Abstract
Protein kinase R (PKR), a sensor of double-stranded RNA, plays an important role in the host response to viral infection. Hepatitis C genotype 2a virus (HCV 2a) has been shown to induce PKR activation to suppress the translation of antiviral interferon stimulated genes (ISGs), suggesting that PKR inhibitor can be beneficial for treating chronically HCV-infected patients in conjunction with interferon alpha and ribavirin. However, in this study, we found that PKR inhibition using siRNA PKR, shRNA PKR or PKR inhibitor enhanced HCV 1a replication and rendered Huh7.5.1 cells more susceptible to HCV1a infection. Additionally, PKR silencing suppressed NF-kB activation and NF-kB mediated STAT1 phosphorylation in Huh7.5.1 cells and HCV1a persistently-infected Huh7.5.1 cells (2HDD4). These effects were accompanied by a reduction of interferon beta response and thereby enhanced HCV1a replication in Huh7.5.1 cells. We conclude that host cells can employ PKR activation to restrict HCV1a replication through regulation of NF-kB expression.
Keywords: PKR silencing, NF-kB activation, IFN-β, HCV1a replication
Introduction
The innate immune system plays a critical role in containing early hepatitis C virus (HCV) infection through the induction of type 1 interferon and the up-regulation of interferon stimulated genes. However, because HCV replicates very rapidly, peak levels of virus are achieved within the first days of infection and the HCV proteins subsequently generated (core, NS2, NS3, NS4B, NS5A) can inhibit many steps in the innate immune response, preventing the activation of NF-kB and facilitating viral replication(Joo et al., 2005; Park et al., 2012). The net effect is that HCV generally circumvents both the innate response and the adaptive immune response resulting in persistent infection in 75%-85% of those infected.
In response to viral infection, the transcription of interferon beta is initiated by enhancesome formation that consists of multiple transcriptional factors including ATF-2/c-Jun, IRF3/IRF7 and NF-kB (Ford and Thanos, 2010; Randall and Goodbourn, 2008). NF-kB is the most critical transcription factor in the regulation of interferon beta and interferon simulated genes (ISGs) and is composed of five elements: RelA, cRel, RelB, p50 and p52. Through the Rel homology domain, these elements can form homodimers and heterodimers. The canonical pathway to NF-kB activation is initiated when upon stimulation by LPS, polyI/C, TNF-alpha or viral infection. Then, IkB is phosphorylated leading to the translocation of the RelA (p65) component of NFkB into the nucleus (Hacker and Karin, 2006; Ting, Duncan, and Lei, 2010). Subsequently, p65 serves as an immediate early transcription activator of interferon beta gene expression and production (Hiscott et al., 2006). In order to antagonize the antiviral effect of interferon beta, viruses have developed many strategies, especially concentrating on inhibiting the activation of NF-kB.
PKR, the sensor of double-stranded RNA (dsRNA), also plays an important role in the antiviral response through phosphorylation of eIF2alpha and inhibition of host cell gene translation and protein synthesis (D’Acquisto and Ghosh, 2001; Robertson and Mathews, 1996; Williams, 2001). DsRNA-activated PKR can induce the degradation of IkB resulting in the activation of NF-kB (Zamanian-Daryoush et al., 2000). PKR has also been shown to be responsible for virus-induced NF-kB activation in engineered vaccinia virus or attenuated Herpes simplex virus-1 infected cells (Lynch et al., 2009; Taddeo et al., 2003). In HCV infection, genetic analysis has shown that the HCV internal ribosomal entry site (IRES), and the core and NS5A proteins contain PKR-binding domains that are critical to PKR function (Gimenez-Barcons et al., 2005; Toroney et al., 2010; Yan et al., 2007). In-vitro inhibition of PKR can increase HCV production 10-fold (Oem et al., 2008), suggesting that PKR is essential to the host’s antiviral response to HCV infection. Thus, HCV infection can lead to a duality of responses and the relative balance can result in either viral clearance or persistence. On the one hand, virus-induced activation of NF-kB can trigger the transcription of IL8, TNF-alpha and interferon beta, which results in restriction of viral replication (Frey et al., 2009; Hassan et al., 2007; Oem et al., 2008), conversely, HCV proteins can diminish NF-kB activation, facilitating viral persistence. Integral to this equation are virus-induced PKR activation and NF-kB activation.
In this study we show that the PKR silencing contributes to HCV1a replication in HCV1a persistently infected Huh7.5.1 cells (2HDD4). Moreover, PKR silencing inhibits the interferon beta response in PKR silenced 2HDD4 cells through NF-kB inactivation. Consistent with this mechanism, shRNA PKR renders cells more susceptible to HCV1a infection in naïve cells. In addition, NF-kB inhibitor improved HCV1a replication in 2HDD4 cells. Hence, host defenses may be attenuated by PKR silencing, enhancing HCV1a replication through the attenuation of the NF-kB mediated interferon beta response.
Results
Inhibition of PKR enhanced the replication of HCV1a in HCV persistently infected Huh-7.5.1 cells
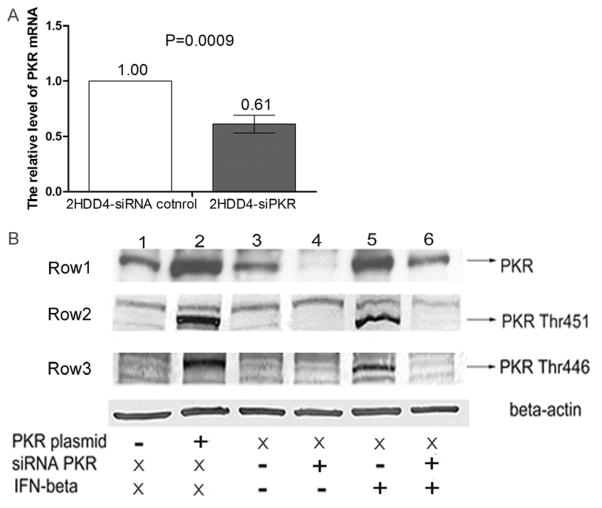
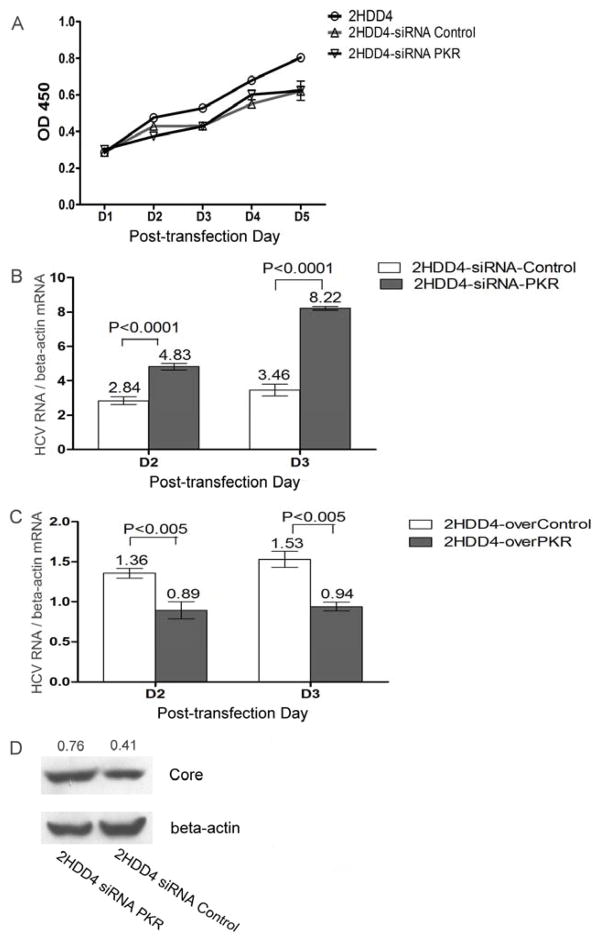
In order to assess the effect of PKR silencing on HCV1a replication and infection, we constructed Huh7.5.1 cells that were persistently infected with HCV1a (2HDD4 cells) and then investigated the effect of PKR protein on HCV1a replication in this cell line. Firstly, we examine the effectiveness of siRNA PKR, siRNA PKR led to a 40% reduction of PKR mRNA in 2HDD4 cells (p=0.0009) (Figure 1A) and a reduction in PKR protein as shown by Western blot (Row1 of Figure 1B, lanes 3 and 4). In addition, IFN-β-induced PKR phosphorylation at Thr451 and Thr446 was inhibited by PKR silencing (Row2 Row3 of Fig 1B lane4 and lane6) confirming that siPKR efficiently suppressed PKR activation in target cells. In contrast, siRNA PKR did not have a significant impact on cell growth (Figure 2A). To examine the impact of PKR silencing on HCV1a replication, we utilized RT-PCR to monitor HCV1a RNA replication in persistently infected 2HDD4 cells at different time intervals after siRNA transfection. The results demonstrated that PKR silencing increased the replication of HCV1a approximately by 1.7-fold at day 2 (p<0.0001) (Figure 2B) and almost a 2.5-fold at day 3 (p<0.0001) and resulted in the increased expression of core protein (Figure 2D). In contrast, over-expression of PKR in 2HDD4 cells inhibited HCV1a replication (p<0.005) (Figure 2C). Combined, these results indicate that PKR knockdown enhances the replication of HCV1a in persistently infected cells.
Figure 1. siRNA PKR efficiently inhibited the activation of PKR.
(A) HCV persistently infected Huh7.5.1 cells (2HDD4 cells) were transfected with siRNA PKR or siRNA control for 6 h. After 24hrs, PKR mRNA level was measured by RT-PCR and normalized by beta-actin mRNA level. Data are represented as mean± SEM; n=3. The data are representative of two independent assays. (B) 2HDD4 cells were transfected with either vector only control plasmid (−) (lane 1), PKR expression plasmid (+) (lane 2), non-targeting siRNA control (−) (lanes 3 and 5), or siRNAs specifically targeting PKR(+) (siRNA PKR, lanes 4 and 6) for 6 h. After overnight incubation, an aliquot of siRNA treated 2HDD4 cells (lanes 3–6) were further stimulated with 200U/ml IFNbeta (+) (lanes 5 and 6) or PBS (−) (lanes 3 and 4). X: no treatment. The expression of PKR protein (Row1) and the phosphorylation of PKR protein at Thr446 (Row2) and Thr451 (Row3) were analyzed by Western blot at 48h post-treatment. Beta-actin was used as the protein loading control.
Figure 2. Inhibition of PKR enhanced the replication of HCV1a in HCV persistently infected Huh7.5.1 cells(2HDD4 cells).
(A) 2HDD4 cells were transfected with siRNA PKR or siRNA control. After transfection, 2000 siPKR cells or 2000 siControl cells and 2HDD4 cells were seeded in 96-well plates. The cell growth was examined by absorbance at 450nm as described in Materials and Methods. Data are represented as mean± SEM; n=6. (B) 2HDD4 cells were transfected with siRNA PKR or siRNA control. After transfection, total cellular RNA was collected at the indicated time points. Intracellular HCV copy numbers were measured by qRT-PCR and normalized by beta-actin mRNA level. Data are represented as mean± SEM; n=6. The data are representative of four independent experiments. (C) 2HDD4 cells were transfected with 2μg PKR plasmid or 2μg control plasmid for 6h. After transfection, total cellular RNA was collected at the indicated time points. Intracellular HCV copy numbers were measured by qRT-PCR and normalized by beta-actin mRNA level. Data are represented as mean± SEM; n=6. The data are representative of four independent experiments. (D) 2HDD4 cells were transfected with siRNA PKR or siRNA control for 6h. After 72 h incubation, the cellular extracts were collected and subjected to immunoblot analysis for HCV core protein. Beta-actin was used as the protein loading control. Each sample represents a pool of three replicates.
shRNA PKR rendered Huh7.5.1 cells more susceptible to HCV1a infection
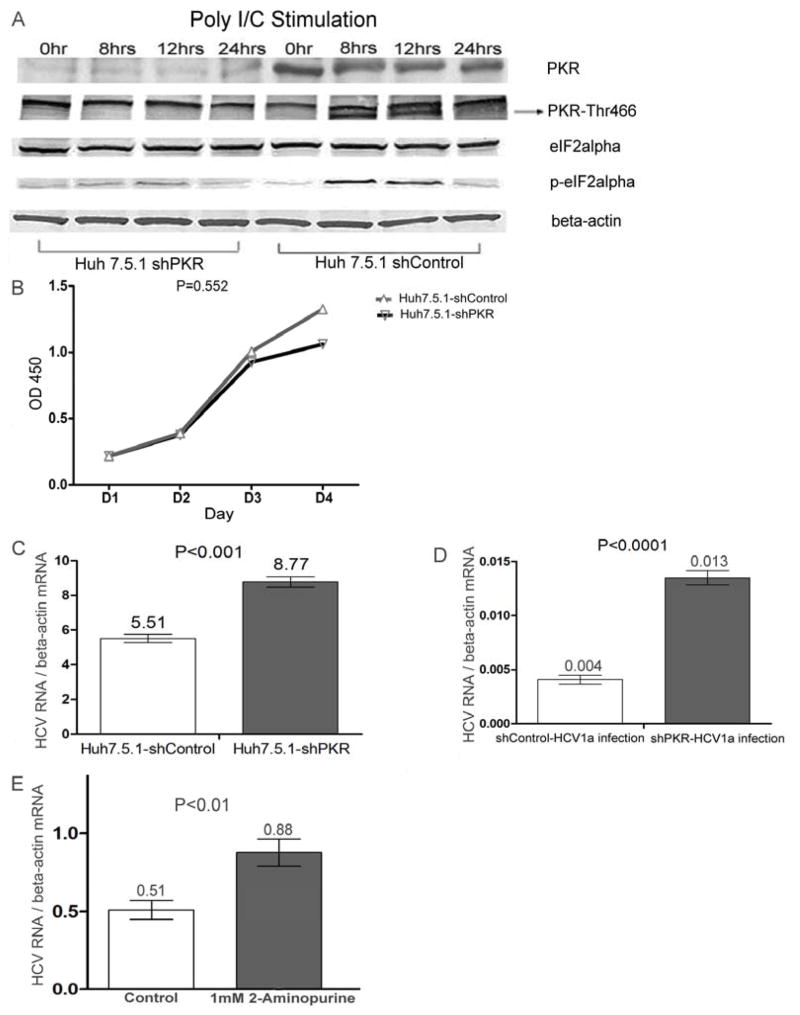
To exclude the potential non-target effect of siRNA and to confirm the observation over a longer incubation period, we developed a stable PKR- silenced single clone from Huh-7.5.1 cells treated with a pool of plasmids containing three PKR-target specific shRNAs. As shown in Figure 3A, PKR silencing significantly suppressed double strand RNA-induced phosphorylation of PKR at residue Thr446 and the phosphorylation of eIF2alpha at residue Ser51, while having no significant impact on cell growth (Figure 3B). Next, HCV1a RNA was transfected into PKR silenced Huh-7.5.1 cells, the results showed that the level of HCV1a RNA replication was higher than that in control cells (p<0.001) (Figure 3C). Furthermore, we used HCV1a virus produced from 2HDD4 cells to infect PKR silenced cells and control cells. The number of HCV copies in PKR silenced cells was significantly (p<0.0001) higher than that in control cells (Figure 3D). To further confirm the positive effect of PKR silencing on HCV1a replication, we used PKR inhibitor, 2-aminopurine, to treat 2HDD4 cells. Similarly, HCV1a RNA replication was enhanced by PKR inhibitor (p<0.01) (Figure 3E).
Figure 3. PKR silencing rendered Huh7.5.1 cells more susceptible for HCV1a infection.
(A) A stably PKR silenced Huh7.5.1 cells (shPKR) and a shRNA control plasmid transfected Huh7.5.1 cells (shControl) were stimulated with 2μg/ml double-strand RNA (polyI/C) by transfection with Lipofectamine 2000. Cellular extracts were collected at the indicated time points and analyzed by Western blot for PKR protein, the phosphorylation of PKR protein at Thr446, eIF2alpha protein, and the phosphorylation of eIF2alpha protein at Ser51. Beta-actin was utilized as internal control. Each sample represents a pool of three replicates. (B) 2000 shPKR cells or 2000 shControl cells were seeded at each well in 96-well plates. The cell growth was examined by absorbance at 450nm as described in Materials and Methods. Data are represented as mean± SEM; n=4. (C) 2 × 105 shPKR cells or 2 × 105 shControl cells were transfected with 2μg HCV1a RNA. 72hrs post- transfection, total cellular RNA was collected. Intracellular HCV copy numbers were measured by qRT-PCR and normalized by beta-actin mRNA level. Data are represented as mean± SEM; n=6. The data are representative of four independent experiments. D) 2 × 104 shPKR cells or 2 × 104 shControl cells in 12-well plates were infected overnight with HCV1a virus produced from the 0.45 μm filtered culture supernatant of HCV1a persistently infected Huh-7.5.1 cells (2HDD4 cells). 48hrs post-infection, intracellular HCV RNA in newly infected cells was analyzed by qRT-PCR and normalized by beta-actin mRNA copies. Data are represented as mean± SEM; n=3. The results are representative of two independent experiments. E) 2HDD4 cells were treated 1mM aminopurine or control. 48h post-treatment, intracellular HCV RNA was analyzed by qRT-PCR and normalized by beta-actin mRNA copies. Data are represented as mean± SEM; n=3. The results are representative of two independent experiments.
NF-kB activation was suppressed in PKR silenced HCV1a infected and transfected Huh7.5.1 cells
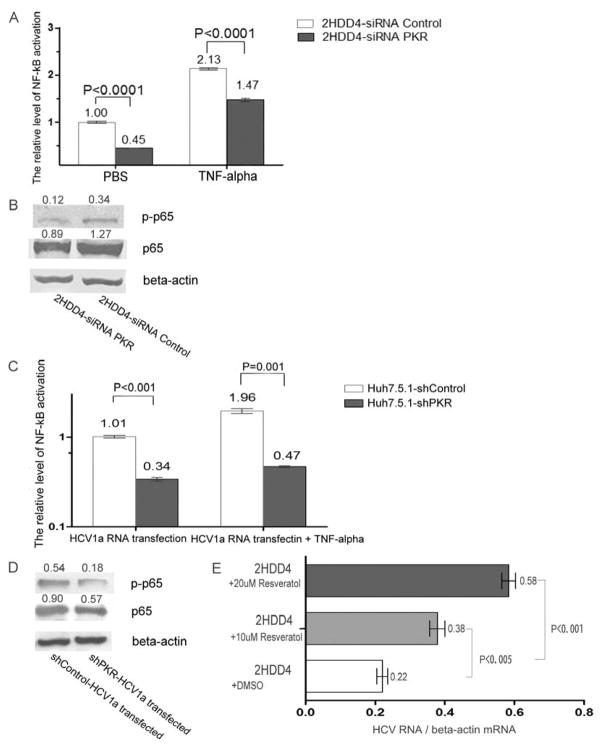
NF-kB activation has been shown to be important for PKR function, so we examined the effects of PKR silencing on the activation of NF-kB in HCV1a persistently infected Huh7.5.1 cells. PKR silencing resulted in a significant decrease in the level of NF-kB activation (p<0.0001), even after stimulation with TNF-alpha, a known NF-kB agonist (Fig. 4A); correspondingly, a lower expression of NF-kB protein and phosphorylated p65 protein were observed in PKR silenced 2HDD4 cells (Figure 4B). Consistent with these results, a significant inhibition of NF-kB activation and a lower expression of NF-kB protein were also found in HCV1a RNA transfected PKR stably-silenced Huh7.5.1 cells (Figure 4C and 4D). To investigate whether the attenuation of NF-kB contributed to the enhancement of HCV1a replication, the NF-kB inhibitor resveratrol was used. The results showed that NF-kB inhibitor significantly improved HCV1a RNA replication in 2HDD4 cells (p<0.005) (Figure 4E). In composite, these results suggest that enhanced HCV1a infection and replication are at least partly mediated by a reduction of NF-kB activity.
Figure 4. NF-kB activation was inhibited in PKR silenced HCV1a infected Huh7.5.1 cells.
(A) 2HDD4 cells were treated by transfection with siRNA Control or siRNA PKR, or treated by a combination of siRNA with TNF-alpha (60 ng/ml) stimulation. 48 h post treatment, the supernatants were collected and examined for NF-kB activation by SEAP activity. Data are represented as mean± SEM; n=3. The results are representative of two independent experiments. (B) 2HDD4 cells were transfected with siRNA PKR or siRNA control for 6h. After 72 h incubation, the cellular extracts were collected and subjected to immunoblot analysis for p65 and phosphorylated p65 proteins. Beta-actin was used as the protein loading control. Each sample represents a pool of three replicates. (C) shPKR cells and shControl cells were treated by transfection with 2μg of HCV1a RNA, or treated by a combination of 2μg HCV1a RNA with TNF-alpha (60 ng/ml) stimulation. 48 h post treatment, the supernatants were collected and examined for NF-kB activation by SEAP activity. Data are represented as mean± SEM; n=3. The results are representative of two independent experiments. (D) shPKR cells and shControl cells were transfected with 2μg of HCV1a RNA for 6h. After 48 hrs incubation, the cellular extracts were collected and subjected to immunoblot analysis for p65 and phosphorylated p65 proteins. Beta-actin was used as the protein loading control. Each sample represents a pool of three replicates. (E) 2HDD4 cells were treated with 10μM resveratrol, 20μM resveratrol or DMSO. 48h post-treatment, intracellular HCV RNA was analyzed by qRT-PCR and normalized by beta-actin mRNA copies. Data are represented as mean± SEM; n=3. The results are representative of two independent experiments.
Inhibition of NF-kB activation attenuated IFN-β response in PKR silenced HCV1a infected Huh7.5.1 cells
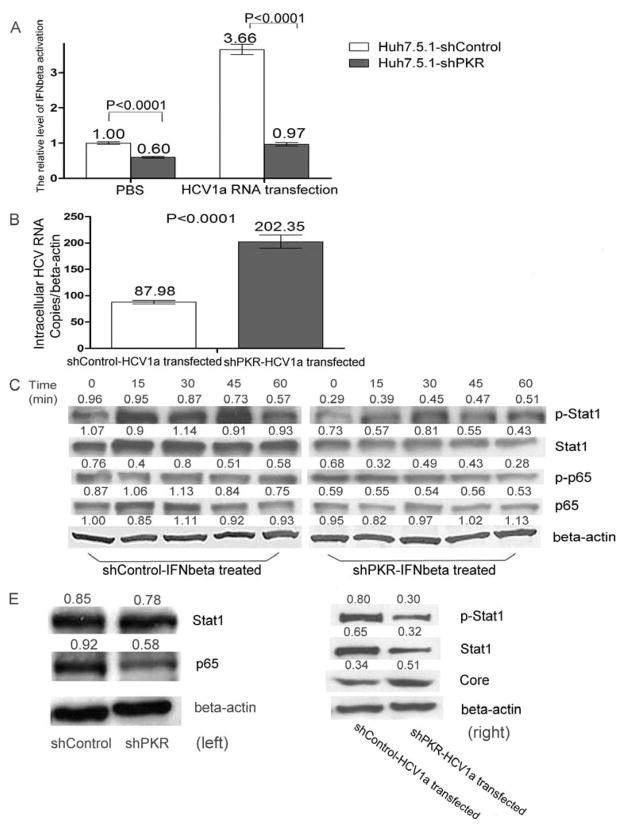
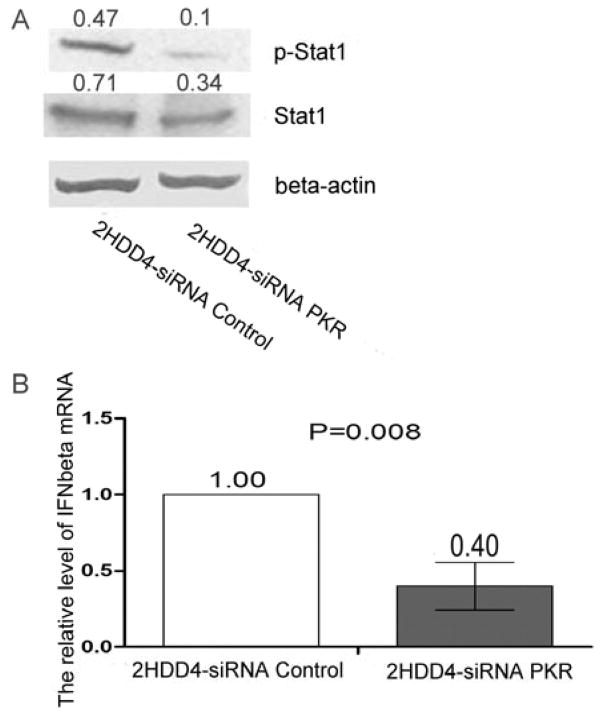
As a downstream effector of NF-kB, the IFN-β response was subsequently investigated in PKR silenced HCV1a infected cells. Examining by a IFN-β reporter gene assay, ablation of PKR suppressed the basic level of IFN-β activity in the absence of HCV1a replication. After cells were transfected with HCV1a RNA, a markedly diminished interferon beta response was found in PKR silenced cells (P<0.0001) as compared to the control cells (Figure 5A) and was accompanied by a significant (p<0.0001) increase in viral replication (Figure 5B). This finding was further elucidated in Huh 7.5.1 cells by examining STAT1 phosphorylation after IFN-β stimulation. As shown in Figure 5C, STAT1 protein was rapidly induced and phosphorylated following IFN-β treatment in control cells. Conversely, inhibited expression of NF-kB, phosphorylated p65 protein, STAT1 protein and phosphorylated STAT1 protein were observed in PKR stably-silenced cells. These results indicate that attenuated NF-kB activation is responsible for the inhibition of IFN-β responses in PKR silenced Huh7.5.1 cells. To confirm that the effect of PKR silencing on interferon beta was mediated by the inhibition of NF-kB, we introduced 2μg of HCV1a RNA into cells and examined STAT1 expression; PKR silencing reduced the expression of STAT1 protein and phosphorylated STAT1 protein (Figure 5D). Consistent with these results, PKR silencing resulted in decreased expression of STAT1 protein and phosphorylated STAT1 protein in HCV persistently infected cells (Figure 6A). The level of IFN-β mRNA was also down-regulated by siRNA PKR in 2HDD4 cells (Figure 6B).
Figure 5. siPKR silencing inhibited IFN-β response in HCV1a transfected Huh7.5.1 cells.
(A) shPKR cells and shControl cells were treated by transfection with 2μg of HCV1a RNA or PBS. After 24hrs, the cells were transfected with IFN-β reporter plasmid. 48 h post-treatment, the supernatants were collected and examined for IFN-β activation by SEAP activity. Data are represented as mean± SEM; n=3. The results are representative of two independent experiments. (B) shPKR cells and shControl cells were transfected with 2μg of HCV1a RNA for 24hrs. After that, the cells were transfected with IFN-β reporter plasmid. 48 h post-treatment, total RNA were isolated. HCV RNA was analyzed by qRT-PCR and normalized by beta-actin mRNA copies. Data are represented as mean± SEM; n=3. The results are representative of two independent experiments. (C) shPKR cells and shControl cells were stimulated with 200u/ml IFN-β. Cellular extracts were collected at the indicated time points (minute) and analyzed by Western blot for p65 protein, the phosphorylation of p56 protein at Serine 596, STAT1 protein, and the phosphorylation of STAT1 protein at Tyr701. Beta-actin was utilized as internal control. Each sample represents a pool of three replicates. (D) shPKR cells and shControl cells were treated by transfection with 2μg of HCV1a RNA. After 48hrs, the cellular extracts of HCV1a transfected cells (right) and control cells (left) were collected and subjected to immunoblot analysis for core protein, STAT1 protein, and the phosphorylation of STAT1 protein at Tyr701. Beta-actin was used as the protein loading control. Each sample represents a pool of three replicates.
Figure 6. siPKR silencing inhibited IFN-β response in 2HDD4 cells.
(A) 2HDD4 cells were transfected with siRNA PKR or siRNA control. After 48hrs, the cellular extracts were collected and subjected to immunoblot analysis for STAT1 protein, and the phosphorylation of STAT1 protein at Tyr701. Beta-actin was used as the protein loading control. Each sample represents a pool of three replicates. (B) 2HDD4 cells were treated with siRNA PKR or siRNA control. After 24hrs, IFN-β mRNA levels were examined with the relative RT-PCR and normalized with beta-actin mRNA level. Data are represented as mean± SEM; n=3. The results are representative of two independent experiments.
Discussion
PKR is an important regulator of innate immunity as a sensor of dsRNA and viral RNA, including HCV. In order to evade this response, HCV utilizes E2 protein and NS5A protein to antagonize PKR activation (Gerotto et al., 2000; Taylor et al., 1999). Recently, the HCV internal ribosomal entry site (IRES) also has been shown to be involved in the inhibition of PKR (Toroney et al., 2010). Previously, the Chisari lab had shown that HCV can hijack PKR activation to suppress the translation of antiviral ISGs and inhibit the IFNβ-induced antiviral response. They emphasized the important role of PKR on the early phase of HCV2a infection (Garaigorta and Chisari, 2009). Based on these findings, it has been inferred that inhibition of PKR activation may be an important determinant of the efficiency of HCV replication. However, the influence of PKR on HCV replicaton is probably complex such that in the HCV2a replicon system, HCV 2a infection has been shown to inhibit the IFN beta response through PKR activation in RIG-I functional Huh7.25 cells. PKR inhibition can lead to the suppression of HCV2a replication (Arnaud et al., 2010).
HCV1a is the most common genotype in the US. In this study, PKR siRNA and PKR shRNA were used to inhibit PKR activation in an HCV1a-replication permissive cell line, Huh7.5.1 and in an HCV1a-persistently infected cell line, 2HDD4. After analysis by qRT-PCR, we found that PKR down-regulation significantly enhanced the replication of HCV1a. This is consistent with a previous report (Chang et al., 2006), which showed that HCV RNA replication was more efficient in PKR null cells than in wild type cells. However, other studies have not confirmed this effect on HCV1a replication in Huh7.5.1 cells. Importantly, Huh7.5.1 cells carry a point mutation in the RIG-I gene which leads to a reduction of host innate immune response to viral RNA and renders cells more permissive to HCV1a infection (Bartenschlager and Pietschmann, 2005; Zeisel and Baumert, 2006). This mutation minimizes the effect of the RIG-I pathway on PKR activation in vivo, a consideration in interpreting the in-vitro data in this study. Moreover, in our study, the effect of PKR silencing on HCV genotype 2a replication was investigated using JFH-1 virus. In contrast to the finding in HCV1a virus, there was no significant difference in virus replication level between JFH-1 virus infected PKR silenced Huh7.5.1 cells and control cells (Supplemental Figure A). This result suggests that the effect of PKR silencing on HCV replication may be genotype specific. Then, we used JFH1/1a chimeric virus to infect Huh-7.5.1 cells with or without PKR knockdown. The results in Supplemental Figure B revealed that PKR silencing enhanced the replication of this chimeric virus. Next, we used PKR inhibitor to treat HCV1a RNA transfected RIG-I functional cells, IMY-4 and IMY-N9 (Date et al., 2004; Ito et al., 2001), respectively, and monitored the replication of HCV1a virus in these cells by RT-PCR. As shown in Supplemental Figure C and D, treatment with PKR inhibitor significantly increased HCV1a replication in these RIG-I functional cells. Based on these results, we concluded that PKR silencing is enhancing HCV1a replication and this enhancement of virus replication may be genotype specific.
In principle, the RIG-I/MVAS pathway is responsible for type I interferon induction and plays an important role in the innate immune response to HCV infection (Vilasco et al., 2006). The study from Foy E et al showed that RIG-I mediated pathway was disrupted by HCV protease NS3/4A in HCV infected cells (Foy et al., 2005). Moreover, HCV NS3/4A can inhibit IFN-beta induction through cleavage of IPS-1 from the membrane of mitochondria (Loo et al., 2006). However, in the RIG-I impaired cell line, Huh7.5.1, it is still difficult to obtain high titers of HCV1a, suggesting a possibility of a RIG-I-independent pathway involved in the host defense against HCV1a viral infection. PKR has been proposed as an important modulator of type I interferon response in viral infection and both Interferon beta transcription and production have been shown to be down-regulated by inhibition of PKR activation in some viral infections (Carpentier, Williams, and Miller, 2007; Fragkoudis et al., 2008). In our study, PKR down-regulation reduced interferon beta transcription in RIG-I nonfunctional cells. Moreover, PKR silencing led to reduction of interferon beta production in HCV1a persistently-infected cells. Although PKR has no direct effect on the interferon beta transcript, PKR protein has been demonstrated to regulate the expression of NF-kB, which is one of the three dominant activators of interferon beta transcription. Direct degradation of IkB by PKR protein has been proposed as an important mechanism whereby PKR regulates NF-kB activity (Kumar et al., 1994). In recent studies, Several viruses have been shown to regulate interferon beta response through PKR-mediated NF-kB activation (McAllister et al., 2010; Iwamura et al., 2001). Hu7.5.1 cells possess a high and constant NF-kB activation, which is an obstacle for HCV infection and replication. In our study, we did not observe any significant differences in the levels of eIF2alpha and phosphorylated eIF2alpha between siRNA PKR treated and untreated cells in a cell line (2HDD4) that was persistently infected with HCV1a (data not shown). Also we did not find any significant differences in the level of eIF2alpha and phosphorylated eIF2alpha between 2HDD4 cells and naive cells, suggesting that the phosphorlyation of eIF2alpha mediated by PKR activation mainly functions on the host against HCV infection at the early phase of viral infection. However, we found that PKR silencing reduced NF-kB activation, leading to enhanced HCV replication and increased expression of core protein. Shavinskaya et al showed that the D2 domain of HCV core is an important determinant of viral production, suggesting that sequence variants in this domain could affect the host immune response to viral infection (Shavinskaya et al., 2007). Also, studies have documented that HCV1a core protein can significantly suppress NF-kB activation (Joo et al., 2005; Ray et al., 2002). Importantly, PKR silencing resulted in the down-regulated expression of STAT1 and the phosphorylation of STAT1. The expression of STAT1 and phosphorylated STAT1 have been implicated in the resistance of Type I interferon pathway to HCV1a infection (Kanda et al., 2007; Raychoudhuri et al., 2010).
Cumulative data suggest that the host immune response can employ PKR activation to facilitate the translation of some ISGs to restrict viral replication and that PKR inactivation can be used by viruses to circumvent innate immunity (Schoggins et al., 2011). Our data reveal the key role played by PKR in HCV1a replication and infection and indicate that PKR silencing can suppress the interferon beta response to HCV1a infection and replication through NF-kB activation. In combination, these data suggest that PKR possesses a double-face in different genotype of HCV infection; on the one side, PKR activation is utilized by HCV2a virus to suppress the immune response, while on the other, it is involved in the host’s innate immune response to inhibit HCV1a virus replication and cell damage by the up-regulation of NF-kB, beta interferon and interferon stimulated genes as well as through the phosphorylation of eIF2alpha. These data support the concept that NF-kB mediated type 1 interferon pathway is an important mechanism underlying the effect of PKR silencing on HCV1a replication. Our findings provide new insight into the role of PKR protein in HCV1a infection and suggest that PKR might be a rational therapeutic target to enhance host immunity to HCV in concert with protease, polymerase and other HCV-specific enzyme inhibitors.
Material and Methods
Cell culture
The human hepatoma cell line, Huh7.5.1 was obtained from Dr. Francis V. Chisari (Zhong et al., 2005). Huh7.5.1 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco BRL) supplemented with 10% heat inactivated fetal bovine serum, 2 mM L-glutamine, 10 mM HEPES, pH7.2, 100 unit/ml penicillin, 100 μg/ml streptomycin, and 1% non-essential amino acids (Gibco BRL) in a humidified 37°C, 5% CO2 incubator. PKR knock-down Huh7.5.1 cells and negative plasmid transfected cells were cultured in the same medium supplemented with 1 μg/ml puromycin. The PKR inhibitor 2-aminopurine (cat. #:tlrl-apr) and the NF-kB inhibitor resveratrol (cat. #:tlrl-resv) were purchased from Invivogen.
Construction of a HCV persistently infected cell line (2HDD4 cells)
Huh7.5.1 cells were transfected with H77S HCV RNA. At two days post-transfection, the cells were serial diluted and seeded in 96-well plates. One clone, expressing a stable high level of HCV1a RNA and producing infectious HCV1a, was selected as HCV persistently infected cells (2HDD4 cells).
Construction of a stable cell line expressing PKR shRNA
Huh7.5.1 cells were transfected with a pool of 3 PKR-target specific shRNAs (Santa Cruz, sc-36263-SH). At two days post-transfection, the cells were cultured in DMEM supplemented with 1 μg/ml puromycin. After 2 weeks, the remaining viable cells were serially diluted and seeded in 96 well plates. Puromycin-resistant clones were expanded and identified by Western blot. One clone, expressing a low level of PKR protein, was selected for further analysis.
In-vitro RNA synthesis
The plasmid pH77-S, which carries a full length HCV1a sequence with five cell culture-adaptive mutations was kindly provided by Dr. Stanley Lemon(Yi et al., 2006). The plasmid was linearized with XbaI restriction enzyme and purified by Qiagen PCR purification kit. In vitro transcribed RNA, using the MEGA-script T7 kit (Ambion, Austin, TX, USA), was purified with Qiagen RNA clean up kit. The RNA pellet was aliquoted and stored at −80°C until use.
HCV RNA transfection
1 μg of HCV1a (H77S) full length RNA was transfected into Huh7.5.1 cells and its derivative cells in a 6-well plate (3×105 cells/well) by using mRNA boost reagent and TranslT-mRNA reagent (Mirus, MIR2250) according to the manufacturer’s instructions.
Small interfering RNA transfection
2×105 cells/well in 6-well plates were transfected with 10 nM of 3 PKR specific-targeting small interfering RNAs (Santa Cruz Biotechnology, Inc, sc-36263) usingLipofectamine 2000 (Invitrogen) for 6 h. Three non-targeting, small interfering RNAs (Santa Cruz Biotechnology, Inc, sc-37007) transfected cells were used as controls. After three days incubation, cells were treated again with specific-targeting siRNA or control siRNA for 3 days and then analyzed.
Plasmid and transfection
Full-length expression plasmid for PKR was purchased from Genecopoeia Inc. (Rockville, MD). 2 μg plasmid DNA was transfected into 2×105 cells/well in 6-well plates using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Cell growth assay
2000 cells/well were inoculated into each well in 96-well plates and 10μl of the cell counting kit-8 solution (Dojindo, CK04) added to each well. Plates were incubated at 37°C for 2hrs. Absorbance was measured at 450nm using a microplate reader.
NF-kB reporter gene assay
After treatment by siRNA PKR or siRNA control, 2×105 cells per well in 6-well plates were transfected with 0.5 μg pNF-kB-SEAP-1 reporter plasmid and 10 ng pMet-Luc plasmid as internal control for 6 h. After overnight incubation, transfected cells were stimulated with TNF-alpha (60ng/ml) or mock treated for 24hrs. SEAP and Met-Luc in the supernatant were analyzed by chemiluminescence as previously described (Monsurro et al., 2010).
Interferon beta reporter gene assays
3×105 cells were transfected with 2ug HCV1a RNA. After overnight incubation, transfected cells were treated with0.5 μg pIFN-β-SEAP-1 reporter plasmid and 10 ng pMet-Luc plasmid as internal control. Supernatant was collected at the indicated time points. Phospha-Light™ SEAP Reporter Gene Assay system for secredted alkaline phosphatase (SEAP) was obtained from Applied Biosystems Ready-To-Glow Secreted Luciferase Reporter system for Metridia secreted luciferase (Met-luc) was obtained from Clontech. SEAP and Met-Luc in the supernatant were analyzed by chemiluminescence according to the previously described method (Monsurro et al., 2010).
Total RNA preparation and cDNA synthesis
Total RNA was extracted from cell culture using Qiagen RNeasy kit according the manufacturer’s instructions. RNA quantity and purity were determined on a NanoDrop spectrophotometer. Following the purification of total RNA, 100 ng of RNA was applied for cDNA synthesis using BluePrint™ RT Reagent Kit (TAK RR737A, Takara Biochemicals). Random primers and total RNA were incubated at 37°C for 15min and inactivated at 85°C for 5 seconds. cDNA was aliquoted and stored at −80°C until utilized.
Sybr-Green Real time PCR
Sybr-Green real-time PCR was carried out with a pair of forward and reverse primers. The reaction mixture (RT-PCR kit, Code RRO43A, Takara Biochemicals) contained 25 μl SYBR Premix Ex Taq (2x) (SYBR® Premix Ex Taq™ Perfect Real Time, RRO41B, Takara Biochemicals), 1 μl of 10 μM PCR forward primer, 1 μl of 10 μM PCR reverse primer, and 100 ng cDNA to give a final reaction volume of 50 μl. The standard cycling conditions were 95°C for 10 sec, followed by 40 cycles at 95°C for 5 sec, 60°C for 30 sec, with the subsequent melting curve analysis by increasing the temperature from 60°C to 95°C. IFN-β was amplified using the primers: 5′-TCC TGT GGC AAT TGA ATG GG-3′ and 5′-TGC TCA TGA GTT TTC CCC TGG-3′; beta-actin was amplified using the primers: 5′-TCACCCTGAAGTACCCCATC-3′ and 5′-TAGCACAGCCTGGATAGCAA-3′. The primers to amplify PKR (sc-36263-PR) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The relative quantitation value of each gene was compared to the relative quantitation value of its matched non-treated sample. The relative quantification values for each gene were normalized against the endogenous housekeeping gene beta-actin.
Quantitative RT–PCR assay
QRT-PCR was performed using the primer set R6-130-S17 (5′CGGGAGAGCCATAGGTGG-3′), R6-290-R19 5′-AGTACCACAAGGCCTTTCG-3′) and Taqman probe R6-148-S21FT (6FAM-5′-CTGCGGAACCGGTGAGTACAC-3′-TAMRA). Samples were quantified in triplicate using a TaqMan EZ RT-PCR core reagent kit (Applied Biosystems, N808-0236). The reactions were carried out on ABI 7900HT system with a program of 50°C for 2 min, 60°C for 30 min, 95°C for 4 min, and then 40 cycles at 95°C for 20 sec and 60°C for 1 min. The copies of HCV RNA were determined by in vitro transcribed HCV1a RNA standards with the Sequence Detector Software (version 2.2; Applied Biosystems) and normalized by beta-actin mRNA level. Beta-actin mRNA level was quantified by in vitro transcribed beta-actin standards with the primers: 5′-AAGTGTGACGTGGACATCCG-3′ and 5′-CACACGGAGTACTTGCGCTC-3′ and Taqma probe: 6FAM-5′-CTGTACGCCAACACAGTGCT-3′-TAMRA.
Western blot
Twenty microgram aliquots of proteins were loaded into each lane in 10% SDS-PAGE and then transferred to nitrocellulose membranes. After probing with primary and second antibody, the target proteins were developed with either TMB 1-component membrane solution (KPL) or chemiluminescent detection. Antibodies to NF-kB (C-20(p65), sc-372), beta-actin (sc-47778), PKR (K17, sc-707), p-stat1(Tyr701)-R (sc-7988R) and stat1α p91 (C-11, sc-417) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); PKR (pT446) (11201) was purchased from Epitomics (Burlingame, CA); PKR (pT451) (ab4818) was purchased from ABcam; P-NF-kB p65 (s536) was purchased from Cell Signaling. The relative expression of protein was normalized beta-actin using soft Image J. The data were the means of three independent experiments.
Statistical analysis
Data from repeated experiments were averaged and expressed as means ± the standard deviations. Statistical analysis was performed by SPSS using the Student t test, or two-way Anova analysis of variance. P values of <0.05 and <0.01 were considered statistically significant, respectively.
Supplementary Material
Highlights.
PKR plays a role in IFN-β response to HCV replication.
PKR silencing enhances HCV 1a replication in Huh7.5.1 cells and HCV1a persistently infected Huh7.5.1 cells.
PKR silencing inhibits NF-kB mediated IFN-β signaling pathway.
Acknowledgments
This study was supported by the Warren G. Magnuson Clinical Center, NIH intramural research program. We thank Dr. Francis Chisari for providing us with Huh-7.5.1 cells, Dr. Stanley Lemon for providing us with H77S plasmids, and Dr. Keijei Mitamura for providing us with IMY-4 and IMY-N9 cell lines. We also would like to appreciate the help from Dr. Liang Jack and Dr. Li Qisheng. They kindly provided JFH-1 virus and JFH1/1a chimeric virus. We also greatly appreciated Dr. Sonia Voiculescu’s assistance in the review of manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, Wakita T, Meurs EF. Hepatitis C virus controls interferon production through PKR activation. PLoS One. 2010;5(5):e10575. doi: 10.1371/journal.pone.0010575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Pietschmann T. Efficient hepatitis C virus cell culture system: what a difference the host cell makes. Proc Natl Acad Sci U S A. 2005;102(28):9739–40. doi: 10.1073/pnas.0504296102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier PA, Williams BR, Miller SD. Distinct roles of protein kinase R and toll-like receptor 3 in the activation of astrocytes by viral stimuli. Glia. 2007;55(3):239–52. doi: 10.1002/glia.20450. [DOI] [PubMed] [Google Scholar]
- Chang KS, Cai Z, Zhang C, Sen GC, Williams BR, Luo G. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J Virol. 2006;80(15):7364–74. doi: 10.1128/JVI.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Acquisto F, Ghosh S. PACT and PKR: turning on NF-kappa B in the absence of virus. Sci STKE. 2001;(89):re1. doi: 10.1126/stke.2001.89.re1. [DOI] [PubMed] [Google Scholar]
- Date T, Kato T, Miyamoto M, Zhao Z, Yasui K, Mizokami M, Wakita T. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J Biol Chem. 2004;279(21):22371–6. doi: 10.1074/jbc.M311120200. [DOI] [PubMed] [Google Scholar]
- Ford E, Thanos D. The transcriptional code of human IFN-beta gene expression. Biochim Biophys Acta. 2010;1799(3–4):328–36. doi: 10.1016/j.bbagrm.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Foy E, Li K, Sumpter R, Jr, Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102(8):2986–91. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkoudis R, Chi Y, Siu RW, Barry G, Attarzadeh-Yazdi G, Merits A, Nash AA, Fazakerley JK, Kohl A. Semliki Forest virus strongly reduces mosquito host defence signaling. Insect Mol Biol. 2008;17(6):647–56. doi: 10.1111/j.1365-2583.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey KG, Ahmed CM, Dabelic R, Jager LD, Noon-Song EN, Haider SM, Johnson HM, Bigley NJ. HSV-1-induced SOCS-1 expression in keratinocytes: use of a SOCS-1 antagonist to block a novel mechanism of viral immune evasion. J Immunol. 2009;183(2):1253–62. doi: 10.4049/jimmunol.0900570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6(6):513–22. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerotto M, Dal Pero F, Pontisso P, Noventa F, Gatta A, Alberti A. Two PKR inhibitor HCV proteins correlate with early but not sustained response to interferon. Gastroenterology. 2000;119(6):1649–55. doi: 10.1053/gast.2000.20230. [DOI] [PubMed] [Google Scholar]
- Gimenez-Barcons M, Wang C, Chen M, Sanchez-Tapias JM, Saiz JC, Gale M., Jr The oncogenic potential of hepatitis C virus NS5A sequence variants is associated with PKR regulation. J Interferon Cytokine Res. 2005;25(3):152–64. doi: 10.1089/jir.2005.25.152. [DOI] [PubMed] [Google Scholar]
- Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;(357):re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- Hassan M, Selimovic D, Ghozlan H, Abdel-Kader O. Induction of high-molecular-weight (HMW) tumor necrosis factor(TNF) alpha by hepatitis C virus (HCV) non-structural protein 3 (NS3) in liver cells is AP-1 and NF-kappaB-dependent activation. Cell Signal. 2007;19(2):301–11. doi: 10.1016/j.cellsig.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Hiscott J, Nguyen TL, Arguello M, Nakhaei P, Paz S. Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene. 2006;25(51):6844–67. doi: 10.1038/sj.onc.1209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Yasui K, Mukaigawa J, Katsume A, Kohara M, Mitamura K. Acquisition of susceptibility to hepatitis C virus replication in HepG2 cells by fusion with primary human hepatocytes: establishment of a quantitative assay for hepatitis C virus infectivity in a cell culture system. Hepatology. 2001;34(3):566–72. doi: 10.1053/jhep.2001.26752. [DOI] [PubMed] [Google Scholar]
- Iwamura T, Yoneyama M, Koizumi N, Okabe Y, Namiki H, Samuel CE, Fujita T. PACT, a double-stranded RNA binding protein acts as a positive regulator for type I interferon gene induced by Newcastle disease virus. Biochem Biophys Res Commun. 2001;282(2):515–23. doi: 10.1006/bbrc.2001.4606. [DOI] [PubMed] [Google Scholar]
- Joo M, Hahn YS, Kwon M, Sadikot RT, Blackwell TS, Christman JW. Hepatitis C virus core protein suppresses NF-kappaB activation and cyclooxygenase-2 expression by direct interaction with IkappaB kinase beta. J Virol. 2005;79(12):7648–57. doi: 10.1128/JVI.79.12.7648-7657.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Steele R, Ray R, Ray RB. Hepatitis C virus infection induces the beta interferon signaling pathway in immortalized human hepatocytes. J Virol. 2007;81(22):12375–81. doi: 10.1128/JVI.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Haque J, Lacoste J, Hiscott J, Williams BR. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc Natl Acad Sci U S A. 1994;91(14):6288–92. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103(15):6001–6. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HE, Ray CA, Oie KL, Pollara JJ, Petty IT, Sadler AJ, Williams BR, Pickup DJ. Modified vaccinia virus Ankara can activate NF-kappaB transcription factors through a double-stranded RNA-activated protein kinase (PKR)-dependent pathway during the early phase of virus replication. Virology. 2009;391(2):177–86. doi: 10.1016/j.virol.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CS, Toth AM, Zhang P, Devaux P, Cattaneo R, Samuel CE. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J Virol. 2010;84(1):380–6. doi: 10.1128/JVI.02630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsurro V, Beghelli S, Wang R, Barbi S, Coin S, Di Pasquale G, Bersani S, Castellucci M, Sorio C, Eleuteri S, Worschech A, Chiorini JA, Pederzoli P, Alter H, Marincola FM, Scarpa A. Anti-viral state segregates two molecular phenotypes of pancreatic adenocarcinoma: potential relevance for adenoviral gene therapy. J Transl Med. 2010;8:10. doi: 10.1186/1479-5876-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oem JK, Jackel-Cram C, Li YP, Kang HN, Zhou Y, Babiuk LA, Liu Q. Hepatitis C virus non-structural protein-2 activates CXCL-8 transcription through NF-kappaB. Arch Virol. 2008;153(2):293–301. doi: 10.1007/s00705-007-1103-1. [DOI] [PubMed] [Google Scholar]
- Park J, Kang W, Ryu SW, Kim WI, Chang DY, Lee DH, Youn Park D, Choi YH, Choi K, Shin EC, Choi C. Hepatitis C virus infection enhances tumor necrosis factor-alpha-induced cell death via suppression of nuclear factor-kappaB. Hepatology. 2012 doi: 10.1002/hep.25726. [DOI] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Ray RB, Steele R, Basu A, Meyer K, Majumder M, Ghosh AK, Ray R. Distinct functional role of Hepatitis C virus core protein on NF-kappaB regulation is linked to genomic variation. Virus Res. 2002;87(1):21–9. doi: 10.1016/s0168-1702(02)00046-1. [DOI] [PubMed] [Google Scholar]
- Raychoudhuri A, Shrivastava S, Steele R, Dash S, Kanda T, Ray R, Ray RB. Hepatitis C virus infection impairs IRF-7 translocation and Alpha interferon synthesis in immortalized human hepatocytes. J Virol. 2010;84(21):10991–8. doi: 10.1128/JVI.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HD, Mathews MB. The regulation of the protein kinase PKR by RNA. Biochimie. 1996;78(11–12):909–14. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem. 2007;282(51):37158–69. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- Taddeo B, Luo TR, Zhang W, Roizman B. Activation of NF-kappaB in cells productively infected with HSV-1 depends on activated protein kinase R and plays no apparent role in blocking apoptosis. Proc Natl Acad Sci U S A. 2003;100(21):12408–13. doi: 10.1073/pnas.2034952100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285(5424):107–10. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327(5963):286–90. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toroney R, Nallagatla SR, Boyer JA, Cameron CE, Bevilacqua PC. Regulation of PKR by HCV IRES RNA: importance of domain II and NS5A. J Mol Biol. 2010;400(3):393–412. doi: 10.1016/j.jmb.2010.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilasco M, Larrea E, Vitour D, Dabo S, Breiman A, Regnault B, Riezu JI, Eid P, Prieto J, Meurs EF. The protein kinase IKKepsilon can inhibit HCV expression independently of IFN and its own expression is downregulated in HCV-infected livers. Hepatology. 2006;44(6):1635–47. doi: 10.1002/hep.21432. [DOI] [PubMed] [Google Scholar]
- Williams BR. Signal integration via PKR. Sci STKE. 2001;(89):re2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- Yan XB, Battaglia S, Boucreux D, Chen Z, Brechot C, Pavio N. Mapping of the interacting domains of hepatitis C virus core protein and the double-stranded RNA-activated protein kinase PKR. Virus Res. 2007;125(1):79–87. doi: 10.1016/j.virusres.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci U S A. 2006;103(7):2310–5. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian-Daryoush M, Mogensen TH, DiDonato JA, Williams BR. NF-kappaB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-kappaB-inducing kinase and IkappaB kinase. Mol Cell Biol. 2000;20(4):1278–90. doi: 10.1128/mcb.20.4.1278-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel MB, Baumert TF. Production of infectious hepatitis C virus in tissue culture: a breakthrough for basic and applied research. J Hepatol. 2006;44(2):436–9. doi: 10.1016/j.jhep.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102(26):9294–9. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.