Abstract
Hypoxia inducible factors (HIFs) are activated in many tumors and show either promoter or suppressor activity depending on the tumor cell biology and background. However, the role of HIF member HIF-2α remains unclear in hepatocellular carcinoma (HCC). Here, HIF-2α expression was measured in HCC and paired peritumoral tissues by qRT-PCR, western blot analysis, and immunofluorescence assays, and the clinical significance was explored in 246 HCC patients. In cell culture, HIF-2α levels were over-expressed or knocked-down by use of expression or short hairpin RNA recombinant plasmid respectively. Cells were analyzed by immunoblot, chromatin immunoprecipitation coupled with microarray, co-immunoprecipitation, and histochemical staining. In vivo tumor growth was analyzed in nude mice. We found that the average expression of HIF-2α was relatively low in HCC tissues, and the decreased level was associated with lower overall survival (p=0.006). High HIF-2α expression in HCC cells induced higher levels of apoptosis and expression of pro-apoptotic proteins, and it inhibited cell and tumor growth. Furthermore, HIF-2α inhibited expression of the novel target gene transcription factor dimerization partner 3 (TFDP3). TFDP3 protein was found to bind with E2F transcription factor 1 (E2F1) and inhibit its transcriptional activity through both p53-dependent and -independent pathways. Re-introduction of TFDP3 expression reversed HIF-2α-induced apoptosis.
Conclusions
Data gathered from cell lines, tumorigenicity studies, and primary HCC samples demonstrate a negative role of HIF-2α in tumors, which is mediated by the TFDP3/E2F1 pathway. Our study provides evidence supporting a possible tumor suppressor role for HIF-2α and has uncovered a mechanism that links HIF-2α to a fundamental biological regulator, E2F1.
Keywords: HIF-2α, apoptosis, TFDP3, E2F1
Hepatocellular carcinoma (HCC) is one of the most common, aggressive malignancies and is the third leading cause of cancer-related deaths. The poor prognosis of HCC is mainly due to the rapid progression of this disease.1–3 Advances in treatment of this disease are likely to stem from a better understanding of its biology and behavior. As most of solid tumors, hypoxic microenvironment exists in HCC as a result of an imbalance between oxygen supply and consumption in proliferating tumors.1 Significant evidences indicate that the hypoxia inducible factors (HIFs) play an important role in the pathogenesis and pathophysiology of HCC. As HIF inhibitors are currently undergoing clinical evaluation as cancer therapeutics, a more thorough understanding of the unique roles performed by HIFs in human tumor is warranted.4, 5
Cellular responses to low oxygen tension are mainly mediated by the activation HIFs, which are consisting of a constitutively expressed subunit (ARNT) and an oxygen-regulated subunit, mainly HIF-1α and HIF-2α. HIF-1α and HIF-2α promote adaptation of tumor cells to hypoxic stress by regulating the expression of genes involved in metabolism, angiogenesis, erythropoiesis, cell proliferation and apoptosis.6–9 HIF-1α is found to be expressed at higher level in dysplastic nodules and implicate a malignant transformation. We and others have previously showed that HIF-1α expression is significantly associated with an advanced stage and aggressive phenotype and indicates a poor prognosis in HCC.2, 3 Approximately 48% amino-acid sequence homology is shared by HIF-1α and HIF-2α;10 However, much less is known about HIF-2α isoform, and there have been inconsistent and even conflicting reports about its role in HCC. Some studies conclude that HIF-2α promotes hypoxic cell proliferation by enhancing c-Myc transcriptional activity,11 whereas others argue that knock-down of HIF-2α increases cell viability and growth by autophagy.1 It is known that HIF-2α correlates with vascular endothelial growth factor (VEGF) expression and indicates a poor prognosis in melanoma and non-small cell lung cancers.12, 13 Other studies show that HIF-2α acts as a tumor suppressor in breast and brain cancer.14, 15
To date, there are limited published reports describing HIF-2α in HCC and the role of this protein is still controversial. 1, 16 Here, we provide evidence for a possible tumor suppressor role for HIF-2α in HCC. Chromatin immunoprecipitation coupled with microarray (ChIP-on-chip) and additional assays indicate that transcription factor dimerization partner 3 (TFDP3) is a novel target of HIF-2α and inhibits the E2F transcription factor 1 (E2F1) transcriptional activity through multiple pathways. This study has uncovered another mechanism connecting HIF-2α to E2F1 in the induction of apoptosis.
Materials and Methods
Patients and Specimens
Tissues were collected and tissue microarray (TMA) analyses performed as previously described.17 The follow-up procedures were described in the supplementary Materials and Methods.
Cell Culture and Recombinant Plasmid DNA
HCC cell lines used in this study, construction of short hairpin RNA (shRNA), and expression of HIF-2α recombinant plasmids are described in the supplementary Materials and Methods. The pcNDA3-TFDP3 plasmid was a gift from Yu Zhang lab, Peking University. To select the stably transformed monoclonal cell lines, selective medium containing neomycin (pcDNA3.1 plasmids) or puromycin (shRNA plasmids) was used.
Real-Time PCR Assay and Western Blot
RNA extraction, cDNA synthesis, quantitative real-time PCR (qRT-PCR) reactions, and western blot were performed as before.18
Immunofluorescence Staining and Apoptosis Analysis
Primary antibodies used were as follows: anti HIF-2α from Novus Biologicals, Littleton, CO, USA; anti Ki67 from DAKO, Carpentaria, CA, USA; and anti caspase-3 from Cell Signaling, Boston, MA, USA.
To quantify the extent of apoptosis, cells were harvested with trypsin without EDTA, and visualized by Annexin V-FITC/propidium iodide staining according to the manufacturer’s protocol (BD Bioscience, Rockville, MD, USA).
ChIP-on-chip, ChIP-PCR and Immunoprecipitation
A complete protocol of ChIP-on-chip provided by NimbleGen Systems (Madison, WI, USA) was included in the supplementary Materials and Methods section. Predicted binding sites were confirmed using ChIP-PCR in MHCC97L cell line. Immunoprecipitation was performed as describe previously19.
Statistical Analysis
Data were analyzed with SPSS 16.0 software as previously described.17 A p value <0.05 was considered statistically significant.
Results
The Expression Level of HIF-2α in HCC Patients and Its Clinical Significance
To explore effects of HIF-2α in HCC, we first detected HIF-2α levels in tumor and peritumoral tissues. HIF-2α showed a mainly cytoplasmic staining in cancer cells as previously indicated (Figure 1A)16 and the average levels of HIF-2α on both protein and mRNA were significantly lower in tumor than peritumoral tissues (Figures 1B, 1C). We further explored the clinical significance of HIF-2α expression in 246 HCC cases. The clinical and pathological characteristics of these patients were listed in Table S1. The percentage and intensity of positively staining cells were largely varying among cases, and tumors were classified as strong in 118 cases (47.9%) and moderately or weakly positive in 128 cases (52.1%) (Figure 1D).13 Duplicate tumor samples showed a good level of agreement with respect to intensity and percentage of positively stained cells. Kaplan-Meier analysis revealed that patients with high HIF-2α expression levels in tissues had significantly longer overall survival (OS) rates than those with low expression (p=0.006) (Figure 1E).
Figure1.
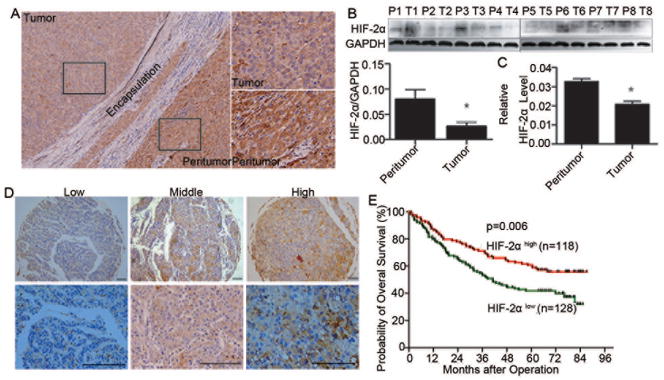
The expression level of HIF-2α in HCC patients and its clinical significance. (A) Representative images of HIF-2α staining in HCC tissue and corresponding peritumoral tissues. (B) The expression levels of HIF-2α in tumor and corresponding peritumoral tissues. Quantitative analysis of protein levels were determined by measuring the ratio of HIF-2α to GAPDH expression. Error bars indicate the standard deviation (SD).*, p=0.02 by two-tailed test. (C) Quantitative RT-PCR analysis of HIF-2α levels in HCC tissues and corresponding peritumoral tissue. GAPDH was used as a control. Error bars indicate standard deviation (SD) (n=60). *, p<0.001 (two-tailed test). (D) Expression of HIF-2α was examined via immunohistochemistry in 246 hepatocellular carcinoma specimens, and representative images are shown. Scale bar, 10 mm. (E) Kaplan-Meier analysis of overall survival (OS) in 246 HCC patients relative to HIF-2α expression.
HIF-2α Inhibits HCC Cell Growth Mainly through Apoptosis
To establish whether HIF-2α plays a suppressor role in HCC tumor growth, human liver cancer cell lines MHCC97H and SMMC-7721 were studied. Both HIF-1α and HIF-2α expression were up regulated in MHCC97H cell line and proliferation rate was lowered under hypoxia condition (Figures S1A, S1B). To explore the role of HIF-2α in overcoming the effects of HIF-1α and other factors, we isolated high- or low- HIF-2α expression clones that had been transfected with expression vector pcDNA3.1-HIF-2α or shRNA inhibition vector pT2sh-HIF-2α, respectively (Figures 2A, 2B). In these clones, HIF-1α levels did not vary significantly (Figure S1C).
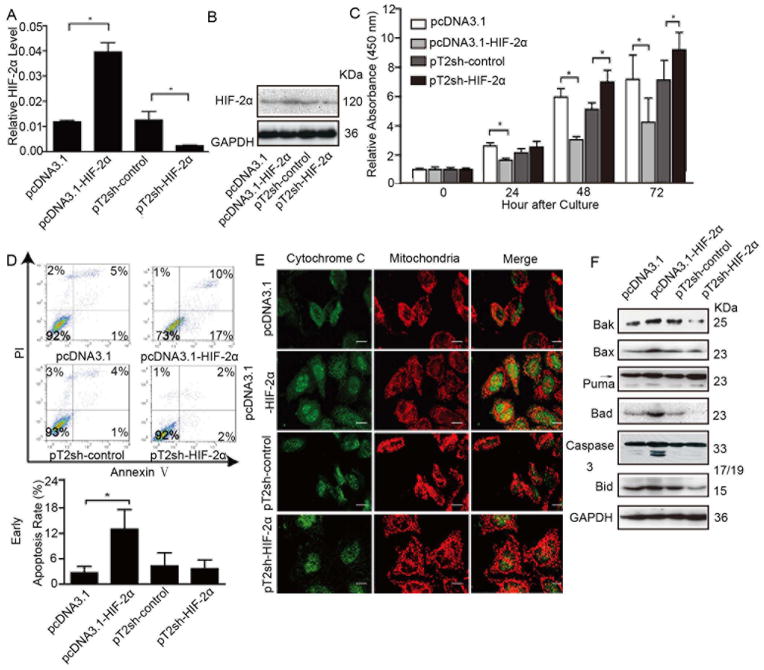
Figure 2.
HIF-2α induces HCC cell growth arrest in vitro mainly through effects on apoptosis. Cells were transfected with pcDNA3.1-HIF-2α or pT2sh-HIF-2α recombinant plasmid or corresponding empty vector (pcDNA3.1, pT2), then monoclones were selected. (A) HIF-2α mRNA levels of each representative monoclone, in which error bars indicate standard deviation (n=3). *, p<0.01 by two-tailed test. (B) Protein levels of HIF-2α and (C) proliferative ability of representative monoclonal cells. The relative absorbance at each time point was normalized to that measured at the first time point (*, p<0.01). (D) Flow cytometry with annexin V/PI plots in different groups represented by an individual monoclone and column bar of mean apoptotic percentage; error bars indicate standard deviation (SD). *, p<0.05. (E) Cells were trypsinized and plated on coverslips. After 48 hours, cells were labeled with mito Tracker™ for 30 minutes at 37°C, subjected to indirect immunofluorescence staining for cytochrome C expression. Representative images with bars =5 μm. (F) Western blot analysis of the expression of Bak, Bax, Puma, Bad, activated caspase 3, Bid, and GAPDH in monoclonal cells.
Initially, viability of transfected cells was investigated. High expression of HIF-2α in MHCC97H cells was associated with a slower growth rate compared to the respective control, while cells transfected with anti-HIF-2α shRNA grew faster than control (Figure 2C). These differences were also observed in the SMMC-7721 cell line (Figures S1D, S1E). HIF-2α also significantly slowed cell growth rate under hypoxic condition no matter with or without HIF-1α RNAi (Figure S1F, S1G). Next, we determined whether the altered growth rate is due to an increased cell death rate. Annexin V/PI assays showed a dramatic apoptosis rate increase in the high-expression HIF-2α clone compared to respective control (Figure 2D). Confocal fluorescence microscopy revealed mitochondrial dysfunction and more cytochrome C released from HIF-2α-overexpressing cells (Figure 2E). Multidomain pro-apoptosis BCL-2 proteins Bax and Bak were expressed at higher levels in cells with high HIF-2α expression relative to controls (Figure 2F), which would facilitate the release of cytochrome C from mitochondria by forming pores in the outer mitochondrial membrane.20, 21 We also observed a higher amount of cleaved caspase 3, a key executer of apoptosis, in over-expressing HIF-2α cells (Figure 2F), which would have been triggered by the cytochrome C release. The BH3-only pro-apoptotic proteins, including the Bcl-2-associated death promoter (Bad), p53 up-regulated modulator of apoptosis (Puma), and Bid were also found to be up-regulated in HIF-2α over-expression cells (Figure 2F). No significant differences were found in Ki67 staining between groups with different HIF-2α levels (Figure S1H). Collectively, these data further confirmed that high levels of HIF-2α in HCC cells caused a cell growth arrest through apoptosis.
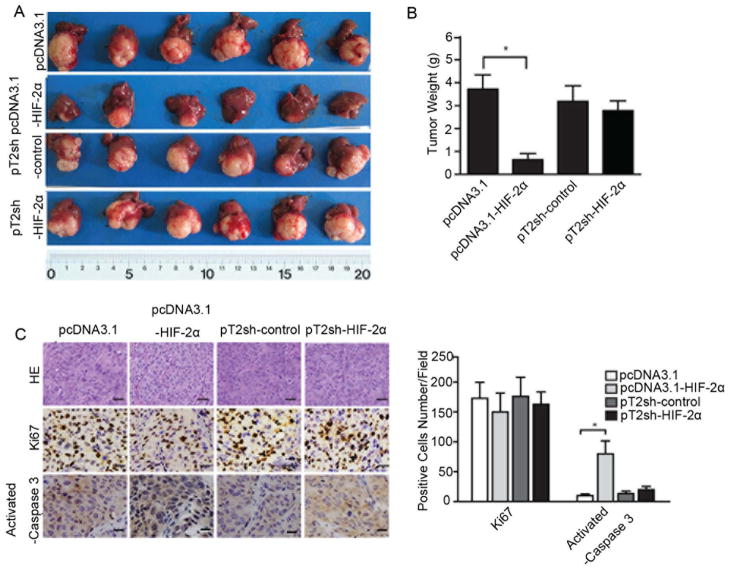
Having established that high expression of HIF-2α in human HCC increases OS rates and induces HCC cell growth arrest in vitro, we further tested the effect of HIF-2α in vivo. Clones with different HIF-2α expression were established (Figure S2A). The tumors formed by cells with high HIF-2α level were smaller than mock controls, and tumors formed by cells with low HIF-2α level showed no significant changes in tumor weight compared to respective controls (Figures 3A, 3B). Cell proliferation and apoptosis measurements indicated that over-expression of HIF-2α induced more cleaved caspase 3 expression in tumor cells compared to mock-treated cells (Figure 3C). These in vivo data support HIF-2α’s role of inducing apoptosis in HCC cells.
Figure 3.
HIF-2α-induced HCC growth arrest and high apoptosis rate in vivo. Monoclonal cells with either high or low HIF-2α expression were implanted into the livers of nude mice (4 weeks old). Tumor tissues were harvested 6 weeks after implantation. (A) Livers bearing tumors formed by implanted cells with different expression levels of HIF-2α. (B) Weights of the tumors in livers of nude mice. Error bars indicate standard deviation (n=6). *, p<0.01, two-tailed test. (C) Representative images of immunohistochemical staining of KI67 and activated caspase 3 in tumor xenografts, with counterstaining by hematoxylin and eosin. Bar = 2 mm.
HIF-2α Augments Apoptosis by Inhibiting Expression of the Target Gene TFDP3
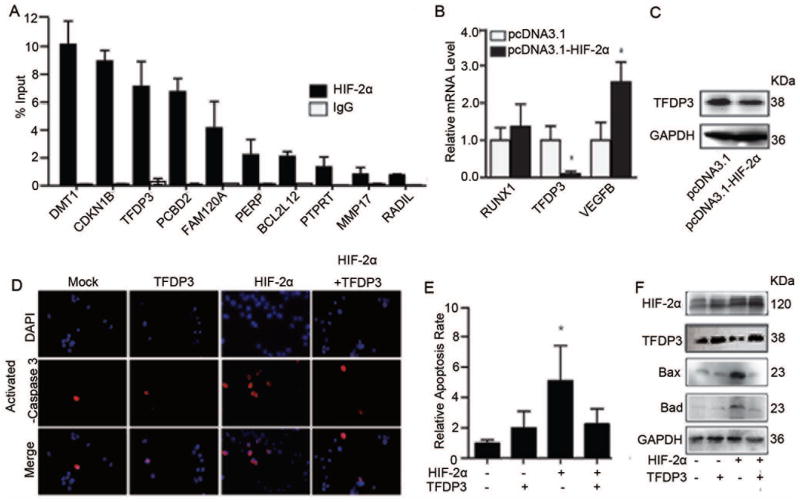
To uncover how HIF-2α affects on apoptosis, we sought to identify possible targets using a ChIP-on-chip screen in MHCC97H cells (GEO accession number GSE37167). With a specific anti-HIF-2α antibody, we identified 470 target genes bound within their promoter regions, spanning 2.2 kb upstream and 500 bp downstream of the transcription start site, by HIF-2α. To validate screening results, we performed an independent ChIP experiment coupled with qRT-PCR (Figure 4A).
Figure 4.
HIF-2α augments cell apoptosis by inhibiting the target gene TFDP3 expression. (A) HIF-2α target genes identified by ChIP-on-chip were further validation by ChIP-PCR. The fold change of HIF-2α content in immunoprecipitates over the input are presented; error bars indicate standard deviation (SD). (B) Relative expression of apoptosis genes in representative monoclonal cells that were stably transfected with empty vector or recombinant HIF-2α expression plasmid (n=3). *, p<0.05, two-tailed test. (C) TFDP3 and GAPDH protein expression in representative monoclonal stable cell lines transfected with empty vector (pcDNA3.1) or recombinant HIF-2α expression plasmid (pcDNA3.1-HIF-2α). (D) Transient expression of HIF-2α by transfection of pcDNA3.1-HIF-2α or TFDP3 (pcDNA3.1-TFDP3) versus control empty vector (pcDNA3.1) in MHCC97H cells. Anti-activated caspase-3 antibody was used to test the rate of apoptosis in each group. (E) Quantification of the relative percentage of activated caspase 3-positive cells relative to varying expression levels of HIF-2α and TFDP3. *, p<0.05. (F) The expression levels of Bax and Bad were determined, and HIF-2α and TFDP3 levels were also confirmed in cells that were transiently transfected with HIF-2α or TFDP3 expression plasmids.
The biological function and cellular component of HIF-2α binding target genes were analyzed according to their ChIP-on-chip assay enrichment score. Genes with the highest enrichment scores were those that regulate cellular processes and are located in the nucleus (Figures S3A, 3B). A substantial number of genes regulating apoptotic processes were found to be HIF-2α-bound genes. The top 10 genes were ranked according to their PeakFDR values and PeakScore values (Table S5). We observed a particularly strong suppression of TFDP3 mRNA (Figure 4B) and protein levels in HIF-2α-overexpressing cells compared to controls (Figure 4C), and these effects were not seen when HIF-1α levels were varied (Figures S3C, S3D).
The TFDP3 gene encodes a member of the dimerization partner (DP) family of transcription factors, 22, 23, 24 which exerts a regulatory function by dimerization with the E2F protein. The TFDP3 protein is known to be highly expressed in HCC but not in normal liver tissues23. We hypothesized that HIF-2α repression of TFDP3 expression could account for the high rate of apoptosis seen in the HIF-2α-overexpressing cell line. To test this idea, we overexpressed TFDP3 or/and HIF-2α in MHCC97H cells (Figure S3E). The number of activated caspase 3-positive cells was decreased when TFDP3 expression was re-established in HIF-2α-overexpressing cells (Figures 4D, 4E). The expression of Bax and Bad was also downregulated in these cells (Figure 4F). TFDP1 and TFDP2 expression levels were tested to exclude the possibility that effects on E2F transcriptional activity (Figure S3F). Collectively, these data show that HIF-2α significantly increases apoptosis by repressing the transcription of TFDP3, and the re-introduction of TFDP3 expression significantly reverses the effect.
HIF-2α Induces Apoptosis through the TFDP3/E2F1 Pathway
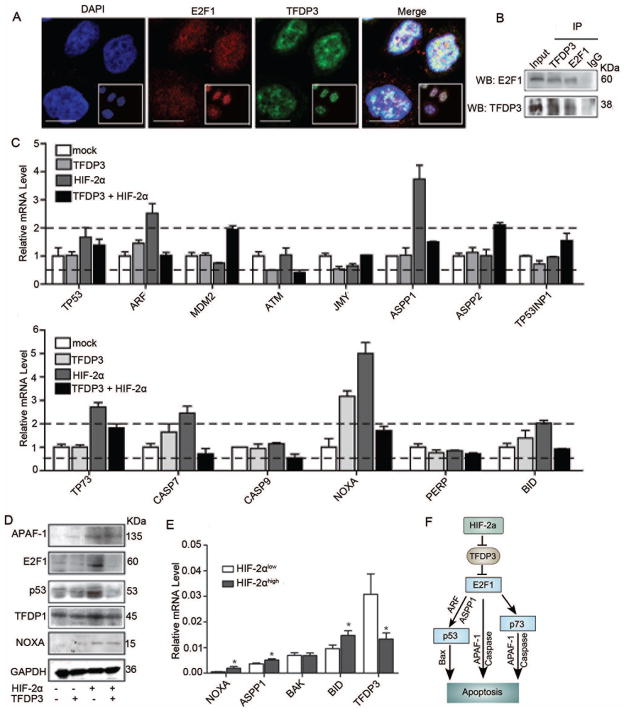
It is well known that DP proteins bind to E2F1, forming heterodimers that are essential for E2F high-affinity DNA binding and efficient transcriptional activity.24, 25 The TFDP3 gene was shown to co-localize with E2F1 by confocal microscopy (Figure 5A). The physical interaction between E2F1 and TFDP3 was further analyzed by co-immunoprecipitation (co-IP). In an E2F1 pull-down assay, TFDP3 bound directly to E2F1. Reciprocally, in an IP assay using anti-TFDP3 antibody, E2F1 was found to be bound (Figure 5B). Knockdown of E2F1 expression by siRNA in HIF-2α-overexpressing cells increased their growth rate and decreased some pro-apoptotic gene expression (Figure S3G, S3H, and S3I). All these data suggest that E2F1 plays an important role in the induction of apoptosis by HIF-2α.
Figure 5.
HIF-2α-TFDP3 controls apoptosis through multiple E2F1 pathways. (A) MHCC97H cells were seeded on coverslips. Representative images of E2F1 and TFDP3 nuclear co-localization were obtained by confocal microscopy. Bar, 10μm. (B) MHCC97H cells were subjected to immunoprecipitation (IP) with anti-TFDP3 or anti-IgG antibody, followed by western blot assay with anti-E2F1 antibody (upper panels). Reciprocal IP was done with anti-E2F1 or anti-IgG antibody, followed by western blot assay with anti-TFDP3 antibody (lower panels). (C) The changes in mRNA expression level of genes involved in the p53-dependent and -independent pathways in cells that were transiently transfected with HIF-2α or TFDP3 expression plasmids. (D) Western blot analysis of proteins involved in the E2F1 pathway in cells with different HIF-2α and TFDP3 expression levels. (E) The expression levels of TFDP3 and select apoptotic genes in sixty human HCC specimens with different HIF-2α levels identified by immunohistochemical staining. (F) Proposed pathway of HIF-2α-induced apoptosis.
E2F1 has been reported to induce apoptosis through both p53-dependent and -independent pathways.26 Therefore, expression levels of key genes in these two pathways were determined. Cells were transiently transfected with both HIF-2α and TFDP3 recombinant expression plasmids, either in combination or individually. Although results showed that mRNA level of TP53 did not change significantly (Figure 5C), its protein level greatly increased in cells transfected with HIF-2α, and it decreased to normal level when cells expressed high HIF-2α and TFDP3 levels (Figure 5D). The p14ARF gene has been reported to be regulated by E2F1 and interacts with Mdm2, thus preventing p53 ubiquitination and subsequent degradation.27 Our results showed that p14ARF levels increased more than 2 fold in the clonal line with high HIF-2α expression and decreased nearly to normal level in cells that expressed both HIF-2α and TFDP3 (Figure 5C). No significant expression changes were found for Mdm2 and p53 kinase ataxia telangiectasia mutated (ATM), which induce p53 phosphorylation28 (Figure 5C). Expression of the apoptosis-stimulating protein of p53 (ASPP1), which directs p53 activity towards apoptosis,29 increased significantly in cells with low TFDP3 expression level, and it decreased to nearly normal level in cells that co-expressed HIF-2α and TFDP3 (Figure 5C). These results suggest that HIF-2α-TFDP3-E2F1 increases apoptosis by increasing p14ARF and ASPP1 transcription, which further increases the p53 protein level.
Expression of some genes in the p53-independent apoptotic E2F1 pathway, such as the p53 homolog p73,30 BID,25 and several caspase 32 were found to be significantly upregulated in cells with high HIF-2α levels, whereas the restoration of TFDP3 expression significantly reversed effects on expression levels of those genes (Figure 5C). Effects on apoptosis protease-activating factor 1 (APAF1),33 Puma, and the BH3-only protein encoding the gene Noxa,34 regulated by both E2F1 and p53, also showed a similar trend (Figures 5C, 5D). Overall, both p53-dependent and -independent pathways play a role in HIF-2α-induced apoptosis. We further measured expression levels of TFDP3 and some representative genes of the E2F1 pathway in HCC specimens, which were with different HIF-2α protein levels determined by IHC staining. We found that patients with high HIF-2α protein expression had low expression levels of TFDP3 but high levels of pro-apoptotic genes (Figure 5E). Collectively, data from in vitro models and HCC patient samples have implicated for the mechanism of HIF-2α-induced apoptosis (Figure 5F).
Discussion
HIFs are detected in most solid tumors and known to correlate with poor patient prognosis. The identification of genes, such as VEGF and CA9, in a HIF-activated pathway reveals a clear mechanism by which HIFs contribute to survival and progression of cancer cells, and clinical therapeutic targeting of HIFs has emerged as a rational approach to treat solid tumors.35 Recently, studies have shown that HIF isoforms differentially regulate gene expression and sometimes have opposing functions during tumor progression. Therefore experiments elaborating the precise roles of each subunit is needed.4, 10 HIF-1α has been widely explored and shown to regulate cell proliferation and angiogenesis and to be correlated with OS in HCC.2, 3, 36 However, studies on HIF-2α function in HCC are limited and these results sometimes are inconsistent.16, 1, 31 Therefore, we explored the clinical significance by measuring gene expression in tumor samples from HCC patients, and putative mechanism by altering cellular HIF-2α expression levels by overexpression or knockdown experiments.
We found that HIF-2α-induced HCC cell growth arrest and knockdown of HIF-2α expression increased cell viability in vitro. These effects have previously been observed in other cell line under hypoxic conditions.1 Our clinical data further confirmed the effect. Through analysis of samples from hundreds of HCC patients, we found that HIF-2α expression in HCC tissues positively correlated with patient OS. Moreover, there was a significant negative correlation between tumor size and relative HIF-2α level (p<0.001). Thus, our results suggest a possible tumor suppressor role of HIF-2α in HCC.
With ChIP-on-chip screening and subsequent confirmatory assays, we showed TFDP3 was a novel transcriptional target of HIF-2α, and its expression was significantly repressed by HIF-2α. Our findings link HIFs transcriptional activity to one of the least understood of the DP family, TFDP3. An important property that distinguishes TFDP3 from other DP members is its ability to inhibit E2F1-induced p53-mediated apoptosis.39 We found that the p53-independent pathway is also modulated by TFDP3. The TFDP3 protein is only expressed in HCC but not in normal liver tissues.23 Further exploring its role as a potential therapeutic target in HCC may uncover new applications.
The E2F family is best known for regulating cell cycle, proliferation, and apoptosis, and is involved in signaling cascades induced by hypoxia.32 The level of E2F1 is up-regulated under hypoxic stimulation, and HIF-2α has been shown to increase E2F1 levels via c-Myc.11 Here, we showed that HIF-2α promotes E2F1 transcriptional activity and expression levels of E2F1-dependent target genes via inhibition of TFDP3 expression (Figure 5F). This property of TFDP3 provides another connection between HIFs and E2F1.
HIF-2α exhibits a mixed nuclear/cytoplasmic localization pattern in HCC samples, and both the percentage and intensity vary greatly between patients.16, 41 HIFs expression is known to be affected by many factors. For example, viral hepatitis infection and liver cirrhosis cause inflammatory cell infiltration and a hypoxic microenvironment, further modulating HIF expression patterns.37, 38 Therefore the rate of viral hepatitis infection and the presence of liver cirrhosis need to be carefully considered when evaluating a study cohort’s HIF expression profile. There were 82.9% HBV-positive and 81.3% cirrhosis-positive patients involved in our study; while these rates were only 67.3% and 45.1%, respectively in another HIF-2α study.16 In addition, there are other epidemiologic features such as geographic regions, racial and ethnic groups, gender, and study methods such as cell type, cultural method, staining method used may also influence HIF-2α expression profile.
The role of HIF-2α in the development of cancer is not clear, and there is some discrepancy in the existing literature. One study showed that HIF-2α was correlated with angiogenesis and a poor outcome in HCC,16 while we and another group found that HIF-2α regulated autophagy and apoptosis, and high HIF-2α expression in HCC correlated with a good outcome.1 Diverse/complex roles of HIF-2α have also been found in non-small cell lung cancer (NSCLC). HIF-2α appears to promote tumor EMT, and it is defined as a promoter of NSCLC.40 On the other hand, deletion of HIF-2α results in an increased NSCLC tumor burden via regulation of tumor suppressor gene secretoglobin 3A1a expression.5
Reconciling these diverse roles is difficult. One possibility is that tumor microenvironment markedly influences the active role of HIFs,1 as has been shown in astrocytoma. Loss of HIF-1α impairs astrocytoma growth subcutaneously, but it increases its proliferative and invasive properties in the brain.14 In HCC, hepatitis virus infection and liver cirrhosis maybe are important factors, which significantly affect tumor microenvironment.37, 38 Another possibility is a tumor may employ a “stop and go” strategy to maintain its growth and survival, as suggested by Acker.15 When oxygen levels are limited, HIF-2α acts negatively on survival by inducing apoptosis, but at the same time, it promotes angiogenesis. Clearly, the mechanisms regulating HIF-2α function need to be further explored.
In conclusion, we have established TFDP3 is a novel transcriptional target of HIF-2α and uncovered another mechanism that links HIFs to a fundamental biologic regulator, E2F1. We have shown that HIF-2α induces HCC cell apoptosis by its effect on the TFDP3/E2F1 pathway, and provided clinical evidences demonstrating a possible tumor suppressor role of HIF-2α.
Supplementary Material
Acknowledgments
Financial Support:
This study was supported through grant support from the National Institute of Health (R01DK080736 and R01DK081417), Michael Rolfe Foundation for Pancreatic Cancer Research, the Major Program of the National Natural Science Foundation of China (No. 81030038), National Key Sci-Tech Project (2012ZX10002011-002, and 2012ZX10002013-005), National Natural Science Foundation of China (No. 81071661, 81071661 and 81000927), Shanghai New Project for Excellent Youth (No.XYQ2011020), Zhongshan Foundation for Youth (No. 2012ZSQN-06) and Research Fund for the Doctoral Program of Higher Education of China (No. 20100071120064).
We thank Dr. Yu Zhang of Peking University for the gift of TFDP3 recombinant plasmid.
Abbreviations
- HCC
hepatocellular carcinoma
- HIFs
Hypoxia inducible factors
- VEGF
vascular endothelial growth factor
- ChIP-on-chip
Chromatin immunoprecipitation coupled with microarray
- TFDP3
transcription factor dimerization partner 3
- TMAs
tissue microarrays
- TNM
tumor-node-metastasis
- BCLC
Barcelona Clinic Liver Cancer
- OS
overall survival
- ALT
alanine aminotransferase
- GGT
gamma-glutamyl transpeptidase
- AFP
alpha-fetoprotein
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HBsAg
hepatitis B surface antigen
- PBS
phosphate-buffered saline
- qRT-PCR
quantitative real-time polymerase chain reaction
- shRNA
small hairpin RNA
Contributor Information
Hai-Xiang Sun, Email: haixiangsun27@hotmail.com.
Yang Xu, Email: drxuyang@gmail.com.
Xin-Rong Yang, Email: yxrdoc@gmail.com.
Wei-Min Wang, Email: wangweimin07@gmail.com.
Haibo Bai, Email: hbai1@jhmi.edu.
Ruo-Yu Shi, Email: shiruoyu2012@163.com.
Suresh K. Nayar, Email: sknayar@yahoo.com.
Ranjan Prasad Devbhandari, Email: ranjandev@hotmail.com.
Yi-zhou He, Email: he_yizhou@163.com.
Qin-Feng Zhu, Email: schxykd@163.com.
Yun-Fan Sun, Email: yunfansun85@gmail.com.
Bo Hu, Email: bohu1120@hotmail.com.
Mehtab Khan, Email: mkhan27@jhmi.edu.
Robert A. Anders, Email: rander54@jhmi.edu.
Jia Fan, Email: jiafan99@yahoo.com.
References
- 1.Menrad H, Werno C, Schmid T, Copanaki E, Deller T, Dehne N, Brune B. Roles of hypoxia-inducible factor-1alpha (HIF-1alpha) versus HIF-2alpha in the survival of hepatocellular tumor spheroids. Hepatology. 2010;51:2183–2192. doi: 10.1002/hep.23597. [DOI] [PubMed] [Google Scholar]
- 2.Dai CX, Gao Q, Qiu SJ, Ju MJ, Cai MY, Xu YF, Zhou J, et al. Hypoxia-inducible factor-1 alpha, in association with inflammation, angiogenesis and MYC, is a critical prognostic factor in patients with HCC after surgery. BMC Cancer. 2009;9:418. doi: 10.1186/1471-2407-9-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie H, Song J, Liu K, Ji H, Shen H, Hu S, Yang G, et al. The expression of hypoxia-inducible factor-1alpha in hepatitis B virus-related hepatocellular carcinoma: correlation with patients’ prognosis and hepatitis B virus X protein. Dig Dis Sci. 2008;53:3225–3233. doi: 10.1007/s10620-008-0296-9. [DOI] [PubMed] [Google Scholar]
- 4.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazumdar J, Hickey MM, Pant DK, Durham AC, Sweet-Cordero A, Vachani A, Jacks T, et al. HIF-2alpha deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci U S A. 2010;107:14182–14187. doi: 10.1073/pnas.1001296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 2001;13:167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 7.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–685. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skuli N, Simon MC. HIF-1alpha versus HIF-2alpha in endothelial cells and vascular functions: is there a master in angiogenesis regulation? Cell Cycle. 2009;8:3252–3253. doi: 10.4161/cc.8.20.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skuli N, Liu L, Runge A, Wang T, Yuan L, Patel S, Iruela-Arispe L, et al. Endothelial deletion of hypoxia-inducible factor-2alpha (HIF-2alpha) alters vascular function and tumor angiogenesis. Blood. 2009;114:469–477. doi: 10.1182/blood-2008-12-193581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci U S A. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giatromanolaki A, Sivridis E, Kouskoukis C, Gatter KC, Harris AL, Koukourakis MI. Hypoxia-inducible factors 1alpha and 2alpha are related to vascular endothelial growth factor expression and a poorer prognosis in nodular malignant melanomas of the skin. Melanoma Res. 2003;13:493–501. doi: 10.1097/00008390-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 13.Giatromanolaki A, Koukourakis MI, Sivridis E, Turley H, Talks K, Pezzella F, Gatter KC, et al. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer. 2001;85:881–890. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blouw B, Song H, Tihan T, Bosze J, Ferrara N, Gerber HP, Johnson RS, et al. The hypoxic response of tumors is dependent on their microenvironment. Cancer Cell. 2003;4:133–146. doi: 10.1016/s1535-6108(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 15.Acker T, Diez-Juan A, Aragones J, Tjwa M, Brusselmans K, Moons L, Fukumura D, et al. Genetic evidence for a tumor suppressor role of HIF-2alpha. Cancer Cell. 2005;8:131–141. doi: 10.1016/j.ccr.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Bangoura G, Liu ZS, Qian Q, Jiang CQ, Yang GF, Jing S. Prognostic significance of HIF-2alpha/EPAS1 expression in hepatocellular carcinoma. World J Gastroenterol. 2007;13:3176–3182. doi: 10.3748/wjg.v13.i23.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY, Xiao YS, Xu Y, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25:2586–2593. doi: 10.1200/JCO.2006.09.4565. [DOI] [PubMed] [Google Scholar]
- 18.Chan N, Pires IM, Bencokova Z, Coackley C, Luoto KR, Bhogal N, Lakshman M, et al. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res. 2010;70:8045–8054. doi: 10.1158/0008-5472.CAN-10-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ke AW, Shi GM, Zhou J, Huang XY, Shi YH, Ding ZB, Wang XY, et al. CD151 amplifies signaling by integrin alpha6beta1 to PI3K and induces the epithelial-mesenchymal transition in HCC cells. Gastroenterology. 2011;140:1629–1641. e1615. doi: 10.1053/j.gastro.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Mignotte B, Vayssiere JL. Mitochondria and apoptosis. Eur J Biochem. 1998;252:1–15. doi: 10.1046/j.1432-1327.1998.2520001.x. [DOI] [PubMed] [Google Scholar]
- 21.Jurgensmeier JM, Xie Z, Deveraux Q, Ellerby L, Bredesen D, Reed JC. Bax directly induces release of cytochrome c from isolated mitochondria. Proc Natl Acad Sci U S A. 1998;95:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao H, Di Stefano L, Tian C, Li YY, Yin YH, Qian XP, Pang XW, et al. Human TFDP3, a novel DP protein, inhibits DNA binding and transactivation by E2F. J Biol Chem. 2007;282:454–466. doi: 10.1074/jbc.M606169200. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Han KJ, Pang XW, Vaughan HA, Qu W, Dong XY, Peng JR, et al. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. J Immunol. 2002;169:1102–1109. doi: 10.4049/jimmunol.169.2.1102. [DOI] [PubMed] [Google Scholar]
- 24.Milton A, Luoto K, Ingram L, Munro S, Thangue NB. A functionally distinct member of the DP family of E2F subunits. Oncogene. 2006;25:3212–3218. doi: 10.1038/sj.onc.1209343. [DOI] [PubMed] [Google Scholar]
- 25.Hitchens MR, Robbins PD. The role of the transcription factor DP in apoptosis. Apoptosis. 2003;8:461–468. doi: 10.1023/a:1025586207239. [DOI] [PubMed] [Google Scholar]
- 26.Putzer BM. E2F1 death pathways as targets for cancer therapy. J Cell Mol Med. 2007;11:239–251. doi: 10.1111/j.1582-4934.2007.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 28.Powers JT, Hong S, Mayhew CN, Rogers PM, Knudsen ES, Johnson DG. E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis. Mol Cancer Res. 2004;2:203–214. [PubMed] [Google Scholar]
- 29.Fogal V, Kartasheva NN, Trigiante G, Llanos S, Yap D, Vousden KH, Lu X. ASPP1 and ASPP2 are new transcriptional targets of E2F. Cell Death Differ. 2005;12:369–376. doi: 10.1038/sj.cdd.4401562. [DOI] [PubMed] [Google Scholar]
- 30.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 31.He C, Sun XP, Qiao H, Jiang X, Wang D, Jin X, Dong X, et al. Downregulating hypoxia-inducible factor-2α improves the efficacy of doxorubicin in the treatment of hepatocellular carcinoma. Cancer Sci. 2012;103:528–534. doi: 10.1111/j.1349-7006.2011.02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, Zhang MQ, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 33.Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, Muller H, et al. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 34.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 35.Patel SA, Simon MC. Biology of hypoxia-inducible factor-2alpha in development and disease. Cell Death Differ. 2008;15:628–634. doi: 10.1038/cdd.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, Wang P, Jiang X, Tan G, Qiao H, Jiang H, Krissansen GW, et al. Antisense hypoxia-inducible factor 1alpha gene therapy enhances the therapeutic efficacy of doxorubicin to combat hepatocellular carcinoma. Cancer Sci. 2008;99:2055–2061. doi: 10.1111/j.1349-7006.2008.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonardi GC, Candido S, Cervello M, Nicolosi D, Raiti F, Travali S, Spandidos DA, et al. The tumor microenvironment in hepatocellular carcinoma (review) Int J Oncol. 2012;40:1733–1747. doi: 10.3892/ijo.2012.1408. [DOI] [PubMed] [Google Scholar]
- 38.Kim KR, Moon HE, Kim KW. Hypoxia-induced angiogenesis in human hepatocellular carcinoma. J Mol Med (Berl) 2002;80:703–714. doi: 10.1007/s00109-002-0380-0. [DOI] [PubMed] [Google Scholar]
- 39.Tian C, Lv D, Qiao H, Zhang J, Yin YH, Qian XP, Wang YP, et al. TFDP3 inhibits E2F1-induced, p53-mediated apoptosis. Biochem Biophys Res Commun. 2007;361:20–25. doi: 10.1016/j.bbrc.2007.06.128. [DOI] [PubMed] [Google Scholar]
- 40.Kim WY, Perera S, Zhou B, Carretero J, Yeh JJ, Heathcote SA, Jackson AL, et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009;119:2160–2170. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talks KL, Turley H, Gatter KC, et al. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–21. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.