Abstract
The nuclear factor κ enhancer binding protein (NF-κB) family of transcription factors regulates the expression of a large array of genes involved in diverse cellular processes including inflammation, immunity and cell survival. Activation of NF-κB requires ubiquitination, a highly conserved and versatile modification that can regulate cell signaling through both proteasome dependent and independent mechanisms. Studies in the past few years have provided new insights into the mechanisms underlying regulation of NF-κB by ubiquitination, including the involvement of multiple linkages of ubiquitin, the essential role of ubiquitin binding, and the function of unanchored polyubiquitin chains. In this review, we will focus on recent advances in understanding the role of ubiquitination in NF-κB regulation in various pathways.
Introduction
Ubiquitin is a 76-amino-acid protein that is highly conserved and ubiquitously expressed in all eukaryotes from yeast to human. The carboxylic acid in the C-terminal glycine of ubiquitin can be covalently attached to the epsilon amine of lysine on another protein through an isopeptide bond. This process, termed ubiquitination, occurs through stepwise enzymatic reactions catalyzed by three classes of enzymes, Ub-activating enzymes (E1), Ub-conjugating enzymes (E2) and Ub protein ligases (E3) [1]. In addition, the C-terminus of ubiquitin can be conjugated to one of the seven lysines (K6, K11, K27, K29, K33, K48, K63) on another ubiquitin, forming polyubiquitin chain of different linkages. Moreover, the C-terminal tail of ubiquitin can be directly attached to the N-terminal methionine of another ubiquitin to form linear polyubiquitin chain. The linkage of polyubiquitin can influence the fate of the substrate, adding another layer of complexity to this modification. For example, K48-linked ubiquitination is usually involved in directing proteins for proteasome-dependent degradation, while K63-linked ubiquitination have nonproteolytic functions such as regulating DNA damage repair, chromatin remodeling, vesicle trafficking and protein kinase activation [2].
Like phosphorylation, which is reversible, ubiquitination can be reversed by deubiquitination, which is carried out by deubiquitinating enzymes (DUBs). Close to 100 DUBs are estimated to be encoded by the human genome [3]. Moreover, more than 20 types of ubiquitin binding domains (UBDs) have been identified [4]. Proteins containing UBDs are able to transduce signals from ubiquitinated substrates to other components in the pathway. Some UBDs prefer to interact with ubiquitin chains of specific linkage, which could be utilized by the cells to regulate specific signaling pathways. This selective affinity of UBDs also make them useful tools for studying the role of distinct ubiquitin linkages in cells (see below).
Ubiquitination plays an essential role in the regulation of NF-κB pathways. NF-κB is a family of heterodimeric transcription factors that is highly involved in a variety of physiological and pathological processes including inflammation, immune response and cell survival. In unstimulated cells, NF-κB binds to inhibitory proteins of κB family (IκB) and is sequestered in the cytoplasm. Upon stimulation, IκB is phosphorylated by the IκB kinase (IKK) complex, which consists of two kinases IKKα and IKKβ, as well as the essential regulatory subunit (NEMO, also known as IKKγ or IKKAP). Phosphorylated IκB is subsequently ubiquitinated and degraded by 26S proteasome, thus allowing NF-κB to translocate to the nucleus, where it regulates the expression of a plethora of genes [5]. In this review, we will highlight some recent progress in dissecting the mechanisms of NF-κB regulation by ubiquitination and deubiquitination in response to multiple stimuli, and discuss some newly developed methods for studying ubiquitin signaling.
Ubiquitination in NF-κB activation by inflammatory cytokines
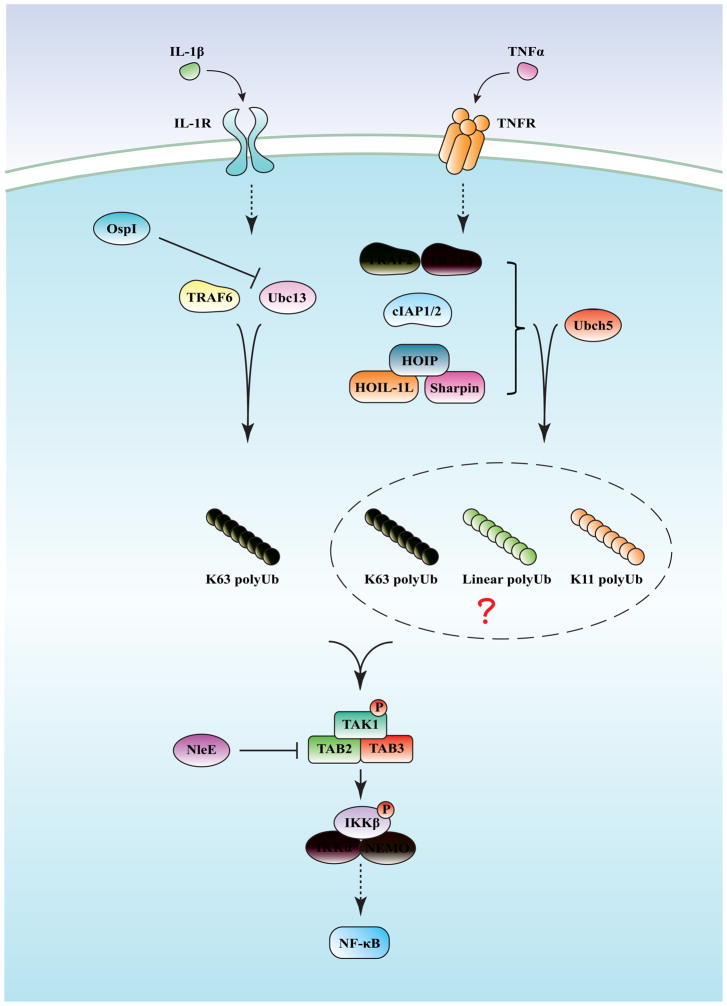
The most well studied pathways of inflammatory cytokine-induced NF-κB activation are those stimulated by Interleukin-1 (IL-1) and Tumor necrosis factor alpha (TNFα). IL-1 and TNFα bind to the receptors IL-1R and TNFR, respectively, and trigger activation of distinct signaling cascades that converge on TGF-β activated kinase 1 (TAK1), which phosphorylates and activates IKK, leading to activation of NF-κB (Figure 1) [5].
Figure 1. NF-κB activation by inflammatory cytokines.
Stimulation of cells with IL-1β leads to the activation of the ubiquitin E3 ligase TRAF6, which catalyzes the synthesis of unanchored K63 polyubiquitin chains that bind to the TAB2 subunit of the TAK1 kinase complex, resulting in TAK1 activation. TAK1 then phosphorylates IKKβ, leading to NF-κB activation. In the TNFα pathway, stimulation of the cells leads to the recruitment of several ubiquitin E3 ligases to the receptor, including TRAF2, TRAF5, cIAP1, cIAP2 and LUBAC (consisting of HOIP, HOIL-1, and Sharpin). These E3s and Ubch5 synthesize ubiquitin chains of different linkages, including K63, K11 and linear (M1). The physiological targets of these ubiquitin chains are still not clear, but it is clear that the ubiquitination events lead to TAK1 and IKK activation. Two bacterial proteins, OspI and NleE, are able to inhibit NF-κB activation by targeting Ubc13 and TAB2/3, respectively.
The first clue that ubiquitination can regulate NF-κB activation through proteasome-independent mechanism came from the discovery that purified IKK complex could be activated by ubiquitination reaction that did not require K48 of ubiquitin or proteasome [6]. The physiological importance of this ubiquitination event became more clear when TRAF6, a protein essential for NF-κB activation by IL-1R and Toll-like receptors (TLRs) [7,8], was shown to function as an E3 ligase to catalyze the synthesis of K63-linked polyubiquitin with the E2 enzyme Ubc13-Uev1A. This polyubiquitination event was shown to activate the TAK1 kinase complex, which in turn phosphorylates and activates IKK (Figure 1) [9,10]. The essential role of K63-linked ubiquitination in IL-1R pathway was further confirmed by using inducible RNAi and rescue to replace endogenous ubiquitin with K63R ubiquitin mutant in human cells [11]. K63R ubiquitin replacement abolished NF-κB activation in response to IL-1β, but not TNFα, suggesting that these two pathways employ distinct mechanisms to activate NF-κB. Interestingly, it has been shown in the same study that IL-1-induced NF-κB activation was dependent on Ubc13, which is highly specific in synthesizing K63-linked polyubiquitin, while TNFα-induced NF-κB activation was dependent on Ubc5, an E2 enzyme that is able to synthesize heterogeneous polyubiquitin chains, suggesting that TNFα-induced NF-κB activation may utilize ubiquitin chains containing different linkages.
In the past few years, growing evidence has revealed that, in addition to K63 ubiquitin chains, other types of ubiquitin chains also play a role in NF-κB activation in response to proinflammatory cytokines. A role for linear ubiquitination in NF-κB activation was suggested through studies of the linear ubiquitin chain assembly complex (LUBAC), which contains two RING finger proteins HOIP (also known as RNF31) and HOIL-1L (also known as RBCK1), and another subunit Sharpin. HOIP knockdown and HOIL-1L knockout partially inhibited NF-κB activation by TNFα [12]. A mouse strain named cpdm (chronic proliferative dermatitis), which carried a spontaneous null mutation in Sharpin developed immune system disorders and multi-organ inflammation. Cells derived from this mouse, including B cells, macrophages and embryonic fibroblasts, exhibited a partial defect in IKK activation in response to TNFα [13–15]. The observation that depletion of HOIP, HOIL or Sharpin always causes a partial rather than complete defect in TNFα-mediated NF-κB activation suggests that LUBAC complex may play redundant roles with other proteins to promote IKK activation. In addition, although evidence has supported a role of LUBAC complex in NF-κB activation by TNFα, more direct evidence for a role of linear polyubiquitination is still lacking.
A recent human genetic study identified three patients harboring loss-of-function mutations of HOIL-1[16]. These patients displayed a paradoxical clinical syndrome that combines immunodeficiency, autoinflammation and amylopectinosis (consisting of intracellular glycogen inclusions). Fibroblasts from these patients were partially defective in IKK activation and had close to 50% reduction in IL6 production in response to IL-1β and TNFα. In contrast, monocytes from the same patients produced elevated levels of inflammatory cytokines, including IL6, IL8, MIP1A and MIP1B, following stimulation with IL-1β. Thus, the role of HOIL-1 in the NF-κB pathway appears to be cell type-specific, which is consistent with increased susceptibility to pyogenic infections and autoinflammation in the patients.
One of the key questions in this field is exactly how ubiquitination promotes IKK activation. As the regulatory subunit of the IKK complex, NEMO has been proposed to be the key factor for transducing ubiquitination signal to IKK activation. NEMO contains two ubiquitin binding domains: the NUB (also known as UBAN or CoZi) domain in the middle region, and the zinc finger (ZF) domain at the C-terminus [17]. Both NUB and ZF domains were able to bind K63 and linear polyubiquitin [18–21], although which linkage is preferred is still controversial due to the variations in the experimental design, including difference in the length of polyubiquitin used and whether full length or truncated NEMO was used. To clarify this issue, it is important to quantitate the amounts of different endogenous polyubiquitin chains associated with NEMO in cells stimulated with NF-κB agonists. In any case, it is clear that ubiquitin binding by NEMO is essential for IKK activation because mutations in the NUB and ZF domain impaired the ability of NEMO to rescue NF-κB activation in NEMO deficient cells [18–22]. Mutations in NEMO that selectively disrupt its binding to linear but not K63-linked ubiquitin chains led to only partial defects in NF-κB activation [21]. This suggests that linear and K63-linked ubiquitin binding to NEMO may play redundant roles in NF-κB activation. NEMO also binds to K11 ubiquitin chains synthesized by cIAP1 and UbcH5 in the TNFα pathway, adding another level of complexity in ubiquitin signaling in this pathway [23].
While there is now a general agreement that non-degradative polyubiquitination and ubiquitin binding by NEMO are important for IKK activation, the physiological ubiquitination targets remain an enigma. Following stimulation of cells with IL-1β, several proteins in the signaling cascade, including IRAK1, TRAF6, TAB2, TAK1 and NEMO, are modified by ubiquitin or polyubiquitin chains. Similarly, multiple proteins in the TNFα signaling pathway, including TNFR1, TRAF2, RIP1, cIAPs, as well as TAB2, TAK1 and NEMO, are ubiquitinated in a signal-dependent manner. It was shown that the mutation of a putative ubiquitination site in human RIP1, K377R, impaired its ability to activate IKK [22,24]. However, there is still no direct evidence yet that K377 of endogenous RIP1 is ubiquitinated in TNF stimulated cells. This is further complicated by the fact that RIP1 knockout does not completely block IKK activation in some cells (i.e., MEFs)[25]. NEMO has been shown to be ubiquitinated at certain lysines, such as K285 (human sequence), but the effects of NEMO lysine mutations on IKK activation were variable, in part because of various expression levels since overexpression of even wild type NEMO could inhibit IKK activation. Overall, the extents of ubiquitination of cellular proteins are very weak, and most studies cannot rule out the possibility that mutations of lysine residues on a protein could affect the structure or function of the protein through effects unrelated to ubiquitination. Moreover, so far there is no direct biochemical evidence that ubiquitination of any target protein could activate IKK in vitro.
Recently, unanchored K63 polyubiquitin chains, which are not conjugated to any other cellular proteins, were found to directly activate the TAK1 kinase complex in vitro through binding to the TAB2 or TAB3 subunit of this complex. The activated TAK1 then phosphorylates IKKβ, leading to IKK activation [26]. Interestingly, the phosphorylation of IKKβ by TAK1 requires NEMO and TAB2 as well as unanchored polyubiquitin chains [27], suggesting that these chains provide a scaffold that recruits both TAK1 and IKK through binding to TAB2 and NEMO, respectively. Stimulation of cells with IL-1β leads to the production of endogenous unanchored polyubiquitin chains associated with TAB2[26]. Curiously, only long K63 polyubiquitin chains synthesized by TRAF6, but not by some other E3s, were able to activate TAK1 and IKK. More work is needed to elucidate the mechanism of protein kinase activation by unanchored polyubiquitin chains and to dissect the roles of unanchored and substrate-anchored ubiquitin chains in IKK activation in vivo.
Ubiquitination in NF-κB activation by microbial pathogens
Microbes, including viruses, bacteria and parasites, are detected by the host immune system that contains and eventually eradicates the infection. Diverse classes of microbial pathogen-associated molecular patterns (PAMP) are detected by distinct pattern recognition receptors (PRR) that trigger different signal transduction pathways that converge on the activation of NF-κB and other transcription factors. PRRs include Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD-like receptors (NLRs) and C-type lectin receptors (CLRs). Remarkably, with the exception of CLRs, for which research is still in its infancy, the other three families of PRRs all extensively engage the ubiquitination machinery to regulate NF-κB activation. Because the role of ubiquitination in TLR and NLR pathways have recently been reviewed elsewhere [1,28,29], here we will discuss some recent advances on the role of ubiquitination in the RLR pathway of antiviral immune responses.
Upon virus infection, viral RNA is released into the cytoplasm, where it is detected by members of the RLR family of RNA sensors, including RIG-I, MDA5, and LGP2. These RLRs then activate the mitochondrial adaptor protein MAVS (also known as IPS-1, VISA, and Cardif), leading to activation of NF-κB and IRF3 (Figure 2). Recent studies have suggested that ubiquitination plays a key role in the RIG-I pathway. Tripartite motif-25 (TRIM25), a RING-domain-containing E3, has been shown to be important for RIG-I antiviral activity [30]. Another ubiquitin E3, Riplet, is also important for RIG-I activation [31]. It was proposed that RIG-I ubiquitination was critical, because TRIM25 and Riplet were able to ubiquitinate RIG-I at the N and C termini, respectively. However, a study using a cell-free system to reconstitute RIG-I pathway in vitro revealed that RIG-I could be activated by the binding of its N-terminal CARD domains to unanchored K63-linked polyubiquitin chains [32]. In addition, endogenous unanchored K63-linked polyubiquitin chains purified from human cells potently activate the RIG-I pathway in vitro. Further study found that both RIG-I and MDA5 are able to bind K63-linked ubiquitin chains [33], and mutations of conserved residues in the CARD domains that disrupt the ubiquitin binding abolished the ability of RIG-I and MDA5 to support downstream activation, highlighting the importance of ubiquitin binding in both pathways.
Figure 2. The role of ubiquitination in the RIG-I antiviral signaling pathway.
After infection by RNA viruses, viral RNA binds to RIG-I and induces a conformational change that exposes the N-terminus of RIG-I which binds to unanchored K63 polyubiquitin chains synthesized by TRIM25 and Riplet. RIG-I then interacts with and activates the mitochondrial membrane protein MAVS, which activates IKK and TBK1 in the cytoplasm. These kinases then activate NF-κB and IRF3, leading to the production of type-I interferons.
Besides RNA viruses, retroviruses like human immunodeficiency virus-1 (HIV-1) can also activate NF-κB through a ubiquitin-dependent mechanism. It is known that the RING-domain E3 ligase TRIM5 restricts the infection of HIV-1 and other retroviruses [34,35]. A recent study showed that TRIM5 functions as a retroviral capsid lattice sensor and restricts retrovirus infection by catalyzing the synthesis of unanchored K63 ubiquitin with Ubc13-Uev1A to activate TAK1 kinase complex and stimulate NF-κB activation [36].
Consistent with a critical role of ubiquitination in NF-κB activation and immune responses, some pathogens have evolved multiple mechanisms to counteract the host defense by hijacking the ubiquitin system (see [29] for a more detailed review). Two recent studies provided excellent examples of how bacteria block NF-κB activation by targeting the production and sensing of K63 polyubiquitin chains. Sanada et al showed that OspI, a virulent effector protein from the bacterial pathogen Shigella flexneri, selectively deamidates the E2 enzyme Ubc13, thereby inhibiting its ability to synthesize K63 polyubiquitin chains, resulting in NF-κB inhibition and suppression of inflammatory responses [37]. Another bacterial protein NleE from Escherichia coli, which is delivered to mammalian cells through the type-III secretion system, was found to possess S-adenosyl-L-methionine-dependent methyltransferase activity [38]. This enzyme specifically methylates two conserved Cysteine residues in the ubiquitin binding domains (known as NZF) of TAB2 and TAB3, two regulatory subunits of the TAK1 complex. Methylation of TAB2 and TAB3 impairs their ability to bind K63-linked polyubiquitin chains, and as a result, the host NF-κB activation is suppressed.
The role of deubiquitinating enzymes in NF-κB activation
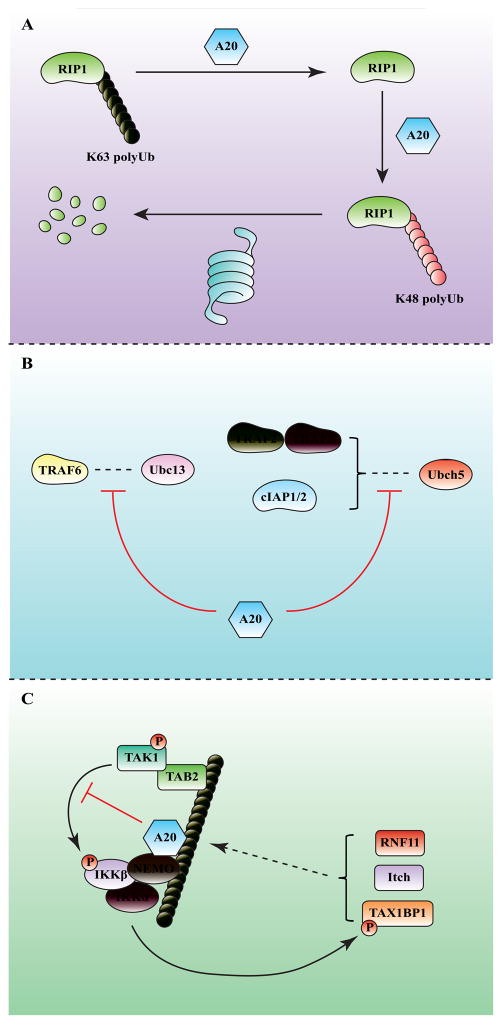
Several deubiquitinating enzymes (DUBs) function as key negative regulators of IKK to allow a tight control of NF-κB activation, which is critical because persistent or excessive activation of NF-κB has been linked to autoimmune disorders and cancer. One of the best studied DUBs in NF-κB pathways is A20 (also known as TNFAIP3), an NF-κB inducible protein that contains an OTU-type DUB domain at the N-terminus, and seven zinc fingers at the C-terminus [39]. A20 knockout mice developed severe inflammation in various tissues including liver, kidney, intestine, joints and bone marrow and died at an early age [40]. Moreover, multiple polymorphisms of the A20 gene have been linked to systemic lupus erythematosus, a human autoimmune disease [41]. A20 deficient cells are hypersensitive to NF-κB activation by multiple stimuli, including TNFα, IL-1β, TLR ligands and RLR ligands [40,42,43], indicating that A20 serves as a crucial negative regulator in NF-κB signaling. It has been proposed that A20 suppress hyperactivation of NF-κB by deubiquitinating K63-linked polyubiquitin attached to RIP1 through the N-terminal OTU domain, as well as promoting K48-linked polyubiquitination of RIP1 for proteasomal degradation [44]. However, A20 cleaves K48-linked polyubiquitin more efficiently than K63-linked polyubiquitin in vitro [45].
Several recent studies have provided evidence that A20 can function through DUB-activity-independent mechanisms. Shembade et al. showed that A20 could antagonize interactions between E3 ligases including TRAF6, TRAF2 and cIAP1 and their E2 partners Ubc13 and Ubch5 (Figure 3) [46]. Skaug et al found that A20 catalytic mutant (C103A) could still inhibit IKK activation both in vitro and in vivo [27]. The same study also showed that A20 directly impaired IKK activation by binding to K63-linked polyubiquitin through its seventh zinc finger (ZnF7) and recruiting NEMO. Mutations in ZnF7 that disrupted ubiquitin binding also abolished A20’s ability to downregulate NF-κB activity in A20 deficient MEF. The role of ubiquitin binding by ZnF7 of A20 in IKK inhibition was confirmed and extended by recent studies showing that this motif also binds to linear ubiquitin chains [47,48].
Figure 3. Mechanisms of IKK inhibition by A20.
A20 inhibits IKK through three proposed mechanisms. (A) A20 functions as an editing enzyme that first utilizes the N-terminal OTU-type deubiquitination enzyme domain to remove K63 polyubiquitin chains from RIP1, followed by K48 polyubiquitination of RIP1 by the C-terminal zinc finger (ZnF4) domain, which functions as a ubiquitin E3 ligase. K48 polyubiquitination of RIP1 targets it for degradation by the proteasome. (B) A20 can inhibit the synthesis of polyubiquitin chains by disrupting the interaction between E2s (Ubc13 and Ubc5) and E3s (TRAFs and cIAPs). (C) A20 can also inhibit IKK through a non-catalytic mechanism that involves its binding to K63 and linear polyubiquitin chains through the zinc finger 7 (ZnF7) domain. This binding facilitates the formation of a complex consisting of A20, NEMO and polyubiquitin chains, resulting in inhibition of IKK phosphorylation by TAK1. A20 also forms a complex with RNF11, ITCH and TAX1BP1, which regulate the inhibitory activity of A20. TAX1BP1 is phosphorylated by IKKα, and this phosphorylation promotes the assembly of the A20 complex to provide a negative feedback inhibition of IKK.
A20 forms a complex with TAX1BP1, immune modulatory protein (ITCH) and RNF11, and formation of this A20 ubiquitin-editing complex is crucial for negative regulation of NF-κB [49–51], although the underlying mechanism remains elusive. Interestingly, IKKα was shown to phosphorylate TAX1BP1 and promote the assembly of A20 ubiquitin-editing assembly complex [52], indicating that IKKα functions as a key factor in coordinating the inhibitory effect of A20 and its binding partners.
The tumor suppressor CYLD is another DUB well known for its role in inhibiting NF-κB. CYLD cleaves K63 as well as linear polyubiquitin chains and its catalytic activity is required for its ability to inhibit IKK. Several mutations that result in the loss-of-function of CYLD have been found in patients afflicted with familial cylindroma and other tumors of the skin appendage [53]. Cells lacking CYLD have elevated NF-κB activation, which likely contribute to the tumor development [54–56]. In cell culture experiments, transfection of CYLD inhibits ubiquitination of several proteins, including RIP1, TRAF2 and NEMO.
However, direct evidence that CYLD could directly remove ubiquitin chains from a protein substrate is still lacking. Rather, it was shown that recombinant CYLD preferentially cleaves unanchored K63 polyubiquitin chains [26]. Further work is needed to clarify the physiological targets of CYLD in the NF-κB pathway. CYLD also promotes cell death through apoptosis and necrosis [56,57], possibly by inhibiting RIP1 ubiquitination or removing ubiquitin chains from RIP1.
Several other DUBs have also been indicated to negatively regulate NF-κB activation, including Ubiquitin-specific peptidase 20 (USP20) [58], USP2a [59], USP21 [60], and USP4 [61]. These DUBs have been shown to deubiquitinate components of NF-κB pathways, such as TRAF6 and RIP1, in vitro. However, rescue experiments with catalytic mutants as well as genetic experiments are required to establish the roles of these DUBs in specific NF-κB pathways.
New methods for studying ubiquitin signaling
Compared to other modifications like phosphorylation and acetylation, ubiquitination is more complicated due to the existence of multiple linkages as well as the difference in length of polyubiquitin chains, which can influence the fates of the modified substrates. As such, it has remained a challenge to study ubiquitination in vivo. During the past two years, however, new methods have emerged as useful tools to elucidate the regulatory role of ubiquitination in cell signaling including NF-κB activation.
Two recent studies have developed ubiquitin sensors to track the dynamics and subcellular localization of polyubiquitin in cells [62,63]. These sensors contained tandem ubiquitin binding domains that rendered them capable of binding to ubiquitin chains of specific linkages with high affinity, and thus can be applied as competitive inhibitor to study the role of particular linkage(s) in signaling pathways. Sims et al. showed that overexpression of K63-linked polyubiquitin sensors inhibited IL-1β-induced NF-κB activity with high efficiency, but it required a much higher concentration of these sensors to obtain an inhibitory effect in the TNF pathway, consistent with the previous observation that K63-linked polyubiquitin plays a more important role in IKK activation by IL-1β than TNFα [11]. These sensors are also anticipated to be useful in live-cell imaging, which will provide important information regarding the dynamics of ubiquitin chain formation and disassembly.
Another important technological development is the rapidly increasing power of mass spectrometry, which is very important for the identification of ubiquitination substrates in cells. The increasingly higher resolution, accuracy, sensitivity, speed and quantitative power of mass spectrometry make it possible to identify low abundant ubiquitinated proteins and map the modification sites on the substrates as well as within the ubiquitin chains. Recently developed antibodies that recognize di-glycine (GlyGly) attached to lysine through an isopeptide bond is a useful tool to enrich the ubiquitinated substrates and help identify ubiquitination substrates [64–66]. These advances, together with the development of ubiquitin-linkage specific antibodies, should significantly advance our understanding of the roles and mechanisms of ubiquitination in the regulation of NF-κB and other cell signaling pathways.
Conclusions
The pace and intensity of research on the role of ubiquitination in NF-kB pathways have increased substantially in the past few years, so have the amounts of information and the degrees of complexity in our understanding of ubiquitin signaling in these pathways. Many proteins involved in the ubiquitin pathway, including ubiquitin conjugating and deconjugating enzymes and ubiquitin binding proteins, have now been shown to regulate IKK activation in different NF-κB signaling pathways. Different types of ubiquitin chains have also been shown to play a role in IKK regulation. The extensive involvement of multiple ubiquitin E2s, E3s, DUBs and different ubiquitin chains in a signaling pathway also presents a challenge in sorting out the mechanism by which ubiquitination regulates IKK activation. For example, stimulation with TNFα leads to the recruitment of multiple E3s and DUBs, including TRAF2, TRAF5, cIAP1, cIAP2, LUBAC, A20 and CYLD, to the receptor. How these proteins, and the ubiquitin chains that they generate, function cooperatively, redundantly or competitively to regulate TAK1 and IKK is still an important issue that remains to be resolved. The physiological ubiquitination targets required for IKK activation have yet to be identified, and the relative contributions of these targets and unanchored ubiquitin chains in IKK activation in vivo require further clarification. Further research that combines modern technological advances with classical biochemical and genetic approaches should bring clarity to these complex issues. A clear understanding of the mechanisms by which ubiquitination and deubiquitination regulate NF-kB is crucial for devising and developing effective therapies of human diseases that are frequently associated with the dysfunction of the ubiquitin network in the NF-kB pathways.
Highlights.
Polyubiquitination activates TAK1 and IKK through proteasome-independent mechanisms.
Unanchored ubiquitin chains have crucial functions in both cytokine- and virus-induced NF-κB activation.
Microbial pathogens can counteract host defense by hijacking the ubiquitin system to repress NF-κB signaling.
A20 is able to regulate NF-κB signaling through noncatalytic mechanisms.
Development of ubiquitin sensors and quantitative assessment of ubiquitination will help advance our understanding of the role of ubiquitination in NF-κB signaling.
Acknowledgments
Research in our laboratory has been supported by grants from the National Institute of Health, the Welch Foundation and Cancer Prevention and Research Institute of Texas. Z.J.C is an Investigator of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu S, Chen ZJ. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 4.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 5.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 6.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 7.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 8.Ishida T, Mizushima S, Azuma S, Kobayashi N, Tojo T, Suzuki K, Aizawa S, Watanabe T, Mosialos G, Kieff E, et al. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J Biol Chem. 1996;271:28745–28748. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- 9.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 10.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 11.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 13.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 14.Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **16.Boisson B, Laplantine E, Prando C, Giliani S, Israelsson E, Xu Z, Abhyankar A, Israel L, Trevejo-Nunez G, Bogunovic D, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nature immunology. 2012;13:1178–1186. doi: 10.1038/ni.2457. This paper shows that humans with HOIL-1 mutations that resulted in a loss of LUBAC complex have both immunodeficiency and autoinflammation, which may be linked to impaired NF-kB activation in some cells but hyper NF-kB activation in other cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwai K. Diverse ubiquitin signaling in NF-kappaB activation. Trends Cell Biol. 2012;22:355–364. doi: 10.1016/j.tcb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 19.Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Veron M, Agou F, Israel A. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 2009;28:2885–2895. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Hadian K, Griesbach RA, Dornauer S, Wanger TM, Nagel D, Metlitzky M, Beisker W, Schmidt-Supprian M, Krappmann D. NF-kappaB essential modulator (NEMO) interaction with linear and lys-63 ubiquitin chains contributes to NF-kappaB activation. J Biol Chem. 2011;286:26107–26117. doi: 10.1074/jbc.M111.233163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 23.Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Kobayashi M, Blonska M, You Y, Lin X. Ubiquitination of RIP is required for tumor necrosis factor alpha-induced NF-kappaB activation. The Journal of biological chemistry. 2006;281:13636–13643. doi: 10.1074/jbc.M600620200. [DOI] [PubMed] [Google Scholar]
- 25.Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J. RIPK1 is not essential for TNFR1-induced activation of NF-kappaB. Cell death and differentiation. 2010;17:482–487. doi: 10.1038/cdd.2009.178. [DOI] [PubMed] [Google Scholar]
- 26.Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *27.Skaug B, Chen J, Du F, He J, Ma A, Chen ZJ. Direct, Noncatalytic Mechanism of IKK Inhibition by A20. Mol Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. This study provides the biochemical evidence that A20 can directly inhibit IKK activation by recruitment of NEMO through binding to ubiquitin, which is independent of the deubiquitinating enzyme activity of A20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tigno-Aranjuez JT, Abbott DW. Ubiquitination and phosphorylation in the regulation of NOD2 signaling and NOD2-mediated disease. Biochimica et biophysica acta. 2012;1823:2022–2028. doi: 10.1016/j.bbamcr.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2011;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- *31.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8:496–509. doi: 10.1016/j.chom.2010.11.008. This study shows that Riplet-deficient mice and cells had impaired interferon production in response to RNA virus infection, confirming the important role of this E3 ligase in the RIG-I pathway. [DOI] [PubMed] [Google Scholar]
- **32.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. This study reconstituted the RIG-I pathway in vitro, revealing that RIG-I activation requires two signals, viral RNA and unanchored K63-linked polyubiquitin chains, which bind to the C- and N-termini of RIG-I, respectively. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-Induced Oligomerization of the RNA Sensors RIG-I and MDA5 Activates Antiviral Innate Immune Response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. This study shows that polyubiquitin binding also plays a critical role in the activation of MDA5, a RIG-I-like receptor. The ubiquitin binding induced oligomerization of RIG-I and MDA5 to activate the downstream signaling cascades. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 35.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- *36.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. The E3 ligase TRIM5 serves as a sensor for the retrovirus capsid lattice and synthesizes unanchored K63-linked polyubiquitin chains with Ubc13-Uev1A to activate NF-κB and AP-1 to restrict retrovirus infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **37.Sanada T, Kim M, Mimuro H, Suzuki M, Ogawa M, Oyama A, Ashida H, Kobayashi T, Koyama T, Nagai S, et al. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature. 2012 doi: 10.1038/nature10894. This paper shows that the bacterial protein OspI selectively deamidates a specific glutamine residue of Ubc13, which inactivates Ubc13’s ability to catalyze K63 polyubiquitination, thereby dampening NF-kB activation. [DOI] [PubMed] [Google Scholar]
- **38.Zhang L, Ding X, Cui J, Xu H, Chen J, Gong YN, Hu L, Zhou Y, Ge J, Lu Q, et al. Cysteine methylation disrupts ubiquitin-chain sensing in NF-kappaB activation. Nature. 2012;481:204–208. doi: 10.1038/nature10690. This study shows that the E. coli protein NleE catalyzes the methylation of a cysteine residue in the ubqiuitin-binding domains of TAB2 and TAB3. This modification blocks ubiquitin binding by TAB2 and TAB3, resulting in inhibition of TAK1 and IKK. [DOI] [PubMed] [Google Scholar]
- 39.Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nature reviews Immunology. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee EG, Boone L, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, Shifrin N, Petri MA, Kamboh MI, Manzi S, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maelfait J, Roose K, Bogaert P, Sze M, Saelens X, Pasparakis M, Carpentier I, van Loo G, Beyaert R. A20 (Tnfaip3) deficiency in myeloid cells protects against influenza A virus infection. PLoS Pathog. 2012;8:e1002570. doi: 10.1371/journal.ppat.1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tavares RM, Turer EE, Liu CL, Advincula R, Scapini P, Rhee L, Barrera J, Lowell CA, Utz PJ, Malynn BA, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 45.Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, Phu L, Phung Q, Maurer B, Arnott D, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokunaga F, Nishimasu H, Ishitani R, Goto E, Noguchi T, Mio K, Kamei K, Ma A, Iwai K, Nureki O. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation. EMBO J. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verhelst K, Carpentier I, Kreike M, Meloni L, Verstrepen L, Kensche T, Dikic I, Beyaert R. A20 inhibits LUBAC-mediated NF-kappaB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 2012;31:3845–3855. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shembade N, Harhaj NS, Liebl DJ, Harhaj EW. Essential role for TAX1BP1 in the termination of TNF-alpha-, IL-1- and LPS-mediated NF-kappaB and JNK signaling. EMBO J. 2007;26:3910–3922. doi: 10.1038/sj.emboj.7601823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shembade N, Harhaj NS, Parvatiyar K, Copeland NG, Jenkins NA, Matesic LE, Harhaj EW. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- 51.Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J. 2009;28:513–522. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shembade N, Pujari R, Harhaj NS, Abbott DW, Harhaj EW. The kinase IKKalpha inhibits activation of the transcription factor NF-kappaB by phosphorylating the regulatory molecule TAX1BP1. Nat Immunol. 2011;12:834–843. doi: 10.1038/ni.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 54.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 55.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 56.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 57.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasunaga J, Lin FC, Lu X, Jeang KT. Ubiquitin-specific peptidase 20 targets TRAF6 and human T cell leukemia virus type 1 tax to negatively regulate NF-kappaB signaling. J Virol. 2011;85:6212–6219. doi: 10.1128/JVI.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He X, Li Y, Li C, Liu LJ, Zhang XD, Liu Y, Shu HB. USP2a negatively regulates IL-1beta- and virus-induced NF-kappaB activation by deubiquitinating TRAF6. J Mol Cell Biol. 2012 doi: 10.1093/jmcb/mjs024. [DOI] [PubMed] [Google Scholar]
- 60.Xu G, Tan X, Wang H, Sun W, Shi Y, Burlingame S, Gu X, Cao G, Zhang T, Qin J, et al. Ubiquitin-specific peptidase 21 inhibits tumor necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J Biol Chem. 2010;285:969–978. doi: 10.1074/jbc.M109.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou F, Zhang X, van Dam H, Ten Dijke P, Huang H, Zhang L. Ubiquitin-specific Protease 4 Mitigates Toll-like/Interleukin-1 Receptor Signaling and Regulates Innate Immune Activation. J Biol Chem. 2012;287:11002–11010. doi: 10.1074/jbc.M111.328187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sims JJ, Scavone F, Cooper EM, Kane LA, Youle RJ, Boeke JD, Cohen RE. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat Methods. 2012;9:303–309. doi: 10.1038/nmeth.1888. See annotation to Ref. [63] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Wijk SJ, Fiskin E, Putyrski M, Pampaloni F, Hou J, Wild P, Kensche T, Grecco HE, Bastiaens P, Dikic I. Fluorescence-Based Sensors to Monitor Localization and Functions of Linear and K63-Linked Ubiquitin Chains in Cells. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.06.017. Refs [62] and [63] developed sensors that recognize polyubiquitin with specific linkages, which are potential powerful tools for studying the localization, dynamics and function of different types of ubiquitin chains in NF-κB signaling as well as other signaling pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. See annotation to Ref. [66] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. See annotation to Ref. [66] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. Refs. [64], [65] and [66] combined the use of a ubiquitin remnant antibody and quantitative mass spectrometry to profile ubiquitination targets and map ubiquitination sites at global scales. [DOI] [PMC free article] [PubMed] [Google Scholar]