Abstract
Many environmental contaminants can disrupt the adaptive immune response. Exposure to the ubiquitous aryl hydrocarbon receptor (AhR) ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and other agonists suppresses the antibody response. The underlying pathway mechanism by which TCDD alters B cell function is not well understood. The present study investigated the mechanism of AhR-mediated pathways and mode of suppression by which TCDD perturbs terminal differentiation of B cells to plasma cells and thereby impairs antibody production. An integrated approach combining computational pathway modeling and in vitro assays with primary mouse B cells activated by lipopolysaccharide was employed. We demonstrated that suppression of the IgM response by TCDD occurs in an all-or-none (binary) rather than graded mode: i.e., it reduces the number of IgM-secreting cells in a concentration-dependent manner without affecting the IgM content in individual plasma cells. The mathematical model of the gene regulatory circuit underpinning B cell differentiation revealed that two previously identified AhR-regulated pathways, inhibition of signaling protein AP-1 and activation of transcription factor Bach2, could account for the all-or-none mode of suppression. Both pathways disrupt the operation of a bistable-switch circuit that contains transcription factors Bcl6, Prdm1, Pax5, and Bach2 and regulates B cell fate. The model further predicted that by transcriptionally activating Bach2, TCDD might delay B cell differentiation and increase the likelihood of isotype switching, thereby altering the antibody repertoire. In conclusion, the present study revealed the mode and specific pathway mechanisms by which the environmental immunosuppressant TCDD suppresses B cell differentiation.
Keywords: TCDD, AhR, all-or-none, bistable, Bach2
Introduction
Activation of the humoral immune response to microbial challenge is an integral component of adaptive immunity. Many environmental pollutants and pharmaceutical compounds can modulate this process (Dooley and Holsapple, 1988; Salazar et al., 2005; Peden-Adams et al., 2008). To understand the toxicity mechanisms of environmental immunotoxicants, it is crucial to examine how these chemicals perturb the signaling dynamics of the molecular pathways and circuits underlying the physiological processes in immune cells. Studying the relevant pathways as dynamic systems and how they are perturbed by exogenous agents is a necessary step toward mechanistically-based interpretation and prediction of dose responses and improved risk assessment for human immune health.
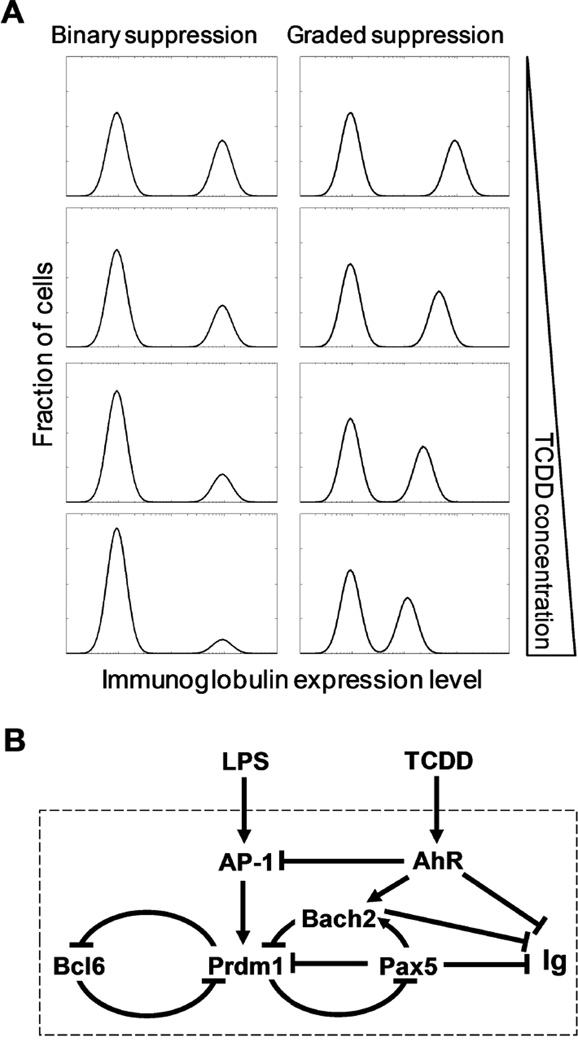
Halogenated aromatic hydrocarbons (HAHs) are a ubiquitous class of environmental immunosuppressants, the most potent of which is 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Kerkvliet, 2002; Sulentic and Kaminski, 2011). It has been well established that mature B cells are a highly sensitive target of TCDD (Sulentic and Kaminski, 2011). By acting via the aryl hydrocarbon receptor (AhR), TCDD suppresses the terminal differentiation of B cells into antibody-secreting plasma cells (Tucker et al., 1986; Sulentic et al., 1998; Vorderstrasse et al., 2001). Although some of the molecular targets of AhR in B cells are known (Suh et al., 2002; Henseler et al., 2009; De Abrew et al., 2011), it is largely unclear how the transcriptional dynamics of the B cell-specific gene circuit is perturbed by these targeted AhR pathways. Differentiation of B cells to plasma cells is an all-or-none phenomenon in individual cells, which express either low or high levels of immunoglobulin, respectively. Consequently, two distinct modes of suppression of the antibody response by TCDD are plausible in theory (Fig. 1A). In an all-or-none (binary) mode, the fraction of B cells that differentiate into antibody-secreting plasma cells would be reduced as the TCDD concentration increases, while the amount of immunoglobulin produced and secreted per plasma cell is unaffected. In a graded mode of suppression, the amount of immunoglobulin produced and secreted per plasma cell would be reduced by TCDD in a concentration-dependent manner while the fraction of plasma cells formed is largely unaffected. Additionally, a hybrid mode exhibiting a mixture of binary and graded suppression is also possible. The binary mode of suppression has been suggested based on earlier studies using Jerne plaque or ELISpot assays (Tucker et al., 1986; Schneider et al., 2008; Lu et al., 2010). However, since neither of these assays provides quantitative measurement of the intracellular immunoglobulin content in individual cells, the exact mode by which TCDD suppresses the antibody response and the responsible AhR signaling mechanism remain to be determined.
Figure 1.
(A) Schematic illustration of modes of suppression of the antibody response by TCDD. In each histogram, left peak represents naïve B cells and right peak represents plasma cells. Intracellular immunoglobulin expression level can be measured by fluorescence intensity using flow cytometry. (B) Schematic illustration of the regulatory network underlying LPS-stimulated B cell terminal differentiation and its disruption by TCDD through AhR-mediated pathways. Arrows and blunted lines indicate activation and repression, respectively.
Terminal differentiation of B cells into plasma cells is believed to be controlled by a bistable gene regulatory network, which ensures all-or-none and irreversible differentiation dynamics (Bhattacharya et al., 2010; Muto et al., 2010; Martinez et al., 2012). The putative bistable-switch circuit is formed by interconnected feedback loops (Fig. 1B) composed of multiple transcription factors (TFs) including B cell lymphoma 6 (Bcl6), PR domain zinc finger protein 1 (Prdm1), paired box 5 (Pax5), BTB and CNC homology 2 (Bach2), and others (Lin et al., 2002; Shaffer et al., 2002; Vasanwala et al., 2002; Sciammas and Davis, 2004; Tunyaplin et al., 2004; Shapiro-Shelef and Calame, 2005; Ochiai et al., 2006; Mora-Lopez et al., 2007). Numerous studies have demonstrated that mature B cells and plasma cells have mutually exclusive expression profiles of these TFs, suggesting that these two cell types represent the two attractor states of the bistable-switch circuit (Barberis et al., 1990; Turner et al., 1994; Cattoretti et al., 1995; Muto et al., 1998; Johnson et al., 2005; Ochiai et al., 2006). A number of signal transduction pathways activated by antigens and cytokines can converge onto the bistable circuit as inputs to trigger the switch, driving B cells to differentiate into plasma cells (Calame, 2008). For the bacterial endotoxin lipopolysaccharide (LPS), a common polyclonal activator, toll-like receptor 4 (TLR4)-mediated activation of activator protein 1 (AP-1) appears to be the primary signaling pathway (Vasanwala et al., 2002; Ohkubo et al., 2005). Pax5, highly expressed in B cells to maintain their identity and subsequently repressed in plasma cells, can be viewed as a primary output of the bistable system, and along with other TFs such as Bach2, it represses the production of immunoglobulin molecules in B cells (Muto et al., 1998; Delogu et al., 2006).
From a dynamic system’s perspective, it could be argued that if TCDD blocks the signal transduction pathways that trigger the bistable gene circuit or interfere with the operation of the circuit itself, it would make the switching of the circuit by an activator more difficult. B cells would then be less likely to differentiate into plasma cells. However, for cells that do manage to switch the circuit on in the presence of TCDD, immunoglobulin genes would still be expressed at full levels because of the all-or-none nature of a bistable switch. This would lead to a binary mode of suppression. Alternatively, if TCDD directly inhibits the transcription and/or secretion of immunoglobulin molecules without interfering with the bistable switching process per se, B cells would differentiate into plasma cells unobstructed, but with each plasma cell producing a smaller amount of immunoglobulin molecules. This would produce a graded mode of suppression. Since TCDD suppresses the plasma cell response by perturbing multiple components of the underlying molecular network (Fig. 1B) including AP-1, Bach2 and immunoglobulin heavy chain (IgH) (Suh et al., 2002; Henseler et al., 2009; De Abrew et al., 2011), the possibility for both binary and graded modes of suppression exists. By using an integrated approach of in vitro experimentation and computational modeling of pathway perturbation, we demonstrate here that TCDD suppresses B cell differentiation in an all-or-none fashion at the level of individual cells. Our modeling study indicates that this all-or-none mode of suppression likely results from perturbations of pathways that interfere with the bistable gene circuit regulating B cell fate.
Materials and Methods
Animals
Virus-free, female B6C3F1 mice (6 weeks of age) were purchased from Charles River (Portage, MI, USA). Mice were randomized, transferred to plastic cages containing bedding (five per cage), and quarantined for 1 week. Mice were given food and water ad libitum and not used until their body weight reached 17–20 g. All experiments were approved by the Michigan State University Institutional Animal Care & Use Committee (East Lansing, MI, USA).
Chemicals
TCDD was purchased from Accustandard (New Haven, CT, USA) and prepared in dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO, USA). Stocks of Salmonella typhosa LPS (Sigma-Aldrich) were prepared in individual aliquots and stored at −20° C until use.
Primary splenocyte isolation and culture
Splenocytes were isolated aseptically from mouse spleens and cultured at 5 × 106 cells/ml in complete media (RPMI 1640 supplemented with 10% bovine calf serum, streptomycin sulfate, and 2-mercaptoethanol). Splenocytes were exposed to either LPS (dissolved in RPMI1640), LPS + vehicle (0.02% DMSO), or LPS + TCDD and incubated in 24-well culture plates at 37°C with 5% CO2 for up to 72 h. No effect on cell viability was observed at any of the TCDD concentrations used throughout the study.
Flow cytometry analysis
At the designated times (0, 24, 36, 48, 60, 72, 120 h), cells were harvested from culture. To identify viable cells LIVE/DEAD Near-IR dye (Invitrogen, Carlsbad, CA, USA) was used according to the manufacturer’s instructions. Proliferation was measured using carboxyfluorescein diacetate succinimidyl ester (CFSE, Invitrogen) following manufacturer’s instructions. Splenocytes at 1 × 107 cells/ml in HBSS were incubated in 5 µM CFSE for 10 min at 37°C with 5% CO2 and washed 3 times with HBSS and re-suspended to 5 × 106 cells/ml in complete media. B cells were identified using anti-CD19 staining (BioLegend, San Diego, CA, USA). Plasma cells were identified using anti-CD138 (BD Biosciences, San Diego, CA, USA). B cell receptors and Fc receptors were blocked in FCS buffer (HBSS with 1% BSA, Sigma-Aldrich) and 0.1% sodium azide (Sigma-Aldrich) before CD19, CD138 and intracellular anti-immunoglobulin M (IgM) staining was preformed. Surface staining of CD19 and CD138 was performed at 4°C for 20 min in FCS buffer and then the cells were washed twice in staining buffer. The stained cells were then fixed with BD Cytofix (BD Biosciences). Intracellular IgM was measured in cells permeablized with 1X BD PERM/WASH followed by intracellular staining with anti-IgM, anti-IgG and anti-IgA (BD Biosciences). Cells were washed twice with PERM/WASH, re-suspended in FCS buffer and analyzed using a Canto II flow cytometer (BD Bioscience). Analysis of data was performed using FlowJo software (Tree Star Inc. Ashland, OR, USA).
Computational model construction and simulation
The computational model was constructed by expanding and updating a previous model of B cell differentiation (Zhang et al., 2010). Recently, we have identified Bach2 as a key target gene of TCDD involved in the suppression of the IgM response (De Abrew et al., 2010; De Abrew et al., 2011). The present study incorporated transcriptional regulations between Bach2 and other factors, including its activation by AhR and Pax5 and repression of Prdm1 by Bach2 (Ochiai et al., 2006; Schebesta et al., 2007), and other related processes. Relevant parameters were estimated and constrained by fitting the model output to experimental data generated in the present study and obtained in the literature. Model details including ODEs, parameter values, steady-state conditions, and simulations tools are provided in Supplemental Tables S1 and S2.Ordinary differentiation equation (ODE)-based deterministic simulation was used for stability analysis, and stochastic simulation, which considers fluctuation of gene expression as normally occurring in cells, was used to model the heterogeneous response among individual B cells. The deterministic version of the model was implemented in PathwayLab (InNetics Inc., Linköping, Sweden), while the stochastic version was implemented in BioNetS (Adalsteinsson et al., 2004) based on Gillespie’s stochastic simulation algorithm (Gillespie, 1977). Both deterministic and stochastic versions of the model were exported into MatLab (The Mathworks, Inc., Natick, MA, USA) for further analysis. Bifurcation diagrams were generated with the XPP-AUT program (Ermentrout, 2002).
Results
LPS-activated all-or-none B cell differentiation
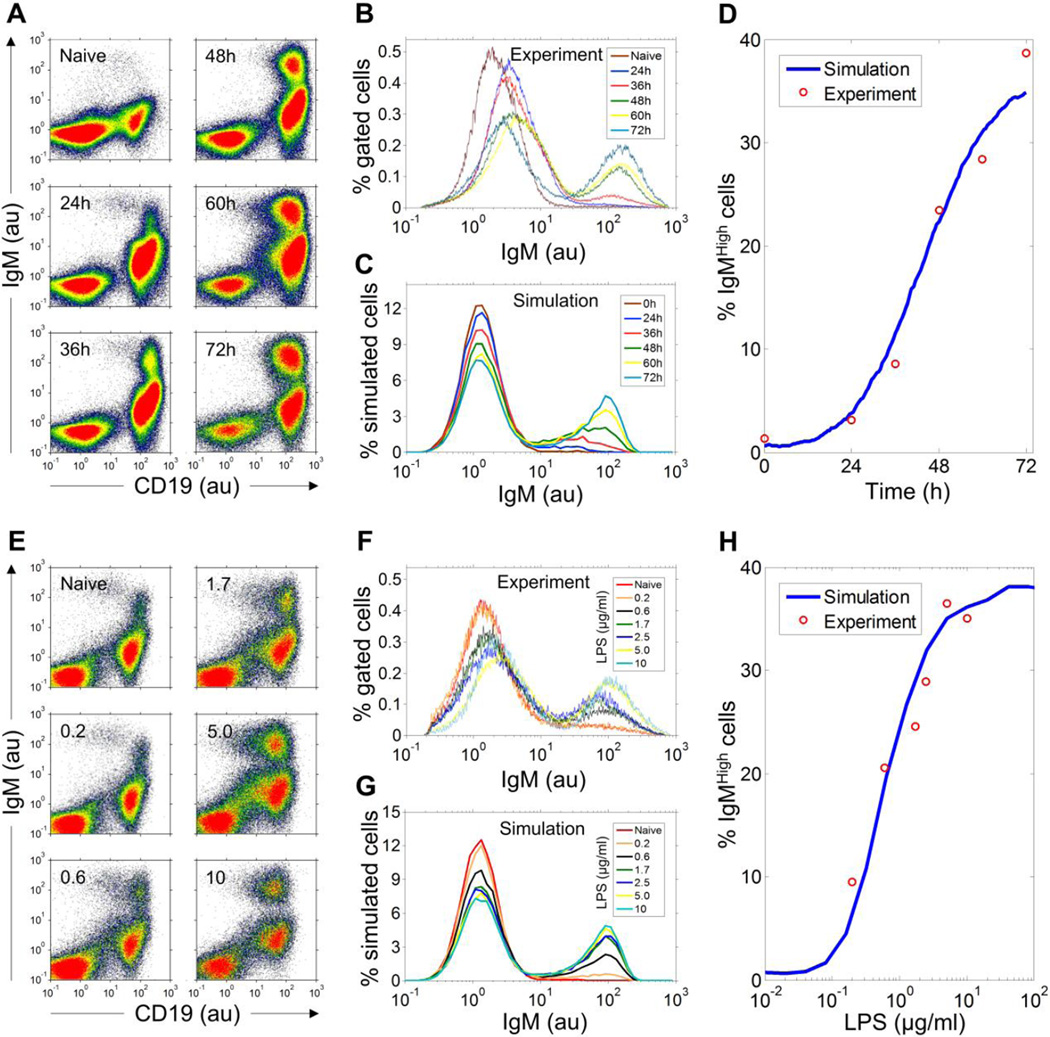
Flow cytometry revealed in the absence of LPS there was a single distinct B cell population (CD19high) expressing low-level intracellular IgM (IgMlow) among freshly isolated mouse splenocytes (Fig. 2A). For cells continuously cultured with 5 µg/ml LPS, a distinct, second CD19high population displaying high levels of intracellular IgM (IgMhigh) gradually emerged over time. Histograms of intracellular IgM fluorescence intensity gated on viable B cells clearly showed two distinct peaks, separated horizontally by about two orders of magnitude (Fig. 2B). The IgMhigh population increased marginally at 24 h, but grew rapidly to ~38% of total viable B cells by 72 h (Fig. 2D).
Figure 2. LPS-stimulated dynamic- and dose-response of differentiation of B cells into IgM-secreting cells.
(A) Co-plots of fluorescence intensity of antibodies against surface CD19 and intracellular IgM in primary mouse splenocytes treated with 5 µg/ml LPS continuously and analyzed at time points indicated. (B) Histograms of intracellular IgM in CD19-gated viable B cells from panel A. For time points of 0, 24, 36, 48, 60, and 72 h, the percentage of IgMhigh cells is 1.30%, 3.12%, 8.56%, 23.41%, 28.37%, and 38.67%, respectively. (C) Histograms of IgM in 20,000 stochastically simulated B cells activated by 5 µg/ml LPS. The scale difference in the Y axis between panels B and C (also for comparable panels in F and G and Fig. 5A, 5B and 5D) was due to the difference in the number of bins used to plot the experimental and simulated histograms. Fractions of IgMlow and IgMhigh cells, which correlate with the area-under-the-curve of associated histogram peaks, are independent of the scale difference. (D) Fitting of the dynamic change of percentage simulated IgMhigh cells in panel C to the experimental data in panel B. (E) Co-plots of fluorescence intensity of surface CD19 and intracellular IgM in primary mouse splenocytes treated continuously for 72 h with LPS of various concentrations indicated (µg/ml). (F) Histograms of intracellular IgM in CD19-gated viable B cells from panel E. For LPS concentrations of 0.2, 0.6, 1.7, 2.5, 5, and 10 µg/ml, the percentage of IgMhigh cells is 9.49%, 20.52%, 24.53%, 28.87%, 36.49%, and 35%, respectively. (G) Histograms of IgM in 20,000 stochastically-simulated B cells stimulated by various concentrations of LPS. (H) Fitting of the concentration-response of percentage simulated IgMhigh cells in panel G to the experimental data in panel F. au: arbitrary unit of fluorescence intensity.
The pattern that B cells split into two distinct sub-populations, IgMlow and IgMhigh, was observed consistently across the entire range of LPS concentrations tested (Fig. 2E). As the LPS concentration was raised, the fraction of cells in the IgMhigh sub-population increased with a concordant decrease of B cells in the IgMlow sub-population (Fig. 2F). The respective mean levels of intracellular IgM in IgMlow and IgMhigh cells, as indicated by the horizontal positions of the two corresponding histogram peaks, remained largely constant irrespective of the LPS concentration, supporting the notion that an underlying all-or-none switch is at work. Co-expression of CD138, a specific plasma cell surface marker, in these IgMhigh cells confirmed their plasma cell identity (Fig. S1).
Simulation of B cell-specific bistable gene circuit recapitulates the all-or-none B cell differentiation stimulated by LPS
The discrete process of cell differentiation is generally underpinned by molecular circuits capable of being bistable (Xiong and Ferrell, 2003). We tested computationally whether the gene circuit in Fig. 1B, consisting of key transcriptional regulations known in B cells, is capable of generating bistability and sufficient to recapitulate the all-or-none IgM response in individual cells. The model, once calibrated by fitting the temporal and dose response data as presented above, would allow us to predict the patterns of response to perturbations by various AhR-mediated pathways.
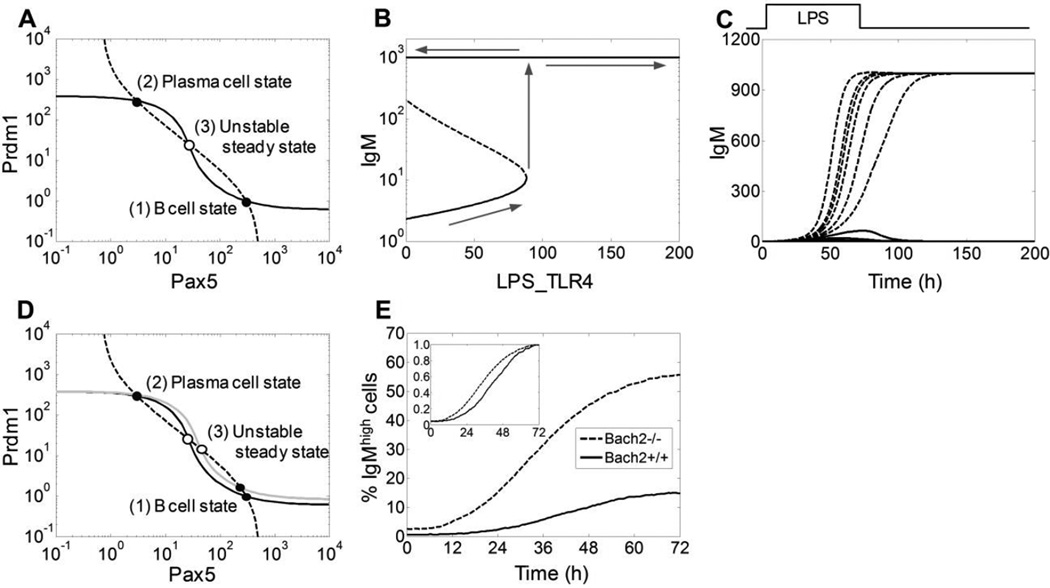
Phase-plane stability analysis (Strogatz, 1994) of the gene circuit, containing coupled feedback loops of Bcl6, Prdm1, Pax5, and Bach2, indicated that the feedback system can be bistable with two mutually exclusive attractor states: one with high Bcl6/Pax5/Bach2 and low Prdm1/IgM, and the other with low Bcl6/Pax5/Bach2 and high Prdm1/IgM. The first state is associated with the naïve B cell phenotype, and the second state with the plasma cell phenotype (Fig. 3A). This mutually exclusive expression profile has been well established experimentally (Barberis et al., 1990; Turner et al., 1994; Cattoretti et al., 1995; Muto et al., 1998; Ochiai et al., 2006). Bifurcation analysis indicated that as the intensity of LPS signaling surpasses a certain threshold, the system irreversibly switches from the low IgM-expressing B cell state to the high IgM-expressing plasma cell state (Fig. 3B). Further, in response to a pulse of LPS lasting 72 h, the system can settle into either a low or high IgM-expressing steady state, depending on the LPS concentration (Fig. 3C). These deterministic analyses demonstrated that the transcriptional feedback system shown in Fig. 1B can function as an irreversible bistable switch that can be triggered by LPS.
Figure 3. Deterministic analysis of the B cell-specific gene circuit and effect of Bach2 deletion.
(A) Reciprocal steady-state stimulus-response curves between Prdm1 and Pax5 in the absence of LPS and TCDD. The dashed curve was obtained by varying Prdm1 as an independent stimulus and the steady-state level of Pax5 as the response; the solid curve was obtained by varying Pax5 as an independent stimulus and the steady-state level of Prdm1 as the response. Intersection points 1 and 2 (filled circles) are stable steady states representing the B cell and plasma cell states respectively, and point 3 (empty circle) represents an unstable steady state. (B) Bifurcation diagram showing that as the signaling intensity of LPS (represented by LPS_TLR4, which is LPS-occupied TLR4 receptor) reaches a certain threshold, the system can irreversibly switch from the B cell state expressing low-level IgM to plasma cell state expressing high-level IgM. (C) Transient LPS signal can irreversibly switch the bistable system from a low IgM-expressing state to a high IgM-expressing state. Trajectories were generated with a 72-h pulse of LPS (square wave on top) at various concentrations (0, 0.05, 0.1, 0.2, 0.3, and 0.4 µg/ml for solid curves; 0.5, 0.6, 0.8, 1.0, 1.2, and 10 µg/ml for dashed curves). (D) Phase-plane stability analysis of deletion of Bach2. Stimulus-response curves were obtained similarly as in panel A except the solid gray curve (Prdm1 vs. Pax5 response) which was obtained in the absence of Bach2 gene (i.e. Bach2−/−). (E) Dynamic changes of percentage IgMhigh cells from 20,000 stochastically-simulated Bach2+/+ (solid curve) and Bach2−/− (dashed curve) B cells stimulated continuously with 0.45 µg/ml LPS. Inset: normalized responses. For panels A–D, the unit of state variables Prdm1, Pax5, IgM, and LPS_TLR4 is number of molecules per cell.
We then simulated the bistable network model stochastically to account for fluctuations in gene expression that are normally expected in single cells, which can produce cell-to-cell variability in responses (Kaern et al., 2005). Stochastic gene expression can render the bistable switching event probabilistic, such that whether and when an individual B cell would differentiate into a plasma cell becomes a random event, with the probability dependent on the stimulation strength (Zhang et al., 2010). A simulation of 20,000 B cells continuously stimulated by 5 µg/ml LPS for 72 h revealed a bimodal distribution of intracellular IgM that closely resembles that observed experimentally (compare Figs. 2B and 2C). The IgMhigh cells appearing at early time points (24 and 36 h) have a somewhat lower intracellular IgM content than those appearing later, as indicated by the left-ward shift of the associated IgMhigh peaks (Fig. 2C, mean fluorescence intensity of IgMhigh peak is 31, 47, 64 ,77, and 87 for 24, 36, 48, 60, and 72 h, respectively). This shift was also observed in the experimental data, albeit to a lesser extent (Fig. 2B). The lower IgM at early times can be attributed to the fact that the switching process of the bistable gene circuit, once initiated, does not complete instantaneously. The inversion of the B cell-specific gene expression profile requires a finite amount of time. Thus the IgMhigh histogram peaks at early time points likely captured those B cells still en route to becoming plasma cells that would eventually produce IgM maximally (Fig. S2A). Importantly, the stochastic model was able to reproduce the experimentally observed temporal change of the IgMhigh population (Fig. 2D), and the IgMhigh cells accumulate at a rate positively correlated with LPS concentration (Fig. S2C).
The IgM histograms of simulated B cells captured at 72 h showed a clear bimodal distribution very similar to those observed experimentally, with LPS concentration affecting largely the percentage of IgMhigh cells rather than the intracellular IgM content in those cells (Fig. 2G, mean fluorescence intensity of IgMhigh peak is 73, 82, 85, 86, 87, and 87 for 0.2, 0.6, 1.7, 2.5, 5, and 10 µg/ml LPS, respectively). The stochastic model was able to reproduce the experimental concentration-response relationship between LPS concentration and the percentage of IgMhigh cells (Fig. 2H). In contrast to the all-or-none IgM response in individual cells, the concentration response at the cell population level was graded.
Bach2 and delayed B cell differentiation
Bach2 plays a key role in promoting class switch recombination (CSR) and somatic hypermutation in B cells (Muto et al., 2004; Watanabe-Matsui et al., 2011). It can be transcriptionally activated by Pax5, and both of these TFs repress the Prdm1 gene, thus forming a coherent feed-forward loop (Mangan et al., 2003; Ochiai et al., 2006; Mora-Lopez et al., 2007; Schebesta et al., 2007). We have recently identified Bach2 as a transcriptional target of AhR, which partially mediates the suppression of B cell differentiation by TCDD (De Abrew et al., 2010; De Abrew et al., 2011). Phase-plane analysis suggested that the gene circuit would still be bistable if the Bach2 gene were deleted (Fig. 3D). However, the B cell attractor state would become less stable by virtue of being situated closer to the unstable steady state that separates the B cell and the plasma cell attractor states, suggesting increased probability of B cells switching to plasma cells. Stochastic simulations showed that LPS at 0.45 µg/ml produced ~15% IgMhigh cells with Bach2 intact in the regulatory circuit, whereas with in silico Bach2 deletion the percentage rose more than three-fold to ~55% (Fig. 3E). These results are quantitatively comparable to the augmented plasma cell response observed with splenic B cells from Bach2−/− mice compared to Bach2+/+ mice (Muto et al., 2010), suggesting the strengths of transcriptional regulations involving Bach2 are reasonably parameterized in the model. Moreover, normalization of the two IgMhigh cell responses (i.e., with and without Bach2) revealed that in addition to suppressing the final fraction of IgMhigh cells formed, Bach2 is also responsible for a relatively delayed response at the cell population level (Fig. 3E inset).
AhR-mediated TCDD pathways and alternative modes of suppression
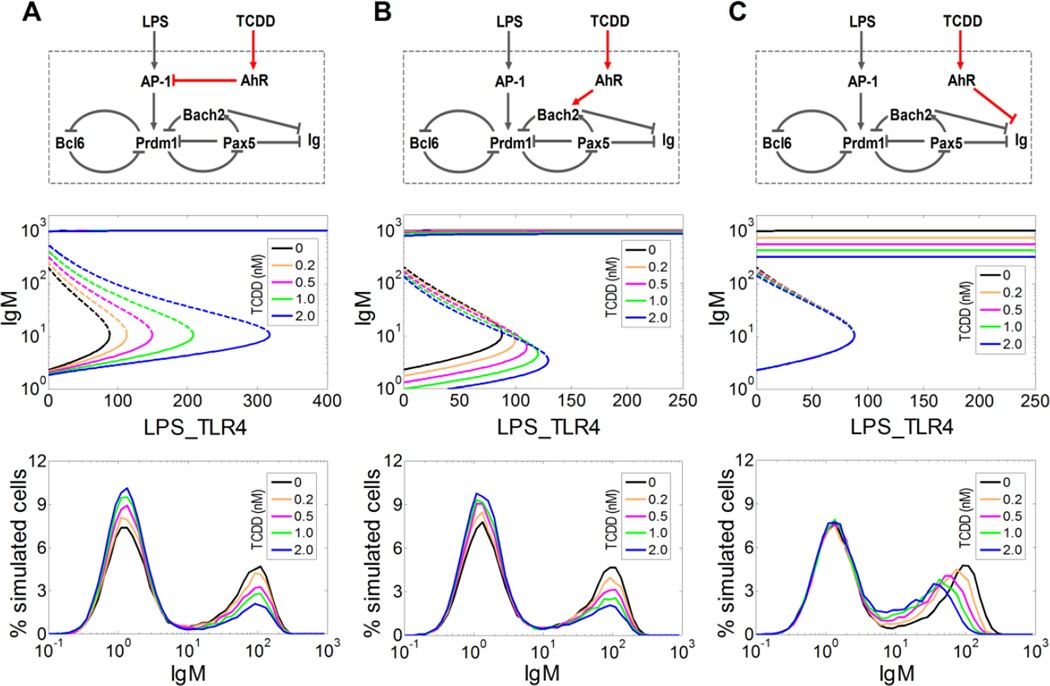
With the stochastic bistable-switch model calibrated with respect to the experimental data on LPS-stimulated IgMhigh cell response and Bach2−/− B cells, we next set out to computationally evaluate the possible modes of suppression of the IgM response by TCDD. In our previous work, we have experimentally identified three molecular targets in B cells that could be regulated by AhR: the signaling complex AP-1, Bach2, and IgH (Suh et al., 2002; Sulentic et al., 2004; Henseler et al., 2009; Schneider et al., 2009; De Abrew et al., 2010; De Abrew et al., 2011). Since these three targets reside in distinct locations with respect to the B cell transcriptional network (Fig. 1B), each of the AhR-mediated pathways may perturb the bistable gene circuit differently, producing different response patterns. We computationally analyzed the effect of each pathway separately, by keeping the other two disabled in turn.
The first pathway involves inhibition by AhR of the AP-1 protein (Suh et al., 2002; Schneider et al., 2009), which transmits the stimulatory signal of LPS to the core B cell transcriptional circuit (Fig. 4A, top panel). Bifurcation analysis showed that through inhibiting AP-1, TCDD raised the threshold of LPS signaling required to trigger the bistable switch, without affecting the IgM level associated with the plasma cell state (Fig. 4A, middle panel). In a stochastic gene expression environment, this increased threshold would lead to a reduced probability of bistable switching. Stochastic simulations supported this prediction: the fraction of IgMhigh cells appearing in response to LPS stimulation was suppressed by TCDD in a concentration-dependent manner, without the IgM level altered in those cells (Fig. 4A, bottom panel). This simulation result is consistent with an all-or-none mode of suppression.
Figure 4. Computational analyses of mode of suppression of B cell differentiation driven by three alternative AhR-mediated TCDD pathways.
(A) Pathway I: AhR represses AP-1 signaling (top panel). The deterministic threshold level of LPS-liganded TLR4 receptors (LPS_TLR4), required to trigger switching from the low IgM-expressing state to the high IgM-expressing state, increases with TCDD concentration, without affecting the level of IgM associated with the IgMhigh plasma cell state (middle panel). This suggests an all-or-none mode of suppression, which is corroborated by stochastic simulation (bottom panel). (B) Pathway II: AhR up-regulates Bach2 (top panel). The threshold level of LPS_TLR4 also increases with TCDD concentration, without tangibly affecting the level of IgM associated with the plasma cell state (middle panel). This again presages an all-or-none mode of suppression, as confirmed by stochastic simulation (bottom panel). (C) Pathway III: AhR represses IgM directly (top panel). The threshold level of LPS_TLR4 is not affected, however, the level of IgM associated with the plasma cell state decreases as TCDD concentration increases (middle panel). This suggests a graded mode of suppression, as confirmed by stochastic simulation (bottom panel). To analyze each pathway illustrated in panels A, B, and C separately, model parameters (Kd31, a71, Kd63) were set to the following sets of values, respectively: (0.75e4, 0, 1.7e9), (2.5e8, 10, 1.7e9), and (2.5e8, 0, 2e3). In all stochastic simulations (bottom panels), 20,000 B cells were stimulated with 10 µg/ml LPS for 72 h in the presence of TCDD at various concentrations indicated.
The second AhR-mediated pathway involves transcriptional activation of Bach2 by TCDD (Fig. 4B, top panel) (De Abrew et al., 2010; De Abrew et al., 2011). Bistable switching of the core circuit requires an inversion of the B cell-specific transcriptional profile (where Bach2 expression is high) to a plasma cell-specific profile (where Bach2 expression is low). Thus by maintaining Bach2 at a high level, TCDD would increase the difficulty with which the bistable circuit is flipped on, thereby increasing the threshold of LPS stimulation. Such an effect is indicated in the bifurcation diagrams, which also show that TCDD has little effect on the IgM level associated with the plasma cell state (Fig. 4B, middle panel). Similar to the AP-1 pathway, stochastic simulations indicated that activation of Bach2 by TCDD also impaired B cell differentiation in an all-or-none manner (Fig. 4B, bottom panel).
The third AhR-mediated pathway involves direct repression by TCDD of the IgH gene (Sulentic et al., 2004; Henseler et al., 2009), a key component of the immunoglobulin molecule (Fig. 4C, top panel). Since IgH is located outside of (a) the core feedback loops that make up the bistable switch and (b) the signaling pathway transducing extracellular stimuli to the switch, suppression of IgH by TCDD is not expected to interfere with the bistable switch itself or its activation. Bifurcation analysis supports this conjecture, showing that TCDD suppresses the IgM level associated with the plasma cell state, without altering the activation threshold of LPS stimulation (Fig. 4C, middle panel). This pathway should therefore lead to suppression of B cell differentiation in a graded manner. Stochastic simulations confirmed that the fraction of IgMhigh cells (which is 1 minus the fraction of IgMlow cells, which remained essentially constant) was unaffected, whereas the mean IgM level in those cells decreased with TCDD, as indicated by the leftward shift of the IgMhigh peak (Fig. 4C, bottom panel).
TCDD impairs B cell differentiation in an all-or-none manner
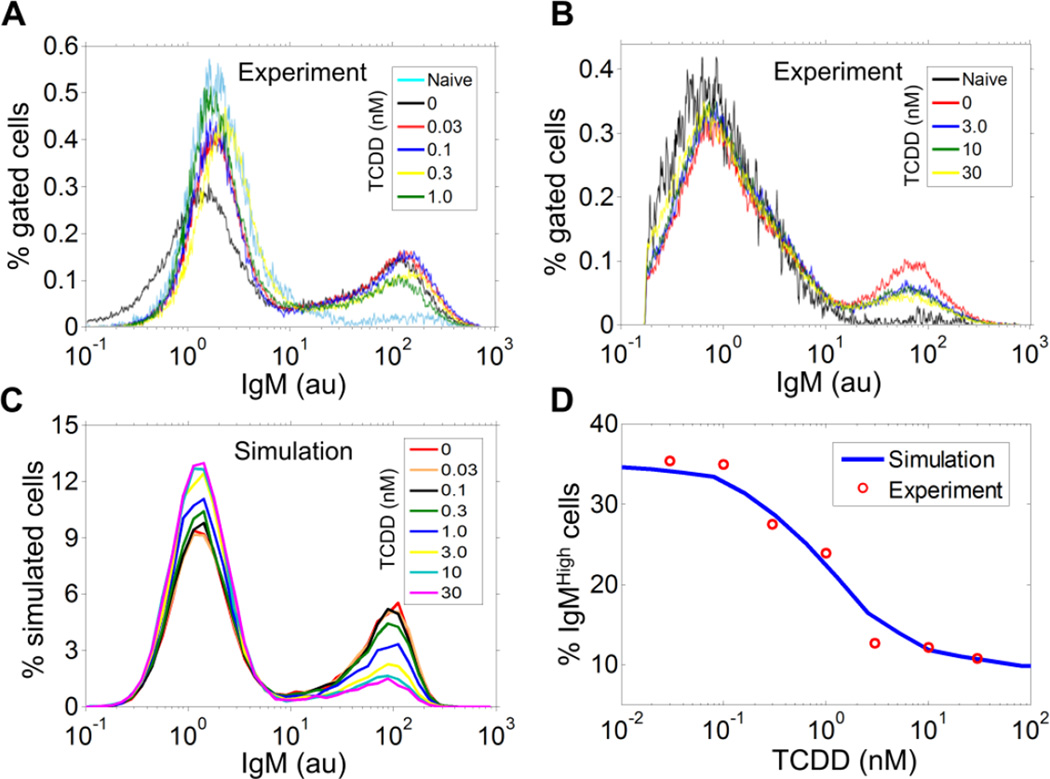
We next experimentally tested whether TCDD impairs B cell differentiation in a binary or graded fashion. Flow cytometry revealed that the fraction of the IgMhigh cell population obtained in the continuous presence of 5 µg/ml LPS for 72 h decreased with increasing TCDD concentration, without any appreciable down-regulation of the intracellular IgM level in these cells (Figs. 5A and 5B). The fraction of IgMhigh cells was maximally suppressed at 30 nM TCDD, reaching only about one third of the vehicle control. It is important to emphasize that consistent with earlier reports (Holsapple et al., 1986; Crawford et al., 2003), TCDD had no effect on LPS-induced proliferation (Fig. S3A). Cell viability was also unaffected (result not shown). Moreover, the suppression of IgMhigh cells observed on day 3 cannot be attributed to a simply shift of the normal response to LPS to a later time as no further increase in IgMhigh cells was observed on day 5 (Fig. S3B). In addition, the decrease in IgMhigh cells by TCDD was not due to increased differentiation of B cells to other isotypes of antibody-producing cells (Fig. S3C). Collectively, these results demonstrated that TCDD suppresses LPS-stimulated B cell differentiation in an all-or-none fashion, by reducing the fraction of IgMhigh cells formed.
Figure 5. All-or-none mode of suppression of B cell differentiation into IgMhigh cells by TCDD.
(A, B) Histograms of intracellular IgM in CD19-gated viable B cells from flow cytometry analysis. Primary mouse splenocytes were simultaneously treated with 5 µg/ml LPS and various concentrations of TCDD for 72 h. For TCDD concentrations of 0.03, 0.1, 0.3, 1, 3, 10, and 30 nM, the percentage of IgMhigh cells is 35.35%, 34.93%, 27.49%, 23.94%, 12.71%, 12.22%, and 10.78%, respectively. (C) Histograms of IgM in 20,000 stochastically simulated B cells simultaneously exposed to 5 µg/ml LPS and various concentrations of TCDD for 72 h, with all three calibrated AhR-mediated TCDD pathways kept active. (D) Fitting of the concentration-response of percentage simulated IgMhigh cells in panel C to the experimental data in panels A and B.
The observed all-or-none mode of suppression suggests that among the three AhR-mediated pathways analyzed above, the pathways of AP-1 inhibition and Bach2 up-regulation by TCDD are likely to be at work while direct repression of IgH transcription may play a modest, if any, role in suppressing the IgM response. We went on to assign an appropriate weight to each of the AhR-mediated TCDD pathways to further constrain the model quantitatively according to the experimental data (see Table S2 for details). Briefly, since direct suppression of IgH appears to have only a negligible effect on the IgM response, we assumed the rate of IgM transcription in simulated plasma cells is maximally reduced by 5% by TCDD. We have recently reported that 10 nM TCDD up-regulated Bach2 gene transcription by about 1.5- to 3-fold in resting CH12.LX mouse B cells (De Abrew et al., 2011). Accordingly, we parameterized the model such that 10 nM TCDD would increase Bach2 protein expression by about 2-fold. Lastly, the parameters controlling the inhibition of the AP-1 pathway by AhR was adjusted such that with all three pathways participating, the maximal suppression of the IgMhigh cell population by TCDD would reach one third of the control, matching the experimental data reported above. When the final calibrated model was stochastically simulated, it was indeed able to reproduce the observed all-or-none mode of suppression by TCDD (Fig. 5C). Moreover, the model quantitatively recapitulated the observed concentration-response relationship between TCDD concentration and the percentage of IgMhigh cells (Fig. 5D). It is clear that although TCDD suppressed the plasma cell response in an all-or-none fashion in individual cells, the population-level response shows a graded behavior due to cell-to-cell heterogeneity, an effect captured by stochastic simulation.
With the calibrated model, it would be intriguing to investigate whether the AP-1 and Bach2 pathways, both of which can mediate an all-or-none mode of suppression by TCDD, can produce outcomes that differ in other aspects, such as the timing of the differentiation process. Stochastic simulations showed that when either of the two AhR-mediated pathways was blocked individually in the final model, suppression of the IgMhigh response was reversed partially and to a comparable extent (Fig. S4A). Normalization of the responses revealed that the appearance of IgMhigh cells was time-delayed when TCDD acts primarily through the Bach2 pathway rather than through the AP-1 pathway (Fig. S4B).
Discussion
Cells can respond to external perturbations with diverse gene expression patterns, which may vary in either an all-or-none or graded fashion (Rossi et al., 2000; Biggar and Crabtree, 2001; Louis and Becskei, 2002; Joers et al., 2004). The all-or-none mode of protein expression may arise from mechanisms including discrete gene promoter dynamics, ultrasensitive switches, and bistability (Huang and Ferrell, 1996; Ozbudak et al., 2004; Pirone and Elston, 2004; Zhang et al., 2006). As an important class of environmental toxicants, TCDD and other AhR ligands have been shown to induce cytochrome P450 1A1 gene expression in a binary, switch-like manner in hepatocytes through an unknown mechanism (Tritscher et al., 1992; Broccardo et al., 2004). In the present study, we demonstrated that TCDD suppresses the differentiation of mature B cells into IgM-secreting cells in an all-or-none fashion. As the concentration of TCDD increases, fewer fractions of B cells can transition into the plasma cell state. However, for those B cells that make into plasma cells, the intracellular IgM level is uncompromised by TCDD exposure.
To mechanistically understand this all-or-none phenomenon, it is necessary to closely examine: (1) the B cell-specific molecular circuit as a dynamic system that underlies the physiological differentiation process; and (2) the TCDD-activated AhR pathways that perturb the dynamic system. The core transcriptional circuit in B cells contains interconnected double-negative feedback loops, consistent with the network structure capable of bistable switching (Ferrell, 2002). Therefore, it is reasonable to postulate that the all-or-none mode of impairment of B cell differentiation by TCDD may be a consequence of interfering with the underlying bistable switch. Based on the gene circuit model which was experimentally-constrained, we predicted that among the three AhR-implicated pathways previously identified in B cells, perturbation through the AP-1 and/or Bach2 pathways is likely to result in an all-or-none mode of suppression by TCDD (Figs. 4A and 4B). In contrast, the pathway involving direct repression of IgH transcription by TCDD can only produce graded suppression as an outcome (Fig. 4C).
The premise that all-or-none suppression by TCDD is achieved through interference with the B cell bistable switch is consistent with our previous findings that TCDD gradually lost its ability to suppress the IgM response as the onset time of its application post B cell activation was pushed back (Tucker et al., 1986; Crawford et al., 2003). A likely explanation for this phenomenon is that for TCDD to disrupt the differentiation process, it must be present in the cellular environment prior to the bistable switch being turned on. Once B cells are committed to the plasma cell fate with the activation of the irreversible bistable switch, TCDD can no longer exert influence on the differentiation program. In contrast, if direct repression of IgH expression were the primary mechanism, TCDD-mediated suppression of the antibody response would not have been as time-sensitive. In that case, TCDD could repress immunoglobulin expression at any time, even after B cells have become plasma cells.
We have previously shown that TCDD treatment of B cells leads to deregulation of AP-1 and Bach2, two critical regulators of Prdm1. Specifically, TCDD inhibited AP-1 DNA binding activity to the Prdm1 promoter, which acts as a positive transcriptional regulator of Prdm1(Schneider et al., 2009). TCDD also transcriptionally up-regulated Bach2, which acts as a repressor of Prdm1 (De Abrew et al., 2010; De Abrew et al., 2011). Although modulation by TCDD of both the AP-1 and Bach2 pathways could mediate the all-or-none suppression of the B cell response to LPS, their relative contributions are unknown. Our experimentally calibrated computational model suggested that both pathways could exert suppression to a comparable extent (Figs. S4A). We have previously demonstrated that the suppression of antibody secretion by TCDD can be reversed by approximately 40% when TCDD-induced up-regulation of Bach2 gene expression is blocked by siRNA interference in CH12.LX B cells (De Abrew et al., 2011). Notably, blocking the up-regulation of the Bach2 pathway in our computational model achieved a similar magnitude of recovery (Fig. S4A). Despite the comparable extent of suppression by the AP-1 and Bach2 pathways, each pathway appears to affect the dynamics of plasma cell accumulation differently, as predicted by our model. When examining the normalized responses, it is evident that when Bach2 is the primary pathway mediating the immunotoxicity of TCDD (i.e. with the AP-1 pathway blocked), the emergence of plasma cells occurred at later times compared with the situation when AP-1 is the primary pathway (i.e., with the Bach2 pathway blocked) (Fig. S4B).
In the B cell regulatory circuit, Pax5, Bach2, and Prdm1 form a coherent feed-forward loop where Pax5 activates Bach2 and both of these TFs repress Prdm1 (Ochiai et al., 2006; Mora-Lopez et al., 2007; Schebesta et al., 2007). This type of feed-forward loop can introduce a time delay in signaling (Mangan et al., 2003). In the context of our model, the feed-forward loop ensures that a decrease in the level of Pax5 would not result in an immediate de-repression of Prdm1, since Bach2, which also represses Prdm1, is unlikely to disappear immediately with Pax5. Thus by providing a time delay within the feedback loop, Bach2 tends to stabilize the B cell state and postpone switching to the plasma cell state. High levels of Bach2 during B cell activation can promote CSR in B cells before they differentiate into the default IgM-secreting plasma cells, thus diversifying the isotype of the antibody response (Muto et al., 2010). Conversely, as our model suggested, absence of Bach2 in the regulatory circuit would destabilize the B cell state, making the switching to the plasma cell state more readily (Fig. 3D). This could explain the more rapid and robust response of B cells harvested from Bach2−/− mice compared to wild-type cells, along with a lower proportion of non-IgM isotypes (Muto et al., 2004; Muto et al., 2010). Deletion of Bach2 in our model recapitulated this augmented plasma cell response and its earlier activation (Fig. 3E).
By maintaining Bach2 gene expression at an elevated level, TCDD could presumably postpone the bistable switching event underlying B cell differentiation and allow more time for CSR to occur. This leads us to predict that while TCDD generally reduces the fraction of activated B cells that eventually differentiate into plasma cells, in the process it may simultaneously alter the proportion of relevant non-IgM-secreting plasma cells as a result of more frequent CSR. Although this prediction remains to be tested experimentally, previous studies have demonstrated that TCDD acting through AhR increased circulating IgA levels while suppressing other types of antibody response in mice infected by influenza virus (Warren et al., 2000; Lawrence and Vorderstrasse, 2004). In B cells, many signal transduction pathways activated by specific antigens, interleukins and cytokines converge to the core bistable circuit containing Bach2 (Calame, 2008). Therefore the all-or-none mode of suppression by TCDD is likely to be conserved in B cells activated by stimuli other than LPS.
Previously, IgH was identified as a direct transcriptional target responsible for TCDD/AhR-mediated suppression of LPS-induced IgM secretion in the CH12.LX mouse B cell line (Sulentic et al., 2004; Henseler et al., 2009). The present study suggests that the AhR-to-IgH pathway likely plays a very modest repressive role in primary mouse B cells. This conclusion is based on the fact that neither a graded nor hybrid (mixed binary/graded) mode of response to TCDD was observed. The inferred lack of involvement of direct IgH suppression in primary mouse B cells is in fact consistent with our previous in vitro findings that the amount of IgM secreted into the cell culture – an endpoint reflecting both the number of plasma cells formed and amount of IgM produced per plasma cell – was suppressed by TCDD to an extent that is not significantly higher than the reduction in the number of plasma cells (Tucker et al., 1986; Sulentic et al., 1998; Crawford et al., 2003; Schneider et al., 2008). However it is also possible that IgH may be produced in excess compared with other immunoglobulin components such as the light and J chains, such that direct repression of IgH by TCDD would not lead to an observable decrease in the amount of the final assembled immunoglobulin molecules.
An integrated approach employing both experimental studies and mathematical modeling is increasingly utilized to understand the gene network dynamics and the roles of constituent transcription factors involved in B cell activation and differentiation (Muto et al., 2010; Sciammas et al., 2011; Martinez et al., 2012). While these studies focused primarily on the physiological B cell activation process, our study examined how the process is perturbed by environmental toxicant TCDD to disrupt humoral immunity. None of these models including our own are inclusive to incorporate all known gene components and interactions implicated in the B cell transcriptional events, owing to insufficient information on molecular regulation details or irrelevance to the model focus. As the wiring diagram of the gene circuit is resolved in greater details, more complete mathematical descriptions of B cell activation, including clonal expansion and cell death, are expected to follow, which will serve as an improved platform to further help investigate how environmental immunotoxicants affect the antibody response.
In conclusion, we have demonstrated that B cell terminal differentiation is disrupted by the environmental contaminant TCDD in an all-or-none manner and that this mode of suppression could result from perturbation of a bistable gene circuit underpinning B cell differentiation. As the field of toxicity testing and risk assessment for environmental toxicants moves steadily toward alternative testing methods, the integrated approach exploited here represents an emerging direction in which computational modeling of pathway perturbation is used to mechanistically interpret in vitro toxicity data and make quantitative predictions.
Supplementary Material
Highlights.
TCDD suppresses B cell differentiation stimulated by LPS in an all-or-none mode.
TCDD reduces the fraction of IgM-secreting cells, not the IgM level in those cells.
A mathematical model indicates deregulation of AP-1 and Bach2 by AhR is involved.
Both pathways interfere with the bistable switch underlying B cell differentiation.
Disruption of the bistable switch leads to all-or-none mode of suppression.
Acknowledgement
We would like to thank the Superfund Research Program of the National Institute of Environmental Health Sciences (P42ES04911 to NEK) for supporting this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors have declared no conflicts of interest. This manuscript has been reviewed by the U. S. Environmental Protection Agency and approved for publication. Approval does not signify that the contents necessarily reflect the views and policies of the agency; nor does the mention of trade names or commercial products constitute endorsement or recommendation for use.
Authors’ Contributions
All authors participated in the design of the in silico and in vitro experiments, and approved the final manuscript. QZ and SB developed the in silico model and performed numerical simulations. DEK and RBC2 performed the in vitro experiments. QZ and NEK wrote the manuscript. SB, RBC3, RST, MEA, and NEK critically reviewed the manuscript.
References
- Adalsteinsson D, McMillen D, Elston TC. Biochemical Network Stochastic Simulator (BioNetS): software for stochastic modeling of biochemical networks. BMC Bioinformatics. 2004;5:24. doi: 10.1186/1471-2105-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis A, Widenhorn K, Vitelli L, Busslinger M. A novel B-cell lineage-specific transcription factor present at early but not late stages of differentiation. Genes Dev. 1990;4:849–859. doi: 10.1101/gad.4.5.849. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Conolly RB, Kaminski NE, Thomas RS, Andersen ME, Zhang Q. A bistable switch underlying B-cell differentiation and its disruption by the environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2010;115:51–65. doi: 10.1093/toxsci/kfq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar SR, Crabtree GR. Cell signaling can direct either binary or graded transcriptional responses. Embo J. 2001;20:3167–3176. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccardo CJ, Billings RE, Chubb LS, Andersen ME, Hanneman WH. Single cell analysis of switch-like induction of CYP1A1 in liver cell lines. Toxicol Sci. 2004;78:287–294. doi: 10.1093/toxsci/kfh077. [DOI] [PubMed] [Google Scholar]
- Calame K. Activation-dependent induction of Blimp-1. Curr Opin Immunol. 2008;20:259–264. doi: 10.1016/j.coi.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, Falini B, Louie DC, Offit K, Chaganti RS, Dalla-Favera R. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- Crawford RB, Sulentic CE, Yoo BS, Kaminski NE. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the regulation and posttranslational modification of p27kip1 in lipopolysaccharide-activated B cells. Toxicol Sci. 2003;75:333–342. doi: 10.1093/toxsci/kfg199. [DOI] [PubMed] [Google Scholar]
- De Abrew KN, Kaminski NE, Thomas RS. An integrated genomic analysis of aryl hydrocarbon receptor-mediated inhibition of B-cell differentiation. Toxicol Sci. 2010;118:454–469. doi: 10.1093/toxsci/kfq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Abrew KN, Phadnis AS, Crawford RB, Kaminski NE, Thomas RS. Regulation of Bach2 by the aryl hydrocarbon receptor as a mechanism for suppression of B-cell differentiation by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Appl Pharmacol. 2011;252:150–158. doi: 10.1016/j.taap.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Dooley RK, Holsapple MP. Elucidation of cellular targets responsible for tetrachlorodibenzo-p-dioxin (TCDD)-induced suppression of antibody responses: I. The role of the B lymphocyte. Immunopharmacology. 1988;16:167–180. doi: 10.1016/0162-3109(88)90005-7. [DOI] [PubMed] [Google Scholar]
- Ermentrout B. Simulating, Analyzing, and Animating Dynamical Systems: A Guide to XPPAUT for Researchers and Students. 1st ed. Philadelphia, USA: SIAM; 2002. [Google Scholar]
- Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- Gillespie DT. Exact stochastic simulation of coupled chemical reactions. Journal of Physical Chemistry. 1977;81:2340–2361. [Google Scholar]
- Henseler RA, Romer EJ, Sulentic CE. Diverse chemicals including aryl hydrocarbon receptor ligands modulate transcriptional activity of the 3'immunoglobulin heavy chain regulatory region. Toxicology. 2009;261:9–18. doi: 10.1016/j.tox.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsapple MP, Dooley RK, McNerney PJ, McCay JA. Direct suppression of antibody responses by chlorinated dibenzodioxins in cultured spleen cells from (C57BL/6 × C3H)F1 and DBA/2 mice. Immunopharmacology. 1986;12:175–186. doi: 10.1016/0162-3109(86)90001-9. [DOI] [PubMed] [Google Scholar]
- Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joers A, Jaks V, Kase J, Maimets T. p53-dependent transcription can exhibit both on/off and graded response after genotoxic stress. Oncogene. 2004;23:6175–6185. doi: 10.1038/sj.onc.1207864. [DOI] [PubMed] [Google Scholar]
- Johnson K, Shapiro-Shelef M, Tunyaplin C, Calame K. Regulatory events in early and late B-cell differentiation. Mol Immunol. 2005;42:749–761. doi: 10.1016/j.molimm.2004.06.039. [DOI] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Kerkvliet NI. Recent advances in understanding the mechanisms of TCDD immunotoxicity. Int Immunopharmacol. 2002;2:277–291. doi: 10.1016/s1567-5769(01)00179-5. [DOI] [PubMed] [Google Scholar]
- Lawrence BP, Vorderstrasse BA. Activation of the aryl hydrocarbon receptor diminishes the memory response to homotypic influenza virus infection but does not impair host resistance. Toxicol Sci. 2004;79:304–314. doi: 10.1093/toxsci/kfh094. [DOI] [PubMed] [Google Scholar]
- Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis M, Becskei A. Binary and graded responses in gene networks. Sci STKE. 2002;2002:pe33. doi: 10.1126/stke.2002.143.pe33. [DOI] [PubMed] [Google Scholar]
- Lu H, Crawford RB, Suarez-Martinez JE, Kaplan BL, Kaminski NE. Induction of the aryl hydrocarbon receptor-responsive genes and modulation of the immunoglobulin M response by 2,3,7,8-tetrachlorodibenzo-p-dioxin in primary human B cells. Toxicol Sci. 2010;118:86–97. doi: 10.1093/toxsci/kfq234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Martinez MR, Corradin A, Klein U, Alvarez MJ, Toffolo GM, di Camillo B, Califano A, Stolovitzky GA. Quantitative modeling of the terminal differentiation of B cells and mechanisms of lymphomagenesis. Proc Natl Acad Sci U S A. 2012;109:2672–2677. doi: 10.1073/pnas.1113019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Lopez F, Reales E, Brieva JA, Campos-Caro A. Human BSAP and BLIMP1 conform an autoregulatory feedback loop. Blood. 2007;110:3150–3157. doi: 10.1182/blood-2007-05-092262. [DOI] [PubMed] [Google Scholar]
- Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, Karasuyama H, Hayashi N, Nakauchi H, Yamamoto M, Groudine M, Igarashi K. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3' enhancer. EMBO J. 1998;17:5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Ochiai K, Kimura Y, Itoh-Nakadai A, Calame KL, Ikebe D, Tashiro S, Igarashi K. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010;29:4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, Ikebe D, Yamamoto M, Igarashi K. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H, Tashiro S, Muto A, Igarashi K. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem. 2006;281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y, Arima M, Arguni E, Okada S, Yamashita K, Asari S, Obata S, Sakamoto A, Hatano M, J OW, Ebara M, Saisho H, Tokuhisa T. A role for c-fos/activator protein 1 in B lymphocyte terminal differentiation. J Immunol. 2005;174:7703–7710. doi: 10.4049/jimmunol.174.12.7703. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- Peden-Adams MM, Keller JM, Eudaly JG, Berger J, Gilkeson GS, Keil DE. Suppression of humoral immunity in mice following exposure to perfluorooctane sulfonate. Toxicol Sci. 2008;104:144–154. doi: 10.1093/toxsci/kfn059. [DOI] [PubMed] [Google Scholar]
- Pirone JR, Elston TC. Fluctuations in transcription factor binding can explain the graded and binary responses observed in inducible gene expression. J Theor Biol. 2004;226:111–121. doi: 10.1016/j.jtbi.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Rossi FM, Kringstein AM, Spicher A, Guicherit OM, Blau HM. Transcriptional control: rheostat converted to on/off switch. Mol Cell. 2000;6:723–728. doi: 10.1016/s1097-2765(00)00070-8. [DOI] [PubMed] [Google Scholar]
- Salazar KD, de la Rosa P, Barnett JB, Schafer R. The polysaccharide antibody response after Streptococcus pneumoniae vaccination is differentially enhanced or suppressed by 3,4-dichloropropionanilide and 2,4-dichlorophenoxyacetic acid. Toxicol Sci. 2005;87:123–133. doi: 10.1093/toxsci/kfi244. [DOI] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Schneider D, Manzan MA, Crawford RB, Chen W, Kaminski NE. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated impairment of B cell differentiation involves dysregulation of paired box 5 (Pax5) isoform, Pax5a. J Pharmacol Exp Ther. 2008;326:463–474. doi: 10.1124/jpet.108.139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Manzan MA, Yoo BS, Crawford RB, Kaminski N. Involvement of Blimp-1 and AP-1 dysregulation in the 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated suppression of the IgM response by B cells. Toxicol Sci. 2009;108:377–388. doi: 10.1093/toxsci/kfp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciammas R, Davis MM. Modular nature of Blimp-1 in the regulation of gene expression during B cell maturation. J Immunol. 2004;172:5427–5440. doi: 10.4049/jimmunol.172.9.5427. [DOI] [PubMed] [Google Scholar]
- Sciammas R, Li Y, Warmflash A, Song Y, Dinner AR, Singh H. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol Syst Biol. 2011;7:495. doi: 10.1038/msb.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- Strogatz SH. Nonlinear dynamics and chaoes with applications to physics, biology, chemistry, and engineering. 1st ed. Cambridge: Perseus Books Group; 1994. [Google Scholar]
- Suh J, Jeon YJ, Kim HM, Kang JS, Kaminski NE, Yang KH. Aryl hydrocarbon receptor-dependent inhibition of AP-1 activity by 2,3,7,8-tetrachlorodibenzo-p-dioxin in activated B cells. Toxicol Appl Pharmacol. 2002;181:116–123. doi: 10.1006/taap.2002.9403. [DOI] [PubMed] [Google Scholar]
- Sulentic CE, Holsapple MP, Kaminski NE. Aryl hydrocarbon receptor-dependent suppression by 2,3,7, 8-tetrachlorodibenzo-p-dioxin of IgM secretion in activated B cells. Mol Pharmacol. 1998;53:623–629. [PubMed] [Google Scholar]
- Sulentic CE, Kaminski NE. The long winding road toward understanding the molecular mechanisms for B-cell suppression by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2011;120(Suppl 1):S171–S191. doi: 10.1093/toxsci/kfq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulentic CE, Zhang W, Na YJ, Kaminski NE. 2,3,7,8-tetrachlorodibenzo-p-dioxin, an exogenous modulator of the 3'alpha immunoglobulin heavy chain enhancer in the CH12.LX mouse cell line. J Pharmacol Exp Ther. 2004;309:71–78. doi: 10.1124/jpet.103.059493. [DOI] [PubMed] [Google Scholar]
- Tritscher AM, Goldstein JA, Portier CJ, McCoy Z, Clark GC, Lucier GW. Dose-response relationships for chronic exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin in a rat tumor promotion model: quantification and immunolocalization of CYP1A1 and CYP1A2 in the liver. Cancer Res. 1992;52:3436–3442. [PubMed] [Google Scholar]
- Tucker AN, Vore SJ, Luster MI. Suppression of B cell differentiation by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol Pharmacol. 1986;29:372–377. [PubMed] [Google Scholar]
- Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169:1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- Vorderstrasse BA, Steppan LB, Silverstone AE, Kerkvliet NI. Aryl hydrocarbon receptor-deficient mice generate normal immune responses to model antigens and are resistant to TCDD-induced immune suppression. Toxicol Appl Pharmacol. 2001;171:157–164. doi: 10.1006/taap.2000.9122. [DOI] [PubMed] [Google Scholar]
- Warren TK, Mitchell KA, Lawrence BP. Exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) suppresses the humoral and cell-mediated immune responses to influenza A virus without affecting cytolytic activity in the lung. Toxicol Sci. 2000;56:114–123. doi: 10.1093/toxsci/56.1.114. [DOI] [PubMed] [Google Scholar]
- Watanabe-Matsui M, Muto A, Matsui T, Itoh-Nakadai A, Nakajima O, Murayama K, Yamamoto M, Ikeda-Saito M, Igarashi K. Heme regulates B-cell differentiation, antibody class switch, and heme oxygenase-1 expression in B cells as a ligand of Bach2. Blood. 2011;117:5438–5448. doi: 10.1182/blood-2010-07-296483. [DOI] [PubMed] [Google Scholar]
- Xiong W, Ferrell JE., Jr A positive-feedback-based bistable 'memory module' that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Andersen ME, Conolly RB. Binary gene induction and protein expression in individual cells. Theor Biol Med Model. 2006;3:18. doi: 10.1186/1742-4682-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bhattacharya S, Kline DE, Crawford RB, Conolly RB, Thomas RS, Kaminski NE, Andersen ME. Stochastic modeling of B lymphocyte terminal differentiation and its suppression by dioxin. BMC Syst Biol. 2010;4:40. doi: 10.1186/1752-0509-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.