Abstract
As important vectors of human disease, phlebotomine sand flies are of global significance to human health, transmitting several emerging and re-emerging infectious diseases. The most devastating of the sand fly transmitted infections are the leishmaniases, causing significant mortality and morbidity in both the Old and New World. Here we present the first global transcriptome analysis of the Old World vector of cutaneous leishmaniasis, Phlebotomus papatasi (Scopoli) and compare this transcriptome to that of the New World vector of visceral leishmaniasis, Lutzomyia longipalpis. A normalized cDNA library was constructed using pooled mRNA from Phlebotomus papatasi larvae, pupae, adult males and females sugar fed, adult females blood fed and fed blood infected with Leishmania major. A total of 47,615 generated sequences were cleaned and assembled into 17,120 unique transcripts. Of the assembled sequences, 50% (8,837 sequences) were classified using Gene Ontology (GO) terms. This collection of transcripts is comprehensive, as demonstrated by the high number of different GO categories. An in depth analysis has revealed 245 sequences with putative homology to proteins involved in blood and sugar digestion, immune response and peritrophic matrix formation. Twelve of the novel genes, including one trypsin, two peptidoglycan recognition proteins (PGRP) and nine chymotrypsins have a higher expression level during larval stages. Two novel chymotrypsins and one novel PGRP are abundantly expressed upon blood feeding. This study will greatly improve the available genomic resources for Ph. papatasi and will provide essential information for annotation of the full genome.
Introduction
Phlebotomine sand flies are important vectors of human disease in both the Old and the New World, transmitting protozoan, bacterial and viral pathogens. These flies are members of the family Psychodidae, which includes a diverse group of vectors that vary widely in geographic distribution, ecology, and the pathogens they transmit. Sand flies serve as vectors for several established, emerging and re-emerging infectious diseases, the most devastating of which are the leishmaniases with 350 million people at risk and approximately two million new cases each year (Cunningham 2002, Desjeux 1996). Human migration, political instability, and warfare is expanding Leishmania-endemic regions and increasing the propensity for epidemics world-wide (Desjeux 2001). In spite of the medical importance of leishmaniasis it is classified as a neglected tropical disease and phlebotomine sand fly species remain understudied.
Approximately 40 different species of Leishmania are transmitted by 35 different sand fly species (Ramalho-Ortigao, Saraiva & Traub-Cseko 2010). Most vectors belong to one of two genera, Phlebotomus and Lutzomyia (Richard P. Lane, Roger W. Crosskey 1993). Phlebotomus species are responsible for transmitting leishmaniasis throughout parts of Africa, southwest Asia, the Middle East, and the Mediterranean basin; Lutzomyia species are vectors throughout the Americas. There is a close ecological relationship between Leishmania species and the vector(s) that transmit that species. For example, Phlebotomus papatasi only transmits Leishmania major whereas Lutzomyia longipalpis transmits Le. infantum chagasi (Killick-Kendrick 1999). Although most vectors are specific under natural conditions, some, such as Lu. longipalpis, can transmit a range of Leishmania species under laboratory conditions.
Genomics approaches enable comprehensive comparisons between diverse organisms, thus facilitating a more complete understanding of their biology. The completed genomes of the more widely studied hematophagous vectors, the malaria vector Anopheles gambiae, the yellow fever mosquito Aedes aegypti, and the West Nile virus vector Culex quinqefasciatus, have already contributed valuable information relative to vectorial capacity, blood-feeding, insect immune system modulation and insecticide resistance (Holt et al. 2002, Christophides et al. 2002, Nene et al. 2007, Arensburger et al. 2010). Previous genomic studies concerning sand flies have focused on specific questions regarding vector-human interactions (analysis of protein expression in salivary glands (Hostomska et al. 2009)) or vector-parasite interactions (analysis of midgut expressed proteins (Ramalho-Ortigao et al. 2007)); only one study has performed a global gene discovery analysis of a sand fly, Lu. longipalpis (Dillon et al. 2006). Here, we expand these studies by characterizing the transcriptome of the Le. major vector, Ph. papatasi. We also similarly reanalyze the Lu. longipalpis Expressed Sequence Tag (EST) dataset (Dillon et al. 2006) for a comparative analysis.
Phlebotomine sand flies along with mosquitoes (family Culicidae) are members of the suborder Nematocera, but these two families are representatives of distinct infraorders within the Nematocera. Ph. papatasi and Lu. longipalpis exhibit distinct geographical distributions, ecology, and vector competence specificity. A comparative approach between these flies will accelerate the discovery of regulatory and biochemical pathways within this family as potential biopharmaceuticals, vaccine candidates, and targets for insecticide development. Moreover, comparative analyses between these and other available vector data sets will elucidate the pathways that lead to arthropod blood-feeding and immunity and inform arthropod phylogenetic relationships.
ESTs represent the expressed portion of mRNA in a cell obtained through single pass sequencing of randomly selected cDNA clones, resulting in about 200-800 bp of sequence information for each clone (Mark Blaxter et al. 2009). EST studies can be used for gene discovery in organisms where sequencing the whole genome is not possible (Lindlof 2003), or in addition to genome information for more accurate gene annotation. A phlebotomine sand fly genome sequencing project was initiated a few years ago to sequence Lu. longipalpis and Ph. papatasi (McDowell et al. 2006). The aim of this study was to increase the availability of EST resources for future sand fly studies, provide useful information regarding the biology of these important vectors, and generate essential data for annotation of the newly sequenced phlebotomine sand fly genomes (McDowell et al. 2006).
Results and Discussion
Assembly
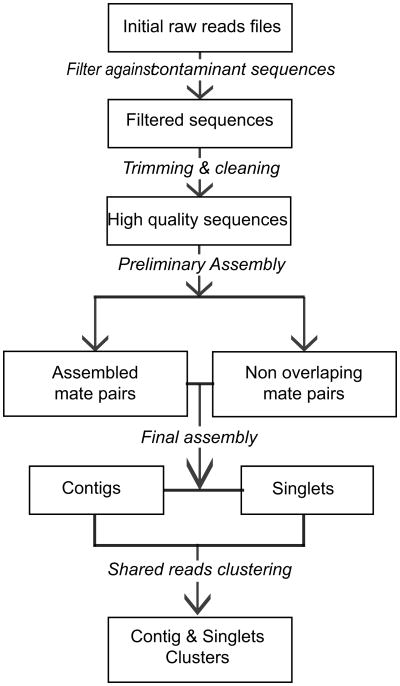
Mate pair information generated during the sequencing process was utilized in a two step assembly process. First, the sequence reads were assembled with their mates using a lower identity criteria based on the assumption that they are opposite ends of the same cDNA clone and thus should assemble. Resulting sequences were assembled using a more stringent identity parameter to avoid over collapsing closely related gene families into a single sequence. There were 47,615 sequences initially generated from the normalized Ph. papatasi library. Of all initial Ph. papatasi reads, 10,128 (21%) failed the screening and filtering steps (see Experimental procedures) (Figure 1). The remaining 37,487 sequences were then assembled into 6,187 contigs and 10,933 singlets (Table 1), representing a total of 11 Mb of the Ph. papatasi transcriptome, average assembled sequence length was 550 bp. Assembled sequences with a length greater than 200 bp were deposited in GenBank (JP539097-JP555361).
Figure 1. Assembly process flowchart.
Boxes indicate a state of the sequences in the pipeline while italic lettering indicates modifications applied to the sequences and arrow indicates the sense of the sequences movement down the pipeline
Table 1.
Assembly results for Phlebotomus papatasi and Lutzomyia longipalpis at all steps of the assembly process presented in Fig. 1.
| Assembly steps | Phlebotomus papatasi | Lutzomyia longipalpis | |
|---|---|---|---|
| Initial sequences | Total | 47,615 | 27,928 |
| Normalized | 47,615 | 26,495 | |
| Non-normalized | N/A | 1,433 | |
| Filtering | Passed | 47,227 | 27,863 |
| Failed | 388 | 65 | |
| Cleaning and trimming | Lucy | 37,708 | 24,102 |
| SeqClean | 37,487 | 24,019 | |
| First tier assembly | Contigs | 7,683 | 6,049 |
| Singlets | 22,121 | 11,101 | |
| Second tier assembly | Contigs | 6,187 | 5,063 |
| Singlets | 10,933 | 4,963 | |
|
| |||
| Total | 17,120 | 10,026 | |
| Sequences longer than 200 nucleotides | 16,265 | N/A | |
A total of 3,909 (14%) Lu. longipalpis sequences failed the screening and filtering steps. The remaining cleaned sequences (24,019) were assembled into 5,063 contigs and 4,963 singlets, average length of 1,041 bp, representing a total of 8 Mb of the Lu. longipalpis transcriptome (Table 1). The differences in assembly results between the current study and the previous assembly by Dillon et al. (Dillon et al. 2006) can be explained by utilization of a different assembly program (Cap3 vs. Phrap) as well as by the different assembly strategy used (Supplementary Figure 1) (Huang, Madan 1999).
For our analyses we defined a read as a sequence that was cleaned and trimmed before the first assembly step. A mate pair represented two reads sequenced from the same cDNA; these reads had the same name with a different ending denoting either the 5′ or 3′ end. A mated read contig referred to mate pairs that co-assembled. A contig was either a mated read or a set of sequences that were not mate pairs but assembled under the conditions indicated above. One contig represented one gene, unless otherwise specified. Singlets are sequences that failed to assemble with any other sequence in the set, including their own mate pairs.
Similarity to known proteins and GO annotation
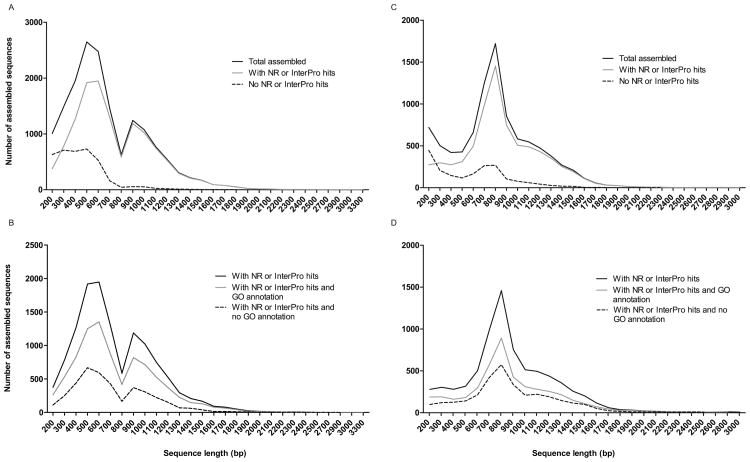
Of the 17,120 Ph. papatasi assembled sequences, 4,286 (25%) had no matches when searched against the NR and InterPro databases using a BLAST search (BLASTX) with an e-value threshold of 10-5, making them potentially unique Ph. papatasi sequences. The average length of assembled sequences with matches against either NR or InterPro database was between 500 and 699 bp long (Figure 2). Of the total assembled sequences, 8,837 (50%) of the Ph. papatasi and 4,411 (44%) of the Lu. longipalpis sequences could be associated with a GO term in at least one of the three main categories (biological process, molecular function or cellular component). The level of annotation is smaller than that for Anopheles mosquito (Anopheles albimanus, 65% (Martinez-Barnetche et al. 2012) and higher than non-model organisms with little or no closely related species in NCBI NR database (∼17- 50%) (Du et al. 2012, Hou et al. 2011, Wang et al. 2010, Shen et al. 2011). The limited annotation may be explained by the lack of sequences for closely related species available in NR and InterPro.
Figure 2. Assembled sequence length distribution.
Distribution of sequence length for sequences with NR or InterPro hits and for sequences with NR or InterPro hits and GO annotation for Ph. papatasi (A,B) and Lu. longipalpis (C,D)
The high number of different GO categories suggests that our cDNA library is representative of the entire organism. The distribution of GO categories was highly similar (Figure S2-4) between the two sand fly species, with Ph. papatasi having 35 (0.2%) sequences in four extra categories (viral reproduction, external encapsulating structure, symbiosis and neurotransmission categories). The smaller number of sequences available for Lu. longipalpis, combined with the small number of sequences annotated in the Ph. papatasi dataset, might account for their absence in the Lu. longipalpis dataset. Some of the differences were probably due to the original source of RNA prepared from the two species (the Lu. longipalpis RNA was extracted from adult females only, while for the Ph. papatasi RNA extraction, immature life stages and males were included) see Experimental procedures and Dillon et all. (Dillon et al. 2006). There were no GO categories present in Lu. longipalpis that were absent in the Ph. papatasi dataset.
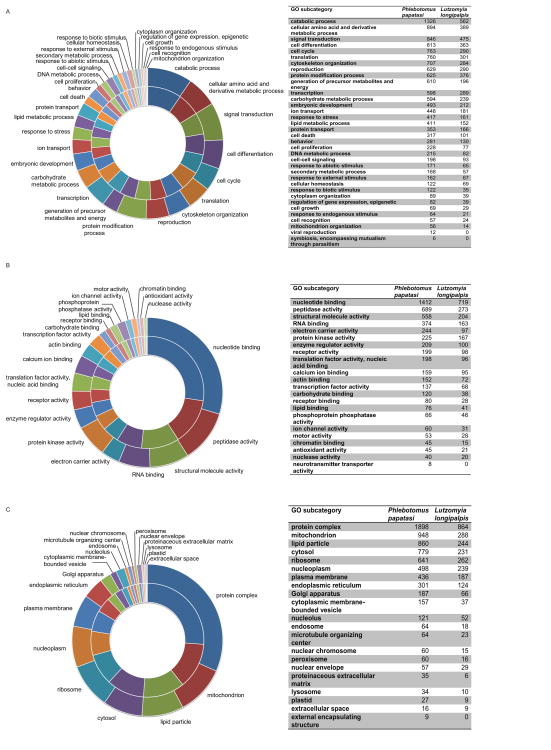
The highest number of sequences in the biological process category for both sand flies were annotated as catabolic process (1,326, 15% - Ph. papatasi; 562, 12.7% - Lu. longipalpis). In the Molecular Function category, the majority of Ph. papatasi and Lu. longipalpis sequences were annotated as nucleotide binding (1,412 or 16%, and 719 or 16.3% respectively). For the Cellular Component category, a high number of sequences were annotated as protein complex (1,898, 21.4%; 864, 19.6%) (Figure 3).
Figure 3. Gene Ontology terms distribution.
Distribution of Ph. papatasi (inner circle) and Lu. longipalpis (outer circle) sequences for the three main GO categories: Biological process (A), Molecular Function(B) and Cellular Component (C).
Digestive proteins
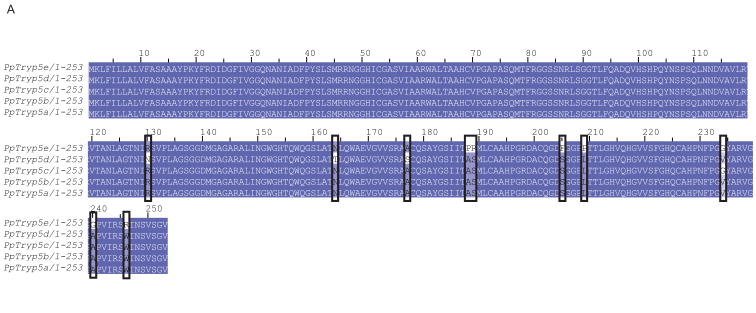
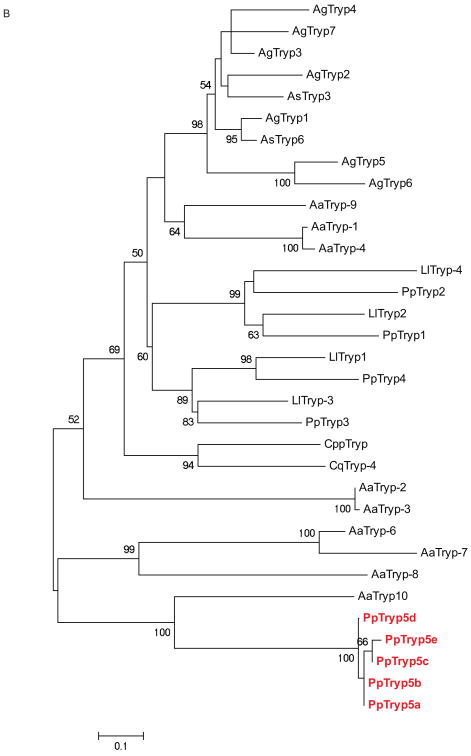
Trypsin and chymotrypsin-like serine endoproteases involved in blood digestion have been characterized in mosquitoes and other blood feeding arthropods, and their expression level was associated with the type and the time elapsed since a blood meal acquisition (Ramalho-Ortigao et al. 2007, Muller et al. 1993, Noriega, Wells 1999, Pitaluga et al. 2009, Telleria et al. 2010, Vizioli et al. 2001). Serine proteases are also involved in many other key processes in insects including complex cascades of proteases involved in immune signaling pathways (Buchon et al. 2009). Serine proteases, like trypsin, are considered important in the interaction of the sand fly host with Leishmania; trypsin activities being modulated in the gut of both Ph. papatasi and Lu. longipalpis. Knockdown of a ‘late’ trypsin in Lu longipalpis enhanced the survival of Le. mexicana (Sant'Anna et al. 2009). Previously, four trypsins and three chymotrypsins in Ph. papatasi (Ramalho-Ortigao et al. 2007, Ramalho-Ortigao et al. 2003) and two trypsins and five chymotrypsins in Lu. longipalpis (Telleria et al. 2010, Jochim et al. 2008) were identified. Here we identify five new trypsin-like sequences including PpTryp5a (JP544502) and PpTryp5b-e (JP542407, JP540627, JP554453, JP544448), that may represent alleles of PpTryp5a because they have an identity over 95% at the amino acid level (Figure 4A). PpTryp5a has a similarity of 32-39% at the amino acid level to known Ph. papatasi trypsins and a 50% identity to the closest related sequence in the NR database –the Ae. aegypti trypsin. The newly identified trypsin sequences (PpTrp5a-e) clustered closest to each other and to the Ae. aegypti trypsin rather than to any of the six previously identified sand fly trypsins (Figure 4B). This grouping may be explained by the difference in cDNA library sources, one possibility is that they are larval or pupal specific genes; previously the trypsin sequences were identified using a library constructed from the midgut of adult females only, while the library described here was constructed using whole sand fly bodies at different life stages that included both sexes. The Ae. aegypti trypsin sequence was identified from the entire genome sequence.
Figure 4. Ph. papatasi novel trypsin sequences.
(A) Novel Ph. papatasi multiple sequence alignment; conserved residues are represented by a darker shading while mismatches between the five sequences are indicated by boxes. Full sequence alignment is available in supplemental materials (S5). (B) Phylogenetic analysis of trypsin amino acid sequences from Ph. papatasi (Pp: PpTry5a-e (JP544502, JP542407, JP540627, JP554453, JP544448), AAM96940.1, AAM96941.1, AAM96942.1, AAM96943.1), Lu. longipalpis (Ll: ABM26904.1, ABM26905.1, ABV60308.1, ABV60300.1), An. gambiae (Ag: CAA80512.1, CAA79328.1, CAA80517.1, CAA80515.1, CAA80514.1, CAA80513.1, CAA80516.1), An. stephensi (As: AAB66878.1, AAA97479.1), Ae. aegypti (Aa: EAT40684.1, EAT42808.1, EAT36350.1, EAT34033.1, EAT42007.1, EAT42008.1, EAT42004.1, EAT37859.1), Cu. quinquefasciatus (Cq: EDS34988.1) and Cu. pipiens palens (Cpp: AAK67462.1). The WAG substitution model was used with variable positions and a bootstrap value of 1000 (only those above 50 are represented on the trees). The scale represents the rate of amino acid substitution per site. The novel Ph. papatasi trypsin sequences (PpTryp5a-e) are indicated in bold.
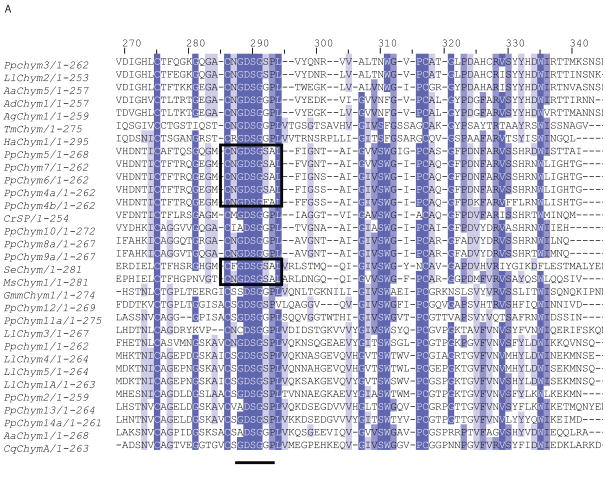
Seventeen novel chymotrypsin-like sequences with an identity of 29-69% to known Ph. papatasi chymotrypsin sequences were identified in the current study. Five putative chymotrypsin sequences PpChym4a,b,5-7 (JP546634, JP554565, JP551370, JP547341, JP554731) contained all the domains identified in other chymotrypsins (Ramalho-Ortigao et al. 2007, Ramalho-Ortigao et al. 2003, Appel 1986, Park, Kwak 2008) except for two amino acid differences in an otherwise highly conserved region. The first substitution, an S to F, is present only in PpChym4b (position 292) while the second substitution, a P to A, was present in all five sequences, PpChym4a,b-7 (position 293) (Figure 5A) . Four sequences representing two putative different chymotrypsins (PpChym14a and PpChym13; PpChym14b and PpChym14c are likely alleles of PpChym14a) cluster close to the previously identified PpChym2 (Ramalho-Ortigao et al. 2003), to which they share a 66, 67 and 69% identity at amino acid level. PpChym11 (including the 2 likely alleles: Ppchym11a and PpChym11b) and PpChym12 cluster closer to midgut chymotrypsin from Glossina morsitans morstans (Alves Silva et al. 2010). The last five of the novel putative chymotrypsin sequences (four of which are likely alleles of PpChym8 and PpChym9) cluster closer to chymotrypsin-like serine protease from Chironomus riparius expressed in the larvae gut (Park, Kwak 2008). The closer clustering of novel Ph. papatasi chymotrypsin sequences to sequences from different species could be explained by a possible difference in expression location (tissue) and/or life stage between the known and the novel chymotrypsin sequences; the known chymotrypsin sequences having been described in adult sand flies (Ramalho-Ortigao et al. 2007, Ramalho-Ortigao et al. 2003) (Figure 5B).
Figure 5. Ph. papatasi novel chymotrypsin sequences.
(A) Multiple sequence alignment fragment depicting the chymotrypsin binding pocket, where the pockets with the P-to-A substitution are boxed; the residue concordance at each position in indicated by the different degree of shading at that position. Full sequence alignment is available in supplemental materials (S6). (B) Phylogenetic analysis of known and novel chymotrypsins from Ph. papatasi (Pp: PpChym4-14 (JP546634, JP554565, JP551370, JP547341, JP554731, JP549601, JP540150, JP554746, JP554644, JP540516, JP543908, JP549141, JP547864, JP543702, JP542007, JP548909, JP549999), AAM96938.1, AAM96939.1, ABV44728.1), Lu. longipalpis (Ll: ABV60294.1, ABV60293.1, ABV60309.1, ABV60291.1, ABV60292.1, ABV60301.1), An. gambiae (Ag: CAA83568, CAA83567), An. darlingi (Ad: ADD17493.1, ADD17494.1), Ae. aegypti (Aa: XP_001663061.1, EAT32679.1, EAT38422.1, AAL93243.1), Cu. quinquefasciatus (Cq: XP_001846630.1, XP_001865429.1, XP_0011863473.1), Ch. riparius (Cr: ACF19792.1), Gl. morisitans morisitans (Gmm:ADD18377.1), He. armigera (Ha: ADI32883.1, ADI32881.1), Ma. sexta (Ms: CAL92020.1, CAM84317.1, CAM84318.1, CAM84319.1), Sp. exigua (Se: AAX35812.1) and Te. molitor (Tm: ABC88746.1). The WAG substitution model was used with variable and invariable positions and a bootstrap value of 1000 (only those above 50 are represented on the trees). The scale represents the rate of amino acid substitution per site. The novel Ph. papatasi chymotrypsin sequences are in bold.
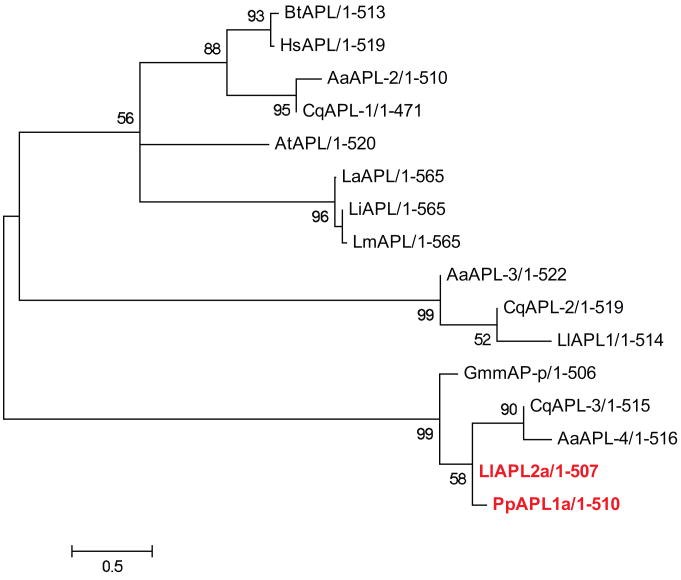
Aminopeptidases catalyze the removal of amino acids from the N-terminus of peptides and proteins. Their expression, along with the expression of exopeptidases, was shown to be modified by the ingestion of infected blood meal in sand flies (Muller et al. 1993, Dillon, Lane 1993). Our analysis identified 45 Ph. papatasi sequences with high identity to different aminopeptidases, including three aminopeptidase P sequences. Of these three sequences, only one (PpAPP1- JP547392) had all the necessary conserved domains (Kulkarni, Deobagkar 2002), with a 73% amino acid identity to Ae. aegypti proline specific aminopeptidase (e-value 10-127). The other two sequences have a high identity to PpAPP1 (JP539747, JP552630) (over 98% identity spanning 100-144 amino acids) but lack one or more conserved domains. There were no aminopeptidase P sequences identified in Lu. longipalpis. This difference between the two species is more likely due to the nature of the datasets rather than the loss of the gene from Lu. longipalpis. We also have identified four sequences (JP41850, JP543532, JP547607, JP546724) with similarity to leucyl aminopeptidase, two of which are unassembled mate reads and all four of them have a higher than 97% identity at the amino acid level making them potentially alleles of the same sequence, PpAPL1 (JP541859). Four sequences with similarity to leucyl aminopeptidase were identified in Lu. longipalpis, three of which are probably allelic variants of LlAPL1a which has 32% identity at the amino acid level to the leucyl aminopeptidase identified by Dillon et al. (Dillon et al. 2006) (Figure 6). It was not possible to confirm aminopeptidase identity (using conserved domain identification) in the remaining 39 Ph. papatasi sequences with homology to known aminopeptidases due to the short sequence length (76-585 AA). These Ph. papatasi sequences are likely to represent aminopeptidase transcript fragments rather than whole sequences (Supplemental Table2).
Figure 6. Phylogenetic analysis of Ph. papatasi and Lu. longipalpis aminopeptidase L.
Philogenetic tree constructed using aminopeptidase L sequences from Ph. papatasi (Pp: PpAPL1a (JP541859)), Lu. longipalpis (Ll: LlAPL1, LlAPL2a), Ar. thaliana (At: CAA45040.1), Bo. taurus (Bt: AAB28170.1), Gl. morisitans morisitans (Gmm: ADD18517.1), Ae. aeygpti (Aa: EAT48532.1, EAT47208.1, EAT45789.1), Cu. quinquefasciatus (Cq: XP_001866727.1, XP_001844372.1, XP_001851548.1), Le. amazonensis (La: AAL16095.1), Le. infantum (Li: CAM68214.1), Le. major (Lm: XP_001683430.1), Homo sapiens (Hs: AAD17527.1) and Bo. mori (Bm: NP_001108470.1). The WAG substitution model was used with variable and invariable positions and a bootstrap value of 1000 (only those above 50 are represented on the trees). The scale represents the rate of amino acid substitution per site. Novel sand fly aminopeptidase L sequences are in bold.
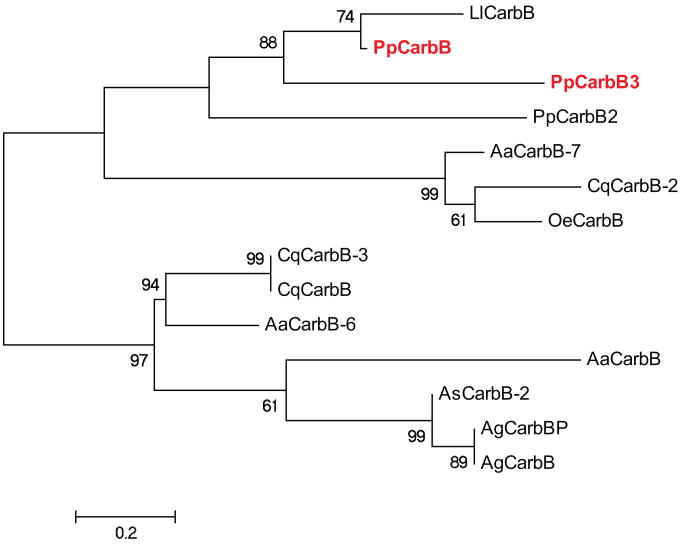
Carboxypeptidases also are enzymes integral to protein digestion in insects (e.g. mosquitoes) (Edwards et al. 2000). In sand flies, five sequences have been isolated from midgut, two carboxypeptidase B (one from each species) and three carboxypeptidase A (one from Ph. papatasi and two from Lu. longipalpis) (Ramalho-Ortigao et al. 2007, Pitaluga et al. 2009). Eighteen sequences (Supplemental Table2) related to carboxypeptidase A were identified in Ph. papatasi, these most likely represent transcript fragments. An additional two Ph. papatasi sequences (JP552630, JP546271) with similarity to known carboxypeptidase B were identified in this study. The new putative carboxypeptidase B sequences share a 38% and 54% identity, respectively, to known sand fly carboxypeptidases (Figure 7). While blood meals are a preferred source of protein for sand flies, they can survive on a sugar diet. Glucosidases are involved in carbohydrate digestion and their expression was shown to be modified by both sugar and blood meals in Ph. langeroni (Dillon, El Kordy 1997). Here we have identified 23 unique Ph. papatasi sequences with high identity (BLASTP, <1e-50) to known glucosidases from mosquitoes.
Figure 7. Phylogenetic analysis of Ph. papatasi and Lu. longipalpis carboxypeptidase B.
Phylogenetic tree constructed using carboxypeptidase B sequences from Ph. papatasi (Pp: PpCarbB2 (JP542144), PpCarbB3 (JP546271), ABV44754.1), Lu. longipalpis (Ll: LlCarbB), An. gambiae (Ag: AAS99341.1, CAF28572.1), An. stephensi (As: ADD31639.1), Ae. aeygpti (Aa: AAT36733.1, AAT36732.1, ABO21077.1), Cu. quinquefasciatus (Cq: XP_001856154.1, EDS34658.1, XP_001856164.1), Oc. epactius (Oe: AAT36738.1). The WAG substitution model was used with variable and invariable positions and a bootstrap value of 1000 (only those above 50 are represented on the trees). The scale represents the rate of amino acid substitution per site. Novel Ph. papatasi carboxypeptidase B sequences are in bold.
Chitin is an insoluble polysaccharide present in the insect cuticle, peritrophic matrix and the lining of the foregut, hindgut and trachea (Zhu et al. 2008). Chitinolytic enzymes are important for the periodical rearrangement and degradation of the exoskeleton and peritrophic matrix. To date, chitinases have been identified in many different arthropod species including fruit flies (Dr. melanogaster) (Zhu et al. 2004), mosquitoes (Ae. aegypti, An. gambiae) (De la Vega et al. 1998), silk moths (Koga et al. 1997), red flour beetles (Tribolium castaneum) (Zhu et al. 2008) and sand flies (Ph. papatasi and Lu. longipalpis) (Ramalho-Ortigao, Traub-Cseko 2003, Ramalho-Ortigao et al. 2005). The two known sand fly chitinases have been identified in the midgut and implicated in Leishmania – sand fly interactions (Ramalho-Ortigao, Traub-Cseko 2003, Ramalho-Ortigao et al. 2005). Seven more sequences with identity to the previously identified chitinase from Ph. papatasi (57%) and from related organisms, including Ae. aegypti and Dr. melanogaster (Ramalho-Ortigao et al. 2005), were identified in Ph. papatasi in the current study. However the search for conserved domains (Zhu et al. 2008) has revealed these sequences to be more likely transcript fragments rather than full-length chitinase sequences. Eight Ph. papatasi sequences related to amylase were identified in this study. Thirty-one sequences with high identity (BLASTP, <1e-50) to lipase also were identified in Ph. papatasi (Supp Table2).
Immune response Proteins
Insect immune responses have been most extensively studied in other dipteran insects including in Dr. melanogaster challenged with bacteria or fungi (Lemaitre, Hoffmann 2007) and in mosquitoes challenged with Plasmodium parasites (Cirimotich et al. 2010, Richman et al. 1997, Tahar et al. 2002) or bacteria (Dimopoulos et al. 1997, Hillyer, Schmidt & Christensen 2004). There have been only a few studies regarding the immune response of sand flies to parasite infection (Ramalho-Ortigao et al. 2007, Pitaluga et al. 2009, Jochim et al. 2008). Here we have identified Ph. papatasi sequences belonging to three of the four immune response pathways (Cirimotich et al. 2010, Tanaka et al. 2008), ranging from recognition receptors to effector proteins. There were no significant matches found to components of the JAK/STAT pathway in either sand fly.
Recognition proteins
Peptidoglycan recognition proteins (PGRPs) are highly conserved (Liu et al. 2001) across species that bind peptidoglycan present in the cell walls of bacteria. To date PGRPs have been identified in several arthropod species, including fruit flies (Werner et al. 2000), moths (Kang et al. 1998), silkworms (Tanaka et al. 2008, Ochiai, Ashida 1999), mosquitoes (Christophides et al. 2002) and sand flies (Pitaluga et al. 2009). Two new PGRP sequences were identified in Ph. papatasi, one is a short singlet with high identify to other sequence (PpPGRP2 –JP540873). PpPGRP2 has 44% identity to Ae. aegypti PGRP-LC and a 41% and 44% identity to known sand fly PGRPs (Lu. longipalpis and Ph. papatasi, respectively (Jochim et al. 2008)). Three additional sequences were identified with high similarity to PGRPs short class (PGRP-SC1-3, JP551327, JP547206, JP539467) (Figure 8).
Figure 8. Phylogenetic analysis of Ph. papatasi PGRP sequences.
Phylogenetic analysis of known and novel Ph. papatasi PGRP sequences from Ph. papatasi (Pp: ABV60369.1, PpPGRP2 (JP540873), PpPGRP-SC1-3 (JP551327, JP547206, JP546057), Lu. longipalpis (Ll: ABV60332.1), Bo. mori (Bm: BAA77209.1), Ca. dromedaries (Cd: CAC19553.1), Dr. melanogaster (Dm: AAF54643.1, AAG32064.1, AAG23735.1, AAG23736.1), Mu. musculus (Mm: AAC31821.1), Ra. norvegicus (Rn: AAF73252.1) and Tr. ni (Tn: AAC31820.1). The WAG substitution model was used with variable positions and a bootstrap value of 1000 (only those above 50 are represented on the trees). The scale represents the rate of amino acid substitution per site. Novel Ph. papatasi PGRP sequences are in bold.
β-glucan recognition proteins (βGRPs) are involved in recognition of Gram-negative bacterial and fungal cell walls and in triggering the prophenoloxidase (PPO) cascade (Yoshida, Ochiai & Ashida 1986, Ochiai, Ashida 2000). The βGRP class is composed of two functionally distinct proteins; one that binds β-1,3 glucan and one that binds Gram-negative bacteria. In the Ph. papatasi dataset we have identified three sequences with high similarity to βGRPs, two (JP543291, JP552580) that likely represent two alleles of the same sequence, PpBGRP1, whereas the third sequence (PpBGRP2- JP544368) is most likely a transcript fragment. These three novel βGRPs have amino acid identity to βGRPs from the pyralid moth Plodia interpunctella (37%) (Fabrick, Baker & Kanost 2003) and from the mosquito Armigeres subalbatus (44%) (Wang et al. 2005). Three sequences (JP542600, JP540290, JP541890) with high similarity to Gram-Negative Binding Proteins (GNBPs) were also identified. No new sequences with similarity to βGRPs were identified in the Lu. longipalpis dataset, LlBGRP1 and LlBGRP2 having previously been described by Dillon et al. (Dillon et al. 2006).
C-type lectins are a large protein family with low conservation at the amino acid level, they have diverse functions (Zelensky, Gready 2005), including pathogen recognition and neutralization (Weis, Taylor & Drickamer 1998). We have identified seven C-type lectins (PpCTL1-3) in the Ph. papatasi dataset with similarity to known C-type lectins from other insects. Two are alleles of PpCTL1a (JP543952, JP44832, JP547369) and one is an allele of PpCTL2a (JP543073, JP55083). PpCTL3a (JP554123) and PpCTL3b (JP540156) share a high identity at amino acid level (over 95%), however PpCTL3b is 139 amino acids longer than PpCTL3a. There were no sequences with high identity to known C-type lectins identified in Lu. longipalpis.
Three classes of the scavenger receptor protein family serve as recognition receptors in immune responses (Peiser, Mukhopadhyay & Gordon 2002, Kiefer et al. 2002, Pierini 2006, Ramet et al. 2001). Dillon et al. (Dillon et al. 2006) previously identified proteins belonging to two of the three classes (B and C) in Lu. longipalpis. Here we added six Ph. papatasi sequences with identity to class B scavenger receptors. Three of the new Ph. papatasi putative scavenger receptors (PpSR1b-d, JP543420, JP543780, JP539722) may represent alleles of PpSRB1a (JP541894). PpSRB1a-d and PpSRB2 (JP543210) share an identity of 29-31% to the known Lu. longipalpis scavenger receptors and 44-49% identity to An. gambiae scavenger receptor class B, while PpSRB3 (JP553019) has an identity of 96% to NSFM-84c11.q1k (SRB) from Lu. longipalpis and 80% to An. gambiae scavenger B receptor.
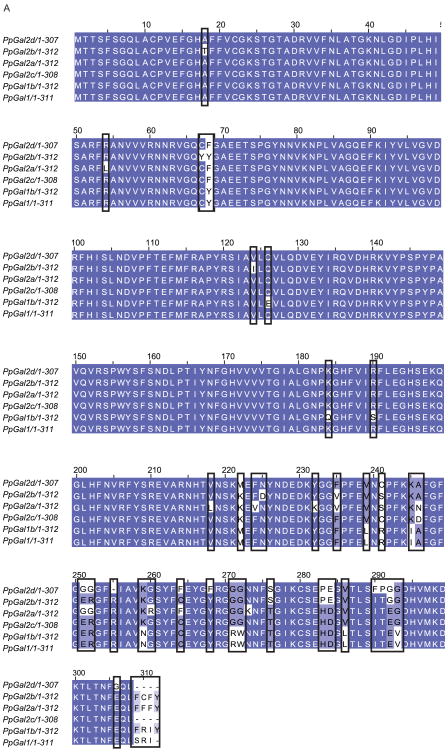
Galectins have been implicated in cell adhesion (Ochieng, Leite-Browning & Warfield 1998), apoptosis (Perillo et al. 1997), and immune response (Tanaka et al. 2008) and, for sand flies, in species-specific binding of Leishmania parasites to the sand fly midgut (Kamhawi et al. 2004). Seven sequences related to galectins were identified in Ph. papatasi. One of these novel sand fly galectins is most likely an allele of the known Ph. papatasi galectin (PpGal1 (JP539532), previously identified as PpGalec [75]), four more sequences (PpGal2a-d, JP540648, JP546602, JP550066, JP540193) have less than 95% identity at the amino acid level to PpGal1, three of which (PpGal2b-d) are alleles of PpGal2a (Figure 9A). The remaining two galectins (PpGal3-4, JP5484429, JP549531, JP543439) share less than 39% identity to PpGal1. The novel galectins share a higher identity to related sequences from Lu. longipalpis (PpGal2a – 56%, PpGal3 – 87%, PpGal4 – 73%), than to mosquito galectins: An. gambiae (36%, 47%, 48%), Ae. aegypti (40%,50%, 60%) and Cu. quinquefasciatus (37%, 45%, 57%). The previously published galectins A-D (Dillon et al. 2006) were identified in our Lu. longipalpis analysis, but no additional galectins were identified (Figure 9).
Figure 9. Ph. papatasi novel galectin sequences.
Multiple sequence alignment of five novel Ph. papatasi galectins (PpGal1b (JP539352), PpGal2a-d (JP540648, JP546602, JP550066, JP540193) and the known galectin sequence (AAT11557.1) from the same organism. Darker shading indicates conservation at each position.
Thioester-containing proteins (TEPs) are homologs of the complement system (Blandin, Levashina 2004) and have been implicated in phagocytosis of gram-negative bacteria in insects (Cirimotich et al. 2010, Tanaka et al. 2008, Blandin, Levashina 2007). Three sequences (JP540471, JP544554, JP555283) with identity to mosquito TEPs were identified in Ph. papatasi. One of these TEP sequences is a contig while two are singlets of short length and likely represent transcript fragments. The contig shares 98% of its amino acids with one singlet and 93% with the other, making them potentially alleles of the contig. In the Lu. longipalpis dataset, two sequences related to mosquito TEPs were identified, one singlet and one contig. The novel Lu. longipalpis TEP sequences share a 72% and 86% identity respectively to the TEP sequence identified by Dillon et al. (Dillon et al. 2006). The novel Ph. papatasi TEP contig has an identity of 85% and 89% to novel Lu. longipalpis and a 60% identity to Ae. aegypti TEPs. The Ph. papatasi TEP singlets have and identity of 89% and 86% respectively for one singlet, 78% and 87% respectively to the novel Lu.longipalpis TEP sequences and 69% and 53% identity to the Ae. aegypti TEP sequence.
Signaling pathways proteins
The Toll signaling pathway plays a key role in the establishment of the dorso-ventral axis of the Drosophila embryo and also is activated in response to microbial infection (Cirimotich et al. 2010, Anderson 2000). Immune responses through this pathway are activated by the recognition of a pathogen by the PGRPs that activates a serine protease cascade culminating with activation of the cytokine-like protein, Spätzle. Subsequent signaling involves Myd88, Tube and Pelle resulting in the degradation of Cactus and the release of Dorsal, a Rel1 protein, from its complex with Cactus (Anderson 2000, Michel et al. 2001). Five sequences with identity to mosquito Spätzle were identified in sand flies; three in Ph. papatasi and two in Lu. longipalpis. Two of the Ph. papatasi Spätzle-like sequences are non-overlapping fragments of the same transcript. The two Lu. longipalpis Spätzle-like have an identity of 99% to each other, making them alleles and 30% identity to the known Lu. longipalpis Spätzle (Dillon et al. 2006). The three novel sand fly Spätzle-like sequences have an identity of 61%, 68% and 68% to Ae. aegypti Spätzle proteins (Nene et al. 2007). The Ph. papatasi sequence identified as MyD88-like has 45% identity to Cu. quinquefasciatus MyD88 and is most likely a fragment, as it is short (122 AA) and a singlet. The absence of MyD88-like sequences from the Lu. longipalpis dataset may be explained by the lower number of sequences available for this sand fly. The four sand fly homologs to Toll-interacting protein (Tollip) (three in Lu. longipalpis and one in Ph. papatasi) have a 97% identity to each other and 68% to Tr. castaneum Tollip protein. Pellino, another component of the Toll pathway, is known to bind phosphorylated Pelle/IRAK and enhance innate immunity in Drosophila fruit flies (Haghayeghi et al. 2010). Here we identified two Ph. papatasi sequences with a 91% identity to the Dr. melanogaster Pellino sequence, however these novel sequences are from a single transcript as they are unassembled mate reads (Supplemental Table 1).
The immunodeficiency (IMD) pathway is similar to the mammalian TNF receptor signaling pathway (Valanne et al. 2007)(Lemaitre, Hoffmann 2007) and is activated by gram-negative bacteria in Dr. melanogaster (Tanaka et al. 2008). Several sequences with homology to components of this pathway were identified in Ph. papatasi including five sequences with identity to inhibitor of apoptosis protein 2 (IAP2), a protein required for antimicrobial peptide expression in fruit flies (Valanne et al. 2007). Four of the novel IAP2 sequences are most likely alleles of one protein, the fifth being rather short (96AA) representing a transcript fragment. The sand fly IAP2 sequences have an identity of 44% to Gl. morsitans morsitans IAP2 sequence (Attardo et al. 2006). One sequence related to IKKβ was also identified in Ph. papatasi (Supp Table 1).
While the first two signaling pathways are well characterized in insect species, there is less information concerning the c-Jun NH2-terminal kinase (JNK) pathway in Diptera (Agaisse, Perrimon 2004). The JNK signaling pathway is formed by genes involved in wound repair, stress repair, and negative feedback control of antimicrobial peptides (Botella et al. 2001, Ramet et al. 2002). A component of the IMD pathway, TAK1, a MAPK, has been implicated in activation of this signaling cascade (Silverman et al. 2003). One sequence related to Hem, a component of the JNK pathway, was identified in each sand fly, but shared no significant identity to each other. Three Fos sequences were identified in the current study; 1 in Lu. longipalpis and two in Ph. papatasi with an identity of 80% between the two sand flies (the 2 Ph. papatasi sequences are alleles). Two sequences with identity to Cu. quinquefasciatus Jun sequence, another JNK pathway component, also were identified in sand flies (one each) (Supp Table 1).
Effector proteins
In insects, prophenoloxidases (PPOs) are involved in melanization, an efficient immune response activated by recognition of lipopolysacharides, peptidoglycan, and β-1,3 glucans (Soderhall, Cerenius 1998, Cerenius, Soderhall 2004). Ten sequences related to PPOs were identified in Ph. papatasi. Only four of these ten sequences have high identity (blastp, 10-50) to related PPO sequences from other organisms. The four PPOs identified here most likely represent three unique transcripts, with PpPPO1 having two alleles (PpPPO1a (JP539467) and PpPPO1b (JP550019)). PpPPO1a,b and PpPPO2 (JP549395) have an identity of 44%, 52% and 63% respectively to Ae. aegypti PPO (XP_001648968.1), while PpPPO3 (JP545596) has an identity of 64% to a different Ae. aegypti PPO (XP_001661891.1). The reduced level of identity of the last six putative PPOs from Ph. papatasi could be due to their short length (the determined ORF encodes a protein less than 300 amino acids). Thirteen sequences with similarity to PPOs were identified in Lu. longipalpis, of these sequences, four are likely alleles of one PPO (LlPPO1) and three are alleles of a second PPO sequence (LlPPO2), with LlPPO2c allele being formed by an unassembled mate pair. The other six PPOs present in Lu. longipalpis share an identity of less than 64% at amino acid level to the previously mentioned Lu. longipalpis PPOs (LlPPO1a-d and LlPPO2a-c). The novel putative Ph. papatasi PPO have a varying degree of identity to their closest Lu. longipalpis counterpart: PpPPO1a -78%, PpPPO2 – 41% and PpPP03 – 37%.
Another type of protein implicated in immune response in mosquitoes has leucine-rich repeats (LRR) and in An. gambiae is involved in bacteria phagocytosis and parasite melanization (Povelones et al. 2009). Here we have identified five LRR-containing sequences in Ph. papatasi, two of which are likely alleles of the same gene, and two Lu. longipalpis sequences with an amino acid identity of 46 and 48% to Ae. aegypti and 68% to Gl. morsitans morsitans LRR- containing proteins. Lysozymes have been implicated in both digestion and immune response in mosquitoes (Ursic Bedoya et al. 2005). We identified two different lysozyme sequences from Ph. papatasi with identity to known insect lysozymes.
Peritrophic matrix (PM)
The PM is a non-membranous extracellular layer that surrounds the food bolus in insects. To date it has been best characterized in insects of medical or agricultural importance. In phlebotomine sand flies, there are two different types of peritrophic matrices depending on the life stage of the fly; PM type 1 (PM1) is present during the adult life stage while PM type 2 (PM2) is present during larval life stages (Marquardt et al. 2005). The synthesis and degradation of PM1 has been related to the ingestion of blood in Lutzomyia sand flies, with the PM being well formed 24h after a blood meal (Secundino et al. 2005). Functionally, PM1 has been implicated in both digestion (Tellam, Wijffels & Willadsen 1999, Shao, Devenport & Jacobs-Lorena 2001) and in Leishmania-sand fly interaction. Glutamine synthetase is involved in chitin synthesis, a major component of PM. We identified nine Ph. papatasi sequences related to glutamine synthetase with identity to sequences from other flies (56-96% Lu. longipalpis; 77-81%, Dr. melanogaster; 77% Gl. morsitans morsitans; 70-75%, Ae. aegypti), six of which may represent alleles of one sequence.
Three sequences with identity to Dr. melanogaster hemomucin, a molecule present in the fruit fly PM and involved in induction of antibacterial effector molecules (Theopold et al. 1996) were identified in Ph. papatasi, these sequences likely represent a single transcript with one sequence containing the signal peptide and the other two containing all the conserved glycosylation sites. Integrins are involved in cell-cell and cell-matrix interactions and have been implicated in phagocytosis of bacteria in An. gambiae (Moita et al. 2006). Five Ph. papatasi sequences were identified with high identity to β-integrins from insects with an amino acid identity between 53% for Ae. aegypti and 91% to Plutella xylostella. The length of these sequences combined with the fact they are either singlets or single mate pairs suggests that they might be fragments of β-integrin present in Ph. papatasi. Peritrophins are important components of insect PM (Lehane 1997), we identified one novel sequence with 71% identity to Dr. melanogaster peritrophin A and 25% identity to known Ph. papatasi peritrophins (Ramalho-Ortigao et al. 2007).
Quantitative mRNA analysis of candidate genes
Fourteen novel Ph. papatasi proteins were selected for an expression analysis. The sequences included the novel putative trypsin and chymotrypsin and PGRP proteins sequences were chosen for their potential role in digestion and immune response. The expression of three previously described transcripts was also analyzed (PpTryp1, PpChym2 and PpPGRP) in order to validate our results.
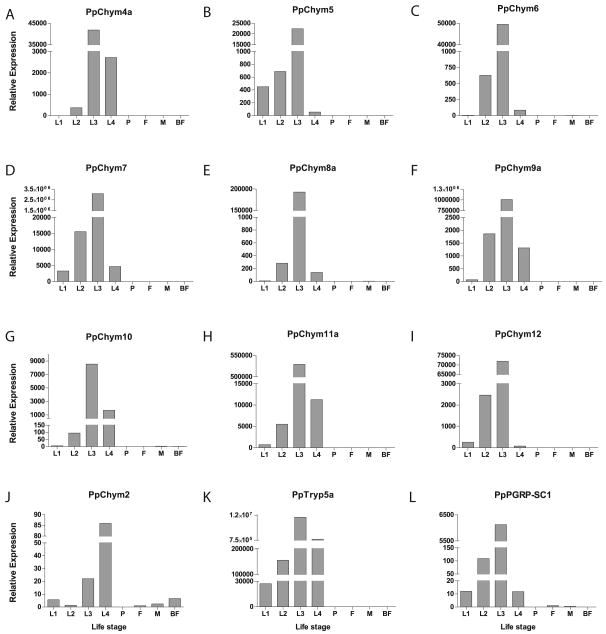
Of the novel putative Ph. papatasi proteins, PpChym4a-12, PpTryp5a and PpPGRP-SC1 (Figure 10) are expressed at a higher level in the immature life stages, indicating that they may be involved in digestion as was shown for larval and pupal trypsin and chymotrypsin in Ae. aegypti (Yang, Davies 1971), where their roles were thought to be in food digestion and immune response, respectively. Previously, the expression of PpChym2 was shown to be influenced by the presence of a blood meal (Ramalho-Ortigao et al. 2003), in this study we demonstrate that while PpChym2 is expressed both in adult and immature life stages (Figure 10) it has a much higher level of expression in late larval stages than upon blood feeding (Figure 10). Unlike PpChym2, PpTryp1 is not expressed in the larval or pupal stages and, as demonstrated by Ramalho-Ortigao et al. (Ramalho-Ortigao et al. 2003) is down regulated upon blood meal (results not shown).
Figure 10. Ph. papatasi novel proteins transcript levels.
qPCR analysis of Ph. papatasi novel putative genes at different life stages, including larvae (L1-L4), pupa (P), 1-3 days past emergence adult females (F) and males (M) and adult females at 24h post blood meal (BF) for (A) PpChym4a (JP546634), (B) PpChym5 (JP551370), (C) PpChym6 (JP547341), (D) PpChym7 (JP554731), (E) PpChym8a (JP549601), (F) PpChym9a (JP554746), (G) PpChym10 (JP540516), (H) PpChym11a (JP543908), (I) PpChym12 (JP547864), (J) PpChym2 (AY128107), (K) PpTryp5a (JP544502) and (L) PpPGRP-SC1 (JP551327).
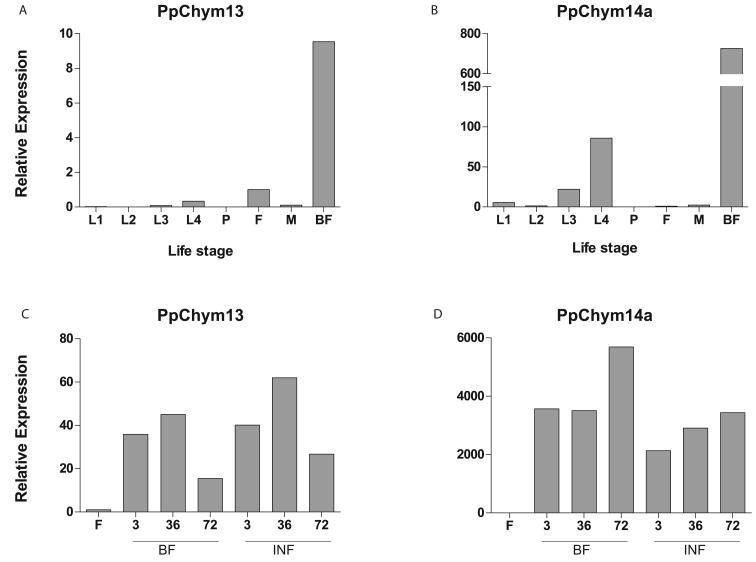
In blood feeding mosquitoes, there are two types of chymotrypsins that have their expression level influenced by taking a blood meal: early chymotrypsin which is highly expressed for the first hours post blood meal in An. gambiae (Shen, Edwards & Jacobs-Lorena 2000) and late chymotrypsin which, in An. gambiae has a peak expression at around 30h post blood meal (Vizioli et al. 2001). One late chymotrypsin has been previously identified in Ph. papatasi – PpChym2, with a peak level of expression at 30h post blood meal (Ramalho-Ortigao et al. 2003). Here we have identified two other novel putative chymotrypsins that have an expression post blood meal – PpChym13 and PpChym14a, indicating a possible implication of these proteins in blood digestion. While PpChym14a is also expressed in immature stages, though at much lower levels, PpChym13 is expressed only upon blood feeding (Figure 11 A&B).
Figure 11. PpChym13 and PpChym14 transcript levels.
qPCR analysis of 2 Ph. papatasi novel putative chymotrypsins at different life stages, including larvae (L1-L4), pupa (P), 1-3 days past emergence adult females (F) and males (M) and adult females at 24h post blood meal (BF) for (A) PpChym13 (JP543702) and (B) PpChym14a (JP542007). Additional blood feeding conditions for adult female sand flies at 3h, 36h and 72h post blood meal (BF) and post Le. major infected blood meal (INF) for (C) PpChym13 and (D) PpChym14a.
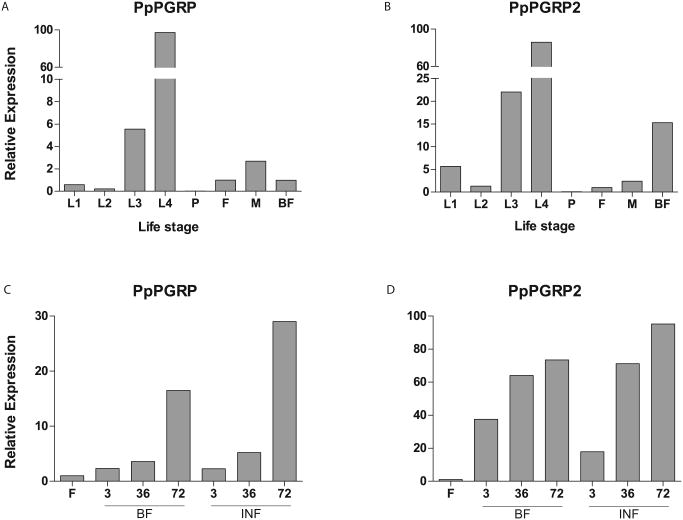
PpPGRP and PpPGRP2 are two other Ph. papatasi proteins potentially involved in immune response whose expression was analyzed in this study. The previously identified PpPGRP (Ramalho-Ortigao et al. 2007, Jochim et al. 2008), is shown here to be expressed in both immature and adult life stages, with a peak of expression during larval stages, suggesting a possible involvement in larval immune response (Figure 12 A&C). PpPGRP2 has an expression profile in the larval stages similar to the PpPGRP with a peak of expression during 4th intar (L4). However, this transcript is also up regulated following a blood meal with or without (Figure 12 B&D).
Figure 12. PpPGRP and PpPGRP2 transcript levels.
qPCR analysis of 2 Ph. papatasi novel putative PGRPs at different life stages, including larvae (L1-L4), pupa (P), 1-3 days past emergence adult females (F) and males (M) and adult females at 24h post blood meal (BF) for (A) PpPGRP (EU130784) and (B) PpPGRP2 (JP540873). Additional blood feeding conditions for adult female sand flies at 3h, 36h and 72h post blood meal (BF) and post Le. major infected blood meal (INF) for (C) PpPGRP and (D) PpPGRP2.
Of the 71 Ph. papatasi sequences addressed in the study, 30 sequences share over 95% amino acid identity with another Ph. papatasi sequence, possibly representing allele variation. The high number of alleles present is suggestive of high genetic heterozygosity within the colony utilized for library construction in spite of the fact that this colony has been maintained under laboratory conditions since the early 1970s with substantial inbreeding. Furthermore, we developed an assembly process that favored a more compacted assembly than otherwise possible, indicating the possibility of gene duplication in Ph. papatasi. The existence of a high degree of polymorphism in the population is possible but further tests are necessary for confirmation.
To estimate colony genetic heterozygosity we quantified the number of SNPs present in each contig, requiring that each qualifying position was covered by at least four ESTs and that the two most common alleles were present in at least two ESTs. Using these criteria, SNPs were discovered in 1285 contigs, comprising 7.1 Mb of sequence. We identified 6312 SNPs and estimated 8.88 SNPs per 1,000 bases. This number is slightly greater than the SNP density for colonized An. funestus mosquitoes (Wondji, Hemingway & Ranson 2007) but lower than that found in the butterflies Melitaea cinxia (Vera et al. 2008) and Papilio Zelicaon (O'Neil et al. 2010), indicating a relatively low level of heterozygosity.
Conclusions
This work represents the first global transcriptome study of the sand fly Ph. papatasi, the principal vector of Le. major in North Africa and the Middle East, and has resulted in the identification of novel sequences involved in parasite-vector interactionthat could be targets for future vector control methods. Furthermore, this EST library will be an essential resource during annotation of the two phlebotomine sand fly genomes currently under way (McDowell et al. 2006).
Experimental procedures
Library construction
A normalized cDNA library (Soares et al. 2009) was constructed from a Ph. papatasi colony (Israeli strain) maintained at the Walter Reed Army Institute of Research (WRAIR). This colony was originally established in the 1970's and has been subjected to several bottlenecks, thus it is thought to display low levels of genetic polymorphism. Using a RNAeasy Mini Kit (Qiagen), total RNA was collected from the four larval instars, pupae, and adult males and females 1, 3, and 10 days post emergence (adults fed only on sugar). Additional RNA was included from females 6, 12, 24, 36, 48, 72, 94 and 120 hours post feeding on uninfected and Le. major (strain Friedlin V1) infected mouse blood. A total of 29 samples were collected with RNA amounts per sample ranging from 700ng (1st instar larvae or L1) to 20μg (48h post blood meal, single females). RNA samples were checked on agarose gel (1% MOPS with 5% formaldehyde). Library construction was completed with pooledRNA, using equal amounts from each of the 29 samples, by Express Genomics (Baltimore, MD) and EST sequencing was performed by the Genome Institute at Washington University (St. Louis, MO).
Sequencing, processing, and assembly
Sequences were generated from the normalized cDNA library using the Sanger-based dideoxy chain termination. Direct colony sequencing using standard high throughput sequencing methods was performed using a DNA track Robot and ABI 3730 sequencers. Ph. papatasi sequencing chromatograms were used to generate sequence and quality score files using Phred/Phrap program suite (Ewing et al. 1998, Ewing, Green 1998). Lu. longipalpis sequences and quality scores were obtained in Fasta format from Dillon et al. (Dillon et al. 2006). Cleaning and filtering of sequence files was performed in three steps. First, contaminant sequences (human, protozoa, bacteria, mouse or rat) were removed using the BLASTX algorithm (Altschul et al. 1990). A sequence was considered to be a contaminant if 12 of the first 20 best hits were any of the organisms listed above. Subsequently low quality regions were trimmed using Lucy2 (Chou, Holmes 2001) with default parameters and the pExpress-1 cloning vector. Lastly, poly(A) tails and small contaminant sequences were removed using the Seqclean algorithm available from The Institute for Genomic Research (TIGR Gene Indices Clustering Tools) (Chen et al. 2007). Only sequences longer than 100 bp were used for further analysis. As a comparison, we also analyzed a previously generated Lu. longipalpis EST data set (Dillon et al. 2006). To eliminate sequence cleaning and assembly biases, the Lu. longipalpis data (first described by Dillon et al. (Dillon et al. 2006) were processed using the same programs and parameters as the Ph. papatasi dataset.
Sequences were assembled into contigs using the Cap3 assembler (Huang, Madan 1999). Overlapping mate pairs were assembled and consensus obtained using relatively relaxed criteria (p 80, o 20, h 95). Consensus sequences obtained above were assembled with remaining ESTs using more stringent parameters (p 95, o 50, h 30) (Figure 1). The resulting sequences are referred to as assembled sequences.
GO annotation and similarity searches
Biological functions for the Ph. papatasi and Lu. longipalpis assembled sequences were assigned using Blast2GO (Conesa et al. 2005). InterProScan analysis also was performed as part of the GO annotation process. Annotated genes were split into the three main GO categories: biological process, molecular function and cellular component.
Similarity to known proteins
Similarity searches to known sequences were performed by using BLAST with an e-value limit of 10-5 against the National Center for Biotechnology Information (NCBI) non-redundant protein database (NR). Protein sequences involved in vector-parasite interactions, specifically sequences involved in blood digestion, immune response, and peritrophic matrix composition were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/protein) and ImmunoDB (http://cegg.unige.ch/Insecta/immunodb/) for available arthropod species. Ph. papatasi sequences with significant similarity (BLASTX, e-value 10-20) to these restricted datasets were further analyzed for presence of conserved domains using ScanPROSITE (Gattiker, Gasteiger & Bairoch 2002) and manual searches. For manual domain searches, sand fly assembled sequences were translated into amino acid sequences using prot4EST pipeline (Wasmuth, Blaxter 2004) and then aligned to known proteins using Muscle (Edgar 2004) with default parameters. Alignments were viewed using Seaview (Gouy, Guindon & Gascuel 2010) and Jalview (Waterhouse et al. 2009). Maximum likelihood trees were built for full sand fly sequences using sequences from related organisms and the MEGA5 algorithm (using the WAG model, bootstrap value: 1000) (Tamura et al. 2011).
Expression analysis of predicted Ph. papatasi proteins
An expression analysis was performed on 14 novel putative Ph. papatasi proteins involved in digestion and immune response (1 trypsin, 11 chymotrypsin, 2 PGRP). Total RNA was isolated from 5 different Ph. papatasi larval stages (L1-P) females and males 1-3 days old and blood fed females at 24h, using the RNeasy Mini Kit (Qiagen). For proteins with a high expression at 24h post blood meal additional samples were extracted from females at 3h, 36h and 72h post blood meal and post Le. major infected blood meal. The DNase (Fermentas) treated RNA was used to generate cDNA using Superscript III (Invitrogen) and oligo (dT12-20). Quantitative PCR was performed using SYBRGreen (ABI), an ABI 7900 RT-PCR system and 20ng of cDNA per sample. Primer sets were designed for each sequence of interest such that, only one sequence was amplified (Supplemental Table 3), however, high level of identity between the potentially different alleles made it impossible to distinguish between them. The 60S Ribosomal protein L10 was used as an internal control. Reactions for each gene and for the control used were carried out in triplicate. Relative expression levels of each gene was determined by the ΔΔCT method, where relative expression is expressed as a fold difference relative to sugar fed females and expressed as 2- ΔΔCT. The following formula was used: ΔΔCT = ΔCT(stage or condition) - ΔCT(Sugar Fed Females) and ΔCT = CT (gene of interest) - CT (60S RNA). Average Ct value for all samples can be found in supplemental tables 4 and 5.
Heterozygosity Analysis
Single nucleotide polymorphisms (SNPs) were generated from the sandfly EST assemblies as described by O'Neil et al. (2010). Specifically, the ACE output of each CAP3 assembly was imported into an AMOS bank (Pop et al. 2004) for programmatic access to underlying read information. SNPs were called using the “loose” criterion, which required that the two most common alleles be found in at least two distinct ESTs (O'Neil et al. 2010). To estimate heterozygosisity, we used AMOS to count both the number of loose criterion SNPs and the number of positions covered by at least four ESTs in each contig. Average heterozygosity was then computed as the total number of SNPs divided by total number of qualifying positions. This simple method was chosen over the Beta statistic (Novaes et al. 2008) because we were not interested in population genetics underlying these two colonies and highly covered arthropod EST contigs tend to be less diverse (O'Neil et al. 2010) justifying the use of more simple SNP criteria.
Supplementary Material
The numbers of sequences were plotted as function of their length. The black line represents sequences obtained in the study using the assembly process detailed in Fig. 1 while the grey line represents sequences obtained by Dillon et al (2006) using the Phrap assembler.
The distribution of the subcategories for the GO Biological process main category were plotted as a function of the percent of sequences annotated for the same main GO category. The Lu. longipalpis dataset is represented by the grey bars while the Ph. papatasi is represented by the black bars.
The distribution of the subcategories for the GO Molecular Function main category were plotted as a function of the percent of sequences annotated for the same main GO category. The Lu. longipalpis dataset is represented by the grey bars while the Ph. papatasi is represented by the black bars.
The distribution of the subcategories for the GO Cellular Component main category were plotted as a function of the percent of sequences annotated for the same main GO category. The Lu. longipalpis dataset is represented by the grey bars while the Ph. papatasi is represented by the black bars.
Acknowledgments
This project was supported by the National Human Genome Research Institute and in part by an award from the Defense Advanced Research Projects Agency (#W911NF0410380) and Telemedicine and Advanced Technology Research Center (#W81XWH-10-1-0085) to Mary Ann McDowell. This project was also supported by VectorBase (NIH/NIAID HHSN72200900039C. The assembly was performed on the Notre Dame Biocomplexity Cluster (University of Notre Dame, Notre Dame IN) supported in part by NSF MRI Grant No. DBI-0420980. BLAST searches were performed on the Biocompute server at Notre Dame.
Footnotes
Authors' Contributions: JA designed and performed all of the bioinformatic analyses. MRO isolated RNA from all samples and coordinated sample shipment to ExpressGenomics for library construction and aided in experimental design. PL maintained the P. papatasi colony and carried out blood feeding and sampling of life-cycle stages. SK organized and performed quality control of the adult and larval samples used for library generation. EDR oversaw the maintenance of the sand fly colony. MRO, MJB, PAB, JGV, FHC, RD and MAM conceived the project, were authors on the white paper that funded the project, and participated in overall experimental design. CT, EA, EM, SC, and RKW were involved in various stages of sequencing and analysis. RD provided L. longipalpis sequences. NFL, FHC, and MAM advised the bioinformatic analysis. SE and SO performed the heterozygosity analysis. DS, GS and JA performed the qPCR analysis. MW and MB maintain the Ph. papatasi colony and collected samples. MAM managed all activities.
Additional Information: The views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Army, Department of Defense, nor the U.S. Government.
Some of the co-authors are employees of the U.S. Government. This work was prepared as part of their official duties. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government’. Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties.
References
- Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunological reviews. 2004;198, no. 1:72. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215, no. 3:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alves Silva J, Ribeiro JMC, Abbeele JVD, Attardo G, Hao Z, Haines LR, Soares MB, Berriman M, Aksoy S, Lemaire HG. An insight into the sialome of Glossina morsitans morsitans. BMC genomics. 2010;11, no. 1:213. doi: 10.1186/1471-2164-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KV. Toll signaling pathways in the innate immune response. Current opinion in immunology. 2000;12, no. 1:13. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- Appel W. Chymotrypsin: molecular and catalytic properties. Clinical biochemistry. 1986;19, no. 6:317. doi: 10.1016/s0009-9120(86)80002-9. [DOI] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Bartholomay L, Bidwell S, Caler E, Camara F, Campbell CL, Campbell KS, Casola C, Castro MT, Chandramouliswaran I, Chapman SB, Christley S, Costas J, Eisenstadt E, Feschotte C, Guigo R, Haas B, Hammond M, Hansson BS, Hemingway J, Hill SR, Howarth C, Ignell R, Kennedy RC, Kodira C, Lobo NL, Mao C, Mayhew G, Michel K, Mori A, Liu N, Naveira H, Nene V, Nguyen N, Pearson MD, Pritham EJ, Puiu D, Qi Y, Ranson H, Ribeiro JMC, Roberston HM, Severson DW, Shumway M, Stanke M, Strausberg RL, Sun C, Sutton G, Tu Z, Tubio JMC, Unger MF, Vanlandingham DL, Vilella AJ, White O, White JR, Wondji CS, Wortman J, Zdobnov EM, Birren B, Christensen BM, Collins FH, Cornel A, Dimopoulos G, Hannick LI, Higgs S, Lanzaro GC, Lawson D, Lee NH, Muskavitch MAT, Raikhel AS, Atkinson PW. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330, no. 6000:86. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo GM, Strickler-Dinglasan P, Perkin SAH, Caler E, Bonaldo MF, Soares MB, El-Sayeed N, Aksoy S. Analysis of fat body transcriptome from the adult tsetse fly, Glossina morsitans morsitans. Insect molecular biology. 2006;15, no. 4:411–424. doi: 10.1111/j.1365-2583.2006.00649.x. [DOI] [PubMed] [Google Scholar]
- Blandin SA, Levashina EA. Phagocytosis in mosquito immune responses. Immunological reviews. 2007;219, no. 1:8–16. doi: 10.1111/j.1600-065X.2007.00553.x. [DOI] [PubMed] [Google Scholar]
- Blandin S, Levashina EA. Thioester-containing proteins and insect immunity. Molecular immunology. 2004;40, no. 12:903–908. doi: 10.1016/j.molimm.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Botella JA, Baines IA, Williams DD, Goberdhan DCI, Proud CG, Wilson C. The Drosophila cell shape regulator c-Jun N-terminal kinase also functions as a stress-activated protein kinase. Insect biochemistry and molecular biology. 2001;31, no. 9:839–847. doi: 10.1016/s0965-1748(01)00029-7. [DOI] [PubMed] [Google Scholar]
- Buchon N, Poidevin M, Kwon H, Guillou A, Sottas V, Lee B, Lemaitre B. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. PNAS: Proceedings of the National Academy of Sciences of the United States of America. 2009;106, no. 30:12442. doi: 10.1073/pnas.0901924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerenius L, Soderhall K. The prophenoloxidase-activating system in invertebrates. Immunological reviews. 2004;198, no. 1:116. doi: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lin C, Wang C, Wu H, Hwang P. An optimized procedure greatly improves EST vector contamination removal. BMC Genomics. 2007;8, no. 1:416. doi: 10.1186/1471-2164-8-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17, no. 12:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller H, Osta MA, Paskewitz SM, Reichhart J, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-Related Genes and Gene Families in Anopheles gambiae. Science. 2002;298, no. 5591:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. Mosquito immune defenses against Plasmodium infection. Developmental comparative immunology. 2010;34, no. 4:387. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21, no. 18:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cunningham AC. Parasitic adaptive mechanisms in infection by Leishmania. Experimental and molecular pathology. 2002;72, no. 2:132. doi: 10.1006/exmp.2002.2418. [DOI] [PubMed] [Google Scholar]
- De la Vega H, Specht CA, Liu Y, Robbins PW. Chitinases are a multi-gene family in Aedes, Anopheles and Drosophila. Insect molecular biology. 1998;7, no. 3:233. doi: 10.1111/j.1365-2583.1998.00065.x. [DOI] [PubMed] [Google Scholar]
- Desjeux P. The increase in risk factors for leishmaniasis worldwide. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2001;95, no. 3:239–243. doi: 10.1016/s0035-9203(01)90223-8. [DOI] [PubMed] [Google Scholar]
- Desjeux P. Leishmaniasis: Public health aspects and control. Clinics in dermatology. 1996;14, no. 5:417–423. doi: 10.1016/0738-081x(96)00057-0. [DOI] [PubMed] [Google Scholar]
- Dillon RJ, El Kordy E. Carbohydrate Digestion in Sandflies:[alpha]-Glucosidase Activity in the Midgut of Phlebotomus langeroni. Comparative biochemistry and physiology B Comparative biochemistry. 1997;116, no. 1:35. doi: 10.1016/s0305-0491(96)00195-2. [DOI] [PubMed] [Google Scholar]
- Dillon RJ, Lane RP. Influence of Leishmania infection on blood-meal digestion in the sandflies Phlebotomus papatasi and P. langeroni. Parasitology research. 1993;79, no. 6:492. doi: 10.1007/BF00931590. [DOI] [PubMed] [Google Scholar]
- Dillon RJ, Ivens AC, Churcher C, Holroyd N, Quail MA, Rogers ME, Soares MB, Bonaldo MF, Casavant TL, Lehane MJ, Bates PA. Analysis of ESTs from Lutzomyia longipalpis sand flies and their contribution toward understanding the insect–parasite relationship. Genomics. 2006;88, no. 6:831–840. doi: 10.1016/j.ygeno.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G, Richman A, Muller H, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proceedings of the National Academy of Sciences. 1997;94, no. 21:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Bao Z, Hou R, Wang S, Su H, Yan J, Tian S, Li Y, Wei W, Lu W, Hu X, Wang S, Hu J. Transcriptome sequencing and characterization for the sea cucumber Apostichopus japonicus (Selenka, 1867) PLoS ONE. 2012;7, no. 3:e33311. doi: 10.1371/journal.pone.0033311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32, no. 5:1792. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MJ, Moskalyk LA, Donelly-Doman M, Vlaskova M, Noriega FG, Walker VK, Jacobs-Lorena M. Characterization of a carboxypeptidase A gene from the mosquito, Aedes aegypti. Insect molecular biology. 2000;9, no. 1:33. doi: 10.1046/j.1365-2583.2000.00159.x. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-Calling of Automated Sequencer Traces UsingPhred. II. Error Probabilities. Genome research. 1998;8, no. 3:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-Calling of Automated Sequencer Traces UsingPhred. I. Accuracy Assessment. Genome research. 1998;8, no. 3:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fabrick JA, Baker JE, Kanost MR. cDNA cloning, purification, properties, and function of a β-1,3-glucan recognition protein from a pyralid moth, Plodia interpunctella. Insect biochemistry and molecular biology. 2003;33, no. 6:579–594. doi: 10.1016/s0965-1748(03)00029-8. [DOI] [PubMed] [Google Scholar]
- Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Applied Bioinformatics. 2002;1, no. 2:107. [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular biology and evolution. 2010;27, no. 2:221. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Haghayeghi A, Sarac A, Czerniecki S, Grosshans J, Schöck F. Pellino enhances innate immunity in Drosophila. Mechanisms of development. 2010;127, no. 5-6:301. doi: 10.1016/j.mod.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Hillyer JF, Schmidt SL, Christensen BM. The antibacterial innate immune response by the mosquito Aedes aegypti is mediated by hemocytes and independent of Gram type and pathogenicity. Microbes and Infection. 2004;6, no. 5:448. doi: 10.1016/j.micinf.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JMC, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chatuverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin J, Lobo NL, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Reiger MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork PBPT, Venter CJ, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298, no. 5591:129. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Hostomska J, Volfová V, Mu J, Garfield M, Rohoušová I, Volf P, Valenzuela JG, Jochim R. Analysis of salivary transcripts and antigens of the sand fly Phlebotomus arabicus. BMC Genomics. 2009;10, no. 282 doi: 10.1186/1471-2164-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou R, Bao Z, Wang S, Su H, Li Y, Du X, Hu J, Wang S, Hu X. Transcriptome Sequencing and De Novo Analysis for Yesso Scallop (Patinopecten yessoensis) Using 454 GS FLX. PLoS ONE. 2011;6, no. 6:e21560. doi: 10.1371/journal.pone.0021560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA Sequence Assembly Program. Genome research. 1999;9, no. 9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochim R, Teixeira C, Laughinghouse A, Mu J, Oliveira F, Gomes R, Elnaiem D, Valenzuela J. The midgut transcriptome of Lutzomyia longipalpis: comparative analysis of cDNA libraries from sugar-fed, blood-fed, post-digested and Leishmania infantum chagasi-infected sand flies. BMC Genomics. 2008;9, no. 1:15. doi: 10.1186/1471-2164-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhawi S, Ramalho-Ortigao JM, Pham VM, Kumar S, Lawyer PG, Turco SJ, Barillas-Mury C, Sacks DL, Valenzuela JG. A role for insect galectins in parasite survival. Cell. 2004;119, no. 3:329. doi: 10.1016/j.cell.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Kang D, Liu G, Lundström A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proceedings of the National Academy of Sciences. 1998;95, no. 17:10078–10082. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C, Sumser E, Wernet MF, von Lintig J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proceedings of the National Academy of Sciences. 2002;99, no. 16:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick-Kendrick R. The biology and control of Phlebotomine sand flies. Clinics in dermatology. 1999;17, no. 3:279–289. doi: 10.1016/s0738-081x(99)00046-2. [DOI] [PubMed] [Google Scholar]
- Koga D, Sasaki Y, Uchiumi Y, Nobuya Hirai N, Arakane Y, Nagamatsu Y. Purification and characterization of Bombyx mori chitinases. Insect biochemistry and molecular biology. 1997;27, no. 8-9:757. doi: 10.1016/s0965-1748(97)00058-1. [DOI] [PubMed] [Google Scholar]
- Kulkarni GV, Deobagkar D. A Cytosolic form of Aminopeptidase P from Drosophila melanogaster Molecular Cloning and haracterization. The Journal of Biochemistry. 2002;131, no. 3:445. doi: 10.1093/oxfordjournals.jbchem.a003120. [DOI] [PubMed] [Google Scholar]
- Lehane MJ. Peritrophic matrix structure and function. Annual Review of Entomology. 1997;42, no. 1:525. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annual Review of Immunology. 2007;25:697. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lindlof A. Gene identification through large scale EST sequence processing. Applied Bioinformatics. 2003;2, no. 3:123. [PubMed] [Google Scholar]
- Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan Recognition Proteins. JBC: the Journal of Biological Chemistry. 2001;276, no. 37:34686. doi: 10.1074/jbc.M105566200. [DOI] [PubMed] [Google Scholar]
- Mark Blaxter, John Parkinson, Yoshihide Hayashizaki B, Franz Lang, Makedonka Mitreva, Gertraud Burger. Expressed Sequence Tags: An Overview. 2009:1. [Google Scholar]
- Marquardt WC, Kondratieff BC, Moore CG, Freier JE, Hagedorn HH, Black IV WC, James AA, Hemingway J, Higgs S. Biology of disease vectors. Second. Elsevier Academic Press; 2005. [Google Scholar]
- Martinez-Barnetche J, Gomez-Barreto R, Ovilla-Munoz M, Tellez-Sosa J, Garcia-Lopez D, Dinglasan R, Mohien C, MacCallum R, Redmond S, Gibbons J, Rokas A, Machado C, Cazarez-Raga F, Gonzales-Ceron L, Hernandez-Martinez S, Rodriguez-Lopez M. Transcriptome of the adult female malaria mosquito vector Anopheles albimanus. BMC genomics. 2012;13, no. 1:207. doi: 10.1186/1471-2164-13-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell MA, Collins FH, Ramalho Ortigao M, Valenzuela J, Shaden K, Dillon RJ, Bates P, Lehane M. Proposal for Sequencing the Genome of the Sand Flies, Lutzomyia longipalpis and Phlebotomus papatasi 2006 [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414, no. 6865:756. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Moita LF, Vriend G, Mahairaki V, Louis C, Kafatos FC. Integrins of Anopheles gambiae and a putative role of a new [beta] integrin, BINT2, in phagocytosis of E. coli. Insect biochemistry and molecular biology. 2006;36, no. 4:282. doi: 10.1016/j.ibmb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Muller HM, Crampton JM, della Torre A, Sinden R, Crisanti A. Members of a trypsin gene family in Anopheles gambiae are induced in the gut by blood meal. EMBO journal. 1993;12, no. 7:2891. doi: 10.1002/j.1460-2075.1993.tb05951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu Z, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NL, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, deBruyn B, DeCaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, LaButti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CFM, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O'Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers Y, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke S, Stinson EO, Tubio JMC, VanZee JV, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Verjovski-Almeida CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316, no. 5832:1718. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega FG, Wells MA. A molecular view of trypsin synthesis in the midgut of Aedes aegypti. Journal of insect physiology. 1999;45, no. 7:613. doi: 10.1016/s0022-1910(99)00052-9. [DOI] [PubMed] [Google Scholar]
- Novaes E, Drost D, Farmerie W, Pappas G, Grattapaglia D, Sederoff R, Kirst M. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC genomics. 2008;9, no. 1:312. doi: 10.1186/1471-2164-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai M, Ashida M. A Pattern-recognition Protein for β-1,3-Glucan. Journal of Biological Chemistry. 2000;275, no. 7:4995–5002. doi: 10.1074/jbc.275.7.4995. [DOI] [PubMed] [Google Scholar]
- Ochiai M, Ashida M. A Pattern Recognition Protein for Peptidoglycan. Journal of Biological Chemistry. 1999;274, no. 17:11854–11858. doi: 10.1074/jbc.274.17.11854. [DOI] [PubMed] [Google Scholar]
- Ochieng J, Leite-Browning ML, Warfield P. Regulation of Cellular Adhesion to Extracellular Matrix Proteins by Galectin-3* 1. Biochemical and biophysical research communications. 1998;246, no. 3:788. doi: 10.1006/bbrc.1998.8708. [DOI] [PubMed] [Google Scholar]
- O'Neil ST, Dzurisin JD, Carmichael RD, Lobo NF, Emrich SJ, Hellmann JJ. Population-level transcriptome sequencing of nonmodel organisms Erynnis propertius and Papilio zelicaon. BMC genomics. 2010;11, no. 1:310. doi: 10.1186/1471-2164-11-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Kwak I. Expression of Chironomus riparius serine-type endopeptidase gene under di-(2-ethylhexyl)-phthalate (DEHP) exposure. Comparative biochemistry and physiology B Comparative biochemistry. 2008;151, no. 3:349. doi: 10.1016/j.cbpb.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Peiser L, Mukhopadhyay S, Gordon S. Scavenger receptors in innate immunity. Current opinion in immunology. 2002;14, no. 1:123–128. doi: 10.1016/s0952-7915(01)00307-7. [DOI] [PubMed] [Google Scholar]
- Perillo NL, Uittenbogaart CH, Nguyen JT, Baum LG. Galectin-1, an endogenous lectin produced by thymic epithelial cells, induces apoptosis of human thymocytes. The Journal of experimental medicine. 1997;185, no. 10:1851. doi: 10.1084/jem.185.10.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierini LM. Uptake of serum-opsonized Francisella tularensis by macrophages can be mediated by class A scavenger receptors. Cellular microbiology. 2006;8, no. 8:1361–1370. doi: 10.1111/j.1462-5822.2006.00719.x. [DOI] [PubMed] [Google Scholar]
- Pitaluga AN, Beteille V, Lobo AR, Ortigao-Farias JR, Davila AMR, Souza AA, Ramalho-Ortigao JM, Traub-Cseko YM. EST sequencing of blood-fed and Leishmania-infected midgut of Lutzomyia longipalpis, the principal visceral leishmaniasis vector in the Americas. Molecular genetics and genomics. 2009;282, no. 3:307. doi: 10.1007/s00438-009-0466-2. [DOI] [PubMed] [Google Scholar]
- Pop M, Phillippy A, Delcher A, Salzberg SL. Comparative genome assembly. Briefings in bioinformatics. 2004;5, no. 3:237. doi: 10.1093/bib/5.3.237. [DOI] [PubMed] [Google Scholar]
- Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-Rich Repeat Protein Complex Activates Mosquito Complement in Defense Against Plasmodium Parasites. Science. 2009;324, no. 5924:258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Ortigao JM, Kamhawi S, Joshi MB, Reynoso D, Lawyer PG, Dwyer DM, Sacks DL, Valenzuela JG. Characterization of a blood activated chitinolytic system in the midgut of the sand fly vectors Lutzomyia longipalpis and Phlebotomus papatasi. Insect molecular biology. 2005;14, no. 6:703. doi: 10.1111/j.1365-2583.2005.00601.x. [DOI] [PubMed] [Google Scholar]
- Ramalho-Ortigao JM, Kamhawi S, Rowton ED, Ribeiro JMC, Valenzuela JG. Cloning and characterization of trypsin-and chymotrypsin-like proteases from the midgut of the sand fly vector Phlebotomus papatasi. Insect biochemistry and molecular biology. 2003;33, no. 2:163. doi: 10.1016/s0965-1748(02)00187-x. [DOI] [PubMed] [Google Scholar]
- Ramalho-Ortigao JM, Traub-Cseko YM. Molecular characterization of Llchit1, a midgut chitinase cDNA from the leishmaniasis vector Lutzomyia longipalpis. Insect biochemistry and molecular biology. 2003;33, no. 3:279. doi: 10.1016/s0965-1748(02)00209-6. [DOI] [PubMed] [Google Scholar]
- Ramalho-Ortigao M, Saraiva EM, Traub-Cseko YM. Sand Fly-Leishmania Interactions: Long Relationships are Not Necessarily Easy. The Open Parasitology Journal. 2010;4:195. doi: 10.2174/1874421401004010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Ortigao M, Jochim R, Anderson J, Lawyer PG, Pham VM, Kamhawi S, Valenzuela J. Exploring the midgut transcriptome of Phlebotomus papatasi:comparative analysis of expression profiles of sugar-fed, blood-fed and Leishmania major-infected sandflies. BMC Genomics. 2007;8, no. 300 doi: 10.1186/1471-2164-8-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramet M, Lanot R, Zachary D, Manfruelli P. JNK Signaling Pathway Is Required for Efficient Wound Healing in Drosophila. Developmental biology. 2002;241, no. 1:145–156. doi: 10.1006/dbio.2001.0502. [DOI] [PubMed] [Google Scholar]
- Ramet M, Pearson A, Manfruelli P, Li X, Koziel H, Göbel V, Chung E, Krieger M, Ezekowitz RAB. Drosophila Scavenger Receptor CI Is a Pattern Recognition Receptor for Bacteria. Immunity. 2001;15, no. 6:1027–1038. doi: 10.1016/s1074-7613(01)00249-7. [DOI] [PubMed] [Google Scholar]
- Lane Richard P, Crosskey Roger W. Medical insects and arachnids. Chapman & Hall; 1993. Chapter 1. General introduction; pp. 1–21. [Google Scholar]
- Richman AM, Dimopoulos G, Seeley D, Kafatos FC. Plasmodium activates the innate immune response of Anopheles gambiae mosquitoes. EMBO journal. 1997;16, no. 20:6114. doi: 10.1093/emboj/16.20.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant'Anna M, Diaz-Albiter H, Mubaraki M, Dillon RJ, Bates PA. Inhibition of trypsin expression in Lutzomyia longipalpis using RNAi enhances the survival of Leishmania. Parasites Vectors. 2009;2, no. 1:62. doi: 10.1186/1756-3305-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secundino NFC, Eger-Mangrich I, Braga EM, Santoro MM, Pimenta PFP. Lutzomyia longipalpis peritrophic matrix: formation, structure, and chemical composition. Journal of medical entomology. 2005;42, no. 6:928. doi: 10.1093/jmedent/42.6.928. [DOI] [PubMed] [Google Scholar]
- Shao L, Devenport M, Jacobs-Lorena M. The peritrophic matrix of hematophagous insects. Archives of Insect Biochemistry and Physiology. 2001;47, no. 2:119. doi: 10.1002/arch.1042. [DOI] [PubMed] [Google Scholar]
- Shen GM, Dou W, Niu J, Jiang H, Yang W, Jia F, Hu F, Cong L. Transcriptome Analysis of the Oriental Fruit Fly (Bactrocera dorsalis) PLoS ONE. 2011;6, no. 12:e29127. doi: 10.1371/journal.pone.0029127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Edwards MJ, Jacobs-Lorena M. A gut-specific serine protease from the malaria vector Anopheles gambiae is downregulated after blood ingestion. Insect molecular biology. 2000;9, no. 3:223–229. doi: 10.1046/j.1365-2583.2000.00188.x. [DOI] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Erlich RL, Hunter M, Bernstein E, Schneider D, Maniatis T. Immune Activation of NF-κB and JNK Requires Drosophila TAK1. Journal of Biological Chemistry. 2003;278, no. 49:48928–48934. doi: 10.1074/jbc.M304802200. [DOI] [PubMed] [Google Scholar]
- Soares MB, Bonaldo MdF, Hackett JD, Bhattacharya D. Expressed Sequence Tags: Normalization and Subtraction of cDNA Libraries. 2009 doi: 10.1007/978-1-60327-136-3_6. [DOI] [PubMed] [Google Scholar]
- Soderhall K, Cerenius L. Role of the prophenoloxidase-activating system in invertebrate immunity. Current opinion in immunology. 1998;10, no. 1:23. doi: 10.1016/s0952-7915(98)80026-5. [DOI] [PubMed] [Google Scholar]
- Tahar R, Boudin C, Thiery C, Bourgouin C. Immune response of Anopheles gambiae to the early sporogonic stages of the human malaria parasite Plasmodium falciparum. EMBO journal. 2002;21, no. 24:6673. doi: 10.1093/emboj/cdf664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28, no. 10:2731. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T, Sunagawa T, Yamaji K, Asaoka A, Mita M, Yamakawa M. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect biochemistry and molecular biology. 2008;38, no. 12:1087. doi: 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Tellam RL, Wijffels G, Willadsen P. Peritrophic matrix proteins. Insect biochemistry and molecular biology. 1999;29, no. 2:87. doi: 10.1016/s0965-1748(98)00123-4. [DOI] [PubMed] [Google Scholar]
- Telleria EL, de Araujo APO, Secundino NF, d'Avila-Levy CM, Traub-Cseko YM. Trypsin-Like Serine Proteases in Lutzomyia longipalpis–Expression, Activity and Possible Modulation by Leishmania infantum chagasi. PLoS ONE. 2010;5, no. 5:188. doi: 10.1371/journal.pone.0010697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theopold U, Samakovlis C, ErdjumentBromage H, Dillon N, Axelsson B, Schmidt O, Tempst P, Hultmark D. Helix pomatia lectin, an inducer of Drosophila immune response, binds to hemomucin, a novel surface mucin. JBC: the Journal of Biological Chemistry. 1996;271, no. 22:12708–12715. doi: 10.1074/jbc.271.22.12708. [DOI] [PubMed] [Google Scholar]
- Ursic Bedoya RJ, Mitzey AM, Obraztsova M, Lowenberger C. Molecular cloning and transcriptional activation of lysozyme-encoding cDNAs in the mosquito Aedes aegypti. Insect molecular biology. 2005;14, no. 1:89–94. doi: 10.1111/j.1365-2583.2004.00534.x. [DOI] [PubMed] [Google Scholar]
- Valanne S, Kleino A, Myllymäki H, Vuoristo J, Ramet M. Iap2 is required for a sustained response in the Drosophila Imd pathway. Developmental comparative immunology. 2007;31, no. 10:991. doi: 10.1016/j.dci.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Vera JC, Wheat C, Fescemyer H, Frilanderm M, Crawford D, Hanski i, Marden J. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Molecular ecology. 2008;17, no. 7:1636. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Vizioli J, Catteruccia F, della Torre A, Reckmann I, Muller HM. Blood digestion in the malaria mosquito Anopheles gambiae. European journal of biochemistry. 2001;268, no. 14:4027. doi: 10.1046/j.1432-1327.2001.02315.x. [DOI] [PubMed] [Google Scholar]
- Wang XW, Luan JB, Li J, Bao Y, Zhang C, Liu S. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC genomics. 2010;11, no. 1:400. doi: 10.1186/1471-2164-11-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fuchs JF, Infanger L, Rocheleau TA, Hillyer JF, Chen C, Christensen BM. Mosquito innate immunity: involvement of β 1,3-glucan recognition protein in melanotic encapsulation immune responses in Armigeres subalbatus. Molecular and biochemical parasitology. 2005;139, no. 1:65–73. doi: 10.1016/j.molbiopara.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Wasmuth JD, Blaxter ML. prot 4 EST: Translating Expressed Sequence Tags from neglected genomes. BMC bioinformatics. 2004;5, no. 1:187. doi: 10.1186/1471-2105-5-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DAM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25, no. 9:1189. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunological reviews. 1998;163, no. 1:19. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proceedings of the National Academy of Sciences. 2000;97, no. 25:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C, Hemingway J, Ranson H. Identification and analysis of single nucleotide polymorphisms (SNPs) in the mosquito Anopheles funestus, malaria vector. BMC genomics. 2007;8, no. 1:5. doi: 10.1186/1471-2164-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Davies DM. Trypsin and chymotrypsin during metamorphosis in Aedes aegypti and properties of the chymotrypsin. Journal of insect physiology. 1971;17, no. 1:117–131. doi: 10.1016/0022-1910(71)90048-5. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Ochiai M, Ashida M. β-1,3-glucan receptor and peptidoglycan receptor are present as separate entities within insect prophenoloxidase activating system. Biochemical and biophysical research communications. 1986;141, no. 3:1177–1184. doi: 10.1016/s0006-291x(86)80168-1. [DOI] [PubMed] [Google Scholar]
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS Journal. 2005;272, no. 24:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Arakane Y, Banerjee D, Beeman RW, Kramer KJ, Muthukrishnan S. Domain organization and phylogenetic analysis of the chitinase-like family of proteins in three species of insects. Insect biochemistry and molecular biology. 2008;38, no. 4:452. doi: 10.1016/j.ibmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Deng Y, Vanka P, Brown SJ, Muthukrishnan S, Kramer KJ. Computational identification of novel chitinase-like proteins in the Drosophila melanogaster genome. Bioinformatics. 2004;20, no. 2:161–169. doi: 10.1093/bioinformatics/bth020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The numbers of sequences were plotted as function of their length. The black line represents sequences obtained in the study using the assembly process detailed in Fig. 1 while the grey line represents sequences obtained by Dillon et al (2006) using the Phrap assembler.
The distribution of the subcategories for the GO Biological process main category were plotted as a function of the percent of sequences annotated for the same main GO category. The Lu. longipalpis dataset is represented by the grey bars while the Ph. papatasi is represented by the black bars.
The distribution of the subcategories for the GO Molecular Function main category were plotted as a function of the percent of sequences annotated for the same main GO category. The Lu. longipalpis dataset is represented by the grey bars while the Ph. papatasi is represented by the black bars.
The distribution of the subcategories for the GO Cellular Component main category were plotted as a function of the percent of sequences annotated for the same main GO category. The Lu. longipalpis dataset is represented by the grey bars while the Ph. papatasi is represented by the black bars.