Abstract
Phospholipase C-β (PLCβ) is directly activated by Gαq, but the molecular basis for how its distal C-terminal domain (CTD) contributes to maximal activity is poorly understood. Herein we present both the crystal structure and cryo-EM 3D reconstructions of human full-length PLCβ3 in complex with murine Gαq. The distal CTD forms an extended, monomeric helical bundle consisting of three anti-parallel segments with structural similarity to membrane-binding bin–amphiphysin–Rvs (BAR) domains. Sequence conservation of the distal CTD identifies putative membrane and protein interaction sites, the latter of which bind the N-terminal helix of Gαq in both the crystal structure and cryo-EM reconstructions. Functional analysis suggests the distal CTD plays roles in membrane targeting and in optimizing the orientation of the catalytic core at the membrane for maximal rates of lipid hydrolysis.
Phospholipase C (PLC) isozymes hydrolyze the inner membrane lipid phosphatidylinositol-4,5-bisphosphate (PIP2) to generate the second messengers diacylglycerol and inositol 1,4,5-triphosphate (IP3), which in turn promote the release of intracellular calcium and activate protein kinase C (PKC)1,2. Among the PLC subfamilies, PLCβ enzymes are unique in their ability to be stimulated via direct interactions with the heterotrimeric G protein subunits Gαq and Gβγ3–5. PLCβ proteins contribute to diverse cellular functions, including chemotaxis6, neural signaling7and opioid sensitivity8. More recently, excess Gαq signaling has been implicated in the development of ocular cancer9. One of the best characterized roles of PLCβ is in the cardiovascular system, where it regulates cardiomyocyte and vascular smooth muscle function10–12. Maladaptive changes in these pathways can result in the onset and maintenance of cardiac arrhythmias13, cardiac hypertrophy14,15, and heart failure16,17.
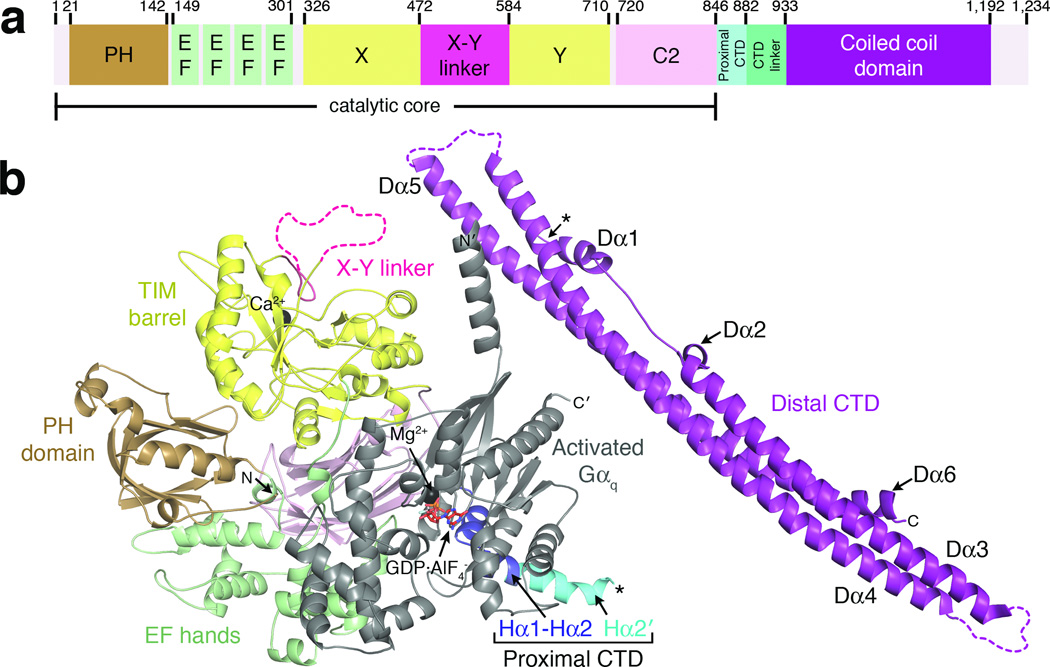
Like most other PLC enzymes, PLCβ contains an N-terminal pleckstrin homology (PH) domain, four tandem EF hand repeats, a triose phosphate isomerase (TIM) barrel-like catalytic domain separated into X and Y halves by the X-Y linker, and a C2 domain1,2, which forms the catalytic core of the enzyme. Unlike other PLC isoforms, PLCβ also has a ~400 amino acid C-terminal extension to the catalytic core that contains the proximal and distal C-terminal domains (CTDs)18,19 separated by a non-conserved, low complexity linker of variable length (Fig. 1a, Supplementary Fig. 1). The proximal CTD is a highly conserved ~40 amino acid segment that contains the principal binding site for activated Gαq (the Hα1-Hα2 hairpin)18 followed by an autoinhibitory helix (Hα2') that binds to the catalytic core19. When Gαq binds to the hairpin, the Hα2' helix is displaced from the catalytic core, leading to enhanced lipid hydrolysis. The distal CTD is less well conserved but numerous studies suggest that it is important for full activity, membrane binding, regulation by Gαq, and GAP activity19–29. However, the molecular basis for how it contributes to these processes remains poorly defined even in light of the structure of a distal CTD from turkey PLCβ 26. To elucidate a structural basis for the functional roles of the distal CTD, we used X-ray crystallography and single particle electron cryo-microscopy (cryo EM) to determine the structure of human full-length PLCβ3 in complex with activated Gαq, revealing the distal CTD in the context of a fully functional signaling complex where it forms unanticipated interactions with Gαq and the catalytic core of PLCβ that likely contribute to regulation.
Figure 1.
Crystal structure of Gαq–PLCβ3 reveals the C-terminal coiled-coil domain (distal CTD) in the context of a fully active signaling complex. (a) Primary structure of human PLCβ3. Numbers above the diagram correspond to domain boundaries. The catalytic core of the enzyme is defined as extending from the N-terminus to the end of the C2 domain. (b) A monomer of Gαq–PLCβ3 from the asymmetric unit. Domains of PLCβ3 are colored as in (a). The observed C-terminus of the proximal CTD and the N-terminus of the distal CTD are marked with black asterisks, although this represents only one possible linkage between the catalytic core and distal CTD in the crystal lattice (see Supplementary Fig. 4). The observed N and C-termini of PLCβ3 and Gαq are labeled N and C, and N' and C', respectively. Ca2+ and Mg2+ are shown as black spheres. Activated Gαq is shown in gray, with bound GDP and AlF4− drawn as red sticks. Dashed lines correspond to disordered loops.
RESULTS
Crystal Structure of Gαq in Complex with Human PLCβ3
The Gαq–PLCβ3 complex crystallized as an asymmetric dimer, with the two-fold interface mediated by the Rac1-binding surface of the PH domains (Table 1, Fig. 1b, Supplementary Fig. 2a). Each Gαq–PLCβ3 catalytic core complex is similar to the structure of Gαq in complex with PLCβ3-Δ887 (PDB entry 3OHM18) (r.m.s.d. of 0.57 Å for 932 Cα atoms from Gαq and residues 11–867 of PLCβ3), although the relative orientation of Gαq with respect to the catalytic core differs from 3OHM by 3° in each Gαq–PLCβ3 catalytic core complex. The greatest conformational differences are observed in theβ1-β2 andβ5-β6 loops of the PH domain, likely due to their contribution to the dimer interface (Supplementary Fig. 2b), and in the Hα2' helix, which adopts a different orientation in each of the Gαq–PLCβ3 complexes in this structure and the 3OHM structure (Supplementary Fig. 2c).
Table 1.
Data collection and refinement statistics
| Gαq–PLCβ3 | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 91.2, 190, 291 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 20.0–4.00 (4.07–4.00)* |
| Rmerge | 0.14 (0.33) |
| I / σI | 8.8 (1.9) |
| Completeness (%) | 76.7 (36.7) |
| Redundancy | 3.3 (2.1) |
| Refinement | |
| Resolution (Å) | 20.0–4.0 |
| No. reflections | 32,433 |
| Rwork / Rfree | 0.214/0.255 |
| No. atoms | |
| Protein | 39,306 |
| Ligand/ion | 98 |
| B-factors (Å2) | |
| Protein | 136 |
| Ligand/ion | 100 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.86 |
Values in parentheses are for highest-resolution shell. Data sets collected from multiple regions of two crystals were merged, and a σ cutoff of 0 was applied during scaling to help limit the effects of radiation damage that occurred over each sweep of data. The data was 85.6% complete at 6.8 Å resolution. Before applying the σ cutoff, there were 41,201 total reflections, corresponding to 91.1% overall completeness.
Only one copy of the distal CTD is observed in the asymmetric unit, despite the preponderance of full length PLCβ3 in the crystals (Supplementary Fig. 3), suggesting the other distal CTD is disordered rather than proteolyzed. The CTD linker (residues 883–933) is not observed in the structure, and thus the distal CTD could be attached to a number of different adjacent catalytic cores in the crystal lattice (e.g. Supplementary Fig. 4). The distal CTD (residues 934–1192) consists of three primarily anti-parallel helical segments that extend nearly the full-length (~140 Å) of the domain (Fig. 1b, 2a). The first segment contains the Dα1 (residues 935–946), Dα2 (954–958) and Dα3 (960–1008) helices. Dα3 forms an extended anti-parallel coiled-coil interaction with the Dα4 helix (residues 1027–1107) in the second segment. Dα4 in turn forms an extended anti-parallel coiled-coil interaction with Dα5 (residues 1115–1183) in the third segment. The Dα1 and Dα2 helices at the beginning of the first segment, and Dα6 (residues 1184–1192) at the end of the third segment, form “arms” that cross the same face of the domain and pack against the principal helices of the other two segments (Fig. 1b, 2a).
Figure 2.
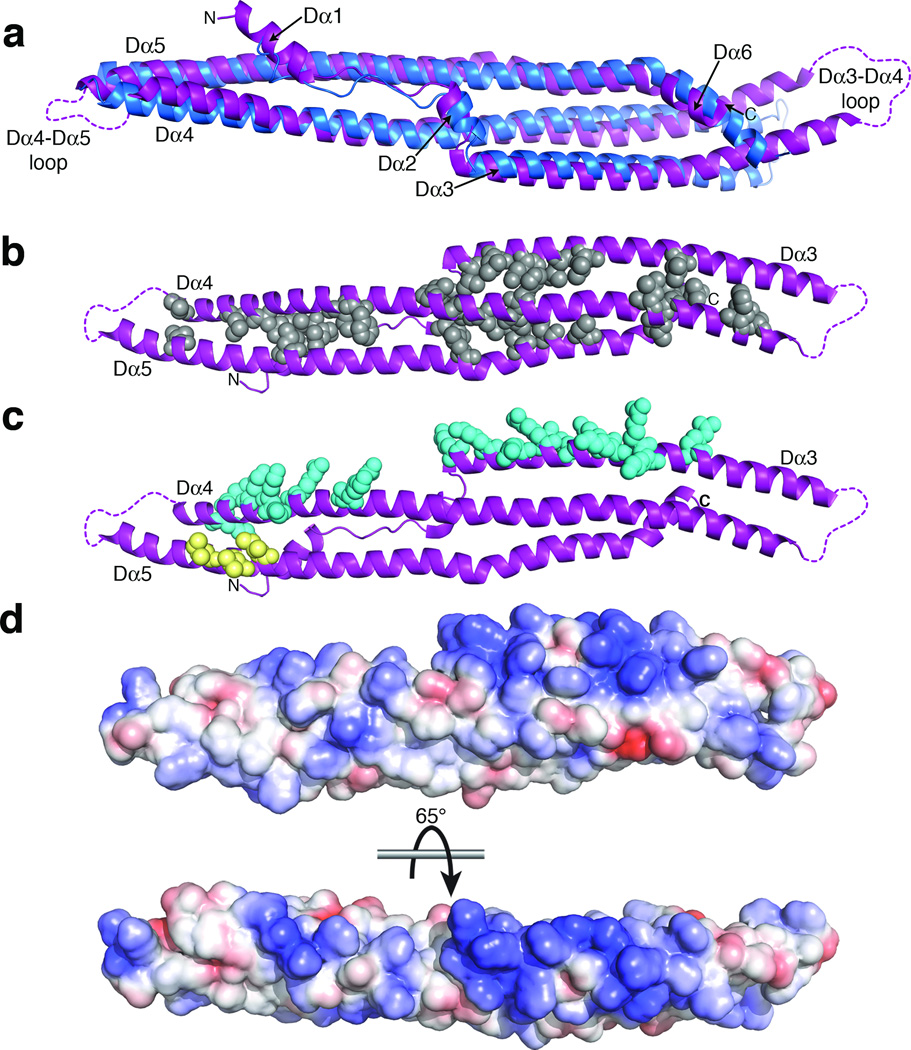
Structural comparison and sequence conservation of the PLCβ distal CTD. (a) Superposition of the distal CTDs from PLCβ3 (purple) and turkey PLCβ (blue)26. Although they have the same fold, they differ substantially in conformation in that the PLCβ3 distal CTD is more curved. Disordered loops within the distal CTD are marked as dashed lines. (b) The distal CTD is stabilized by conserved hydrophobic residues (gray spheres) that pack between the three segments of the domain (see Supplementary Fig. 1). The view is from the opposite side of the domain as shown in panel (a), rotated around a horizontal axis. (c) The distal CTD has two conserved, solvent-exposed regions: a basic ridge along Dα3 and Dα4 (blue spheres), and a hydrophobic patch on Dα5 (yellow spheres). In our structure the hydrophobic patch interacts with the N-terminus of Gαq (see Fig. 1b and Fig. 4a, c). (d) Electrostatic surface of the distal CTD. The top image corresponds to the same view as in (b). The surface is colored from −4 kT·e−1 (red) to +4 kT·e−1 (blue).
Although clearly sharing the same fold, the PLCβ3 distal CTD superimposes poorly with that of turkey PLCβ (PDB entry 1JAD26) (Fig. 2a), with an r.m.s.d. of 4.4 Å for the Dα2-Dα6 helices (197 Cα atoms), much higher than the expected 1.6 Å r.m.s.d. based on sequence identity alone (27%)30. One difference lies in the Dα5 helix, which is kinked in the PLCβ3 CTD structure by Pro1166. A second difference is that the PLCβ3 CTD is more curved than the turkey CTD, such that when the two domains are superimposed, the ends of the PLCβ3 domain are bent more towards the side of the domain with the “arms”. This could be due to differences in the crystal packing of each structure, or to the fact that the turkey CTD crystallized as a homodimer, or to the multiple protein-protein interactions formed by the PLCβ3 distal CTD within its crystal lattice, or to a combination of all these factors. Although the turkey CTD contained a deletion in the Dα3-Dα4 loop at one end of the domain, this deletion corresponds to residues that are disordered in the PLCβ3 CTD and are therefore unlikely to be responsible for global differences in domain conformation. A homology search using Dali31 confirmed the structural similarities between the distal CTD and the turkey CTD structure (Z-scores of 15.4 and 8.6 for the B and A chains of the turkey CTD homodimer, respectively) as well as homology to bin-amphiphysin-Rvs (BAR) domains, with Z-scores between 6.7–8.3. BAR domains are also extended helical bundles composed of three helical segments and have a similar topology (Supplementary Fig. 5). BAR domains interact nonspecifically with negatively charged phospholipids, as has been reported for the PLCβ isozymes25,28,29,32, and can sense and/or induce membrane curvature33,34.
Conservation of the Distal CTD Reveals Putative Interaction Surfaces
Most of the highly conserved residues in the distal CTD (Supplementary Fig. 1) are hydrophobic and pack between the three segments of the domain to form a hydrophobic core, with the largest cluster located at the interface of the Dα2, Dα4, and Dα5 helices at the center of the domain (Fig. 2b). Two other clusters of conserved residues are on the surface of the domain and likely serve other functions. A ridge of highly conserved basic residues runs along the same face of the Dα3 and Dα4 helices (Fig. 2c) and constitutes a potential membrane-interaction site. Indeed, previous studies found that deletion of the distal CTD abolishes particulate association21,23,25,28, and mutation of conserved basic residues within Dα3 of PLCβ1 impairs maximal Gαq-stimulated activity21,27. The electrostatic surface of the domain (Fig. 2d) is also consistent with the ability to bind negatively charged membranes. Some BAR domains also have basic patches located at similar topological positions to those observed in Dα3 and Dα4, suggesting a functional relationship with the PLCβ distal CTD33–35. A second patch of conserved surface residues is formed by residues Ile1129, Val1133, and Ile1136 in the Dα5 helix (Fig. 2c). This region is orthogonal to the putative membrane-binding surface, and may therefore represent a protein-protein interaction site involved in regulation of PLCβ at the membrane.
Solution Architecture of the Gαq–PLCβ3 Complex
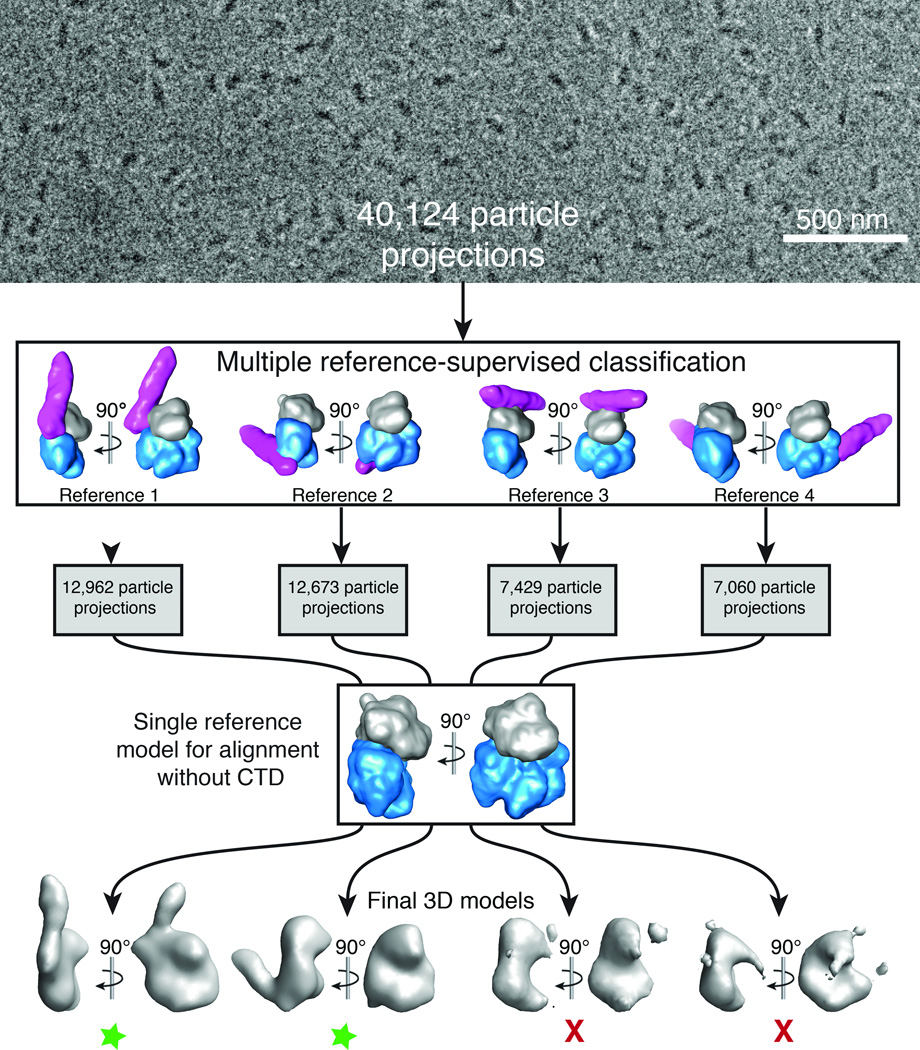
The distal CTD makes several direct contacts with Gαq and the catalytic core of PLCβ3 in the crystal lattice. Although of all these interactions could not be formed at the same time by a single distal CTD in the context of a PLCβ monomer, each one represents an interaction that could contribute to activity and/or regulation. To identify which of the interactions made by the distal CTD also occur in solution, we used single particle cryo-EM to determine the solution structure of the Gαq–PLCβ3 complex. A combination of thin vitrified ice and CCD image acquisition under an electron accelerating voltage of 120 kV allowed adequate contrast for alignment of the ~180 kDa protein particle projections (Fig. 3). We initially calculated a preliminary 3D reconstruction from our entire dataset of 40,124 projections by using the Gαq–PLCβ3 structure without the distal CTD as initial reference. The reconstruction was characterized by relatively poor definition for the Gαq–PLCβ3 core, and lacked clear density for the distal CTD. Instead, several noise-like densities appeared at various positions about the core complex (Supplementary Fig. 6).
Figure 3.
Cryo-EM projection classification and 3D reconstruction scheme. 40,124 particle projections were subjected to multiple 3D reference-supervised classification against volumes of four Gαq–PLCβ3 core complexes observed in the crystal lattice. The density corresponding to Gαq is shown in gray, the PLCβ3 core in blue, and the distal CTD in purple. Each group of particles was used to generate a unique 3D reconstruction using the Gαq–PLCβ3 structure, omitting the distal CTD, as the initial reference. Two final 3D reconstructions were of good quality as judged by the definition of the Gαq–PLCβ3 core complex (green stars). In the first model the distal CTD is docked with the N-terminal helix of Gαq, and in the second with the hydrophobic ridge of the catalytic core of PLCβ3. The third represents a contact between the Dα3-Dα4 loop of the distal CTD and the second EF hand (Eα2 and Fα2 helices) of the catalytic core. The fourth model represents an interaction between the distal CTD and the Ras-like domain of Gαq (Supplementary Fig. 2d). Based on functional analysis (Table 2) and failure to obtain reliable reconstructions, the last two interactions (models 3 and 4) likely represent simple lattice contacts. A representative cryo-EM image of purified Gαq–PLCβ3 complexes is shown at the top of the figure.
To test whether the observed peripheral densities corresponded to flexibility or multiple distinct conformations of the distal CTD with respect to the catalytic core, we employed multiple 3D reference-supervised classification to categorize the cryo-EM projections. The categorization was based on the similarities of projections to reprojections of four 20 Å-filtered volumes representing different direct Gαq–PLCβ3 interactions with the distal CTD that were observed in the crystal lattice. We then used the cryo-EM projections in each category to generate four independent 3D reconstructions, all using the same starting reference, which corresponded to a 20 Å-filtered volume of the Gαq–PLCβ3 catalytic core without the distal CTD (Fig. 3). The omission of the CTD density in the reference volume ensured that we eliminated any reference bias for its position with respect to the catalytic core.
The majority of cryo-EM projections showed good agreement with two of the initial reference structures (>30% of projections each, Fig. 3). These two subgroups provided 3D reconstructions with very good definition for the core complex and additional elongated density attached at two different positions (Fig. 4a, b). These non-biased reconstructions (for the CTD), with estimated resolutions of 19 Å and 21 Å (at FSC=0.5, Supplementary Fig. 7), showed excellent agreement with the two corresponding models used for particle categorization with a cross-correlation greater than 0.93. In contrast, the other subgroups (with ~18% of projections each, Fig. 3) not only failed to reveal a clear density for the distal CTD, but also showed very poor volumes for the Gαq–PLCβ3 core complex. This may indicate that the orientation of the distal CTD within these subgroups is highly variable, resulting in overall projection misalignment and consequently poor quality 3D reconstructions.
Figure 4.
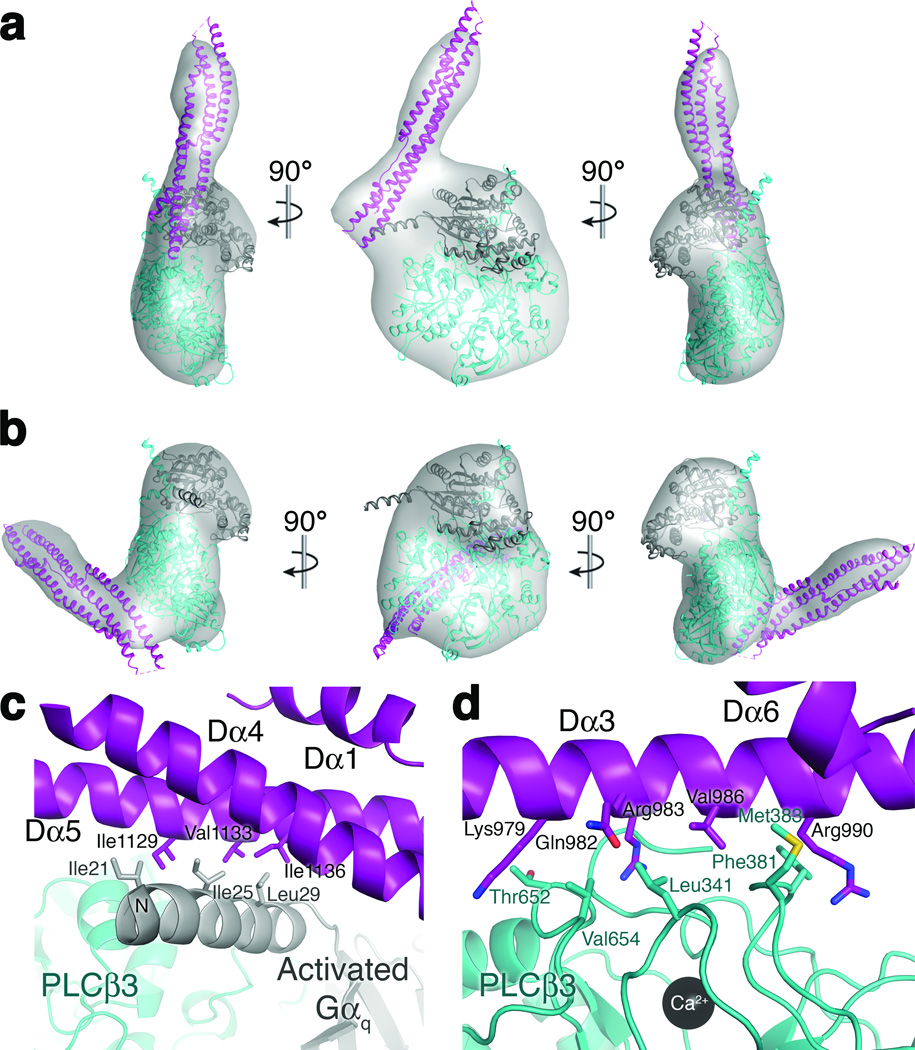
The distal CTD interacts with the N-terminal helix of Gαq and the PLCβ3 catalytic core in solution and in crystals. (a) A 19 Å 3D EM reconstruction with docked crystal structure of the distal CTD in complex with the N-terminal helix of Gαq. The distal CTD is shown in purple, the PLCβ3 core in teal, and Gαq in gray. (b) A 21 Å 3D reconstruction with docked crystal structure of the distal CTD in complex with the hydrophobic ridge of the catalytic core. (c) The interface between the distal CTD hydrophobic patch and the N-terminal helix of Gαq, burying ~850 Å2 of accessible surface area. (d) The interface between the Dα3 helix and the hydrophobic ridge of the catalytic core, burying ~1000 Å2 of accessible surface area.
Therefore, under our experimental conditions, two of the interactions of the distal CTD are favored in solution and recapitulated in the crystal lattice (Fig. 4). Both involve conserved residues. The first interaction is between the distal CTD and the N-terminal amphipathic helix of Gαq (Fig. 4a, c). The second is between the distal CTD and the hydrophobic ridge of the PLCβ3 catalytic core (Fig. 4b, d), a cluster of conserved and solvent exposed hydrophobic residues adjacent to the active site that are thought to insert into the membrane and promote catalysis36,37. The latter interaction would therefore be predicted to simultaneously hinder access of substrates into the active site and prevent the conserved basic patch on Dα3 from interacting with membranes.
Functional Analysis of the Gαq N-terminus–Distal CTD Interaction
To assess the significance of the interaction between the distal CTD and the N-terminal helix of Gαq (Fig. 4a, c) we expressed a truncated Gαq in which the first 36 residues, corresponding to its N-terminal helix, were deleted (Gαq-ΔN), and a PLCβ3 I1129E V1133E I1136E triple mutant (PLCβ3 EEE), which eliminates the hydrophobic patch on Dα5. We used a liposome-based assay to measure the rates of basal and Gαq-stimulated PIP2 hydrolysis. The maximum activities of PLCβ3 and PLCβ3 L876A, which contains a mutation that abrogates Hα2' autoinhibition, were significantly lower when stimulated by Gαq-ΔN than wild-type Gαq (Table 2, Supplementary Fig. 8a, b). The Gαq-stimulated activities of PLCβ3 EEE and PLCβ3 L876A EEE were also significantly reduced compared to wild-type PLCβ3 and PLCβ3 L876A, respectively. Conversely, the activities of PLCβ3-Δ892, which lacks the distal CTD, and PLCβ3 EEE were the same for Gαq and Gαq-ΔN, indicating these PLCβ3 variants were insensitive to the presence of the N-terminal helix of Gαq (Table 2).
Table 2.
Functional analysis of interactions of the distal CTD
| PLCβ3 variant | Basal specific activity ± SEM (nmol IP3/min/ nmol PLCβ3) (n)b |
Gαq variant | Ki ± SEM (nM) (n)b |
EC50 ± SEM (nM) |
Max specific activity ± SEM (nmol IP3/min/nmol PLCβ3) (n)b |
|---|---|---|---|---|---|
| PLCβ3 | 54 ± 2.7* | Gαq | 220 ± 30 (24) | 120 ± 30 | 2900 ± 130 (32) |
| -ΔN | 270 ± 110 (9) | 39 ± 1 | 1600 ± 100 (6) | ||
| -K311A | 260 ± 39 (4) | 130 ± 21 | 2900 ± 360 (10) | ||
| -D296A/P298D | 270 ± 23 (3) | 430 ± 110 | 3400 ± 400 (12) | ||
| -C9S/C10S | NDa | 90 ± 11 | 1800 ± 190 (6) | ||
| PLCβ3 L876A | 950 ± 69* | Gαq | 20 ± 3.2 (11) | 19 ± 2 | 6500 ± 460 (6) |
| -ΔN | 24 ± 7.7 (7) | 5 ± 1 | 2800 ± 250 (6) | ||
| -K311A | 16 ± 3.9 (3) | 14 ± 2 | 4400 ± 150 (4) | ||
| -D296A/P298D | 12 ± 3.4 (4) | 27 ± 3 | 6800 ± 640 (5) | ||
| -C9S/C10S | NDa | 40 ± 4 | 3900 ± 440 (6) | ||
| PLCβ3-Δ892 | 4.7 ± 0.6* | Gαq | 490 ± 66 (14) | 59 ± 7 | 39 ± 2 (6) |
| -ΔN | 490 ± 130 (6) | 59 ± 7 | 31 ± 1 (6) | ||
| -K311A | 380 ± 70 (3) | 150 ± 20 | 48 ± 3 (6) | ||
| -D296A/P298D | 420 ± 88 (6) | 540 ± 250 | 40 ± 4 (8) | ||
| -C9S/C10S | NDa | 150 ± 20 | 29 ± 2 (6) | ||
| PLCβ3 EEE | 46 ± 7 (9) | Gαq | 93 ± 23 (5) | 280 ± 130 | 1900 ± 170 (5) |
| -ΔN | 78 ± 26 (3) | 120 ± 30 | 2000 ± 150 (6) | ||
| PLCβ3 L876AEEE | 730 ± 63 (10) | Gαq | 26 ± 9.1 (4) | 32 ± 20 | 3900 ± 1300 (4) |
| -ΔN | 42 ± 30 (4) | 9 ± 3 | 3400 ± 680 (6) | ||
| PLCβ3 T652E | 5.0 ± 0.5 (6) | Gαq | 160 ± 38 (5) | 1150 ± 790 | 460 ± 170 (6) |
| PLCβ3-Δ892 T652E | 1.0 ± 0.1 (7) | Gαq | 250 ± 88 (3) | 250 ± 80 | 13 ± 2 (4) |
| PLCβ3-Δ882-937 | 200 ± 8 (7) | Gαq | 230 ± 100 (3) | NDa | 200 ± 20 (6) |
| PLCβ3-Δ882-937 L876A | 500 ± 95 (9) | Gαq | 12 ± 2.3 (3) | NDa | 450 ± 25 (5) |
Previously reported19.
Not Determined.
Number (n) of independent experiments conducted in duplicate.
We also tested whether perturbation of the Gαq interfaces formed by the distal CTD would decrease binding affinity for activated Gαq. We used a flow cytometry-based bead-binding assay to measure the ability of PLCβ3 variants to displace a fluorescently labeled mutant of PLCβ3 (PLCβ3-Δ892 R872A L876A L879A)19 from bead-bound Gαq. In this assay, PLCβ3 bound Gαq with higher affinity than PLCβ3-Δ892 (Ki values of 220 ± 30 nM and 490 ± 66 nM, respectively), but with significantly less affinity than PLCβ3 L876A (Ki= 20 ± 3.2 nM) (Table 2, Supplementary Fig. 8c), consistent with the L876A mutation abrogating the autoinhibitory interaction of Hα2' with the catalytic core19. Although these Ki values are overall higher than reported in prior studies19, they maintain the same trends. The values are also higher than the EC50 values we measured for activation. This is likely because the bead-binding assay lacks liposomes, which likely enhance the apparent affinity of Gαq and PLCβ3. We next tested whether the interaction between the distal CTD and the N-terminal helix of Gαq was responsible for the difference in binding affinities between PLCβ3 and PLCβ3-Δ892, using the Gαq-ΔN and PLCβ3 EEE variants to abolish the interface. However, neither variant showed a significant binding defect (Table 2, Supplementary Fig. 8d), consistent with their similar EC50 values for activation. Thus, our results support the conclusion that a direct interaction between the N-terminus of Gαq and the hydrophobic patch of Dα5 in the distal CTD occurs during catalysis that is required for full efficacy, but not potency.
Gαq Palmitoylation Also Contributes to Full Efficacy
The Gαq proteins used in these experiments were purified from the soluble fraction of baculovirus-infected insect cells, but some fraction of these proteins may retain palmitoyl groups at Cys9 and Cys10 (ref.38), which could account for the higher efficacy observed for full-length Gαq relative to Gαq-ΔN. Acyl-biotin exchange chemistry confirmed at least a fraction of the purified Gαq was palmitoylated (Supplementary Fig. 9). Thus, we assessed the ability of palmitoylation-defective Gαq (Gαq C9S C10S) to activate PLCβ3 variants (Table 2, Supplementary Fig. 8b). Gαq-ΔN and Gαq C9S C10S activated PLCβ3 and PLCβ3 L876A with similarly low efficacies, indicating that palmitoylation does contribute to the higher efficacy observed for wild-type Gαq in vitro. However, palmitoylated Gαq likely increases PLCβ3 activity through a mechanism other than increased localization of the complex to vesicles, as it does not seem to increase association of PLCβ3 with membranes25,38,39. PLCβ3-Δ892 was activated to similar extents by Gαq, Gαq-ΔN and Gαq C9S C10S (Table 2), indicating that the palmitoylation status of Gαq is irrelevant in the absence of the distal CTD.
PLCβ3 and the Gαq–PLCβ3 Complex Are Monomeric
The residues that comprise the hydrophobic patch in Dα5 contribute to a homodimer interface in the turkey distal CTD structure26. Therefore, defects in specific activity exhibited by mutants such as PLCβ3 EEE could be attributed to disruption of the dimer interface. In addition, dimerization of the distal CTD, though structurally incompatible with the Gαq N-terminal helix-distal CTD interaction (Fig. 4a, c), could occur simultaneously with the PLCβ3 core-distal CTD interface (Fig. 4b, d). We therefore tested whether full-length PLCβ3 or Gαq–PLCβ3 formed higher order complexes using multi-angle light scattering. PLCβ3 and Gαq–PLCβ3 elute from a WTC-050S5 silica-based column with molecular weights of 152 kDa and 182 kDa, respectively, very close to the expected molecular weights for monomeric PLCβ3 (139 kDa) and Gαq–PLCβ3 species (181 kDa). Furthermore, cryo-EM images of the Gαq–PLCβ3 complex incubated on the surface of PIP2 lipid monolayers did not reveal higher order assemblies of the complex (data not shown), suggesting membrane association does not induce oligomerization. Thus, the differences in efficacy measured for mutants such as PLCβ3 EEE do not seem to be linked to changes in the oligomeric state of the enzyme.
Analysis of Other Interfaces of the Distal CTD Formed in Crystals
The interface formed between the distal CTD and the PLCβ3 hydrophobic ridge (Fig. 4b, d) could represent a mechanism by which the distal CTD contributes to the regulation of basal activity. We therefore generated Leu341, Phe381, Met383, Thr652, and Val654 point mutants in both PLCβ3 and PLCβ3-Δ892. Of these, only T652E could be expressed and purified in the background of the PLCβ3 construct, suggesting these residues are essential for protein folding and/or stability. PLCβ3 T652E and PLCβ3-Δ892 T652E showed significant decreases in basal activity, higher EC50 values, and greatly reduced efficacies upon Gαq activation, confirming the importance of the hydrophobic ridge in PLCβ3 for overall activity (Table 2). The activity of the T652E mutant was the same regardless of the presence of the distal CTD, suggesting that the distal CTD does not influence basal activity. However, our inability to confirm this with other mutations in this region renders interpretation difficult.
The distal CTD also contacts the Ras-like domain of Gαq, burying ~850 Å2 of surface area (Supplementary Fig. 2d). In this interaction, the Gαq α3-α4 loop and α4 helix interact with residues in Dα3 and Dα4 of the distal CTD. Gαq Pro298 forms a hydrophobic contact with the side chains of Arg1062 and Gln1066 in αB, and electrostatic interactions are formed between Gαq Asp296 and K311A and Dα3 residues Arg969, Asp973, and Glu976. To test this interaction, we expressed D296A P298D and K311A variants of Gαq and tested their ability to bind and activate PLCβ3 variants. All were able to activate PLCβ3 variants with EC50 values and maximum activity comparable to wild-type Gαq, although EC50 values for D296A P298D were elevated. However, none had a significant effect on binding affinity (Table 2).
Another interaction observed in crystals is between the Dα3-Dα4 loop of the distal CTD and the second EF hand (Eα2 and Fα2 helices) of the catalytic core. Although this interaction is sufficient to cause a conformational change in a loop of the EF hand relative to the 3OHM structure and the other chain of PLCβ3 in our structure, the residues involved in this interaction are poorly conserved and not well ordered. Indeed, Dα3-Dα4 loop residues 1009–1026 could not be modeled. Thus, this interaction most likely represents a simple crystal contact.
The CTD Linker Is Required for Gαq Activation
Point mutants of residues in the hydrophobic ridge that interact with the distal CTD suggest that these positions are essential for folding and basal activity, consistent with their proposed role in catalysis. We therefore indirectly tested this interface by deleting the CTD linker, which should prevent the distal CTD from forming any of the interactions observed in the crystal lattice or by cryo-EM. PLCβ3-Δ882–938 showed a significant increase in basal activity compared to PLCβ3 (Table 2). This increase could be attributed to a loss of autoinhibition mediated by the distal CTD or to an increase in the local concentration of the catalytic core at the membrane by virtue of a shorter linker. However, deletion of the CTD linker had no significant effect on basal activity in the background of PLCβ3 L876A (Table 2). Thus the mechanism by which basal activity is increased is not clear. Strikingly, Gαq failed to activate PLCβ3-Δ882–938 or PLCβ3-Δ882–938 L876A at all concentrations tested (Table 2, Supplemental Fig. 8a) even though both proteins bound Gαq with comparable affinity to PLCβ3 and PLCβ3 L876A, respectively (Table 2, Supplementary Fig. 8c). Because the sequence of the CTD linker is not conserved among PLCβ isozymes, its length and/or flexibility seems to be required for the proximal and distal CTDs to make all the interactions necessary for activation by Gαq.
DISCUSSION
The structure of Gαq in complex with full-length human PLCβ3 provides new insights into the structure and potential functions of the enigmatic distal CTD of PLCβ isozymes. One edge of this domain bears highly conserved basic residues that likely directly interact with negatively charged membranes. Structural similarity between the distal CTD and BAR domains (Supplementary Fig. 5) is also consistent with membrane targeting being a key role for the distal CTD, and raises the possibility that the distal CTD may likewise sense or influence membrane curvature33,34. Orthogonal to this surface is a conserved hydrophobic patch that interacts with the amphipathic N-terminal helix of Gαq in both the crystal structure and cryo-EM reconstructions. Our in vitro functional analysis supports a role for this interaction in promoting full efficacy of activation by Gαq. However, oligomerization does not appear to play a role in this process26 as PLCβ3 and the Gαq–PLCβ3 complex are monomeric by structural analysis and multi-angle light scattering.
Our structural and functional analysis of the distal CTD in the context of a fully functional signaling complex allows re-interpretation of prior functional studies. Ilkeava et al27 identified five regions in the distal CTD of rat PLCβ1 that caused significant defects in Gαq activation and GAP activity (“Group III” mutants; Supplementary Fig. 1). The first three regions perturbed the conserved basic residues in Dα3 and Dα4 that we propose to be involved in membrane binding. The fourth region perturbed the hydrophobic patch that interacts with the N-terminus of Gαq. Site-directed mutation of the conserved basic residues (particularly within Dα3) reduced Gαq-stimulated activity and greatly reduced association with the particulate fraction of cells21. Considered along with our data, substitution of the conserved basic residues most likely decreases Gαq-stimulated activity because the enzyme is impaired in its ability to associate with the membrane where its substrate is located. Defects in GAP activity resulting from perturbations of the distal CTD could likewise be explained as defects in membrane binding, as this property was measured in steady state assays using receptor and heterotrimeric G proteins reconstituted in lipid vesicles27.
However, membrane binding is clearly not the sole function of the distal CTD. Perturbation of the hydrophobic patch in Dα5 leads to a significant decrease in maximum Gαq-stimulated activity, despite the presence of palmitoylated Gαq and an intact membrane-binding surface on the distal CTD. Furthermore, deletion of the CTD linker leads to complete loss of Gαq activation in our assays (Table 2, Supplementary Fig. 8a). The simplest explanation for these observations is that the interaction between the distal CTD and the palmitoylated N-terminus of Gαq optimizes the orientation of the catalytic core at the membrane (Fig. 5)1. Thus, activation of the enzyme by Gαq is at least a combination of allosteric control (e.g. release of autoinhibition by Hα2')19 and proper positioning of the catalytic core. Either or both of these processes could also influence displacement of the X-Y linker from the active site through a process of interfacial activation18,40.
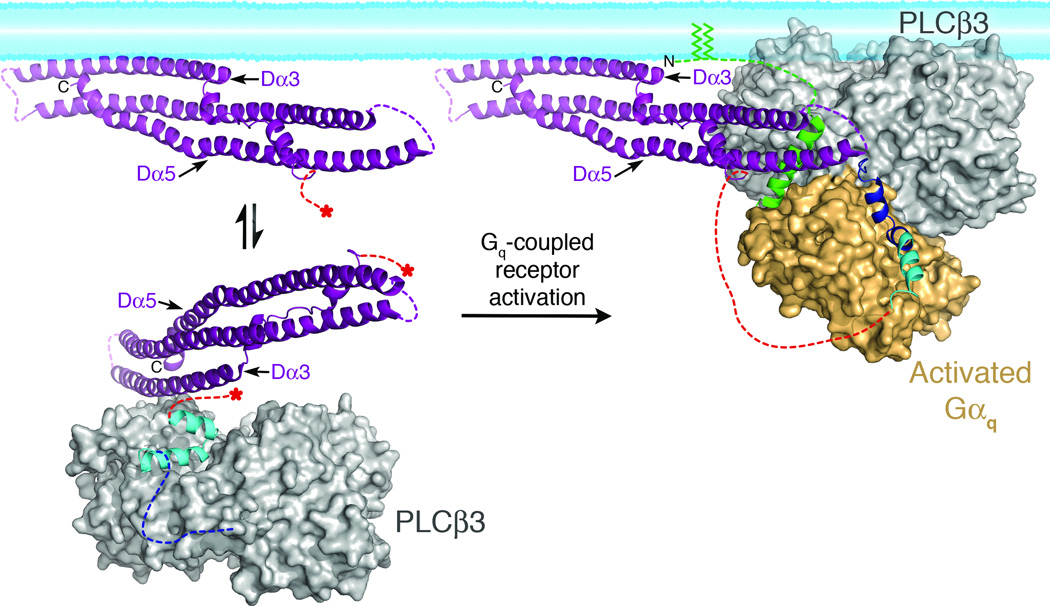
Figure 5.
Model of PLCβ3 regulation by the distal CTD and Gαq. In the absence of Gαq (left), the Hα2' inhibitory helix from the proximal CTD (cyan), is bound to the catalytic core (gray surface), stabilizing the enzyme in a more quiescent conformation (based on PDB entry 3QR0)19. The primary Gαq binding site (dashed blue line) is disordered in this state. The distal (purple) and proximal CTDs are connected by the disordered CTD linker (dashed red lines). Red asterisks mark the start and end of the linker in the basal state. The distal CTD could help partition the enzyme between membrane-associated and cytosolic populations by forming a transient interaction with the hydrophobic ridge of the catalytic core (see Fig. 4b, d). Upon activation, Gαq binds to the Hα1-Hα2 helical hairpin (blue), displacing Hα2' from the catalytic core. The hydrophobic patch of the distal CTD engages the N-terminal helix of Gαq (green), and helps orient the catalytic core of PLCβ with respect to the lipid bilayer. The palmitoylated cysteines of the Gαq N-terminus (green lines) also help anchor the complex at the membrane. This model was created by independently docking the N-terminal helix of Gαq in complex with the distal CTD, and the remainder of Gαq in complex with the PLCβ3 core at a common membrane surface, then translating the distal CTD complex such that the Gαq N-terminal helix makes a reasonable connection to the rest of Gαq. The model also brings the PH domain in close proximity to the membrane, consistent with studies indicating this domain also contributes to membrane binding44,47.
Adding to the complexity is the regulation of PLCβ by Gβγ subunits4,41 and small GTPases42,43. Gβγ and small GTPases are typically geranylgeranylated, resulting in their constitutive association with the cell membrane. Activation by Gβγ and Rac1 has also been proposed to involve the optimization of the orientation of the PLCβ active site at the membrane43,44, but the role of the distal CTD in response to these activators is less clear. Future structural studies will be essential in understanding how Gβγ interacts with and activates PLCβ, in some cases synergistically with Gαq45,46.
ONLINE METHODS
Protein expression, purification, and mutagenesis
N-terminally His-tagged human PLCβ3 (residues 10–1234) and variants were expressed in baculovirus infected insect cells and purified as described previously19, with the modification that resuspended cells were lysed by douncing on ice. Murine Gαq (residues 7–359) and variants were expressed and purified with a TEV-cleavable N-terminal His-tag as described previously19, with some modifications. Following elution from an Ni-NTA column, the protein was concentrated and applied to two tandem Superdex S200 columns equilibrated with 20 mM HEPES pH 8, 200 mM NaCl, 2 mM DTT, 1 mM MgCl2, and 10 µM GDP. Gαq used for complex crystallization had the His-tag removed by the addition of TEV protease at 2% (w/w) of Gαq, then dialyzed overnight at 4 °C in Buffer B (20 mM HEPES pH 8, 100 mM NaCl, 10 mM βME, 3 mM MgCl2, and 10 µM GDP). Following dialysis, the proteins were applied to a Ni-NTA column pre-equilibrated with Buffer B. The flow-through, containing TEV-cleaved Gαq, was collected, passed through the column three times, then concentrated and applied to two tandem Superdex S200 columns. Point mutations and internal deletions in the coding regions of PLCβ3 and Gαq were introduced using QuikChange Site-Directed Mutagenesis (Stratagene), and were confirmed by sequencing over the entire open reading frame.
Purification of the Gαq–PLCβ3 Complex
TEV-cleaved Gαq was incubated on ice with Buffer C (20 mM HEPES pH 8, 200 mM NaCl, 2mM DTT, 0.9 mM CaCl2, 5 mM MgCl2, 10 mM NaF, 30 µM AlCl3, and 50 µM GDP) for 15–20 min. Purified PLCβ3 was added to Gαq in a 1:1.2 molar ratio, and incubated for an additional 30 min on ice. The reaction was loaded on two tandem Superdex S200 columns equilibrated with Buffer C. Peak fractions containing Gαq–PLCβ3 were pooled and concentrated in a 100 kDa Amicon concentrator.
Crystallization and Structure Determination of the Gαq–PLCβ3 Complex
Gαq–PLCβ3 was suspended in hanging drops containing 1 µl Gαq–PLCβ3 at 6.2 mg ml−1 mixed with 1 µl well solution. The well solution contained 100 mM MES pH 6.75, 100–200 mM NaCl, and 11–12% PEG 3350, with crystals appearing in 4–7 d at 4 °C. Crystals were harvested in 20 mM HEPES pH 8, 400–500 mM NaCl, 2 M DTT, 0.9 mM CaCl2, 5 mM MgCl2, 10 mM NaF, 30 µM AlCl3, 50 µM GDP, 15% (w/v) PEG 3350, and 30% (v/v) 1,3-butanediol, then frozen on nylon loops in liquid nitrogen for data collection.
Diffraction data was collected at the Advanced Photon Source at LS-CAT Beam line 21-ID-D and GM/CA-CAT beam line 23-ID-D from crystals maintained at 110 K at wavelengths of 1.13 Å and 1.03 Å respectively. Although initial diffraction to 3.3 Å spacings was observed, rapid decay limited useful data to 4.0 Å spacings (Table 1). Data sets were reduced using HKL200048, and initial phases were derived by molecular replacement using the 3OHM structure18 as a search model. Models were refined via restrained refinement with TLS using the program REFMAC549. Five TLS groups were used: one for each Gαq chain, its ligands (Mg2+, GDP, and AlF4− molecules) and its bound PLCβ3 proximal CTD (residues 863–881), one for each PLCβ3 catalytic core and its associated Ca2+ atom, and one for the distal CTD. NCS restraints were imposed on the two Gαq chains (residues 39–353 and its ligands) and the two PLCβ3 chains (residues 16–194, 202–862, and the Ca2+ atoms). Stereochemical correctness of the final model was assessed with MolProbity50. In the final model, 94.7% of residues were in favored regions of the Ramachandran plot, 4.9% of residues in allowed regions, and 0.4% in disallowed regions. Residues in disallowed regions were also observed the 2.7 Å Gαq–PLCβ3-Δ887 structure18. Electron density was observed for residues 11–470 and 573–881 in chain A of PLCβ3, and 10–196, 199–470, and 575–881 in the second. Residues 471–572 correspond to a region of the X-Y linker that has low sequence homology and is typically poorly ordered18,19,40,43. In the distal CTD, the Dα3-Dα4 (residues 1009–1025) and Dα4-Dα5 (residues 1108–1114) loops and C-terminal residues 1193–1234 lacked electron density, consistent with being disordered (Supplementary Fig. 3a). In Gαq, electron density was observed for residues 18–354 in the first molecule and 35–355 in the second.
Cryo-EM Sample Preparation and Imaging
3 µl of purified Gαq–PLCβ3 was adsorbed on glow-discharged Quantifoil R2/2 200 mesh grids, and vitrified using a Vitrobot (FEI Mark IV). The specimen was imaged on a Tecnai F20 transmission electron microscope (FEI) equipped with a field emission gun and operated at 120kV. Images were recorded at a magnification of 71,138x on a Gatan US4000 CCD camera and defocus values ranging from –2 to –3 µm. All images were binned (2 × 2 pixels) to obtain a pixel size of 4.48 Å on the specimen level.
Cryo-EM 3D reconstructions and molecular modeling
40,124 particle projections from Gαq–PLCβ3 images were interactively selected and excised using Boxer (EMAN 1.9 software suite)51. The CTF parameters for each micrograph were determined using ctfit, and CTF correction was applied accordingly using the program Applyctf (part of EMAN 1.9 package). Multiple 3D reference-supervised classification was applied to the data set, using four possible models of the complex observed in the crystal lattice. These unique particle datasets were used to calculate four independent 3D reconstructions using the same initial reference of the Gαq–PLCβ3 core but lacking the distal CTD. 20 Å-filtered volumes of Gαq–PLCβ3 structures observed in the crystal lattice were computationally fitted as rigid bodies in the EM maps and cross-correlation values obtained using Chimera52. For a more detailed description, see Supplementary Note.
PLCβ3 activity assays
PLCβ3 basal and Gαq-stimulated activity was determined by measuring the amount of free [3H]-IP3 released from [3H]-PIP2-containing liposomes, as previously described19,53. PLCβ3 variants were assayed at a final concentration that resulted in activity within the linear range over the time course of the experiment. PLCβ3, PLCβ3 EEE, and PLCβ3-Δ882–937 were assayed at 0.3 ng µl−1 and PLCβ3 T652E at 3 ng µl−1. PLCβ3-L876A, PLCβ3 L876A EEE, and PLCβ3-Δ882–937 L876A were assayed at 0.05 ng µl−1. PLCβ3- Δ892 was assayed at 10 ng µl−1 and PLCβ3-Δ892 T652E at 20 ng µl−1.
Flow Cytometry Protein Interaction Assay (FCPIA)
Equilibrium binding of PLCβ3 variants to Gαq variants was measured using FCPIA as described previously19, wherein a PLCβ3 variant with high affinity for Gαq (PLCβ3-Δ892 R872A L876A L879A)19 was fluorescently labeled with Alexa-Fluor-488 (AF488) C5-maleimide (Invitrogen). Gαq proteins were biotinylated (b-Gαq)19,54 and linked to SPHERO Streptavidin Coated Particles (Spherotech). Unlabeled PLCβ3 variants were added to the bead-bound b-Gαq proteins over a range of concentrations, after which AF488-labeled PLCβ3-Δ892 R872A L876A L879A was added at its measured KD (40 nM) and incubated for 1 h prior to being processed with an Accuri C6 flow cytometer19.
Detection of Palmitoylated Gαq
Purified His-tagged Gαq variants at identical concentrations were diluted to a final volume of 400 µl in 50 mM HEPES pH 7.5, 150 mM NaCl, 1% Triton X-100, 25 mM N-ethylmaleimide (USB), protease inhibitors, and 0.1% SDS, then rotated overnight at 4 °C with 35 µl Ni-NTA beads. Samples were divided in half, with controls incubated with 0.5 M HEPES pH 7.5, and all other samples with 0.25 M HEPES pH 8 and 0.25 M hydroxylamine (Sigma-Aldrich). Samples were incubated and rocked for 4 h at room temperature. Ni-NTA beads were washed twice with PBS, and resuspended in 225 µl PBS. Samples were treated with 0.4 mM N-[6-(biotinamido)hexyl]-3'-(2'-pyridyldithio)propionamide (HPDP-biotin, Thermo Scientific) and incubated and rocked for 2 h at room temperature. Beads were washed once with PBS, and resuspended in SDS loading dye. Protection of cysteines via palmitoylation was then detected by western blot using a biotin antibody (StrepTactin-HRP Conjugate at 1:10,000 dilution, Biorad)55,56.
Multi-Angle Light Scattering
Purified PLCβ3 or Gαq–PLCβ3 were filtered and applied to a 50S size exclusion column with a molecular weight range of 15,000–5,000,000 Da connected to multi-angle wavelength detector (Wyatt Technology). The system was equilibrated with either PLCβ3 Buffer A (20 mM HEPES pH 8, 200 mM NaCl, 2 mM DTT, 0.1 mM EGTA, and 0.1 mM EDTA), or Gαq–PLCβ3 Buffer C. The molecular weights of PLCβ3 and Gαq–PLCβ3 were determined by fitting the elution peak of the protein using ASTRA software.
Statistical Methods
Statistical analyses used analysis of variance (ANOVA) with a Tukey’s post test as implemented in Prism (version 5.0a). All comparisons deemed significant had P values of at least < 0.01.
Figures. Structure images were generated using Pymol Version 1.5.0 (Schrödinger, LLC), and electrostatic surfaces were calculated using APBS57. Figures were created using Adobe® Photoshop and Illustrator.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Southworth (Life Sciences Institute, University of Michigan) for help with light scattering experiments. This work was supported by National Institutes of Health (NIH) grants HL086865, HL071818 and GM081655 (J.J.G.T), NIH grant DK090165 and the University of Michigan Biological Sciences Scholars Program (G.S.) and an American Heart Association Post-Doctoral Fellowship (A.M.L.). G.S. is a Pew Scholar of Biomedical Sciences. This work used the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center, supported by DK20572. This research was supported in part by the National Institutes of Health through the University of Michigan Cancer Center Support Grant (P30 CA046592). Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and Michigan’s Technology Tri-Corridor (grant 085P100817). Use of GM/CA at the Advanced Photon Source at Argonne National Laboratory has been funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Services (Y1-GM-1104). Use of the Advanced Photon Source at Argonne National Laboratory was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357.
Footnotes
ACCESSION CODES
Coordinates and diffraction intensities for the Gαq–PLCβ3 complex are deposited with the Protein Data Bank as entry 4GNK.
AUTHOR CONTRIBUTIONS
A.M.L., G.S., and J.J.G.T. designed the overall experimental approach. A.M.L. and C.A.B. cloned, expressed and purified all Gαq and PLCβ3 variants and conducted FCPIA assays. A.M.L. carried out all activity-based assays. A.M.L. and C.A.B. crystallized the Gαq–PLCβ3 complex, and A.M.L. determined the structure. S.D. and G.S. carried out all cryo-EM experiments. A.M.L., S.D., G.S., and J.J.G.T. co-wrote the manuscript.
REFERENCES
- 1.Gresset A, Sondek J, Harden TK. The phospholipase C isozymes and their regulation. Subcell Biochem. 2012;58:61–94. doi: 10.1007/978-94-007-3012-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadamur G, Ross EM. Mammalian phospholipase C. Annu Rev Physiol. 2012 doi: 10.1146/annurev-physiol-030212-183750. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 3.Taylor SJ, Chae HZ, Rhee SG, Exton JH. Activation of the β1 isozyme of phospholipase C by α subunits of the Gqclass of G proteins. Nature. 1991;350:516–518. doi: 10.1038/350516a0. [DOI] [PubMed] [Google Scholar]
- 4.Smrcka AV, Sternweis PC. Regulation of purified subtypes of phosphatidylinositol-specific phospholipase Cβ by G protein α and βγ subunits. J Biol Chem. 1993;268:9667–9674. [PubMed] [Google Scholar]
- 5.Smrcka AV, Hepler JR, Brown KO, Sternweis PC. Regulation of polyphosphoinositide-specific phospholipase C activity by purified Gq. Science. 1991;251:804–807. doi: 10.1126/science.1846707. [DOI] [PubMed] [Google Scholar]
- 6.Jiang H, et al. Roles of phospholipase C β2 in chemoattractant-elicited responses. Proc Natl Acad Sci U S A. 1997;94:7971–7975. doi: 10.1073/pnas.94.15.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D, et al. Phospholipase C isozymes selectively couple to specific neurotransmitter receptors. Nature. 1997;389:290–293. doi: 10.1038/38508. [DOI] [PubMed] [Google Scholar]
- 8.Xie W, et al. Genetic alteration of phospholipase C β3 expression modulates behavioral and cellular responses to μ opioids. Proc Natl Acad Sci U S A. 1999;96:10385–10390. doi: 10.1073/pnas.96.18.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Raamsdonk CD, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Descorbeth M, Anand-Srivastava MB. High glucose increases the expression of Gq/11α and PLCβ proteins and associated signaling in vascular smooth muscle cells. Am J Physiol Heart Circ Physiol. 2008;295:H2135–H2142. doi: 10.1152/ajpheart.00704.2008. [DOI] [PubMed] [Google Scholar]
- 11.Descorbeth M, Anand-Srivastava MB. Role of growth factor receptor transactivation in high glucose-induced increased levels of Gq/11α and signaling in vascular smooth muscle cells. J Mol Cell Cardiol. 2010;49:221–233. doi: 10.1016/j.yjmcc.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Ushio-Fukai M, Griendling KK, Akers M, Lyons PR, Alexander RW. Temporal dispersion of activation of phospholipase C-β1 and -γ isoforms by angiotensin II in vascular smooth muscle cells. Role of αq/11, α12, and βγ G protein subunits. J Biol Chem. 1998;273:19772–19777. doi: 10.1074/jbc.273.31.19772. [DOI] [PubMed] [Google Scholar]
- 13.Woodcock EA, Kistler PM, Ju Y-K. Phosphoinositide signalling and cardiac arrhythmias. Cardiovasc. Res. 2009;82:286–295. doi: 10.1093/cvr/cvn283. [DOI] [PubMed] [Google Scholar]
- 14.Grubb DR, Vasilevski O, Huynh H, Woodcock EA. The extreme C-terminal region of phospholipase Cβ1 determines subcellular localization and function; the "b" splice variant mediates α1-adrenergic receptor responses in cardiomyocytes. FASEB J. 2008;22:2768–2774. doi: 10.1096/fj.07-102558. [DOI] [PubMed] [Google Scholar]
- 15.Filtz TM, Grubb DR, McLeod-Dryden TJ, Luo J, Woodcock EA. Gq-initiated cardiomyocyte hypertrophy is mediated by phospholipase Cβ1b. FASEB J. 2009;23:3564–3570. doi: 10.1096/fj.09-133983. [DOI] [PubMed] [Google Scholar]
- 16.Dent MR, Dhalla NS, Tappia PS. Phospholipase C gene expression, protein content, and activities in cardiac hypertrophy and heart failure due to volume overload. Am J Physiol Heart Circ Physiol. 2004;287:H719–H727. doi: 10.1152/ajpheart.01107.2003. [DOI] [PubMed] [Google Scholar]
- 17.Woodcock EA, Grubb DR, Iliades P. Potential treatment of cardiac hypertrophy and heart failure by inhibiting the sarcolemmal binding of phospholipase Cβ1b. Curr Drug Targets. 2010;11:1032–1040. doi: 10.2174/138945010791591331. [DOI] [PubMed] [Google Scholar]
- 18.Waldo GL, et al. Kinetic scaffolding mediated by a phospholipase C-β and Gqsignaling complex. Science. 2010;330:974–980. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyon AM, et al. An autoinhibitory helix in the C-terminal region of phospholipase C-β mediates Gαqactivation. Nat Struct Mol Biol. 2011;18:999–1005. doi: 10.1038/nsmb.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, Shin S, Hepler J, Gilman A, Rhee S. Activation of phospholipase C-β2 mutants by G protein αqand βγ subunits. J Biol Chem. 1993;268:25952–25957. [PubMed] [Google Scholar]
- 21.Kim CG, Park D, Rhee SG. The role of carboxyl-terminal basic amino acids in Gqα-dependent activation, particulate association, and nuclear localization of phospholipase C-β1. J Biol Chem. 1996;271:21187–21192. doi: 10.1074/jbc.271.35.21187. [DOI] [PubMed] [Google Scholar]
- 22.Park D, Jhon DY, Lee CW, Ryu SH, Rhee SG. Removal of the carboxyl-terminal region of phospholipase C-β1 by calpain abolishes activation by Gαq. J Biol Chem. 1993;268:3710–3714. [PubMed] [Google Scholar]
- 23.Wu D, Jiang H, Katz A, Simon MI. Identification of critical regions on phospholipase C-β1 required for activation by G-proteins. J Biol Chem. 1993;268:3704–3709. [PubMed] [Google Scholar]
- 24.Schnabel P, Camps M, Carozzi A, Parker PJ, Gierschik P. Mutational analysis of phospholipase C-β2. Identification of regions required for membrane association and stimulation by guanine-nucleotide-binding protein βγ subunits. Eur J Biochem. 1993;217:1109–1115. doi: 10.1111/j.1432-1033.1993.tb18343.x. [DOI] [PubMed] [Google Scholar]
- 25.Jenco JM, Becker KP, Morris AJ. Membrane-binding properties of phospholipase C-β1 and phospholipase C-β2: role of the C-terminus and effects of polyphosphoinositides, G-proteins and Ca2+ Biochem J. 1997;327((Pt 2)):431–437. doi: 10.1042/bj3270431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singer AU, Waldo GL, Harden TK, Sondek J. A unique fold of phospholipase C-β mediates dimerization and interaction with Gαq. Nat Struct Biol. 2002;9:32–36. doi: 10.1038/nsb731. [DOI] [PubMed] [Google Scholar]
- 27.Ilkaeva O, Kinch LN, Paulssen RH, Ross EM. Mutations in the carboxyl-terminal domain of phospholipase C-β1 delineate the dimer interface and a potential Gαqinteraction site. J Biol Chem. 2002;277:4294–4300. doi: 10.1074/jbc.M109612200. [DOI] [PubMed] [Google Scholar]
- 28.Adjobo-Hermans MJ, Goedhart J, Gadella TW., Jr Regulation of PLCβ1a membrane anchoring by its substrate phosphatidylinositol (4,5)-bisphosphate. J Cell Sci. 2008;121:3770–3777. doi: 10.1242/jcs.029785. [DOI] [PubMed] [Google Scholar]
- 29.Adjobo-Hermans MJ, et al. PLCβ isoforms differ in their subcellular location and their CT-domain dependent interaction with Gαq. Cell Signal. 2013;25:255–263. doi: 10.1016/j.cellsig.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 30.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romoser V, Ball R, Smrcka AV. Phospholipase C β2 association with phospholipid interfaces assessed by fluorescence resonance energy transfer. G protein betagamma subunit-mediated translocation is not required for enzyme activation. J Biol Chem. 1996;271:25071–25078. doi: 10.1074/jbc.271.41.25071. [DOI] [PubMed] [Google Scholar]
- 33.Qualmann B, Koch D, Kessels MM. Let's go bananas: revisiting the endocytic BAR code. EMBO J. 2011;30:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 35.Peter BJ, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 36.Essen LO, Perisic O, Cheung R, Katan M, Williams RL. Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature. 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- 37.Ellis MV, et al. Catalytic domain of phosphoinositide-specific phospholipase C (PLC). Mutational analysis of residues within the active site and hydrophobic ridge of PLCδ1. J Biol Chem. 1998;273:11650–11659. doi: 10.1074/jbc.273.19.11650. [DOI] [PubMed] [Google Scholar]
- 38.Hepler JR, et al. Functional importance of the amino terminus of Gqα. J Biol Chem. 1996;271:496–504. doi: 10.1074/jbc.271.1.496. [DOI] [PubMed] [Google Scholar]
- 39.Nomura S, Fukaya M, Tsujioka T, Wu D, Watanabe M. Phospholipase Cβ3 is distributed in both somatodendritic and axonal compartments and localized around perisynapse and smooth endoplasmic reticulum in mouse Purkinje cell subsets. Eur J Neurosci. 2007;25:659–672. doi: 10.1111/j.1460-9568.2007.05334.x. [DOI] [PubMed] [Google Scholar]
- 40.Hicks SN, et al. General and versatile autoinhibition of PLC isozymes. Mol Cell. 2008;31:383–394. doi: 10.1016/j.molcel.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyer JL, Waldo GL, Harden TK. βγ-subunit activation of G-protein-regulated phospholipase C. J Biol Chem. 1992;267:25451–25456. [PubMed] [Google Scholar]
- 42.Illenberger D, et al. Stimulation of phospholipase C-β2 by the Rho GTPases Cdc42Hs and Rac1. EMBO J. 1998;17:6241–6249. doi: 10.1093/emboj/17.21.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jezyk MR, et al. Crystal structure of Rac1 bound to its effector phospholipase C-β2. Nat Struct Mol Biol. 2006;13:1135–1140. doi: 10.1038/nsmb1175. [DOI] [PubMed] [Google Scholar]
- 44.Han DS, Golebiewska U, Stolzenberg S, Scarlata SF, Weinstein H. A dynamic model of membrane-bound phospholipase Cβ2 activation by Gβγ subunits. Mol Pharmacol. 2011;80:434–445. doi: 10.1124/mol.111.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Philip F, Kadamur G, Silos RG, Woodson J, Ross EM. Synergistic activation of phospholipase C-β3 by Gαqand Gβγ describes a simple two-state coincidence detector. Curr Biol. 2010;20:1327–1335. doi: 10.1016/j.cub.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rebres RA, et al. Synergistic Ca2+responses by Gαi- and Gαq-coupled G-protein-coupled receptors require a single PLC β isoform that is sensitive to both Gβγ and Gαq. J Biol Chem. 2011;286:942–951. doi: 10.1074/jbc.M110.198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang T, Pentyala S, Rebecchi MJ, Scarlata S. Differential association of the pleckstrin homology domains of phospholipases C-β1, C-β2, and C-δ1 with lipid bilayers and the βγ subunits of heterotrimeric G proteins. Biochemistry. 1999;38:1517–1524. doi: 10.1021/bi982008f. [DOI] [PubMed] [Google Scholar]
Methods-Only-References
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 50.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 52.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Ghosh M, Smrcka AV. Assay for G protein-dependent activation of phospholipase C β using purified protein components. Methods Mol Biol. 2004;237:67–75. doi: 10.1385/1-59259-430-1:67. [DOI] [PubMed] [Google Scholar]
- 54.Shankaranarayanan A, et al. Assembly of high order Gαq-effector complexes with RGS proteins. J Biol Chem. 2008;283:34923–34924. doi: 10.1074/jbc.M805860200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 56.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 57.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.