601601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 244.Miller G, Panov KI, Friedrich JK, Trinkle-Mulcahy L, Lamond AI, Zomerdijk JC. hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J. 2001;20:1373–1382. doi: 10.1093/emboj/20.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hirschler-Laszkiewicz I, Cavanaugh A, Hu Q, Catania J, Avantaggiati ML, Rothblum LI. The role of acetylation in rDNA transcription. Nucleic Acids Res. 2001;29:4114–4124. doi: 10.1093/nar/29.20.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Pelletier G, Stefanovsky VY, Faubladier M, Hirschler-Laszkiewicz I, Savard J, Rothblum LI, Cote J, Moss T. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell. 2000;6:1059–1066. doi: 10.1016/s1097-2765(00)00104-0. [DOI] [PubMed] [Google Scholar]
- 247.Meraner J, Lechner M, Loidl A, Goralik-Schramel M, Voit R, Grummt I, Loidl P. Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res. 2006;34:1798–1806. doi: 10.1093/nar/gkl101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kong R, Zhang L, Hu L, Peng Q, Han W, Du X, Ke Y. hALP, a novel transcriptional U three protein (t-UTP), activates RNA polymerase I transcription by binding and acetylating the upstream binding factor (UBF) J Biol Chem. 2011;286:7139–7148. doi: 10.1074/jbc.M110.173393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Chi YH, Haller K, Peloponese JM, Jr, Jeang KT. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J Biol Chem. 2007;282:27447–27458. doi: 10.1074/jbc.M703098200. [DOI] [PubMed] [Google Scholar]
- 250.Muth V, Nadaud S, Grummt I, Voit R. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001;20:1353–1362. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Gonugunta VK, Nair BC, Rajhans R, Sareddy GR, Nair SS, Vadlamudi RK. Regulation of rDNA transcription by proto-oncogene PELP1. PLoS One. 2011;6:e21095. doi: 10.1371/journal.pone.0021095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zhang Y, Smith ADt, Renfrow MB, Schneider DA. The RNA polymerase-associated factor 1 complex (Paf1C) directly increases the elongation rate of RNA polymerase I and is required for efficient regulation of rRNA synthesis. J Biol Chem. 2010;285:14152–14159. doi: 10.1074/jbc.M110.115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zhang Y, Sikes ML, Beyer AL, Schneider DA. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci U S A. 2009;106:2153–2158. doi: 10.1073/pnas.0812939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Drygin D, Siddiqui-Jain A, O'Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 255.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 256.Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics. 2010;9:457–470. doi: 10.1074/mcp.M900429-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Boisvert FM, Lamond AI. p53-Dependent subcellular proteome localization following DNA damage. Proteomics. 2010;10:4087–4097. doi: 10.1002/pmic.201000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hein N, Sanij E, Quin J, Hannan KM, Ganley A, Hannan RD. The nucleolus and ribosomal genes in aging and senescence. Senescence. 2011 [Google Scholar]
- 260.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem Soc Trans. 2008;36:619–624. doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 264.van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci U S A. 2009;106:17829–17834. doi: 10.1073/pnas.0906811106. [DOI] [PMC free article] [PubMed] [Google Scholar]