Abstract
Mechanical stimuli can trigger intracellular calcium (Ca2+) responses in osteocytes and osteoblasts. Successful construction of bone cell networks necessitates more elaborate and systematic analysis for the spatiotemporal properties of Ca2+ signaling in the networks. In the present study, an unsupervised algorithm based on independent component analysis (ICA) was employed to extract the Ca2+ signals of bone cells in the network. We demonstrated that the ICA-based technology could yield higher signal fidelity than the manual region of interest (ROI) method. Second, the spatiotemporal properties of Ca2+ signaling in osteocyte-like MLO-Y4 and osteoblast-like MC3T3-E1 cell networks under laminar and steady fluid flow stimulation were systematically analyzed and compared. MLO-Y4 cells exhibited much more active Ca2+ transients than MC3T3-E1 cells, evidenced by more Ca2+ peaks, less time to the 1st peak and less time between the 1st and 2nd peaks. With respect to temporal properties, MLO-Y4 cells demonstrated higher spike rate and Ca2+ oscillating frequency. The spatial intercellular synchronous activities of Ca2+ signaling in MLO-Y4 cell networks were higher than those in MC3T3-E1 cell networks and also negatively correlated with the intercellular distance, revealing faster Ca2+ wave propagation in MLO-Y4 cell networks. Our findings show that the unsupervised ICA-based technique results in more sensitive and quantitative signal extraction than traditional ROI analysis, with the potential to be widely employed in Ca2+ signaling extraction in the cell networks. The present study also revealed a dramatic spatiotemporal difference in Ca2+ signaling for osteocytic and osteoblastic cell networks in processing the mechanical stimulus. The higher intracellular Ca2+ oscillatory behaviors and intercellular coordination of MLO-Y4 cells provided further evidences that osteocytes may behave as the major mechanical sensor in bone modeling and remodeling processes.
Keywords: Osteocytic and Osteoblastic Networks, Fluid Flow, Calcium Signaling, Independent Component Analysis, Spatiotemporal Properties
Introduction
Bone, acting as an adaptive system, can remodel its structure in response to the external mechanical environment [1]. Understanding the mechanism of bone remodeling is of critical importance in osteoporosis, age-related fractures and bone loss in prolonged space flight. Osteocytes, accounting for almost 95% of the bone cell population, are encapsulated in a fluid-filled mineralized matrix and interconnected with each other through long dendritic processes to form extensive networks in the lacunae-canaliculi system. It has been long conjectured that this elaborate osteocytic network behaves as the central mechanosensory system in bone adaptation and bone homeostasis [2]. Osteocytic networks can transduce physical stimuli into biochemical signals to further regulate the behaviors of osteoblasts and osteoclasts, thus coordinating bone modeling and remodeling processes [3]. Several studies have shown that osteoblasts themselves could respond to mechanical stimuli with a cascade of cellular activities, such as calcium (Ca2+) signaling, and release of nitric oxide (NO) and prostaglandin E2 (PGE2) [4–6]. Mechanical loading triggers Ca2+ release in both osteocytes and osteoblasts [7–11], which is a pivotal and ubiquitous second messenger regulating many downstream cellular activities, including cell proliferation, differentiation, and apoptosis [12]. Hence, a comprehensive understanding of Ca2+ signaling in osteocytes and osteoblasts under mechanical stimuli is of tremendous value in deciphering the mechanisms by which bone processes the biophysical stimulus.
Ca2+ signaling in osteocytes and osteoblasts can propagate between neighboring cells and generate Ca2+ waves in the bone cell populations [10, 13]. However, most studies in bone cell mechanotransduction have been performed on confluent or sub-confluent uncontrolled cell monolayers. In our previous studies, a two-dimensional patterned bone cell network was successfully constructed to mimic the elaborate in vivo bone cell network topology using microcontact printing and self-assembled monolayers (SAMs) techniques [14]. Our recent findings demonstrated that the osteocytic network showed repetitive spike-like Ca2+ peaks under fluid flow induced shear stress. These oscillations were dramatically different from those found in the osteoblastic network regardless of the magnitude of shear stress [15]. However, there are still two major obstacles in studying Ca2+ signaling in these cellular networks. First, bone cells patterned in the topologic network are spatially connected with their neighboring cells, so the time course of Ca2+ dynamics neglected the important spatial and temporal information embedded in the network responses. This information is critical to help provide essential insights into Ca2+ dynamics of individual cells and Ca2+ wave propagation in the cell network, which has attracted extensive attention in the signal analysis of neuronal cell types, such as astrocytes, glial cells and Purkinje cells [16–18]. Therefore, it necessitates more elaborate and systematical analysis of the spatiotemporal characteristics of Ca2+ signaling in bone cell networks. Second, most previous extraction methods for Ca2+ signaling in bone cells have been mainly based on a manual region of interest (ROI) analysis, which can be laborious and subjective, requiring users to select the target boundary manually according to the cell morphology. The manual ROI was subject to the constraints of image qualities, and the large number of cells in our bone cell network data also further increased the difficulty for manual extraction. Therefore, an unsupervised signal extraction technique is needed to reduce the workload and minimize the artificial errors.
Independent component analysis (ICA) is an unsupervised blind source separation procedure that transforms signal mixtures into a corresponding set of statistically independent source signals [19]. ICA has also been successfully applied for identifying and characterizing physiological signals in many research areas, such as electroencephalography (EEG), electrocardiography (ECG), magnetocardiography (MCG), and functional magnetic resonance imaging (fMRI) [20–23]. Isolating the individual Ca2+ signals in a bone cell network, sharing a number of similarities with separating the electrophysiological signals from the recorded mixtures, may also benefit from the unsupervised ICA technique. However, no study to date has employed this technique to extract the intracellular Ca2+ signaling of bone cells and systematically investigated the spatiotemporal properties of Ca2+ signaling in a cell network pattern.
In the present study, osteocyte-like MLO-Y4 and osteoblast-like MC3T3-E1 cell networks were respectively stimulated under physiological related fluid shear stress (0.5–4 Pa) and Ca2+ responses were extracted and analyzed using a set of novel unsupervised techniques. An ICA-based algorithm was used to separate the individual Ca2+ signals from the cell networks. Spike rate and power spectrum density (PSD) analysis were then employed to evaluate the temporal dynamics of Ca2+ signaling, and cell-cell spike synchronization and signal correlation were analyzed to reveal the spatial intercellular communications of Ca2+ signaling in the networks. This study represents the first effort to systematically study and compare the spatiotemporal characteristics of Ca2+ signaling in osteocytic and osteoblastic networks.
Materials and Methods
Bone Cell Network
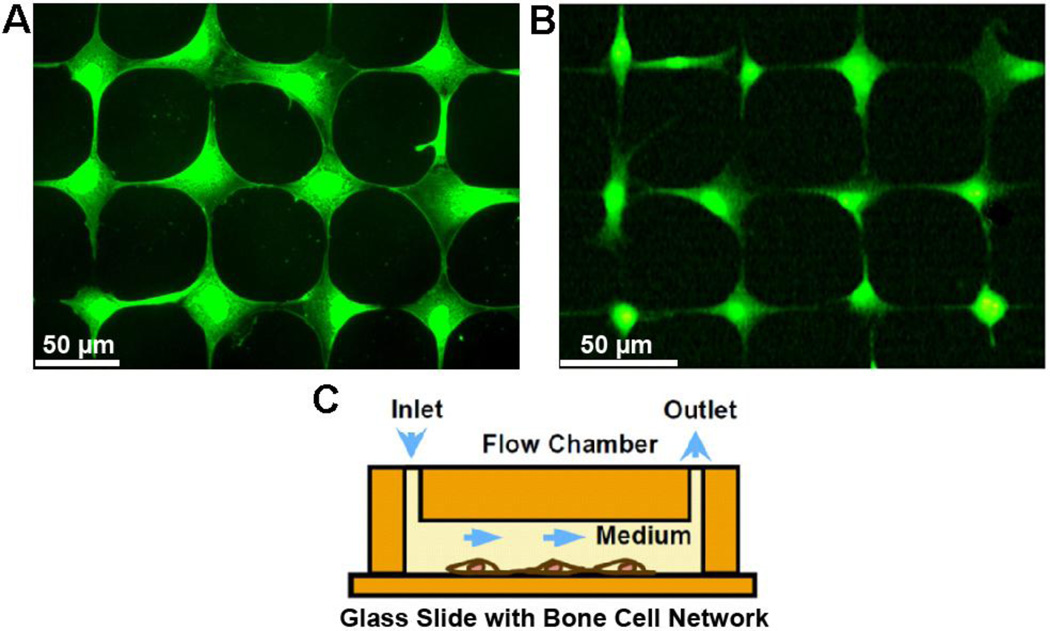
Osteocyte-like MLO-Y4 cells (a gift obtained from Lynda Bonewald, University of Missouri) were cultured on type I rat tail collagen (BD Biosciences, San Jose, CA, USA) coated Petri-dish in α-MEM containing 5% FBS, 5% CS and 1% P/S [24]. MC3T3-E1 cells were maintained in α-MEM supplemented with 10% FBS and 1% P/S. Microcontact printing and SAMs surface chemistry technologies were employed to construct the in vitro bone cell networks, which have been described in detail in our previous studies [14–15]. The diameter of the round island for a cell to reside was 20 µm for MC3T3-E1 cells and 15 µm for MLO-Y4 cells, while the edge-to-edge distances between neighboring islands were 50 µm and 35 µm respectively. These geometric parameters were optimized based on cell morphology to obtain well-formed networks. A grid cell pattern was printed on a chromium mask, and then the pattern was replicated to a master made of positive photoresist (Shipley 1818, MicroChem Corp, Newton, MA). Polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning, Midland, MI) stamps were made using the master. The stamp was coated with an adhesive SAM (octadecanethiol; Sigma–Aldrich Co., St. Louis, MO), and pressed onto a gold-coated glass slide for 1 min. Ethylene glycol terminated SAM solution (HS-C11-EG3; Prochimia, Sopot, Poland) was then added onto the slide for 3 h, and the slide was then incubated in a fibronectin solution for 1 h to facilitate cell adhesion. Cells were then seeded onto the slide and cultured in α-MEM medium supplemented with 2% charcoal-stripped fetal bovine serum (CSFBS, Hyclone Laboratories Inc., Logan, UT) for 24 h before fluid flow stimulation. CSFBS was used here to control the growth of cells and to avoid double-cell-occupancy on a single circular spot on the pattern [10, 15]. The fluorescence images of MC3T3-E1 and MLO-Y4 cell networks are shown in Fig. 1A&B.
Figure 1.
Fluorescent images of micro-patterned bone cell networks and flow chamber setup for fluid flow stimulation. Osteoblast-like MC3T3-E1 (A) and osteocyte-like MLO-Y4 (B) cell networks were established using microcontact printing and SAMs surface chemistry technologies. Cells were loaded with Ca2+ indicator Fura-2 AM. A laminar flow chamber (C) was employed to apply fluid flow on the cell surface.
Fluid Flow Stimulation and Ca2+ Imaging
The patterned cells were loaded with 10 µM Fura-2 AM (Molecular Probes, Eugene, OR) for 45 min. The glass slide was then mounted into a parallel plate flow chamber (Fig. 1C). The flow chamber was fixed on an inverted fluorescence microscope (Olympus IX71, Melville, NY) with a 10X objective. Steady, laminar fluid flow at different shear stress (0.5, 1, 2, and 4 Pa) on the cell surface was applied through the chamber by varying the power output of a magnetic gear pump connected to the flow chamber. α-MEM supplemented with 2% CSFBS was used as the flow medium for the experiment [15, 31]. The Ca2+ responses of cells were recorded for a 10-min period: 1 min for baseline and 9 min after the onset of fluid flow. MetaFluor Imaging Software 7.0 (Molecular Devices, Downingtown, PA) was used to acquire and record the Fura-2 340 nm/380 nm ratio images every 3 sec during the course of flow stimulation.
ICA-based Signal Extraction Algorithm
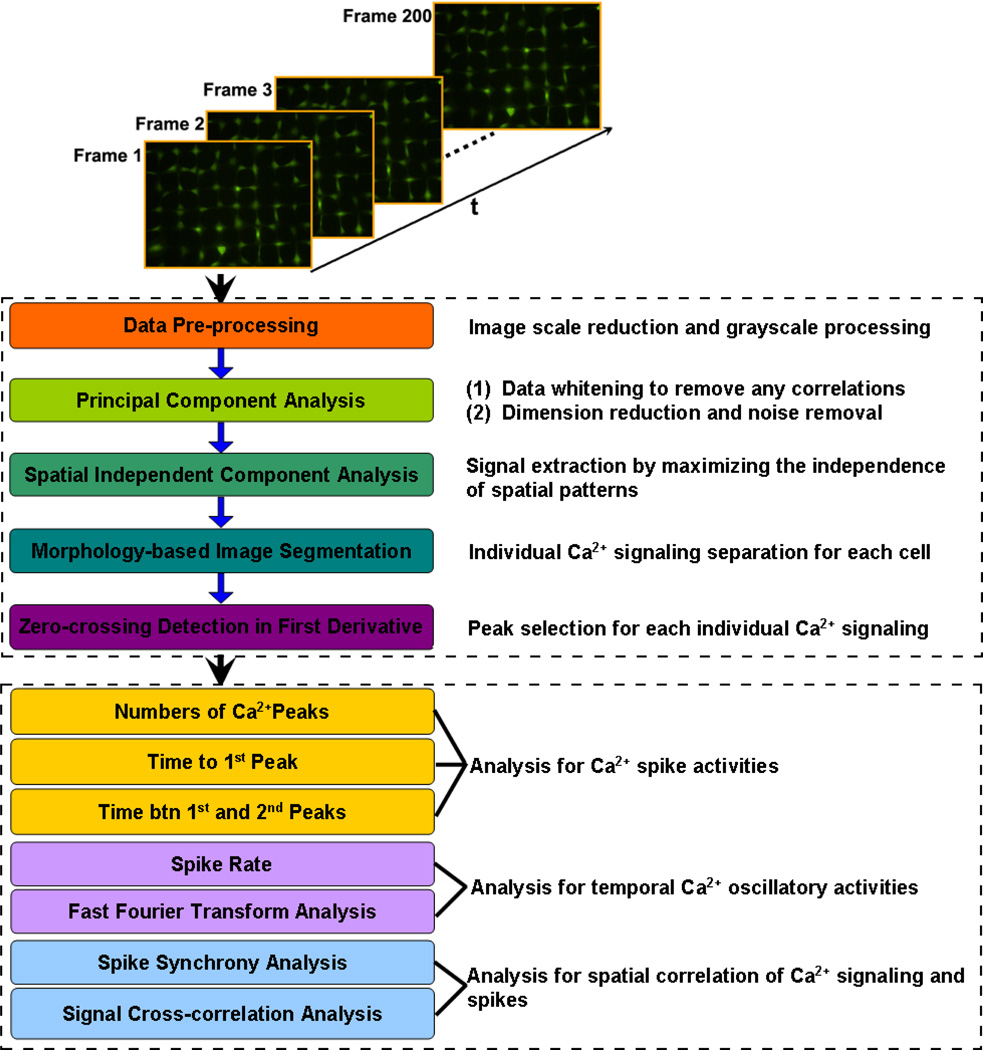
Flow chart for the unsupervised signal extraction algorithm and spatiotemporal analysis strategy for Ca2+ signaling in bone cell networks is shown in Fig. 2. All procedures were performed using Matlab (R2010, The MathWorks Inc., Natick, MA). First, the pseudocolor fluorescence time-lapse images were transformed into grayscale images and resized from 1,334×1,024 pixels to 336×256 pixels for a total of 200 frames with the bilinear interpolation algorithm to reduce the computation load. Second, the signal in each pixel was normalized by the mean value over all time frames at that pixel. Then, principal component analysis (PCA) was used to serve as a preprocessing procedure prior to ICA by whitening the original data matrix. The combination between the PCA and ICA algorithm can simplify the separation of the underlying independent source signals and also improve the robustness of ICA [25–27]. The major aim of PCA in the present study is to transform the original image sequence data X (Mxy × Nt) into a whitened matrix Xwhite, in which components are uncorrelated with each other and covariance matrix equals the identity matrix. As shown in Equation 1, the original image sequence data X was decomposed into a series of spatial patterns U (Mxy × R) and their corresponding time series V (Nt × R) using singular value decomposition.
| (1) |
R is the rank of X, R ≤ min (Mxy, Nt), while D is the R × R non-negative diagonal matrix of singular values in decreasing order. If R=Nt, the approximation will be exact. V and D can be obtained by computing XTX. V are the eigenvectors of XTX, and D2 are the eigenvalues of XTX. U can be computed as:
| (2) |
The smaller the eigenvalue, the less energy along the corresponding eigenvector there will be. Therefore, a small eigenvalue often corresponds to noise signals. In this study, the largest 40~60 singular values in matrix D were selected to ensure that the components encoding the random noise signals were excluded. After filtering the principal components, the whitened matrix Xwhite with reduced dimension was obtained:
| (3) |
Figure 2.
Flow chart for the unsupervised signal extraction algorithm and spatiotemporal analysis strategy for Ca2+ signaling in bone cell networks.
ICA applied in the present study was based on the whitened data Xwhite generated by PCA. Spatial ICA (sICA) was used to extract the signals to identify spatial components that maximized the independence of the spatial patterns, together with their corresponding time courses. Some well-developed ICA algorithms in the literature could be directly used [27–28]. We employed the FastICA algorithm, which is a fixed-point iteration algorithm by measuring the non-Gaussianity to seek the statistically independent sources from their mixtures [29]. The FastICA algorithm has been widely used in previous studies to separate physiological signals [18, 20].
After performing sICA, the spatial patterns between each pair of components are statistically independent. In practice, however, one single IC sometimes contains several spatially separated cell regions. ICA tends to sort these disconnected regions to be one single IC, since they may have strongly high correlations in the spatial patterns with each other, which can be hardly separated by ICA. Therefore, a morphology-based segmentation algorithm was used to split each IC into spatially separated sub-ICs to ensure that each sub-IC corresponds to a target cell region. The spatial patterns were first transformed to a binary matrix. Then, the binary image segmentation procedure was implemented using function ‘bwlabel’ in Matlab to distinguish the spatially disconnected regions [30]. After this procedure, any IC that contains multiple spatially separated cell regions can be split into the corresponding sub-ICs. Then, a threshold value of the minimal cell area (20 pixels in practice) was set to eliminate the ICs encoding the noise signals or background fluctuation. To identify the cell region occupied by multiple sub-ICs, the distance between the centroids of each two sub-IC regions was computed, and a threshold of minimal distance value (10 pixels in practice) was set to select the overlapped sub-ICs. The sub-IC with maximal area value among the overlapped sub-ICs was selected as the target spatial component. All these procedures ensured a one-to-one correspondence between each sub-IC and a cell region.
After signal extraction, a peak selection algorithm was developed to detect the peaks in each cellular signal. The signal was first normalized using the standardized z-score, and then the first derivative of curve was smoothed using the triangular smooth method. The downward zero-crossings in the smoothed first derivative were then detected. The points were selected as the peak points by setting the threshold values for the amplitude of the selected zero-crossings as:
| (4) |
where Stest is the value of the test point, Smin is the minimal value of the signal, Smax is the maximal value of the signal, and Pthreshold is the threshold value. In this study, Pthreshold was set to 1/4 [15].
Validation for the Signal Extraction Algorithm
Simulation movies were generated to mimic the typical cell shapes and Ca2+ signal curves in bone cell networks. The signals were artificially contaminated by generating Gaussian noise with different variances. The signal-to-noise ratio ranged from 10 to 30 dB, which covers the normal range of the experimental fluorescence images. The signal fidelity, defined as the Pearson’s correlation coefficient between the true signal and extracted signal, was compared between the unsupervised ICA method and manual ROI method.
Analysis for the Spatiotemporal Characteristics of Ca2+ Signaling
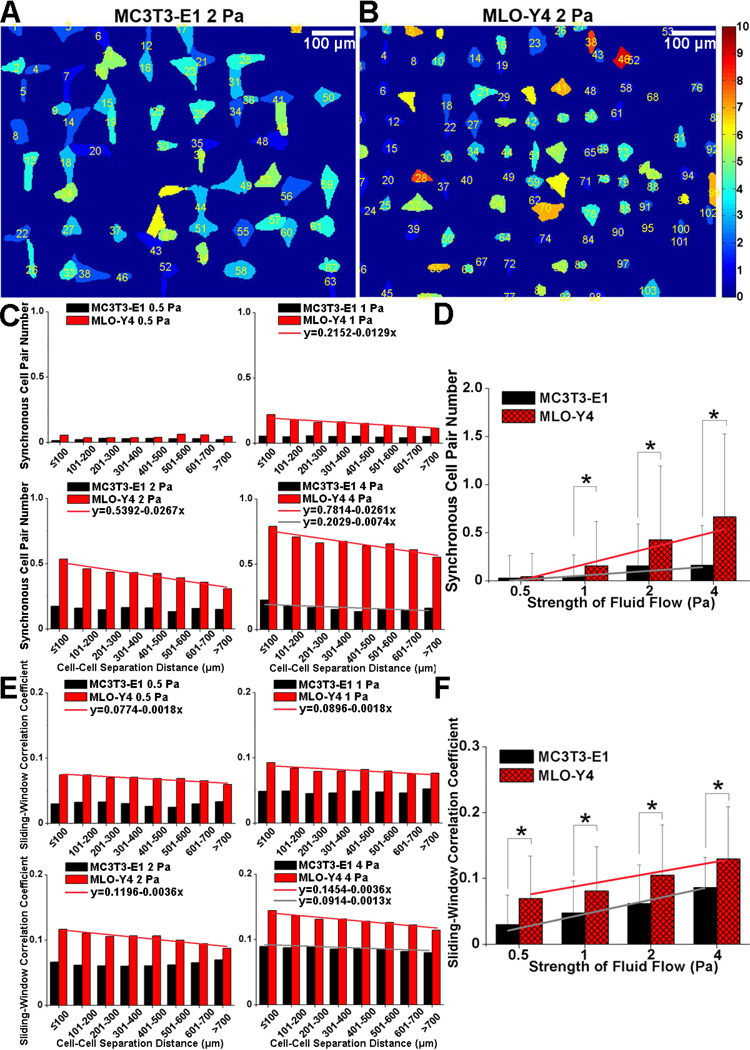
After signal extraction, information relating to spikes was automatically recorded, including the total responsive cell number, Ca2+ spike number in each cell, and time of spike occurrence in each cell. A color-code image with different color intensities corresponding to cells with different spike numbers was also generated automatically to facilitate describing the spatial distribution of Ca2+ signaling (Fig 6A&B). The average Ca2+ spike number, occurrence time of the 1st spike and time between the 1st and 2nd spikes were compared between MC3T3-E1 and MLO-Y4 cells in various magnitudes of shear stress (0.5, 1, 2 and 4 Pa) [15, 31]. Since the present ICA-based Ca2+ signaling extraction algorithm is only sensitive to the cells which exhibited Ca2+ response, the total cell number in each slide was manually counted to compare the responsive rate between MC3T3-E1 and MLO-Y4 cells. The spike rate was then systematically analyzed to elucidate the temporal distributions of spike activities of Ca2+ signals. In brief, a time bin (10 frames in practice) was set so that the time range was segmented into several sub-ranges with equal length (20 sub-ranges in practice). All the spikes of cells were mapped into the corresponding sub-range, and the spike number was determined in each sub-range. The spike rate in each sub-range was calculated as:
| (5) |
where Rspike is the spike rate, Nspike is the total spike number in each sub-range, Δ (t) is the bin size, and Ncell is the total cell number in each sub-range.
Figure 6.
Comparison of the spatiotemporal characteristics of spike synchrony and sliding-window cross correlation coefficients of Ca2+ signaling at 0.5, 1, 2 and 4 Pa shear stress for MC3T3-E1 and MLO-Y4 cell networks. After ICA-based signal extraction, a color-coded image with different color intensities corresponding to cells with different spike numbers was generated automatically to describe the spatial distribution of Ca2+ signaling. Fig. A&B show the typical color-code images at 2 Pa shear stress for MC3T3-E1 and MLO-Y4 cell networks. The spike synchrony (C&D) and signal correlation (E&F) were analyzed between each cell pair in the networks. Cell pairs were excluded from the analysis with intercellular distance lower than 60 µm for osteoblasts and 40 µm for osteocytes. Values are all expressed as mean ± SD. Trend line in C&E represents the statistically negative correlation between the analyzed parameter and cell-cell separation distance based on linear regression analysis. Trend line in D&F represents the statistically negative correlation between the analyzed parameter and fluid flow strength. * represents the significant difference between MC3T3-E1 and MLO-Y4 cell networks at a specific fluid flow strength with P<0.05.
The centroid of each cell was also automatically extracted in the signal extraction program for the spike synchrony and signal correlation analysis. All cells in the field of view from each movie were compared in a pair-wise manner. Since the center to center distance between neighboring round islands for cells to reside was 70 µm for osteoblasts and 50 µm for osteocytes, the cell pair was excluded for the spatial correlation analysis with intercellular distance lower than 60 µm for osteoblasts and 40 µm for osteocytes, given that the possible non-coincidence between the cell centroid and the island center. Because most cells’ 1st spikes occurred almost synchronously when fluid flow was applied, the spikes after the 1st main peak were analyzed to investigate the synchronization of multiple responses of Ca2+ signaling in bone cell networks. From our previous study, the 1st spike was resulted from Ca2+ entry into the cell while the subsequent spikes resulted from ATP-related signal propagation. Spikes between two cells were defined as synchronous ones if the time offset between the spike pair was less than 5 frames (15 sec). In this study, the sliding-window cross correlation analysis (window width: 60 sec; step size: 60 sec) was employed to evaluate the signal correlation. This method can provide enhanced localization analysis in the temporal domain by splitting the signals with the sliding window, and thus quantitate the Ca2+ signal correlation more precisely compared with the normal correlation analysis [32–34]. We selected the maximum value over the lag range from −5 to 5 frames (−15 to 15 sec) in each sub-range. The correlation value between two signals was calculated by averaging the values over all the sub-ranges.
To compare the periodicities and frequency distribution characteristics of Ca2+ signaling between MC3T3-E1 and MLO-Y4 cells, the normalized Ca2+ signals were analyzed using fast Fourier transform (FFT)-based power spectrum density (PSD) processing routines in Matlab. All the frequency curves of cellular signals in each group were averaged to identify the difference in frequency distribution between MC3T3-E1 and MLO-Y4 cells.
Statistical Analysis
All data were presented as the mean ± standard deviation (SD). Statistical analyses were performed using a SPSS version 10.0 for Microsoft Windows (SPSS, Chicago, IL, USA). At each fluid stimulation strength, differences of each parameter (the number of Ca2+ peaks, time to the 1st peak, time between the 1st and 2nd peak, synchronous cell pair number, and sliding-window correlation coefficient) between MC3T3-E1 and MLO-Y4 cells were examined using a Student t-test. To determine the correlation between a parameter and the stimulation strength, linear regression analysis was performed. Linear regression analysis was also used to evaluate the correlation of the spike synchrony and signal correlation with the cell-cell spatial separation distance. P<0.05 was considered statistically significant.
Results
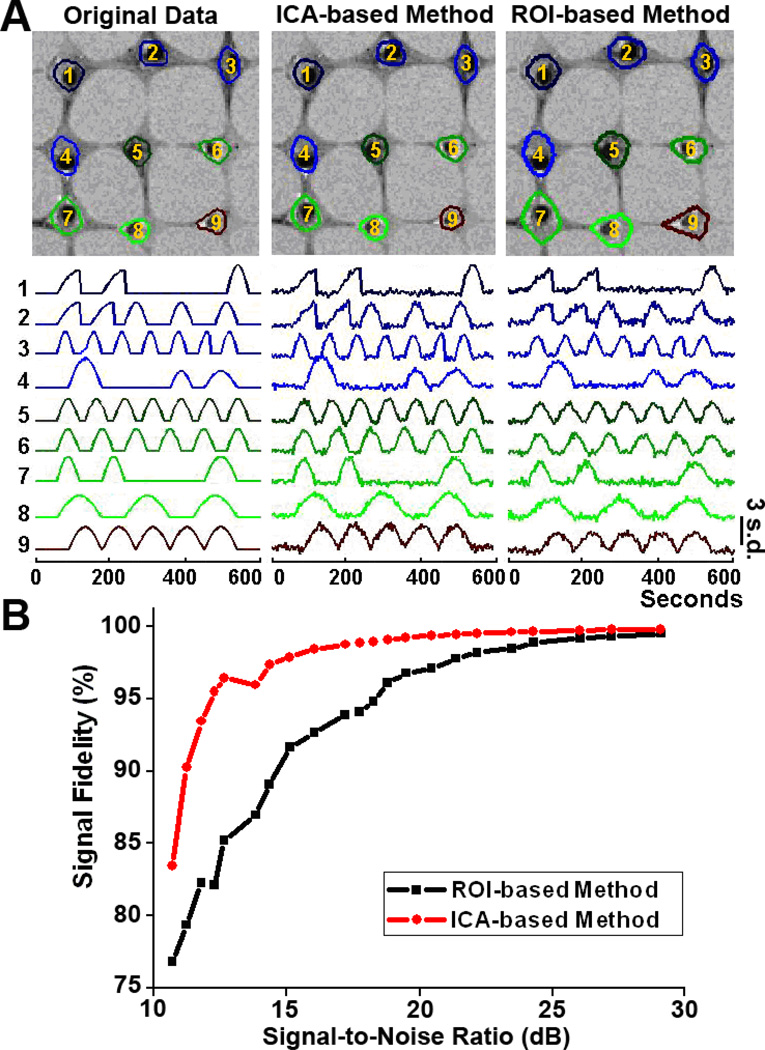
The spatial patterns and temporal signals of the original simulated movies are shown in Fig. 3A (Left). Gaussian noise was artificially introduced to the movies (SNR ranged from 10 to 30 dB). The unsupervised ICA-based technique was used to extract the signals in the noise-contaminated movies. To compare the extraction capacity between the ICA-based and ROI-based methods, signals in the movies were manually extracted by three analysts using the traditional ROI method. Fig. 3A (Middle & Right) show the results of spatial separation and signal extraction by ICA-based and ROI-based methods when the SNR in the movie was 20 dB. These results demonstrated that the unsupervised ICA-based method achieved better performance than the manual ROI method in both spatial target and temporal signal extraction at 20 dB SNR. Moreover, the signal fidelity extracted by the ICA-based method was obviously superior to that identified by the manual ROI method as shown in Fig. 3B for SNR ranging from 10 to 20 dB. As SNR increased from 20 dB to 30 dB, the signal fidelity by the ROI method gradually approached that by the ICA-based method.
Figure 3.
Validation of the ICA-based Ca2+ signal extraction technique. Simulated time-lapse images were created to mimic typical cell shapes and curves of Ca2+ signals in bone cell networks (A, Left). Signals were artificially contaminated by generating Gaussian noise with different variances to create movies with signal-to-noise ratio (SNR) ranging from 10 to 30 dB, which covered the normal SNR range of our experimental fluorescence time-lapse images. (A, Middle) and (A, Right) showed the signal extraction in the noise-contaminated movie with 20 dB SNR using unsupervised ICA-based technique and manual ROI method, respectively. The signal fidelity, defined as the Pearson’s correlation coefficient between true signal and extracted signal, was compared between ICA-based and ROI-based signal extraction methods (B).
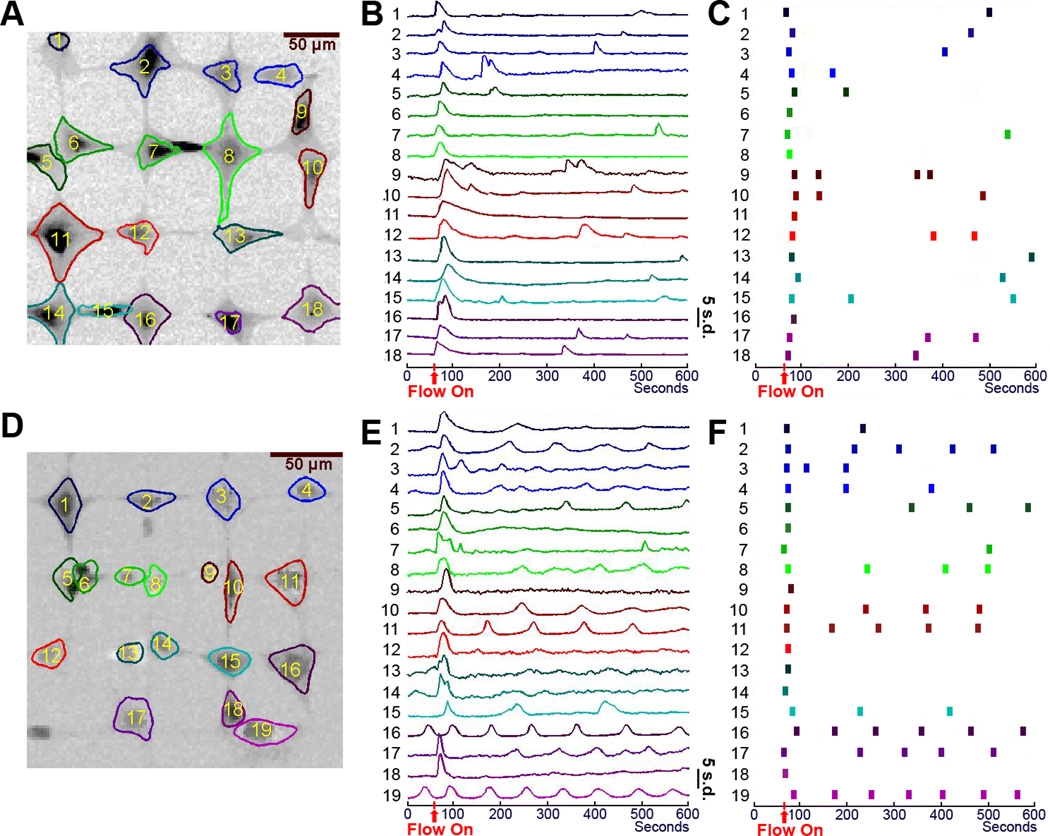
Signals extracted by the ICA-based technique from experimental data of typical MC3T3-E1 and MLO-Y4 cell networks under 2 Pa shear stress are shown in Fig. 4. Most MC3T3-E1 cells and MLO-Y4 cells released 1st spikes at the onset of flow. After the 1st main spike, most MC3T3-E1 cells exhibited a few lower spikes during the 9-min fluid flow stimulation period (Fig. 4B). MLO-Y4 cells showed significantly more frequent Ca2+ oscillatory activities than MC3T3-E1 cells, and some of them exhibited repetitive and robust multiple peak responses in addition to the 1st peak (Fig. 4E). Each individual dot in Fig. 4C&F represents an identified spike. These two plots clearly showed that MLO-Y4 cells exhibited many more Ca2+ peaks under fluid flow than MC3T3-E1 cells.
Figure 4.
Signal extraction based on the ICA-based technique for the experimental data of typical MC3T3-E1 and MLO-Y4 cell networks under 2 Pa shear stress. The spatial patterns and temporal signals from MC3T3-E1 (A&B) and MLO-Y4 (D&E) cell networks are shown. Each spike in one cellular signal is depicted as a dot (C for MC3T3-E1 cells and F for MLO-Y4 cells).
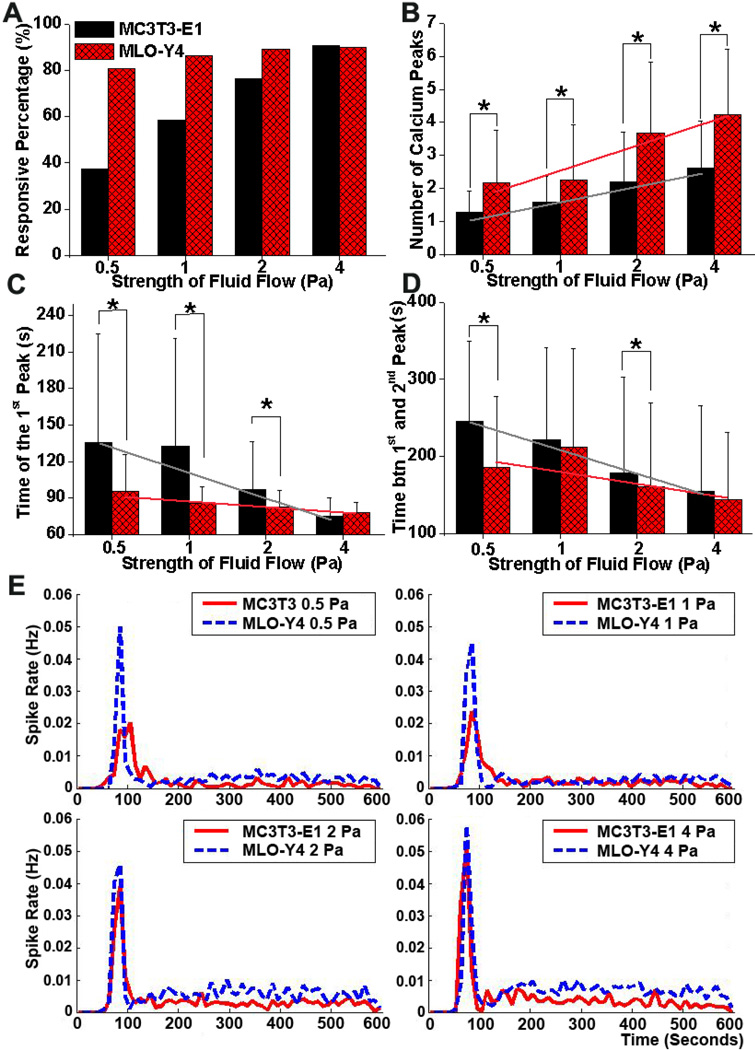
With an increase in fluid shear strength, the average number of Ca2+ peaks increased in both cell types. MLO-Y4 cells exhibited significantly more Ca2+ peaks than MC3T3-E1 cells at all levels of shear stress (P<0.01, Fig. 5B). The percentage of responsive MC3T3-E1 cells increased with the shear stress levels, and the responsive percentage in MLO-Y4 cells was higher than that in MC3T3-E1 cells at 0.5, 1 and 2 Pa shear stress (Fig. 5A). MLO-Y4 cells also took significantly less time than MC3T3-E1 cells to release the 1st spike in response to the fluid flow under 0.5, 1 and 2 Pa shear stress, and recovered faster from previous response to release the next Ca2+ peak under 0.5 and 2 Pa shear stress (Fig. 5C&D). Linear regression indicated that Ca2+ peak number was positively correlated, and the time to the 1st peak and time between the 1st and 2nd peaks were negatively correlated for both cell types with fluid flow levels. These results are consistent with our previous findings using the manual extraction method [15]. As shown in Fig. 5E, the spike rate of both MC3T3-E1 and MLO-Y4 cells reached a peak at the onset of flow. MLO-Y4 cells also showed relatively higher spike rate in the whole flow experimental period under all loading levels, which further suggested that MLO-Y4 cells had more frequent spike activities and more obvious features of multiple spikes than MC3T3-E1 cells (Fig. 5E).
Figure 5.
Comparison of responsive percentage (A), number of Ca2+ peaks excluding non-responsive cells (B), time to the 1st peak (C), time between the 1st and 2nd peaks (D), and spike rate (E) at 0.5, 1, 2 and 4 Pa shear stress for MC3T3-E1 and MLO-Y4 cell networks. After ICA-based signal extraction, information relating to spikes was automatically recorded, including the total responsive cell number, spike number of each cell, and spike times of each cell. The total cell number in each slide was manually counted. Statistical analysis was then applied and values are all expressed as mean ± SD. Trend line represents the statistically negative correlation between the analyzed parameter and fluid flow strength based on linear regression analysis. * represents the significant difference between MC3T3-E1 and MLO-Y4 cell networks at a specific fluid flow strength with P<0.05.
As shown in Fig. 6C&E, MLO-Y4 cell networks had significantly higher spike synchrony and signal correlation than MC3T3-E1 cell networks in all sub-groups with different cell-cell separation distance at all fluid flow levels. Linear regression showed that the spike synchrony and signal correlation decreased with the increase of cell-cell separation distance at 1, 2 and 4 Pa shear stress for MLO-Y4 cell networks, indicating that cells with shorter separation distance exhibited higher cross-talk than those with farther separation distance. However, the cross-talk between cells in MC3T3-E1 cell networks only exhibited minor negative correlation with the cell-cell separation distance in the 4 Pa shear stress group. Fig. 6D&F revealed significantly higher spike synchrony in MLO-Y4 cell networks under 1, 2 and 4 Pa, and also higher signal correlation in MLO-Y4 cell networks under all fluid flow levels (P<0.01). Linear regression further showed an increasing trend for both spike synchrony and signal correlation in the two types of cells with the increase of the fluid flow levels.
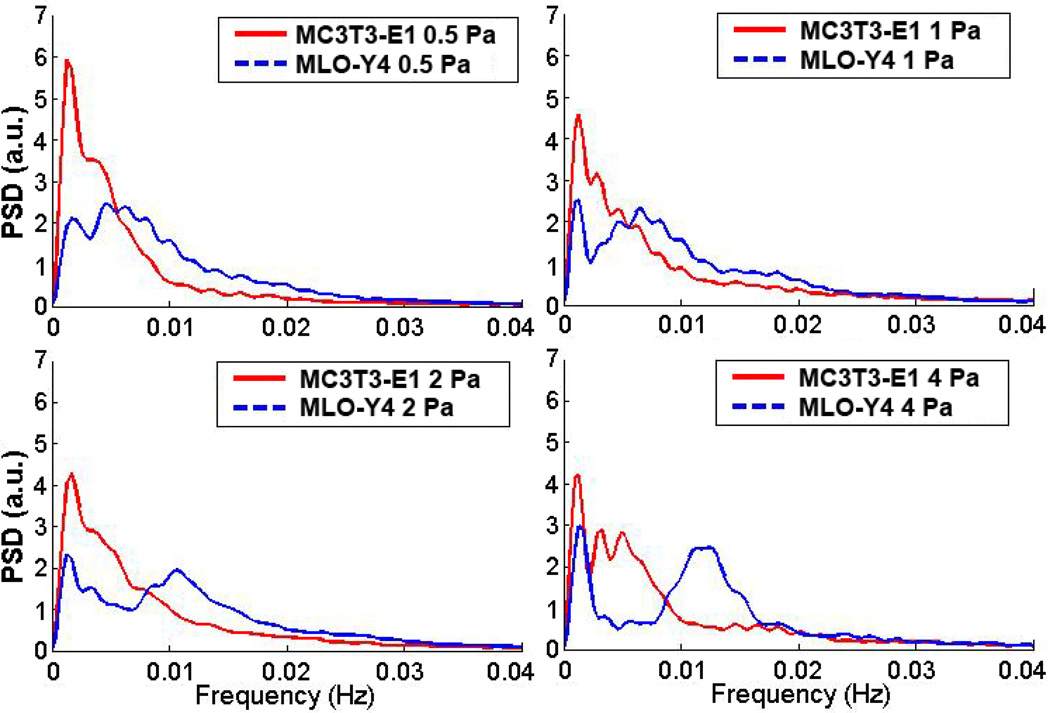
In the frequency spectral analysis (Fig. 7), both MC3T3-E1 and MLO-Y4 cells had a peak in the lower frequency band (0.001~0.002 Hz) at all loading levels, and the energy distribution of MC3T3-E1 cells was much more prominent than MLO-Y4 cells in this frequency range. However, MLO-Y4 cells exhibited striking 2nd peak in the higher frequency band and the frequency corresponding to the peak was increased with the fluid flow rate (0.0047 Hz at 0.5 Pa, 0.0068 Hz at 1 Pa, 0.0106 Hz at 2 Pa, and 0.0124Hz at 4 Pa). This indicated that the Ca2+ oscillations in MLO-Y4 cells were more dramatic than MC3T3-E1 cells, and also positively correlated with the stimulation strength.
Figure 7.
Comparison of frequency domain characteristics using fast Fourier transform (FFT)-based power spectrum density (PSD) analysis at 0.5, 1, 2 and 4 Pa shear stress for MC3T3-E1 and MLO-Y4 cell networks.
Discussion
In the present study on Ca2+ signaling in bone cell networks, a signal processing technique allowing for a cell’s spatial target separation, temporal signal extraction and spike detection were implemented based on PCA followed by sICA algorithm. The PCA followed by ICA algorithm has been extensively applied in ECG, MCG and fMRI studies for separating target signals from the background, which can increase the robustness and simplify the implementation complexity than the single ICA method [21–22, 26–27]. Previous optical imaging studies in neuroscience fields have used a combined PCA-ICA method to identify the distinct functional region and to explore the spatiotemporal relationship between vessels and surrounding tissue in the cerebral cortex [35–36]. Mukamel et al. employed the PCA followed by spatiotemporal ICA method to identify Purkinje cells or Bergmann glial signals in two-photon images [18]. In our bone cell fluorescence imaging data, one IC sometimes contained multiple spatially separated target regions after sICA. ICA tends to sort these disconnected regions to be one IC, since they may have strongly high correlations with each other. Therefore, morphology-based image processing techniques were necessary to further segment each IC into spatially separated sub-ICs and ensure the one-to-one correspondence between each sub-IC and cellular signal. Validation studies proved that the ICA-based technique was more accurate than the manual ROI method for our image data captured with a fluorescence microscope, which had high image quality with the SNR ranging from 10 to 30 dB. Moreover, the large cell number in our bone cell network data (100~200 cells for each set of data) also greatly increased the difficulty and artificial errors during manual ROI selection. In our studies of Ca2+ signaling in bone cell networks, it took an experienced analyst 1~2 hours to analyze one set of data using commercial analysis software, including manually selecting the cell regions, extracting the signals, and identifying the spikes of each signals. The current ICA-based signal extraction technique greatly reduced the workload and also facilitated the analysis of Ca2+ signal spatiotemporal characteristics. Therefore, this study suggests that the unsupervised ICA-based algorithm has great potential in analyzing Ca2+ signals of bone cell networks.
Under fluid flow stimulation with the same levels, more frequent Ca2+ spike activities and vigorous temporal Ca2+ oscillations were demonstrated in MLO-Y4 cell networks. The osteocyte-like MLO-Y4 cells, exhibiting unique repetitive Ca2+ firing activities stimulated by fluid flow, act similar with the action potentials of a neural network. This further supported our previous hypothesis that the osteocyte network holds the key to cellular memory of mechanical loading history in bone [14–15, 37]. The prominent difference in temporal Ca2+ oscillatory activities implied different mechanisms of Ca2+ responses between the two types of cells. Previous studies indicated that the [Ca2+]i elevation under fluid flow stimulation was mainly from two sources: extracellular calcium from the environment and Ca2+ stores (e.g. endoplasmic reticulum, ER) [38–39]. The influx of extracellular calcium was the major source for the rapid initial Ca2+ spike, and the ER Ca2+ stores and ATP pathways played a critical role in the subsequent multiple Ca2+ peaks in osteoblasts [15, 31]. T-type voltage gated calcium channels (VGCC), which regulate the pacemaking and repetitive firing activities in cardiac and neuronal cells [40–41], have also proven to be specifically expressed in osteocytes, but not in osteoblasts [42]. Therefore, the mechanism mediating more dramatic temporal oscillatory activities in osteocytic networks might be attributed to multiple channels and their relevant downstream signal pathways, including ATP-gated P2R ion channels and T-type VGCC, etc. The exact cellular and molecular mechanisms are being studied by our group.
In the present study, MLO-Y4 cell networks possessed more robust intercellular spike synchronization and signal correlation than MC3T3-E1 cells under all fluid flow levels. The synchronous activities in MLO-Y4 cells also negatively correlated with the cell-cell separation distance, but tended to be flat in MC3T3-E1 cells. These observations share similarities with many previous in vivo Ca2+ dynamics studies on cells of nervous system. Hirase et al. found the coordination of Ca2+ events were low and similar between nearby and distant astrocytes in the cerebral cortex in the absence of provoking conditions. After bicuculline treatment, a significantly negative correlation of synchronization with the cell-cell distance was found, which revealed more obvious Ca2+ wave propagation between cells in the astrocytic networks [16]. Studies also found that the Purkinje cells in cerebellar microzones also exhibited faster Ca2+ wave propagation when mice arose from rest to locomotion, evidenced by the increase of synchronous activities and negative correlation of cell crosstalk with the cell-cell distance [18]. Therefore, the results in our present study, which were similar with these findings in neural cells, indicated that the Ca2+ wave propagation might be much more obvious and faster in MLO-Y4 cell networks than that in MC3T3-E1 cell networks. Our previous study demonstrated that ATP played a dominant role in mediating the intercellular Ca2+ wave propagation from the single nanoindented bone cell to its neighboring cells over gap junctions in the network pattern [10], and inhibition for the ATP-related signal pathway can also reduce Ca2+ oscillation to one single spike in both osteoblasts and osteocytes under fluid flow [15, 31]. Therefore, a possible faster extracellular ATP diffusion between neighboring cells in osteocytic network may act as a critical factor to dominate the dramatic difference of spatial intercellular coordination of Ca2+ events between these two kinds of cells. Extracellular ATP can activate the membrane-bound P2 nucleotide receptors, leading to sequential activation of phospholipase C (PLC), generation of IP3, and thus triggering the release from the IP3-sensitive ER calcium stores [43–44]. Our previous studies have shown that depletion of the ER calcium stores could severely hamper both the intracellular calcium response and intercellular calcium wave propagation in bone cells [10, 15]. The exact cellular and molecular mechanisms underlying the spatiotemporal differences in Ca2+ signaling between osteocyte and osteoblasts are still unknown. We believe that the governing mechanisms depend on unique fast kinetics of membrane Ca2+ channels and the refilling of the ER, as well as faster extracellular ATP diffusion between neighboring cells in osteocytes to generate much more frequent and synchronous Ca2+ events, and also promote many more nearby cells to generate intracellular Ca2+ spikes more synchronously than the distant cells. Ongoing studies on bioluminescence trace for extracellular ATP, and also simultaneous visualization for the cytoplasmic and ER Ca2+ dynamics using FRET-based approach by our group will help further understand the exact cellular and molecular mechanisms.
Conclusion
The present study clearly demonstrated that the ICA-based signal extraction technique exhibited more accurate signal extraction capacity than traditional ROI analysis for Ca2+ signaling in bone cell networks. This unsupervised technique can dramatically reduce our effort to extract the Ca2+ signals and also facilitate more elaborate spatiotemporal analysis for Ca2+ signaling in bone cell networks. Our results showed that MLO-Y4 cell networks exhibited more frequent temporal Ca2+ oscillatory activities than MC3T3-E1 cell networks. The spatial intercellular synchronous activities of Ca2+ events in MLO-Y4 cell networks were higher than MC3T3-E1 cell networks and also negatively correlated with the intercellular distance, revealing much more obvious and faster Ca2+ wave propagation in MLO-Y4 cell networks. The findings highlight that cells in osteocytic networks, possessing more sensitive and interactive spatiotemporal characteristics in detecting and processing mechanical signals than those in osteoblastic networks, are qualified as the major mechanical sensor in bone.
Table 1.
Number of cell networks (slides) and analyzed cells (responsive cells) in different experimental groups.
| Shear Stress (Pa) | 0.5 | 1 | 2 | 4 | |
|---|---|---|---|---|---|
| MLO-Y4 | Slides | 3 | 5 | 4 | 5 |
| Responsive Cells | 397 | 625 | 446 | 509 | |
| MC3T3-E1 | Slides | 7 | 6 | 6 | 4 |
| Responsive Cells | 219 | 324 | 459 | 290 | |
Highlights.
-
➢
The unsupervised technique based on independent component analysis exhibits more accurate signal extraction than the region of interest method
-
➢
The independent component analysis-based technique significantly reduces our workload and facilitates more elaborate spatiotemporal analysis of calcium signaling
-
➢
Osteocytic networks exhibit higher spike rate and temporal oscillating frequency of calcium signaling than osteoblastic networks under fluid flow
-
➢
Osteocytic networks demonstrate higher spatial intercellular synchronous activities of calcium signaling than osteoblastic networks
-
➢
The spatial synchronous activities are negatively correlated with the intercellular distance in osteocytic networks, but not in ostoblastic networks
Acknowledgments
This work was supported by NIH grant R21 AR052417, R01 AR052461, and RC1 AR058453 (XEG). The first author was thankful to the China Scholarship Council (CSC) for its partial financial supports. The authors also sincerely thank Genevieve Brown for her help in drafting the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
All authors state that they have no conflicts of interest.
References
- 1.Duncan RL, Turner CH. Mechanotransduction and the functional response of bone to mechanical strain. Calcif Tissue Int. 1995;57:344–358. doi: 10.1007/BF00302070. [DOI] [PubMed] [Google Scholar]
- 2.Tatsumi S, Ishii K, Amizuka N, Li M, Kobayashi T, Kohno K, Ito M, Takeshita S, Ikeda K. Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab. 2007;5:464–475. doi: 10.1016/j.cmet.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Bonewald LF. The Amazing Osteocyte. J Bone Miner Res. 2011;26:229–238. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang GL, White CR, Stevens HY, Frangos JA. Temporal gradients in shear stimulate osteoblastic proliferation via ERK1/2 and retinoblastoma protein. Am J Physiol Endocrinol Metab. 2002;283:E383–389. doi: 10.1152/ajpendo.00547.2001. [DOI] [PubMed] [Google Scholar]
- 5.Reich KM, Frangos JA. Effect of flow on prostaglandin E2 and inositol trisphosphate levels in osteoblasts. Am J Physiol Cell Physiol. 1991;261:C428–C432. doi: 10.1152/ajpcell.1991.261.3.C428. [DOI] [PubMed] [Google Scholar]
- 6.Bakker AD, Soejima K, Klein-Nulend J, Burger EH. The production of nitric oxide and prostaglandin E(2) by primary bone cells is shear stress dependent. J Biomech. 2001;34:671–677. doi: 10.1016/s0021-9290(00)00231-1. [DOI] [PubMed] [Google Scholar]
- 7.Adachi T, Aonuma Y, Ito S, Tanaka M, Hojo M, Takano-Yamamoto T, Kamioka H. Osteocyte calcium signaling response to bone matrix deformation. J Biomech. 2009;42:2507–2512. doi: 10.1016/j.jbiomech.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Charras GT, Horton MA. Single cell mechanotransduction and its modulation analyzed by atomic force microscope indentation. Biophys J. 2002;82:2970–2981. doi: 10.1016/S0006-3495(02)75638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs CR, Yellowley CE, Davis BR, Zhou Z, Cimbala JM, Donahue HJ. Differential effect of steady versus oscillating flow on bone cells. J Biomech. 1998;31:969–976. doi: 10.1016/s0021-9290(98)00114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huo B, Lu XL, Costa KD, Xu Q, Guo XE. An ATP-dependent mechanism mediates intercellular calcium signaling in bone cell network under single cell nanoindentation. Cell Calcium. 2010;47:234–241. doi: 10.1016/j.ceca.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung CT, Pollack SR, Reilly TM, Brighton CT. Real-time calcium response of cultured bone cells to fluid flow. Clin Orthop Relat Res. 1995;313:256–269. [PubMed] [Google Scholar]
- 12.Papachristou DJ, Papachroni KK, Basdra EK, Papavassiliou AG. Signaling networks and transcription factors regulating mechanotransduction in bone. Bioessays. 2009;31:794–804. doi: 10.1002/bies.200800223. [DOI] [PubMed] [Google Scholar]
- 13.Adachi T, Aonuma Y, Taira K, Hojo M, Kamioka H. Asymmetric intercellular communication between bone cells: propagation of the calcium signaling. Biochem Biophys Res Commun. 2009;389:495–500. doi: 10.1016/j.bbrc.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Guo XE, Takai E, Jiang X, Xu Q, Whitesides GM, Yardley JT, Hung CT, Chow EM, Hantschel T, Costa KD. Intracellular calcium waves in bone cell networks under single cell nanoindentation. Mol Cell Biomech. 2006;3:95–107. [PubMed] [Google Scholar]
- 15.Lu XL, Huo B, Chiang V, Guo XE. Osteocytic network is more responsive in calcium signaling than osteoblastic network under fluid flow. J Bone Miner Res. 2012;27:563–574. doi: 10.1002/jbmr.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirase H, Qian L, Barthó P, Buzsáki G. Calcium dynamics of cortical astrocytic networks in vivo. PLoS Biol. 2004;2:E96. doi: 10.1371/journal.pbio.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agulhon C, Platel JC, Kolomiets B, Forster V, Picaud S, Brocard J, Faure P, Brulet P. Bioluminescent imaging of Ca2+ activity reveals spatiotemporal dynamics in glial networks of dark-adapted mouse retina. J Physiol. 2007;583:945–958. doi: 10.1113/jphysiol.2007.135715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 2009;63:747–760. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung TP, Makeig S, Lee TW, McKeown MJ, Brown G, Bell AJ, Sejnowski TJ. Independent component analysis of biomedical signals, 2nd International workshop on independent component analysis and signal separation. Helsinki. 2000:633–644. [Google Scholar]
- 20.Vigário R, Särelä J, Jousmäki V, Hämäläinen M, Oja E. Independent component approach to the analysis of EEG and MEG recordings. IEEE Trans Biomed Eng. 2000;47:589–593. doi: 10.1109/10.841330. [DOI] [PubMed] [Google Scholar]
- 21.Zarzoso V, Nandi AK, Bacharakis E. Maternal and foetal ECG separation using blind source separation methods. IMA J Math Appl Med Biol. 1997;14:207–225. [PubMed] [Google Scholar]
- 22.DiPietroPaolo D, Müller HP, Nolte G, Erné SN. Noise reduction in magnetocardiography by singular value decomposition and independent component analysis. Med Biol Eng Comput. 2006;44:489–499. doi: 10.1007/s11517-006-0055-z. [DOI] [PubMed] [Google Scholar]
- 23.McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ. Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. Establishment of an osteocyte-like cell line, MLO-Y4. J Bone Miner Res. 1997;12:2014–2023. doi: 10.1359/jbmr.1997.12.12.2014. [DOI] [PubMed] [Google Scholar]
- 25.Bai P, Shen HP, Huang XM, Truong Y. A supervised singular value decomposition for independent component analysis of fMRI. Stat Sinica. 2008;18:1233–1252. [Google Scholar]
- 26.Stone JV, Porrill J, Porter NR, Wilkinson ID. Spatiotemporal independent component analysis of event-related fMRI data using skewed probability density functions. Neuroimage. 2002;15:407–421. doi: 10.1006/nimg.2001.0986. [DOI] [PubMed] [Google Scholar]
- 27.Hyvarinen A, Oja E. Independent component analysis: algorithms and applications. Neural Netw. 2000;13:411–430. doi: 10.1016/s0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 28.Roberts S, Everson R. Independent component analysis: principles and practice. Cambridge University Press; Cambridge: 2001. [Google Scholar]
- 29.Hyvarinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 1999;10:626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- 30.Boland MV, Murphy RF. A neural network classifier capable of recognizing the patterns of all major subcellular structures in fluorescence microscope images of HeLa cells. Bioinformatics. 2001;17:1213–1223. doi: 10.1093/bioinformatics/17.12.1213. [DOI] [PubMed] [Google Scholar]
- 31.Huo B, Lu XL, Hung CT, Costa KD, Xu Q, Whitesides GM, Guo XE. Fluid flow induced calcium response in bone cell network. Cell Mol Bioeng. 2008;1:58–66. doi: 10.1007/s12195-008-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurent G, Wehr M, Davidowitz H. Temporal representations of odors in an olfactory network. J Neurosci. 1996;16:3837–3847. doi: 10.1523/JNEUROSCI.16-12-03837.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castelo-Branco M, Neuenschwander S, Singer W. Synchronization of visual responses between the cortex, lateral geniculate nucleus, and retina in the anesthetized cat. J Neurosci. 1998;18:6395–6410. doi: 10.1523/JNEUROSCI.18-16-06395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beierlein M, Gibson JR, Connors BW. A network of electrically coupled interneurons drives synchronized inhibition in neocortex. Nat Neurosci. 2000;3:904–910. doi: 10.1038/78809. [DOI] [PubMed] [Google Scholar]
- 35.Reidl J, Starke J, Omer DB, Grinvald A, Spors H. Independent component analysis of high-resolution imaging data identifies distinct functional domains. Neuroimage. 2007;34:94–108. doi: 10.1016/j.neuroimage.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 36.Siegel RM, Duann JR, Jung TP, Sejnowski T. Spatiotemporal dynamics of the functional architecture for gain fields in inferior parietal lobule of behaving monkey. Cereb Cortex. 2007;17:378–390. doi: 10.1093/cercor/bhj155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner CH, Robling AG, Duncan RL, Burr DB. Do bone cells behave like a neuronal network? Calcif Tissue Int. 2002;70:435–442. doi: 10.1007/s00223-001-1024-z. [DOI] [PubMed] [Google Scholar]
- 38.Hung CT, Allen FD, Pollack SR, Brighton CT. Intracellular Ca2+ stores and extracellular Ca2+ are required in the real-time Ca2+ response of bone cells experiencing fluid flow. J Biomech. 1996;29:1411–1417. doi: 10.1016/0021-9290(96)84536-2. [DOI] [PubMed] [Google Scholar]
- 39.Chen NX, Ryder KD, Pavalko FM, Turner CH, Burr DB, Qiu J, Duncan RL. Ca2+ regulates fluid shear-induced cytoskeletal reorganization and gene expression in osteoblasts. Am J Physiol Cell Physiol. 2000;278:C989–997. doi: 10.1152/ajpcell.2000.278.5.C989. [DOI] [PubMed] [Google Scholar]
- 40.Talavera K, Nilius B. Biophysics and structure-function relationship of Ttype Ca2+ channels. Cell Calcium. 2006;40:97–114. doi: 10.1016/j.ceca.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Nilius B, Talavera K, Verkhratsky A. T-type calcium channels: The never ending story. Cell Calcium. 2006;40:81–88. doi: 10.1016/j.ceca.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Thompson WR, Majid AS, Czymmek KJ, Ruff AL, García J, Duncan RL, Farach-Carson MC. Association of the α(2)δ(1) subunit with Ca(v)3.2 enhances membrane expression and regulates mechanically induced ATP release in MLO-Y4 osteocytes. J Bone Miner Res. 2011;26:2125–2139. doi: 10.1002/jbmr.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 44.Iqbal J, Zaidi M. Molecular regulation of mechanotransduction. Biochem Biophys Res Commun. 2005;328:751–755. doi: 10.1016/j.bbrc.2004.12.087. [DOI] [PubMed] [Google Scholar]