Abstract
Photocrosslinking approaches can be used to map interactome networks within the context of living cells. Photocrosslinking methods rely on use of metabolic engineering or genetic code expansion to incorporate photocrosslinking analogs of amino acids or sugars into cellular biomolecules. Immunological and mass spectrometry techniques are used to analyze crosslinked complexes, thereby defining specific interactomes. Because photocrosslinking can be conducted in native, cellular settings, it can be used to define context-dependent interactions. Photocrosslinking methods are also ideally suited for determining interactome dynamics, mapping interaction interfaces, and identifying transient interactions in which intrinsically disordered proteins and glycoproteins engage. Here we discuss the application of cell-based photocrosslinking to the study of specific problems in immune cell signaling, transcription, membrane protein dynamics, nucleocytoplasmic transport, and chaperone-assisted protein folding.
Introduction
A network of protein-protein interactions (interactome) underlies all biological processes. While the human genome consists of only approximately 20,000 protein-encoding genes, these gene products are estimated to engage in hundreds of thousands of protein-protein interactions [1, 2]. The interactome is dynamic; every significant cellular event – growth, motility, division – is accompanied by changes in the interactome. Moreover, at the organismal level, alterations in the protein interactome are clearly fundamental to physiological processes, such as organ development, and to pathophysiological events, such as cancer [3, 4]. However, our maps of the interactome remain incomplete and poorly validated. In addition, functional insight is often lacking, even for known interactions. More comprehensive knowledge of the interactome will provide mechanistic insight into myriad normal and pathological processes.
Traditional methods for mapping the interactome include the yeast 2-hybrid method (Y2H), affinity purification, and microscopy [4]. While these approaches have provided a wealth of information on protein complexes, each suffers from limitations. Y2H studies and affinity purification interrogate interaction formation in non-native environments; as a result, spurious interactions may be identified, while authentic complexes can be overlooked. Fluorescence resonance energy transfer (FRET) experiments are effective in assessing protein-protein proximity but may be difficult to initiate, often require structural knowledge of proteins being examined, and are not well-suited to discovery efforts. In contrast, photo-activated crosslinking technology offers a powerful way to discover and characterize interactions in the setting of living, intact cells. Photocrosslinking groups can be incorporated into cellular biomolecules and then used in native environments to identify specific, direct protein-protein interactions [5]. Along with the benefit of interrogating native interactions, a light-activated crosslinking process offers the possibility of monitoring interaction events with temporal resolution [6]. Furthermore, while the Y2H method and affinity purification are biased toward detection of strong protein-protein interactions, the covalent nature of photocrosslinking enables detection of low-affinity interactions such as those in which intrinsically disordered proteins and glycoproteins engage.
Metholodology for Interactome Analysis by In-Cell Photocrosslinking
Multiple photocrosslinking functionalities are known and three classes of crosslinkers have achieved recent popularity – diazirines, aryl azides, and benzophenone (Bpa). These functional groups are stable in the absence of ultraviolet (UV) radiation, a feature that allows them to be appropriately incorporated and localized within living cells. UV radiation activates the photocrosslinking group, resulting in production of a highly reactive intermediate that can react with a neighboring functional group, forming a new covalent bond. This new bond is termed a “crosslink” and serves to covalently capture an otherwise non-covalent interaction. The high reactivity of photocrosslinking intermediates has several significant consequences: first, any nearby molecule, irrespective of functional group composition, will likely be covalently crosslinked; second, high reactivity corresponds to a short half-life (ns – µs) of the intermediate, minimizing the probability of non-specific crosslinking; and third, if an interaction partner is not present, the intermediate can either relax back into the ground state, in the case of Bpa, or react with water or even itself, in the case of diazirines and aryl azides, thereby reducing the incidence of nonspecific crosslinking [5].
Photocrosslinking groups are non-native functionalities that must be introduced into biomolecules of interest. Early work in the photocrosslinking field relied on chemical methods to add the necessary photocrosslinking functional groups to the target molecule. As a result, photocrosslinking studies typically could not be used to study proteins in native, cellular settings. Over the past decade, multiple methods have been developed to incorporate photocrosslinkers into biomolecules in living cells. Now, photocrosslinking analogs of amino acids can be incorporated into proteins in place of naturally occurring amino acids. Photocrosslinking analogs of sugars can be added to both glycoproteins and glycolipids in place of the naturally occurring sugars. These photocrosslinking molecules can be activated in intact cells to achieve photochemical capture of native interactions.
The ability to selectively and strategically incorporate photocrosslinking groups into cellular biomolecules provides advantages over other, more traditional forms of crosslinking. The most familiar form of crosslinking relies on the use of formaldehyde, a non-selective reagent that can be added to either intact cells or lysates [7]. While the non-selectivity of formaldehyde crosslinking aids in unbiased interaction discovery, it can also present a challenge, by resulting in extensive and complex crosslinking, which hinders the ability to isolate and characterize crosslinked complexes. Moreover, formaldehyde crosslinking has extremely slow kinetics, adding to the potential for off-target crosslinking events. In contrast, photo-activated crosslinking is highly specific and capable of identifying protein-protein interactions in physiologically relevant contexts. Here we summarize methods commonly used for introducing photocrosslinking groups into cellular biomolecules and for analysis of crosslinked complexes.
Incorporating photocrosslinkers into cellular biomolecules
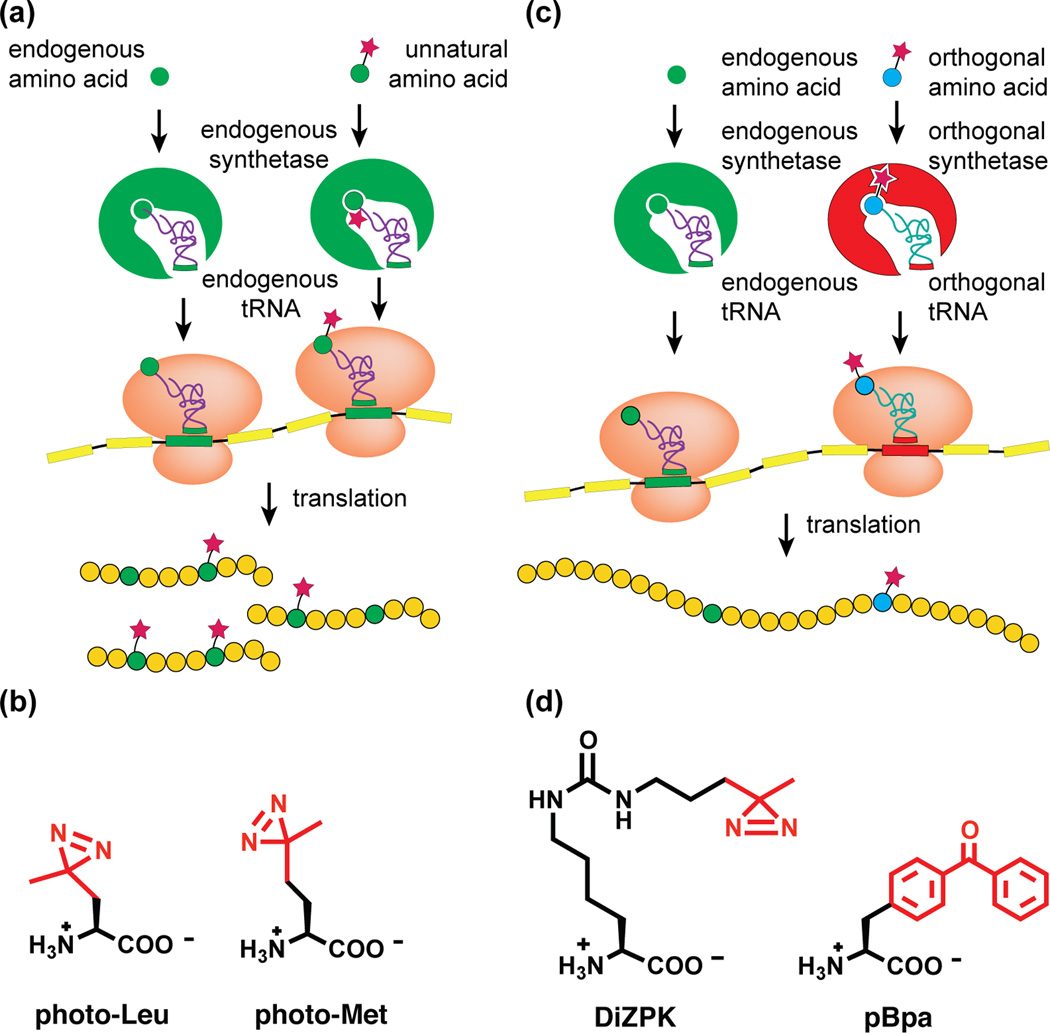
Photocrosslinking amino acids may be incorporated either residue-specifically or site-specifically. Residue-specific incorporation is achieved by metabolic labeling methods in which cells are cultured with an unnatural amino acid analog that resembles a naturally occurring amino acid. The endogenous translational machinery recognizes the photocrosslinking amino acid and incorporates it into proteins in place of the similar natural amino acid [8] (Figure 1a). In this way, photo-active analogs of leucine (photo-Leu) and methionine (photo-Met) can be incorporated into cellular proteins (Figure 1b). Site-specific incorporation of photocrosslinking amino acids is accomplished by genetic code expansion [9] (Figure 1c). In general, genetic code expansion methods rely on modification of an existing aminoacyl tRNA synthetase to create a mutant that is capable of transferring the unnatural amino acid of interest to a cognate tRNA. Cells engineered to express these mutant tRNA/aminoacyl-tRNA synthetase pairs are cultured with photocrosslinking amino acid analogs (Figure 1d). These variant amino acids are sites-pecifically incorporated into proteins via stop codon suppression. The toolkit of the expanded genetic code includes a selection of photocrosslinking amino acids containing benzophenone, aryl azide, alkyl diazirine, and trifluoromethylphenyldiazirine functional groups.
Figure 1. Methods for incorporating photocrosslinking amino acids.
(a) Residue-specific incorporation of photocrosslinking amino acids relies on endogenous tRNA synthetases that accept an unnatural amino acid whose shape is similar to a natural amino acid. tRNA synthetases charge their cognate tRNAs with unnatural amino acids, which then are incorporated into proteins in place of the natural amino acid. (b) Structures of metabolically incorporated photocrosslinking amino acids. Both contain an alkyl diazirine photocrosslinking group (red). (c) Genetically encoded photocrosslinkers are site-specifically incorporated into proteins using stop codon suppression. This method requires an exogenous, engineered orthogonal tRNA/tRNA synthetase pair. (d) Structures of genetically encoded photocrosslinking amino acids. DiZPK contains an alkyl diazirine (red) and pBpa contains a benzophenone (red).
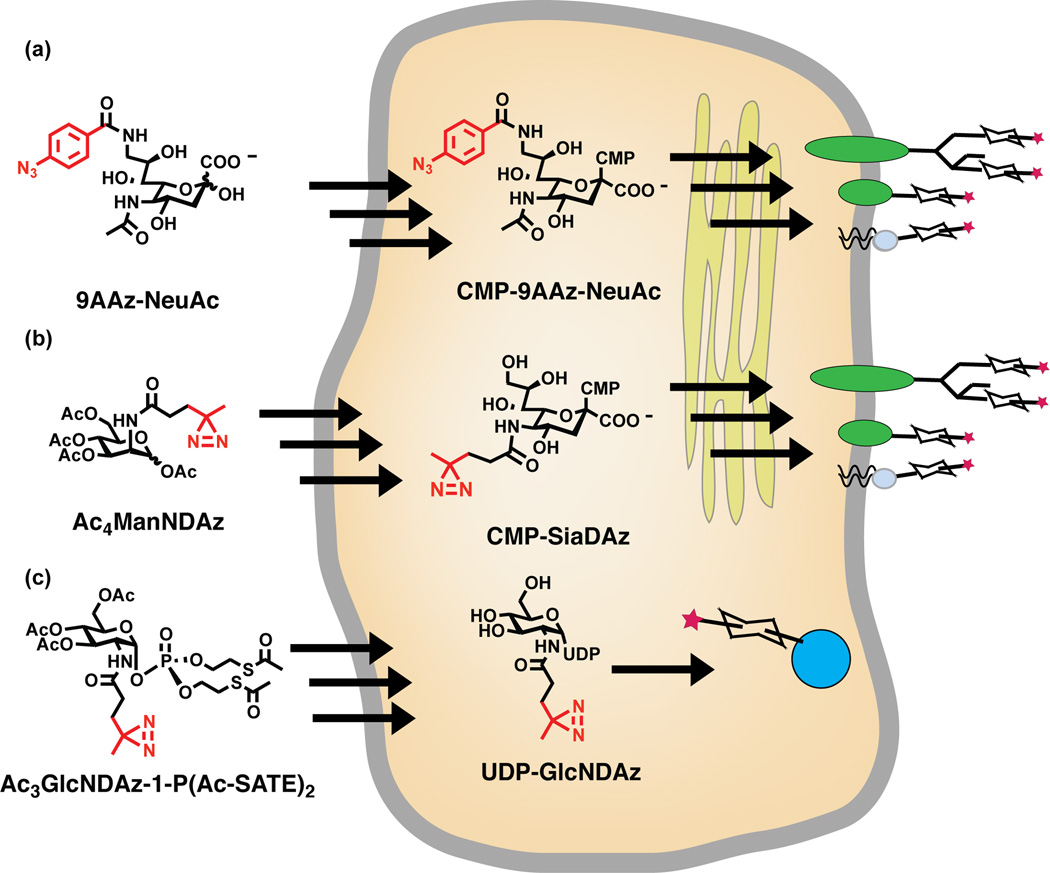
Photocrosslinking sugar analogs are incorporated by metabolic engineering methods, which take advantage of the promiscuity of endogenous glycosylation machinery [10]. In the simplest case, cells are cultured in media that contains the unnatural analog. For example, a sialic acid analog modified with an aryl azide at the C9 position (9AAzNeuAc) can enter cells, be activated to a nucleotide sugar donor, and be transferred to glycoconjugates in place of naturally occurring sialic acids [11] (Figure 2a). While unprotected sugars are sometimes used, polar sugar analogs typically exhibit poor cell permeability; entry into the cell can be facilitated by a pro-drug strategy relying on acylation of hydroxyl groups [12] (Figure 2b). Inside the cell, the acyl protecting groups are removed by endogenous esterases. Different photocrosslinking sugars display dramatically different efficiencies of metabolism. In some cases, the relevant metabolic and glycosylation enzymes are tolerant of unnatural modifications, allowing a metabolic precursor of the sugar of interest to be employed: N-acetylmannosamine (ManNAc) analogs can be metabolized to their sialic acid counterparts, provided that the appended photocrosslinking group is correctly positioned and not too large [13]. In other cases, the endogenous enzymes are remarkably non-permissive, necessitating complex engineering efforts: the diazirine functional group can be incorporated onto O-linked β-N-acetylglucosamine (O-GlcNAc)-modified proteins by combined use of a cell-permeable, phospho-sugar analog and an engineered pyrophosphorylase, as described below [••14] (Figure 2c). To date, cell-based photocrosslinker modification of glycoconjugates includes incorporation of photocrosslinking sialic acid residues onto both glycoproteins and glycolipids and incorporation of photocrosslinking O-GlcNAc onto intracellular proteins [11, ••14, 15, •16].
Figure 2. Metabolic incorporation of photocrosslinking sugars.
(a) 9AAzNeuAc is a photocrosslinking analog of sialic acid. The aryl azide crosslinker is shown in red. Cells are cultured with 9AAzNeuAc, which enters cells and is activated to CMP-9AAzNeuAc by endogenous enzymes. CMP-9AAzNeuAc is transported into the secretory pathway where endogenous sialyltransferases transfer 9AAzNeuAc onto glycoproteins. Although there is currently no direct evidence, it is likely that 9AAzNeuAc is also transferred onto glycolipids. (b) SiaDAz is another photocrosslinking analog of sialic acid. The alkyl diazirine crosslinker is shown in red. Cells cultured with a cell-permeable precursor to SiaDAz, Ac4ManNDAz, use endogenous enzymes to metabolize this compound to CMP-SiaDAz. CMP-SiaDAz is transported into the secretory pathway where endogenous sialyltransferases transfer SiaDAz to glycoproteins and glycolipids. (c) GlcNDAz is a photocrosslinking analog of GlcNAc, containing an alkyl diazirine photocrosslinker (red). Cells are cultured with a cell-permeable analog of GlcNDAz-1-P, which is converted to UDP-GlcNDAz by intracellular enzymes. The endogenous O-GlcNAc transferase (OGT) transfers GlcNDAz to serines and threonines of nucleocytoplasmic proteins. More detail given in Figure 5.
Analyzing photocrosslinked complexes
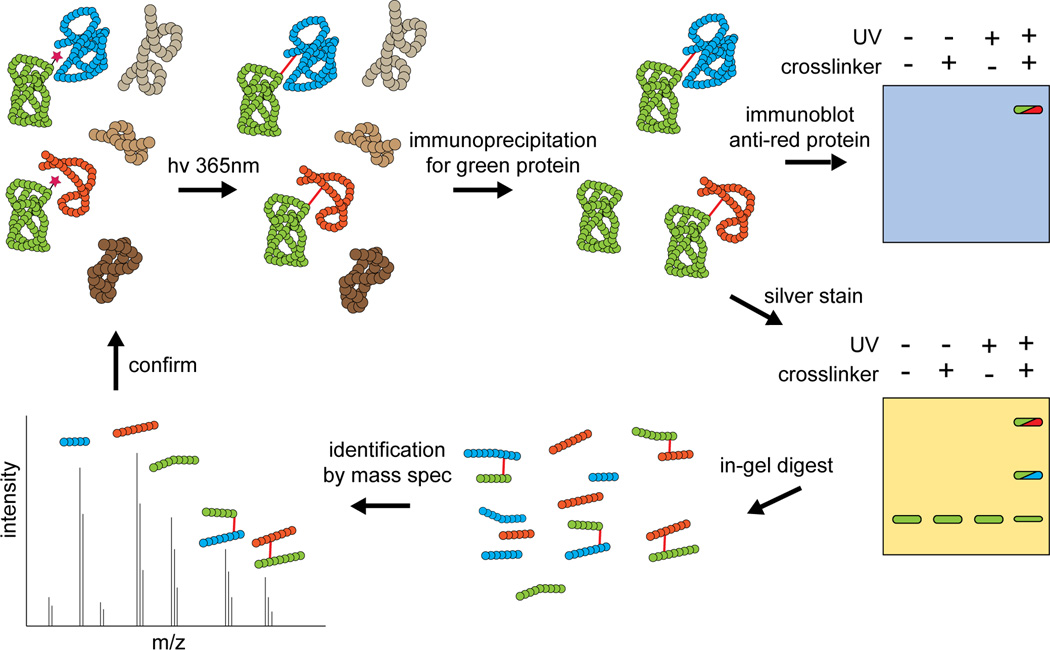
Once photocrosslinkers have been incorporated, cells are exposed to UV radiation to induce crosslinking of the interactome in a native setting. Because photocrosslinking results in covalent linkages between interacting molecules, interactions are effectively frozen in time and cells can be lysed for further analytical steps (Figure 3) [17, 18]. Typically, molecules of interest are isolated for further characterization. Immunoaffinity purification can be used to isolate a particular (glyco)protein, along with all crosslinked complexes that contain this molecule. If certain candidate interacting partners are under consideration, the immunopurified material can be analyzed by immunoblot, using antibodies that recognize potential interaction partners. In this experimental approach, a putative interaction is confirmed by the appearance of a new species that appears only under crosslinking conditions and whose apparent molecular weight is consistent with the size of the crosslinked complex. When possible, identification of the crosslinked complex can be further solidified by a reciprocal experiment, in which immunoprecipitation is carried out using an antibody against potential interaction partner, followed by immunoblot to detect the target protein.
Figure 3. Interactome discovery by mass spectrometry-assisted photocrosslinking analysis.
Following UV-induced crosslinking, a target protein of interest and its crosslinked complexes are isolated by immunopurification. The covalent bond crosslink maintains interaction complexes, even under stringent washing conditions. Crosslinking to candidate interaction partners can be assessed by immunoblot. Alternatively, a candidate list can be generated by performing mass spectrometry on crosslinked material. Candidates identified by mass spectreometry must be verified by additional crosslinking experiments.
If a candidate list is not yet available, photocrosslinked samples can be subjected to mass spectrometry analysis to identify potential interaction partners. As described above, the (glyco)protein of interest and its crosslinked complexes must first be isolated by immunopurification. In this discovery-oriented mode, the immunopurified sample is subjected to tryptic digest, followed by tandem mass spectrometry (MS/MS) to characterize individual tryptic peptides from both the protein of interest and its crosslinked binding partners. Resultant MS/MS data are searched against an appropriate database to identify protein components of the immunopurified sample. While simple MS/MS analysis of crosslinked, immunopurified samples can sometimes deliver a suitable candidate list, more sensitive detection of crosslinked species can be obtained by comparing crosslinked and non-crosslinked samples. The comparison can be performed computationally, using normalized spectral index quantification to compare MS/MS data obtained from crosslinked or non-crosslinked samples [19]. Alternatively, one of the samples can be labeled isotopically (stable isotope labeling by amino acids in cell culture; SILAC) allowing accurate comparison of peptide abundance in the two samples [20]. Enrichment of peptides in the crosslinking sample over the non-crosslinking sample provides insight into potential interaction partners. Regardless of what method is employed, data obtained through mass spectrometry analysis should be viewed as a candidate list and must be validated by immunoprecipitation and immunoblot, as described above.
Obviously, high quality antibodies, suitable for both immunoprecipitation and immunoblot, are critical tools for the analyses described above. However, in some cases, such antibodies are not readily available, or crosslinked complexes may be poorly recognized by antibodies. In these situations, it is useful to introduce a gene encoding the target protein of interest, modified with an affinity or epitope tag. Tagged proteins and their complexes can typically be efficiently immunopurified, facilitating further analysis by either immunoblot or mass spectrometry. However, tagging does negate one advantage of photocrosslinking, which is that it can be performed with endogenous levels of proteins.
Context Matters: Interactome Analysis in Native and Non-Native Settings
As described above, one of the drawbacks of common interaction detection methods is that they interrogate binding interactions outside of their native context. For example, two proteins may localize to distinct subcellular locations and thus never meet in nature, yet may incidentally interact in the context of an affinity purification experiment. Cell-based photocrosslinking approaches capture interactions in their native context, avoiding detection of spurious binding events. Indeed, as described below, studies conducted in non-native settings can deliver a list of candidate interaction partners, while cell-based photocrosslinking studies can determine the subset of those candidates that function in the native context.
CD22 is a member of the siglec (sialic acid binding, immunoglobulin-like lectins) family of proteins that interact with sialylated molecules displayed on the same cell (cis interactions) or opposing cells (trans interactions) [21]. Siglec-ligand interactions mediate B-cell receptor signaling, homing of immune cells, and T-cell signaling. CD22 recognizes a simple glycan structure and therefore interacts with many sialylated glycoproteins in lysates, but a smaller number of specific binding partners likely function in a physiologically relevant context [11, ••22]. The Paulson group determined the specific cis binding partners of CD22 by photocrosslinking experiments that relied on metabolic incorporation of the 9-aryl azide derivative of sialic acid (9AAzNeuAc) [23]. CD45, PMCA, and IgM had been identified previously as candidate CD22 interacting partners by traditional techniques. However, photocrosslinking experiments conducted in living cells showed no evidence for crosslinking of these proteins to CD22 and revealed only large homomultimeric complexes of CD22. In addition, CD22 was the only cis interacting partner identified by mass spectrometry, furthering confirming its identity as the primary cis interacting partner. Homomultimerization of CD22 has been confirmed by a second photocrosslinking approach, which used a cell-permeable, diazirine-modified ManNAc analog to generate diazirine-modified sialylated glycoconjugates [24].
Experiments to identify the CD22 trans ligand exemplify one the most significant challenges associated with the use of photocrosslinking to identify binding partners – the production of sufficient crosslinked material for mass spectrometry analysis. To surmount this challenge, photocrosslinking was first conducted using excess soluble CD22 to generate the candidate list [••22]. Then, photocrosslinking of cell co-cultures produced enough photocrosslinked CD22 trans complex for analysis by subsequent immunoprecipitation and immunoblot, in which individual candidates could be confirmed or invalidated. Only three of nineteen candidate proteins (IgM, CD45, and Basigin) survived the validation process. Of these three, IgM crosslinking was the most robust. Thus, CD22 is the primary cis ligand and the B-cell receptor IgM is the primary trans ligand for CD22. In both cases, photocrosslinking was critical to determining that only a subset of candidate interacting partners functioned in the native context. Analogous approaches may prove useful for other context-dependent interactions.
Mapping Binding Interfaces Within an Immense Macromolecular Complex
Photocrosslinking approaches can offer insight into the topology of large macromolecular machines [25, •26, 27, 28]. In many cases, structures of individual proteins or even complexes are known, but the intact and complete macromolecular machinery eludes structural biology efforts. The eukaryotic transcriptional machinery is one such complex. Transcription is facilitated by two massive multi-protein complexes – RNA polymerase II (Pol II) and Mediator [29, 30]. Pol II consists of ~12 subunits and has a molecular mass of >500 kDa while the Mediator complex consists of ~25 subunits with an overall mass of 1 MDa. Direct interactions between the two complexes have been suggested by genetic, immunoprecipitation, and electron microscopy (EM) experiments. Nonetheless, the subunits that form the exact binding surface remained unknown. To solve this problem, the Werner group used a metabolic labeling approach to nonselectively incorporate photo-Leu and photo-Met into proteins within living cells [••31]. After subsequent UV irradiation, hemagluttinin (HA)-tagged subunits of Mediator were immunoprecipitated and crosslinking to specific Pol II subunits was determined by immunoblotting. Of the eighty potential Pol II-Mediator interactions tested, only one direct interaction – between Med17 and Rpb3 – was identified. Thus, the photocrosslinking results provided specific detection of a distinct interaction interface. This approach was superior to EM, which only narrowed the potential interacting proteins to sixteen Mediator subunits and five Pol II subunits [••31, 32], and to formaldehyde crosslinking, which identified seventeen Mediator subunits that interacted (both directly and indirectly) with Rpb1 and Rpb3 [33]. The identification of the Pol II-Mediator interface aptly demonstrates the power of photocrosslinking in identifying specific, direct interactions even within massive multi-protein complexes.
Probing Dynamic Interfaces with Strategically-Incorporated Photocrosslinkers
Protein complexes are not static structures. The presence of a small molecule ligand or protein partner may induce conformational changes that dramatically alter the interfaces and topology of the complex. Furthermore, the function of a protein complex may require short-lived, yet significant changes in local protein-protein interactions. Photocrosslinkers can be deployed to illuminate these interaction dynamics in living cells (for example, see [25]). The Kahne group used such an approach to gain insight into the dynamics of membrane protein interactions in bacteria [••34]. Lipopolysaccharide (LPS) is transported to the outer membrane (OM) of gram-negative bacteria by a poorly understood mechanism. Implicated in the transit process are two essential proteins, LptD and LptE, which associate, forming a stable complex. LptD had been proposed to form a β-barrel, with LptE providing a complementary plug, but the interaction interface was not well characterized. The Kahne group introduced pBpa at 27 distinct positions throughout LptE via genetically encoded unnatural amino acid mutagenesis. Subsequent UV irradiation of living cells covalently captured native LptE-LptD interactions. Some, but not all, of the pBpa-containing LptE variants were able to crosslink LptD, highlighting the specificity of photocrosslinking. In addition to mapping specific residues of LptE that interacted with LptD, the Kahne group coupled mass spectrometry with photocrosslinking and identified the specific peptide region (predicted to be an extracellular loop) of LptD that was contacted by residue 150 of LptE. Furthermore, while LptD and LptE form a stable complex, the specific interaction detected in this work may be more transient. Indeed, deletion of the LptD loop did not affect association of LptD and LptE in vitro, but did impair OM assembly in vivo, suggesting that dynamic interactions at this particular site are critical for complex assembly. Thus, photocrosslinking was able to identify dynamic interactions between LptD and LptE that are critical for complex assembly in a physiologically relevant context.
One of the more remarkable aspects of this work is that the authors were able to use mass spectrometry to identify the LptD-LptE crosslinked peptide, thereby delineating the regions of the proteins that physically associate. To date, this type of mass spectrometry analysis has not been routine, due to the complexity of crosslinked samples and the computational effort necessary to search mass spectrometry datasets for all potential crosslinked products. New mass spectrometry methods and software tools are now becoming available and the area promises to mature in coming years. Routine identification of crosslinked peptides will be an essential step toward comprehensive interactome analysis (Figure 3).
Capturing Ephemeral Binding Events
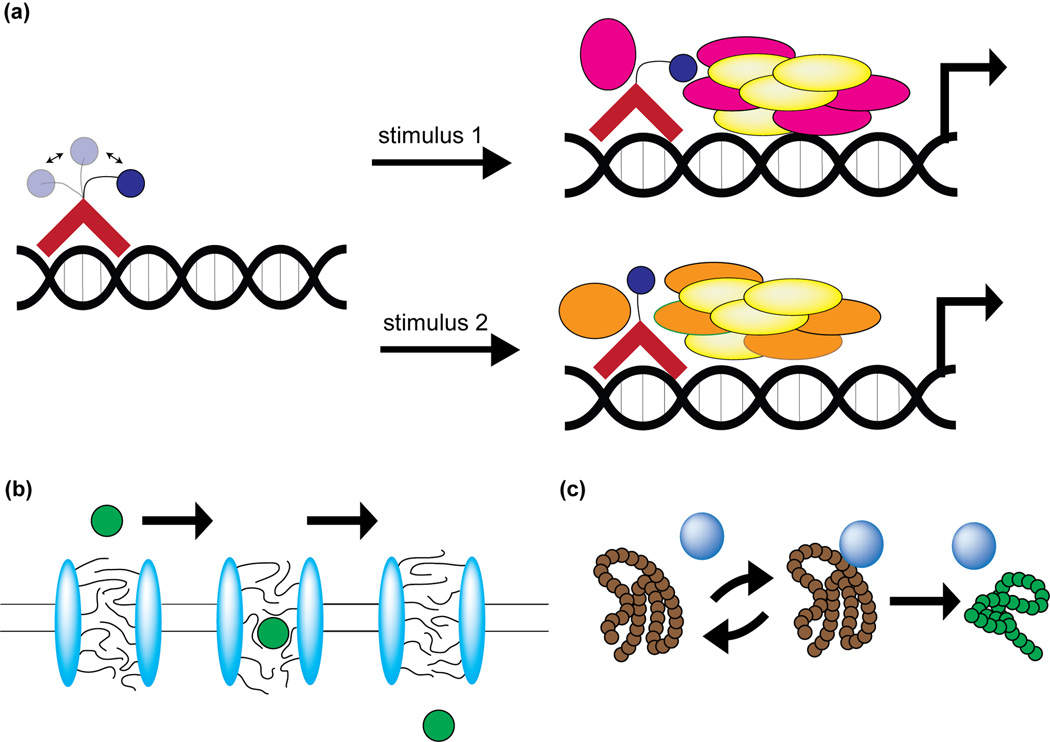
The true power of cell-based photocrosslinking technology lies in its ability to resolve low-affinity, transient interactions. Low-affinity interactions are often dismissed as non-specific, but, in fact, these transient interactions facilitate critical cellular processes such as cell adhesion, nucleocytoplasmic transport, protein folding, and transcriptional activation [35–39] (Figure 4). The intrinsically disordered proteins (IDPs) that commonly mediate low-affinity binding events share certain similarities [40]. Upon protein binding, disordered proteins undergo a partial or full disordered to order transition resulting in a large decrease in conformational entropy [41]. This uncouples specificity and binding affinity, allowing for specific reversible interactions. Furthermore, the disordered nature of IDPs allows for multiple binding partners to exist. Thus, IDPs can engage in reversible, transient, yet biologically-relevant interactions with multiple binding partners. Similar principles may hold for recognition of certain glycoproteins [42]. Both IDPs and glycoproteins are abundant and biologically significant, yet relatively understudied [43, 44]. Proteins containing disordered regions often fill promiscuous “hub” positions in signaling networks, while glycosylated proteins mediate diverse extracellular communication events [45]. For both classes of proteins, photocrosslinking is an ideal tool to determine which binding partner is relevant in a particular physiological context.
Figure 4. Transient, dynamic interactions are fundamental to cellular biology.
(a) Activation domains of transcription factors tend to be intrinsically disordered. This flexibility allows the activation domain to interact with different co-activators under different stimuli. (b) FG repeats of the nuclear pore complex form a dynamic sieve that acts as a gateway between the nucleoplasm and the cytoplasm. (c) Molecular chaperones undergo multiple interaction cycles with unfolded proteins. This process is critical for proper folding of nascent proteins and also of misfolded proteins that appear during times of stress.
Transcriptional Activation
Transcriptional activation relies on weak yet specific contacts that occur between activators and coactivators (Figure 4a). Most transcriptional activators are believed to contain at least one intrinsically disordered segment [46]. Thus, the interactions in which activators engage are often low-affinity and difficult to study using traditional techniques. Indeed, while there have been extensive reports of in vivo and in vitro recruitment of the Swi/Snf chromatin-remodeling complex during transcriptional activation, direct contacts between Swi/Snf subunits and transcriptional activators remain poorly characterized. Adding to the challenge, coactivator complexes can be very large, containing multiple subunits. For instance, the vertebrate Swi/Snf chromatin remodeling complex consists of about eleven subunits encoded by twenty genes, implying that ~288 assemblies of this complex are possible [47]. Additionally, the composition of the coactivator complex varies in response to cellular stimuli and specific contacts between activator and coactivator may also vary in different physiologic contexts.
Recently, the Mapp group used a photocrosslinking approach to identify the subunits of the Swi/Snf complex that directly interact with the activation domains of two transcriptional activators, VP16 and Gal4 [••48]. Using genetically encoded photocrosslinkers, they incorporated pBpa site-specifically into the activation domains of VP16 and Gal4. UV-induced crosslinking in live cells suggested that Snf2 of the Swi/Snf complex interacts with these activation domains. Furthermore, Snf2 did not crosslink the activation domain of Gcn4, highlighting the specificity of this technique. Taken together, photocrosslinking identified Snf2 as a subunit of the Swi/Snf complex that directly binds activation domains of certain transcription factors. Knowledge of the direct binding partners of transcriptional activators could provide a route for development of inhibitors to prevent pathologic transcriptional activation.
Transport through the nuclear pore
Transient interactions are essential to the process of transport across the nuclear membrane (Figure 4b). Molecules must be recognized specifically in order for transport to be selective, yet the recognition must occur with low affinity so that transit is not impeded. Nucleoporins (Nups) containing phenylalanine-glycine (FG) repeats are essential players in the transport process. The FG regions of FG Nups are large, highly disordered segments that pack densely in the nuclear pore, creating a hydrogel-like phase that acts as a dynamic molecular sieve [36]. In vertebrates, the FG regions are further modified by O-GlcNAc on serines/threonines [49]. Given the size, complexity, and unique biophysical properties of the nuclear pore, direct protein interactions that occur during nuclear transport are extremely difficult to study. Taking advantage of the presence of O-GlcNAc on FG Nups, the Kohler lab used a novel photocrosslinking approach to identify proteins that directly interact with certain FG Nups [••14].
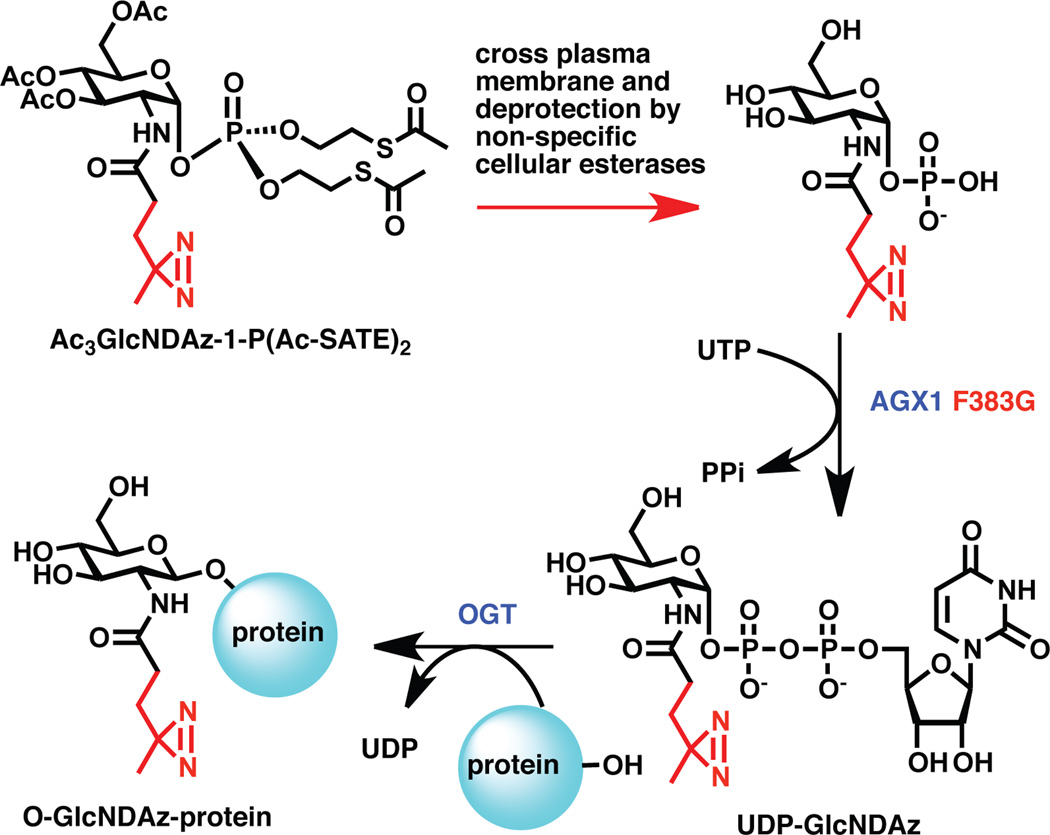
To incorporate the diazirine photocrosslinker directly onto O-GlcNAc residues, the Kohler group employed a metabolic engineering approach that relied on chemical synthesis and enzyme engineering (Figure 5). To produce diazirine-modified O-GlcNAc (O-GlcNDAz), cells were engineered to express a mutant form of the GlcNAc-metabolizing enzyme AGX1 and cultured with a cell-permeable, diazirine-modified derivative of GlcNAc-1-phosphate (Ac3GlcNDAz-1-P(Ac-SATE)2). Intact O-GlcNDAz-producing cells were UV irradiated to photochemically crosslink O-GlcNAc-modified proteins to neighboring molecules. Multiple FG Nups (Nup153, Nup214, Nup358) were immunoprecipitated using a single monoclonal antibody and several crosslinked species were observed. Molecules crosslinked to the FG Nups were identified by mass spectrometry. Identified proteins included known transport factors exportin-1, transportin-1, transportin-2, importin β-1, and nuclear RNA export factor 1. The Kohler lab then characterized the specific FG Nups that transportin-1 crosslinked, identifying Nup358 and Nup153, but not Nup214, as interaction partners. Thus, photocrosslinking was used to specifically identify transient interactions that occur during the process of nuclear transport.
Figure 5. Production of O-GlcNDAz requires chemical synthesis and enzyme engineering.
Diazirine-modified, cell-permeable GlcNAc-1-P (Ac3GlcNDAz-1-P(Ac-SATE)2) is prepared synthetically and added to cultured cells, which remove the protecting groups. The deprotected compound is converted to UDP-GlcNDAz by the action of a mutant form of AGX1. OGT transfers GlcNDAz from UDP-GlcNDAz to substrate proteins.
Chaperone-assisted protein folding
Photocrosslinking is an ideal method for identifying and characterizing the transient interactions that occur during chaperone-assisted protein folding. Numerous chaperone systems have evolved to maintain the integrity of cellular proteins [37, 50]. Chaperones must transiently interact with nascent or damaged proteins, releasing them when they are properly folded (Figure 4c). Furthermore, a critical role of chaperones is to protect cells from proteins that misfold in times of stress. Therefore, the chaperone interactome is likely altered under conditions of stimulus or stress. Previous in vitro work with photocrosslinkers elucidated how chaperones interact with nascent proteins, aggregated proteins, and subunits of large, multiprotein complexes [51–55]. A more recent report from the Chen group used crosslinking to define the interactome of E. coli HdeA in a unique in vivo environment [••56].
The Chen group illuminated the interactome of E. coli chaperone HdeA by genetically incorporating a diazirine-containing amino acid site specifically into HdeA [••56]. Photocrosslinking experiments conducted in live E. coli at neutral pH confirmed previous work suggesting that HdeA is a dimer under non-stressed conditions [57]. Next, they placed the bacteria in an acidic environment to mimic the stress that occurs when pathogenic bacteria traverse the human stomach en route to the small intestine. At pH 2.3, UV irradiation of live cells resulted in numerous crosslinked HdeA species, which were isolated by immunopurification and analyzed by mass spectrometry. After identifying the proteins that interact directly with HdeA under acidic conditions, the Chen group further characterized the mechanism of HdeA action. They demonstrated that HdeA binds to two major perisplasmic chaperones (SurA and DegP), but not another (Skp), highlighting the specificity of HdeA photocrosslinking. Thus, SurA and DegP are protected from aggregation during acid stress and upon neutralization assist HdeA in facilitating refolding. In this study, photocrosslinking was used to identify transient interactions that occur between proteins in a unique environment that would be extremely difficult to study using traditional techniques. Furthermore, knowledge of direct protein interactions provided clear insight into biological mechanism.
Conclusions and Outlook
All cellular processes are governed by protein-protein interactions that range from strong and effectively permanent to weak and transient. Due to the limitations of traditional techniques however, the vast majority of research has focused on strong interactions and neglected weak transient contacts. Photocrosslinking has demonstrated ability to identify low affinity interactions and to illuminate the biological pathways that they control. Indeed, novel insights into the function of intrinsically disordered proteins and glycoproteins, two particularly difficult classes of proteins to study, have been gained by the use of photocrosslinkers.
A significant advantage of photocrosslinking is that it enables detection of interactions that occur in intact cells. To date, most work in this area has focused on interactions that occur under resting conditions. But the use of photoactivation also allows for exquisite temporal control. UV-induced crosslinking enables the covalent capture of transient interactions that occur in response to cellular stimuli. Indeed, the Chen group utilized photocrosslinking to determine differences in the HdeA acid chaperone interactome under neutral versus acidic conditions. Analogous strategies could be employed to discover how other interactomes change in response to stimuli. For example, CD22 binding partners could be surveyed under B-cell stimuli or in co-culture with different immune cells. The contacts between transcriptional activator and coactivator could be assessed under different growth stimuli. The interactions that underlie nuclear transport could be evaluated under various types of stress. In this way, we predict that photocrosslinking will provide unprecedented resolution of interactome dynamics under a variety of cellular conditions.
In addition to genetic code expansion and metabolic engineering, other methods for introducing photocrosslinkers onto cellular proteins exist, such as intein-mediated protein ligation [58, 59], co-opting the activity of lipoic acid ligase [60, 61], and use of ligand-directed tosylation [62, •63]. Future application of these methods will expand the scope of photocrosslinking applications. In the same vein, we expect that new developments in protein engineering, particularly of the ribosome and of glycosyltransferases, will enhance our ability to selectively incorporate photocrosslinkers into specific sites within proteins and glycoproteins [•64]. Futhermore, other classes of biomolecules can be targeted with photocrosslinking approaches. Photocrosslinking sugar analogs can already be incorporated into cellular glycolipids [•16, 65], and may be deployed to investigate the roles of these molecules in cell signaling [66–70].
Finally, we note that the use of photocrosslinkers has thus far been restricted to cells in tissue culture. However, the methods commonly used to introduce photocrosslinking groups – metabolic labeling and genetic code expansion – can also be applied in multicellular organisms [71, 72]. In the future, it may be possible to incorporate photocrosslinking amino acids or sugars in a living animal and determine the nature of the interactome in vivo. The major challenge associated with these experiments would be achieving effective photoactivation, perhaps by using a transparent animal or by surgically implanting a light emitter.
Highlights.
Photocrosslinking provides insight into context-dependent interactions
Interactome dynamics can be determined by photocrosslinking
Use of photocrosslinkers defines specific interaction interfaces within large complexes
Crosslinking captures transient interaction complexes containing IDPs and glycoproteins
Acknowledgements
We acknowledge financial support from the National Institutes of Health (F30AG040909, T32GM007062, and R01GM090271) and thank all members of the Kohler lab for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, Wiuf C. Estimating the size of the human interactome. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat. Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charbonnier S, Gallego O, Gavin AC. The social network of a cell: recent advances in interactome mapping. Biotechnol. Annu. Rev. 2008;14:1–28. doi: 10.1016/S1387-2656(08)00001-X. [DOI] [PubMed] [Google Scholar]
- 4.Garner AL, Janda KD. Protein-protein interactions and cancer: targeting the central dogma. Curr. Top. Med. Chem. 2011;11:258–280. doi: 10.2174/156802611794072614. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka Y, Bond MR, Kohler JJ. Photocrosslinkers illuminate interactions in living cells. Mol. Biosyst. 2008;4:473–480. doi: 10.1039/b803218a. [DOI] [PubMed] [Google Scholar]
- 6.Gorostiza P, Isacoff EY. Optical switches for remote and noninvasive control of cell signaling. Science. 2008;322:395–399. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutherland BW, Toews J, Kast J. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J. Mass Spectrom. 2008;43:699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- 8.Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods. 2005;2:261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 9.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat. Rev. Mol. Cell. Bio. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 10.Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VDP, Yarema KJ. Metabolic glycoengineering: Sialic acid and beyond. Glycobiology. 2009;19:1382–1401. doi: 10.1093/glycob/cwp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat. Chem. Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 12.Luchansky SJ, Goon S, Bertozzi CR. Expanding the diversity of unnatural cell-surface sialic acids. Chembiochem. 2004;5:371–374. doi: 10.1002/cbic.200300789. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka Y, Kohler JJ. Photoactivatable crosslinking sugars for capturing glycoprotein interactions. J. Amer. Chem. Soc. 2008;130:3278–3279. doi: 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- 14. Yu SH, Boyce M, Wands AM, Bond MR, Bertozzi CR, Kohler JJ. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4834–4839. doi: 10.1073/pnas.1114356109. A diazirine photocrosslinker was incorporated onto O-GlcNAc residues in cells and used to provide direct evidence that O-GlcNAc residues are intimately associated with protein-protein recognition events.
- 15.Yu SH, Bond MR, Whitman CM, Kohler JJ. Metabolic labeling of glycoconjugates with photocrosslinking sugars. Methods Enzymol. 2010;478:541–562. doi: 10.1016/S0076-6879(10)78026-5. [DOI] [PubMed] [Google Scholar]
- 16. Bond MR, Whitman CM, Kohler JJ. Metabolically incorporated photocrosslinking sialic acid covalently captures a ganglioside-protein complex. Mol. Biosyst. 2010;6:1796–1799. doi: 10.1039/c0mb00069h. A diazirine-modified sialic acid was metabolically incorporated into glycolipids. These modified glycolipids were covalently crosslinked to a protein binding partner in a cellular setting.
- 17.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinette D, Neamati N, Tomer KB, Borchers CH. Photoaffinity labeling combined with mass spectrometric approaches as a tool for structural proteomics. Expert Rev. Proteomic. 2006;3:399–408. doi: 10.1586/14789450.3.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundgren DH, Hwang S-I, Wu L, Han DK. Role of spectral counting in quantitative proteomics. Expert Rev. Proteomic. 2010;7:39–53. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- 20.Mann M. Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell. Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 21.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 22. Ramya TN, Weerapana E, Liao L, Zeng Y, Tateno H, Yates JR, 3rd, Cravatt BF, Paulson JC. In situ trans ligands of CD22 identified by glycan-protein photocross-linking-enabled proteomics. Mol. Cell. Proteomics. 2010;9:1339–1351. doi: 10.1074/mcp.M900461-MCP200. The publication provides an outstanding example of the use of metabolically incorporated photocrosslinking in combination with mass spectrometry to provide a candidate list of potential interaction partners. The authors go on to determine that only a subset of the identified candidates are physiologically relevant.
- 23.Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat. Chem. Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 24.Bond MR, Zhang H, Vu PD, Kohler JJ. Photocrosslinking of glycoconjugates using metabolically incorporated diazirine-containing sugars. Nat. Protoc. 2009;4:1044–1063. doi: 10.1038/nprot.2009.85. [DOI] [PubMed] [Google Scholar]
- 25.Mori H, Ito K. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16159–16164. doi: 10.1073/pnas.0606390103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. The authors use photocrosslinking to examine the dynamics of bacterial RNA polymerase.
- 27.Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H-T, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wild T, Cramer P. Biogenesis of multisubunit RNA polymerases. Trends Biochem. Sci. 2012;37:99–105. doi: 10.1016/j.tibs.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451–1454. doi: 10.1126/science.1200188. The authors explore vast combinatorial possibilities for possible interactions between RNA polymerase II and mediator. Using cell-based photocrosslinking, they were able to identify a specific bimolecular interaction with high confidence.
- 32.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 33.Tardiff DF, Abruzzi KC, Rosbash M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19948–19953. doi: 10.1073/pnas.0710179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freinkman E, Chng SS, Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2486–2491. doi: 10.1073/pnas.1015617108. The authors used photocrosslinking to probe the interface of an essential membrane protein complex in bacteria. Their impressive mass spectrometry analysis revealed the specific regions of the each protein that engaged in crosslinking.
- 35.Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr. Opin. Chem. Biol. 2004;8:617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Hulsmann BB, Labokha AA, Gorlich D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell. 2012;150:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 37.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 38.Khan SH, Kumar R. An overview of the importance of conformational flexibility in gene regulation by the transcription factors. J. Biophysics. 2009;2009:210485. doi: 10.1155/2009/210485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat. Chem. Biol. 2008;4:728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 42.Taylor ME, Drickamer K. Structural insights into what glycan arrays tell us about how glycan-binding proteins interact with their ligands. Glycobiology. 2009;19:1155–1162. doi: 10.1093/glycob/cwp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Informatics. 2000;11:161–171. [PubMed] [Google Scholar]
- 44.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 45.Singh GP, Ganapathi M, Dash D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins. 2007;66:761–765. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krishnamurthy M, Dugan A, Nwokoye A, Fung YH, Lancia JK, Majmudar CY, Mapp AK. Caught in the act: covalent cross-linking captures activator-coactivator interactions in vivo. ACS Chem. Biol. 2011;6:1321–1326. doi: 10.1021/cb200308e. This paper tackled the challenge of identifying the direct binding partner(s) of an intrinsically disordered activation domain. The authors were able to capture a transient interaction between an activator and a single subunit of the Swi/Snf complex.
- 49.Hanover JA, Cohen CK, Willingham MC, Park MK. O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J. Biol. Chem. 1987;262:9887–9894. [PubMed] [Google Scholar]
- 50.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 51.Liu C, Young AL, Starling-Windhof A, Bracher A, Saschenbrecker S, Rao BV, Rao KV, Berninghausen O, Mielke T, Hartl FU, et al. Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature. 2010;463:197–202. doi: 10.1038/nature08651. [DOI] [PubMed] [Google Scholar]
- 52.Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, et al. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- 53.Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol. Cell. 2007;25:247–260. doi: 10.1016/j.molcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 54.Lakshmipathy SK, Tomic S, Kaiser CM, Chang HC, Genevaux P, Georgopoulos C, Barral JM, Johnson AE, Hartl FU, Etchells SA. Identification of nascent chain interaction sites on trigger factor. J. Biol. Chem. 2007;282:12186–12193. doi: 10.1074/jbc.M609871200. [DOI] [PubMed] [Google Scholar]
- 55.Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM. Real-time observation of trigger factor function on translating ribosomes. Nature. 2006;444:455–460. doi: 10.1038/nature05225. [DOI] [PubMed] [Google Scholar]
- 56. Zhang M, Lin S, Song X, Liu J, Fu Y, Ge X, Fu X, Chang Z, Chen PR. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nature Chem. Biol. 2011;7:671–677. doi: 10.1038/nchembio.644. The full power of photoactivated crosslinking is demonstrated in this paper. The authors used genetically incorporated diazirine to compare the interactomes of a disordered protein in specialized cellular conditions. The information obtained from these experiments providing insight into the biological mechanisms of acid resistance.
- 57.Gajiwala KS, Burley SK. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 2000;295:605–612. doi: 10.1006/jmbi.1999.3347. [DOI] [PubMed] [Google Scholar]
- 58.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shah NH, Vila-Perello M, Muir TW. Kinetic control of one-pot trans-splicing reactions by using a wild-type and designed split intein. Angew. Chem. 2011;50:6511–6515. doi: 10.1002/anie.201102909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nature Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baruah H, Puthenveetil S, Choi YA, Shah S, Ting AY. An engineered aryl azide ligase for site-specific mapping of protein-protein interactions through photo-cross-linking. Angew. Chem. 2008;47:7018–7021. doi: 10.1002/anie.200802088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsukiji S, Miyagawa M, Takaoka Y, Tamura T, Hamachi I. Ligand-directed tosyl chemistry for protein labeling in vivo. Nature Chem. Biol. 2009;5:341–343. doi: 10.1038/nchembio.157. [DOI] [PubMed] [Google Scholar]
- 63. Tamura T, Tsukiji S, Hamachi I. Native FKBP12 engineering by ligand-directed tosyl chemistry: labeling properties and application to photo-cross-linking of protein complexes in vitro and in living cells. J. Amer. Chem. Soc. 2012;134:2216–2226. doi: 10.1021/ja209641t. The paper presents proof-of-principle experiments demonstrating the use of a new method to add photocrosslinking functionality to endogenous proteins.
- 64. Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. A tour-de-force of genetic evolution yields an engineered ribosome capable of decoding quadruplet codons. This work has the potential for far-reaching applications, including the simultaneous incorporation of multiple unnatural amino acids. For example, a protein containing both a photocrosslinkers and a purification handle could be encoded.
- 65.Whitman CM, Yang F, Kohler JJ. Modified GM3 gangliosides produced by metabolic oligosaccharide engineering. Bioorg. Med. Chem. Lett. 2011;21:5006–5010. doi: 10.1016/j.bmcl.2011.04.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fra AM, Masserini M, Palestini P, Sonnino S, Simons K. A photo-reactive derivative of ganglioside GM1 specifically cross-links VIP21-caveolin on the cell surface. FEBS Lett. 1995;375:11–14. doi: 10.1016/0014-5793(95)95228-o. [DOI] [PubMed] [Google Scholar]
- 67.Ono M, Handa K, Sonnino S, Withers DA, Nagai H, Hakomori S. GM3 ganglioside inhibits CD9-facilitated haptotactic cell motility: coexpression of GM3 and CD9 is essential in the downregulation of tumor cell motility and malignancy. Biochemistry. 2001;40:6414–6421. doi: 10.1021/bi0101998. [DOI] [PubMed] [Google Scholar]
- 68.Wendeler M, Hoernschemeyer J, Hoffmann D, Kolter T, Schwarzmann G, Sandhoff K. Photoaffinity labelling of the human GM2-activator protein. Mechanistic insight into ganglioside GM2 degradation. Eur. J. Biochem. 2004;271:614–627. doi: 10.1111/j.1432-1033.2003.03964.x. [DOI] [PubMed] [Google Scholar]
- 69.Vodovozova EL. Photoaffinity labeling and its application in structural biology. Biochemistry (Moscow) 2007;72:1–20. doi: 10.1134/s0006297907010014. [DOI] [PubMed] [Google Scholar]
- 70.Haberkant P, van Meer G. Protein-lipid interactions: paprazzi hunting for snap-shots. Biol. Chem. 2009;390:795–803. doi: 10.1515/BC.2009.074. [DOI] [PubMed] [Google Scholar]
- 71.Kayser H, Zeitler R, Kannicht C, Grunow D, Nuck R, Reutter W. Biosynthesis of a nonphysiological sialic-acid in different rat organs, using N-propanoyl-D-hexosamines as precursors. J. Biol. Chem. 1992;267:16934–16938. [PubMed] [Google Scholar]
- 72.Greiss S, Chin JW. Expanding the genetic code of an animal. J. Amer. Chem. Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]