Abstract
Recent surveillance data of multidrug-resistant tuberculosis (MDR-TB) reported the highest rates of resistance ever documented. As further amplification of resistance in MDR strains of Mycobacterium tuberculosis occurs, extensively drug-resistant (XDR) and totally drug resistant (TDR) TB are beginning to emerge. Whilst for the most part, the epidemiological factors involved in the spread of MDR-TB are understood, insights into the bacterial drivers of MDR-TB have been gained only recently, largely owing to novel technologies and research in other organisms. Herein, we review recent findings on how bacterial factors such as persistence, hypermutation, the complex interrelationship between drug resistance and fitness, compensatory evolution, and epistasis affect the evolution of multidrug resistance in M. tuberculosis. Improved knowledge of these factors will help better predict the future trajectory of MDR-TB, and contribute to the development of new tools and strategies to combat this growing public health threat.
Keywords: drug resistance, persistence, hypermutation, compensatory mutations, epistasis
The rise of MDR-TB
Tuberculosis (TB), which is caused by Mycobacterium tuberculosis, is one of humankind’s deadliest diseases. The World Health Organization (WHO) estimated 8.7 million new cases and 1.4 million deaths due to TB in 2011, including 430,000 deaths estimated to have occurred among HIV co-infected patients [1]. Although most recent models suggests that the absolute number of TB cases has decreased since 2006 [1], multidrug-resistant (MDR) TB is threatening disease control efforts throughout the globe [2]. MDR-TB is defined by the resistance to isoniazid and rifampicin, the two most potent anti-TB drugs (Box 1), and requires treatment with much more expensive second-line drugs, which, in addition to being costly, cause more adverse events and show lower cure rates [2, 3]. MDR-TB with additional resistance to any fluoroquinolone and one of the second-line injectable drugs kanamycin, amikacin, or capreomycin is referred to as extensively drug-resistant (XDR) TB [2, 3]. Recently, cases of vaguely defined totally drug-resistant (TDR) TB were reported [4, 5], suggesting that human TB is joining the growing list of bacterial diseases entering the post-antibiotic era [6].
Box 1. Treatment of tuberculosis.
Treatment of TB consists of a standardized six or eight month chemotherapy for new or previously treated cases, respectively. In countries where DST is routinely performed, MDR-TB treatment is individualized. Otherwise, MDR-TB is often treated using standardized drug regimens; however, these may vary between countries.
New TB cases
Two months of isoniazid, rifampicin, pyrazinamide and ethambutol (intensive phase).
Four months of isoniazid and rifampicin (continuation phase).
Previously treated TB cases
Two months of isoniazid, rifampicin, pyrazinamide, ethambutol and streptomycin (intensive phase).
One month of isoniazid, rifampicin, pyrazinamide and ethambutol (intensive phase).
Five months of isoniazid, rifampicin and ethambutol (continuation phase).
MDR-TB cases
At least four drugs likely to be effective must be included.
Any first-line oral agents likely to be effective should be included (e.g. pyrazinamide or ethambutol).
One effective injectable aminoglycoside or polypeptide drug should be included (kanamycin, amikacin, capreomycin or streptomycin).
One fluoroquinolone should be included.
Intensive phase therapy including the injectable drug should last at least 6 months. Total duration of therapy should be at least 18 months.
Drug-resistant TB can be “acquired” de novo during treatment, or it can occur through the transmission of already resistant strains; the latter being referred to as “primary resistance” [2]. Primary resistance poses a particular challenge as less than 20% of the estimated MDR-TB cases in the world are believed to be properly diagnosed, largely due to the lack of appropriate laboratory infrastructure in low-income areas that bear the highest disease burden [1]. Disturbingly, while disease control has improved in many areas, MDR-TB incidence rates have increased considerably in other regions [7]. The prevalence of MDR-TB varies significantly across the globe (Figure 1) as a result of many contributing factors. These include variably effective control programs [7], the presence of co-morbidities, which may influence the emergence and spread of MDR-TB (e.g., HIV co-infection) [7, 8], numerous patient-related factors, including societal [2, 9], immunological [10] and genetic factors [11], and many less well understood bacterial properties such as bacterial physiology, genetics, and population biology (Figure 1 and 2) [12, 13]. Ecological theory predicts that bacterial fitness plays an important role in the emergence of drug-resistant bacteria [14]. Based on the dogma that drug-resistant strains are less fit than their drug-sensitive counterparts (see below) [15], early mathematical models suggested that MDR-TB would remain a localized problem [16, 17]. However, subsequent models allowed for heterogeneous fitness in drug-resistant strains and indicate that the former views were too optimistic [18, 19]. Importantly however, attempts to measure the relative fitness of drug-resistant strains of M. tuberculosis using epidemiological approaches have been highly inconclusive (reviewed in [20]). This highlights the need to better understand the underlying biology of M. tuberculosis and the evolutionary forces driving the emergence and spread of drug-resistant bacteria. For many years, the bacterial factors involved in the emergence of MDR-TB remained elusive, but recently, an increasing number of studies investigating how bacterial biology influences the evolution of MDR-TB have begun to provide new insights into this important facet of MDR-TB. The purpose of this review is to summarize these findings and place them in the broader context of our current understanding of MDR-TB. We then discuss how these new insights could be used to improve the treatment and prevention of this deadly disease.
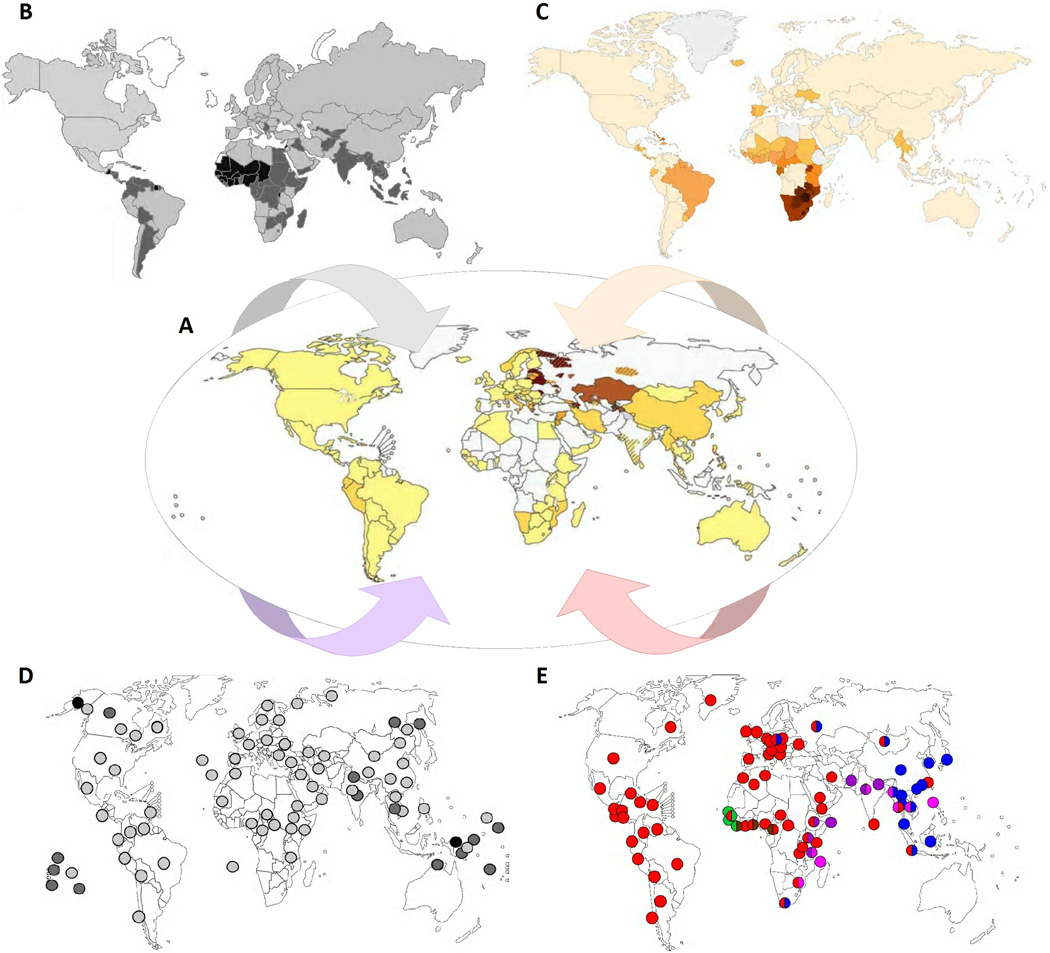
Figure 1. The heterogeneity of MDR-TB.
A. The proportion of new TB cases with MDR-TB (centre; darker shadings indicate an increasing proportion) [7] is heterogeneously distributed and influenced by a multitude of factors including: B. Variably effective control programs (here, represented by the estimated global case detection rates; darker shadings indicate lower case detection rates) [87]; C. The variable presence of co-morbidities (here, represented by the HIV prevalence among incident TB cases; darker shadings indicate a higher prevalence) [1]; D. Different host-related factors (here, represented by the presence of the HLA II allele DQB1*0503, which was the first HLA allele associated with increased TB risk; darker shading indicates a higher abundance) [88]; and E. The global distribution of different phylogenetic lineages of M. tuberculosis (the six main lineages of human TB-associated strains of M. tuberculosis known to date and their geographical distribution are represented by circles of different colour; the colours correspond to the clades indicated in the phylogenetic tree shown in Figure 4). Figure adapted from www.Globalhealthfacts.org and [7, 89, 90].
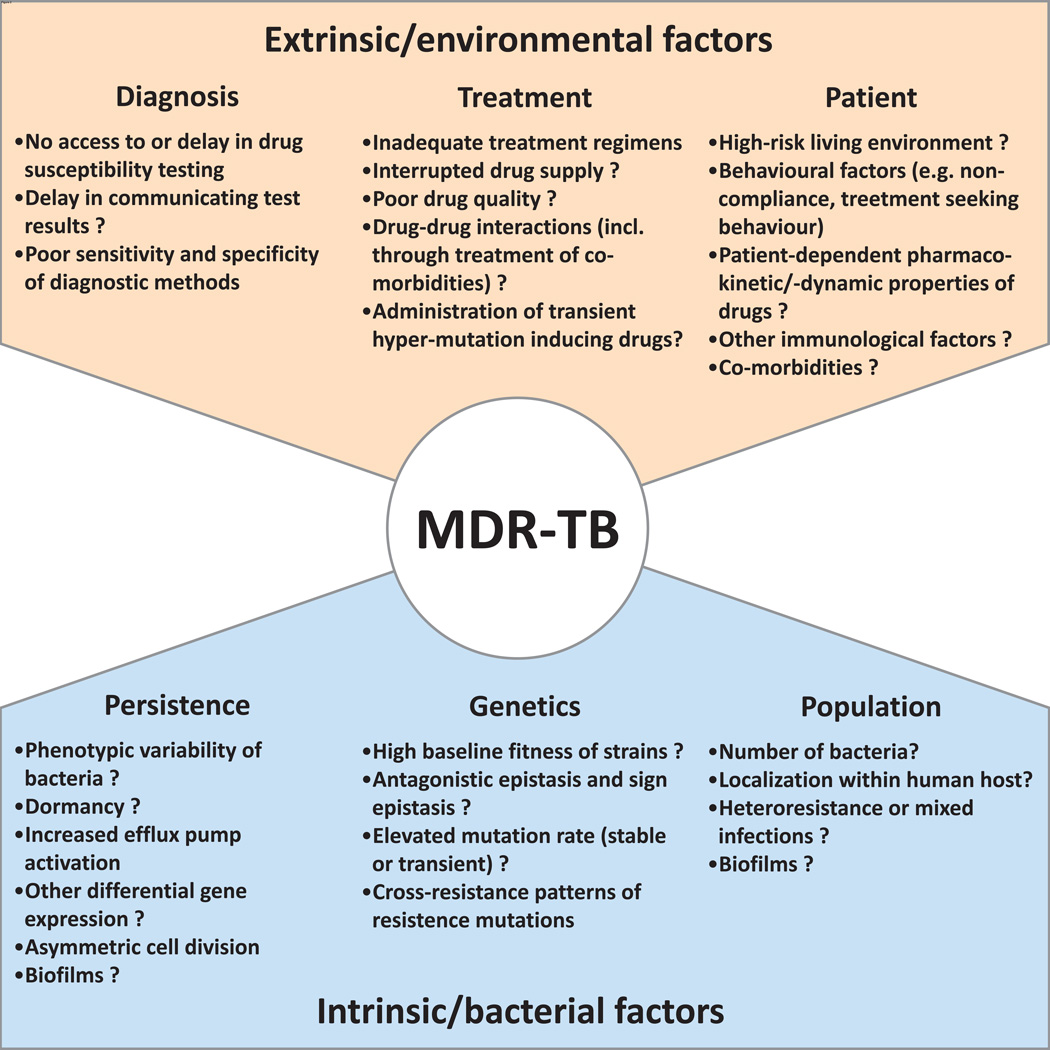
Figure 2. A selection of extrinsic and intrinsic factors contributing to the emergence of MDR-TB.
A non-exhaustive list of extrinsic, non-bacterial, and intrinsic bacterial factors referred to in the text that modulate the evolution of MDR-TB. Question marks indicate factors that might be involved, but for which there is no direct evidence for in M. tuberculosis.
The development of drug resistance in M. tuberculosis
M. tuberculosis is intrinsically resistant to numerous antibiotics, and only a handful of drugs are effective for treatment [21]. This intrinsic resistance is partially due to the thick, lipid-rich cell wall, which is an important characteristic of mycobacteria, limiting the penetration of antibiotics. Moreover, mycobacteria possess various defence systems against the activity of antibiotics, including potent β-lactamases and other drug-neutralizing enzymes [21]. Herein, we will focus on the de novo development of resistance to the drugs that wild-type M. tuberculosis is usually susceptible to, i.e., the drugs used for standard TB treatment (Box 1). Clinically relevant drug resistance in TB is defined as the genetically encoded, increased capacity to tolerate the presence of a specific drug compared to drug-susceptible bacilli [2]. Unlike many other bacteria, M. tuberculosis harbours no resistance plasmids, and drug resistance arises through the acquisition of specific chromosomal mutations [22]. The population structure of M. tuberculosis is essentially clonal, suggesting a limited role for horizontal gene transfer (HGT) in the biology of M. tuberculosis [23]. A recent report has challenged this view [24], but a putative role of HGT in the emergence of drug resistance in M. tuberculosis remains to be demonstrated. Drug resistance-conferring mutations in M. tuberculosis have been described in genes encoding enzymes directly targeted by the antibiotics, in regulatory regions of these genes, or in gene products involved in the activation of pro-drugs (Table 1) [25]. Classical Darwinian models suggest that resistance-conferring mutations appear stochastically within bacterial populations and independently of drug exposure [26]. The average rate of emergence of spontaneous resistance mutations to isoniazid and rifampicin in M. tuberculosis was estimated at 10−8 and 10−9 mutations/bacterium/cell division, respectively [27], suggesting that during mono-therapy, the appearance of resistant bacteria will occur almost inevitably, given an average number of approximately 108 bacilli present in individual TB lesions [28]. Consequently, TB has to be treated with a combination therapy including at least four drugs (Box 1) [2, 3]. Nevertheless, selection of drug-resistant bacteria can occur during intermittent exposure to sub-therapeutic drug levels, initially producing hetero-resistant and finally fully resistant bacterial populations. Hetero-resistance refers to the situation where patients harbour different subpopulations of bacteria, some drug-resistant, and others still drug-susceptible [29]. The de novo emergence of drug resistance in an individual patient can happen because of patient non-adherence [2], poor drug quality, and patient-dependent pharmacodynamic and -kinetic properties of the drugs administered [11, 30]. Additionally, treatment of co-morbidities (e.g., HIV) can influence pharmacodynamic properties of anti-TB drugs and increase the likelihood of resistance development [31]. Importantly, resistance-conferring mutations to a given drug may confer cross-resistance to other drugs targeting related metabolic pathways [32], and the anti-TB drugs themselves can interact in ways that could promote drug resistance [33].
Table 1.
Anti-TB drugs and mechanisms of drug resistance.
| Drug | Genetic region involved in resistance formation* |
Natural function of gene | Role in resistance formation when mutated |
|---|---|---|---|
| Isoniazid | ahpC | Alkyl hyperperoxide reductase | Compensatory mutations |
| fabG | 3-oxoacyl-thioester reductases | Unknown | |
| fadE24 | Involved in fatty acid β-oxidation | Unknown | |
| inhA | Enoyl reductase | Alteration of drug target | |
| inhA promoter** | Regulation of expression of InhA | Overexpression of drug target | |
| iniA | Efflux pump associated | Altered efflux pump activity | |
| katG** | Catalase/peroxidase | Elimination of pro-drug conversion | |
| Rifampicin | rpoA | α subunit of RNA polymerase | Compensatory mutations |
| rpoB** | β-subunit of RNA polymerase | Alteration of drug target | |
| rpoC | β'-subunit of RNA polymerase | Compensatory mutations | |
| Pyrazinimide | pncA** | Nicotinamidase | Elimination of pro-drug conversion |
| Streptomycin | gidB | 7-methylguanosine methyltransferase | Alteration of drug target |
| rpsL** | S12 ribosomal protein | Alteration of drug target | |
| rrs** | 16S rRNA | Alteration of drug target | |
| Ethambutol | embA | Arabinosyl transferase | Alteration of drug target |
| embB** | Arabinosyl transferase | Alteration of drug target | |
| embC | Arabinosyl transferase | Alteration of drug target | |
| embR | Regulator of embCAB operon expression | Overexpression of drug target | |
| iniA | Efflux pump associated | Altered efflux pump activity | |
| rmlD | dTDP-4-dehydrorhamnose reductase | Unknown | |
| Fluoroquinolones | gyrA** | DNA gyrase | Alteration of drug target |
| gyrB** | DNA gyrase | Alteration of drug target | |
| Kanamycin/amikacin | rrs** | 16S rRNA | Alteration of drug target |
| rrs | 16S rRNA | Compensatory mutations | |
| Capreomycin/viomycin | tlyA | rRNA methyltransferase | Alteration of drug target |
| rrs** | 16S rRNA | Alteration of drug target | |
| Ethionamide | inhA | Enoyl reductase | Alteration of drug target |
| inhA promoter | Regulation of expression of inhA | Overexpression of drug target | |
| para-amino salicylic acid | thyA | Thymidylate synthase A | Elimination of pro-drug conversion |
| PA-824 and OPC-67683 | Rv3547 | Hypothetical 16.4 kDa | Alteration of drug target |
| TMC207 | atpE | ATP synthase | Alteration of drug target |
A selection of genetic regions, previously associated with drug resistance according to www.TBDreamDB.com or previously identified as potential compensatory mutations (see text).
High-confidence drug resistance-associated genetic regions according to www.TBDreamDB.com
Among the bacterial factors promoting drug resistance, recent findings highlight a possible role for “persisters” [34]. These are subpopulations of cells that can phenotypically tolerate increased concentrations of drugs, however, they are not genetically resistant (i.e., drug resistance will not be inherited by daughter cells). This phenomenon is also referred to as “phenotypic drug tolerance”. Following the classical understanding, persistence requires phenotypic differentiation into persister cells and is often linked to a state of slow growth or dormancy. This process can be triggered by various factors such as starvation, quorum sensing, intracellular signals, and antibiotic treatment itself [34, 35]. However, persistence is not strictly associated with dormancy; for example, active mycobacterial growth inside macrophages can induce efflux pumps and confer persistence to bacteria upon exposure to anti-TB drugs [36]. Moreover, phenotypic variation among genetically identical bacterial cells can also involve stochastic processes, including spontaneous phenotypic switches, which are thought to facilitate adaptation to fluctuating environments [37]. In addition, deterministic phenotypic variation among genetically identical bacteria can contribute to persistence. For example, it was recently shown that cell division occurs asymmetrically in mycobacteria, which results in an unusual, unipolar growth and a differential ability of the two daughter cells to tolerate anti-TB drugs [38]. Although the phenotypic resistance of persisters is not heritable, persistence prolongs the average lifetime of bacteria exposed to drugs and could therefore increase the likelihood for the subsequent acquisition of a high-level resistance mutation.
A recent study has shown that mutations in M. tuberculosis can occur during latent infections, when bacilli are thought to be replicating very slowly or not at all [39]. These mutations are presumably the result of oxidative DNA damage rather than replication errors, and indicate that M. tuberculosis might have the capacity to acquire drug resistance during latency [39]. This process could be further facilitated in bacteria with an intrinsically elevated mutation rate (mutator genotypes), such as ones with mutations in the mismatch repair system [40]. An association between the acquisition of drug resistance and heritable hypermutation has been described for various bacterial species, including Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa (reviewed in [40]). In M. tuberculosis, a particular phylogenetic lineage known as the “Beijing family” has been repeatedly associated with MDR-TB [reviewed in [20]]. It was hypothesized that “Beijing” strains might exhibit hypermutation as this strain family harbours various nonsynonymous substitutions in putative mutator genes [41]. However, subsequent attempts to measure differences in mutation rates across M. tuberculosis lineages have yielded contradictory results [42, 43].
In addition to heritable hypermutation, the emergence of transient mutators has been described. For example, in M. tuberculosis, increased expression of dnaE2, a gene encoding an additional copy of the major replicative DNA polymerase DnaE2, was linked to transient error-prone replication under stress and to the emergence of drug resistance in vivo [44]. Moreover, the use of fluoroquinolones and streptomycin, both important anti-TB drugs (Box 1, Table), was shown to induce mutator phenotypes in E. coli [45, 46]. Fluoroquinolones are being evaluated as new first-line antibiotics for TB [47]. Whether the wider use of these drugs could promote the development of drug resistance in the future is unknown, but it is a disturbing possibility that should be carefully considered. In summary, many bacterial factors contribute to the acquisition of drug resistance-conferring mutations in M. tuberculosis. Below we discuss the effect of these mutations on bacterial fitness, and the evolutionary forces that determine the fate of these mutations in bacterial populations.
Fitness cost and compensatory mechanisms
Studies from the 1950s revealed a reduced pathogenicity of isoniazid-resistant tubercle bacilli compared to drug-sensitive bacteria when inoculated into guinea pigs [48]. These and other observations led to the dogma that drug-resistant M. tuberculosis show reduced “fitness”, as they are presumably less transmissible and therefore unlikely to spread successfully in immunocompetent human populations [15]. However, this rather simplistic model is contradicted by numerous recent reports describing community outbreaks of MDR-TB, and the finding that in some areas, MDR-TB is more frequently caused by patient-to-patient transmission of drug-resistant strains than through de novo acquisition of resistance during patient treatment [2, 7]. Experimental studies in various bacterial species have shown that not all resistance-conferring mutations inflict the same level of fitness-cost in absence of the drug, with some showing mild or no detrimental effects (reviewed in [49]). In M. tuberculosis, similar studies have identified such “low/no-cost mutations” conferring resistance to various anti-TB drugs (Figure 3) [50–55]. Intriguingly, these studies also found that these mutations account for the majority of the resistance detected in clinical isolates [50, 51, 53–55], suggesting that strains harbouring low/no-cost mutations are positively selected among drug-resistant strains, either within the bacterial population infecting a host and/or as a result of variable transmission efficiency. In support of the latter, several studies have shown that among isoniazid-resistant strains of M. tuberculosis, those carrying low/no-cost resistance-conferring mutations were more successfully transmitted than strains harbouring other mutations [56–58].
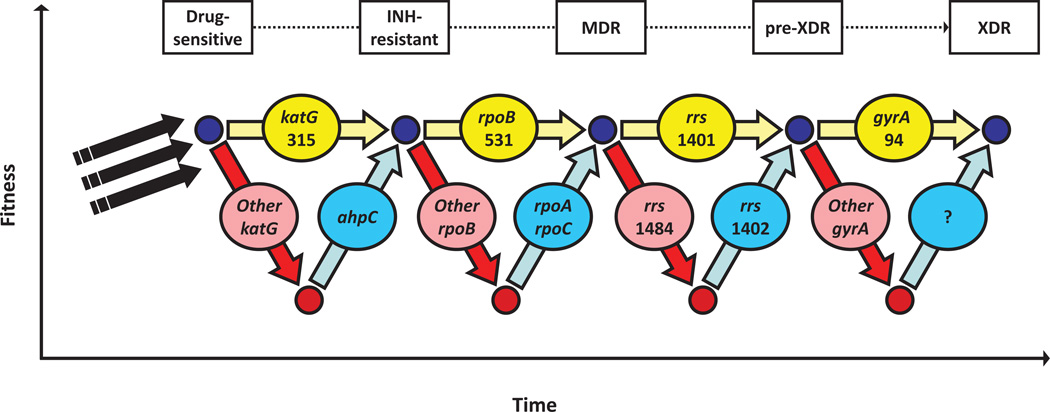
Figure 3. The influence of Darwinian fitness on the emergence of MDR and XDR-TB.
In the absence of drugs, the acquisition of resistance mutations is often associated with a fitness-cost. The most prevalent resistance mutations in clinical isolates show low or no fitness cost. However, high fitness costs incurred by other generally less common mutations can be alleviated by compensatory mutations. Higher and lower fitness levels of bacteria are indicated by small blue or red circles, respectively. Yellow and red arrows and ovals indicate low-cost and high-cost resistance mutations, respectively. Blue arrows and ovals indicate compensatory mutations. Black arrows refer to baseline genomic characteristics of a particular strain genetic background. Figure based on data from [22, 25, 63–65].
In addition to the occurrence of low/no-cost mutations, research in different bacterial species has shown that the deleterious effects of drug resistance mutations can be mitigated by compensatory mutations (reviewed in [59]). In M. tuberculosis, reversions of resistance mutations are rarely observed, indicating a higher likelihood for the occurrence of compensatory evolution, consistent with the view that many more mutational targets exist for compensation compared to reversion [60]. Compensatory mutations impact the disease epidemiology and treatment practices for bacterial infections by preventing drug-resistant bacteria from being out-competed by drug-susceptible strains, even if the respective antibiotics are withdrawn [59]. Many mechanisms of compensatory evolution have been described in other bacteria (reviewed in [61]). These involve intragenic or intergenic mutations, which may reduce the need for, or restore the functionality of an impaired enzyme. Alternatively, the need of a particular enzyme may be by-passed by an alternative enzymatic pathway. Evidence for compensatory evolution has also been reported for M. tuberculosis (Figure 3). KatG, a catalase-peroxidase involved in the degradation of harmful reactive oxygen intermediates, is required to convert the pro-drug isoniazid into its bioactive form (Table 1). Whilst the most frequently detected resistance mutations in katG at amino-acid position 315 only confer a low fitness-cost and do not abolish the function of KatG [53], other, more deleterious mutations in this gene were shown to be associated with compensatory mutations in the regulatory region of ahpC (Figure 3) [56, 62]. The resulting over-expression of the ahpC gene product, another peroxidase, is believed to partly compensate for the loss of KatG activity [63]. Similarly, mycobacterial resistance to aminoglycoside is mediated by mutations in the 16S rRNA. Some of these mutations destabilize the rRNA secondary structure, which can be compensated by a secondary mutation in the same gene (Figure 3) [64]. Compensatory mutations in the ahpC promoter and in the 16S rRNA are rarely observed in clinical isolates of M. tuberculosis [22], indicating that these mutations might have only limited epidemiological relevance. By contrast, recently described compensatory mutations in the RNA polymerase of M. tuberculosis were detected in more than 30% of MDR clinical isolates from high-burden countries [65, 66]. These mutations are situated within rpoA and rpoC, encoding the α- and β’-subunits of the RNA polymerase, respectively, and alleviate the fitness-cost associated with resistance mutations in rpoB, the β-subunit of the RNA polymerase and target of rifampicin (Figure 3) [65]. Recent genetic reconstructions in Salmonella enterica confirmed that similar compensatory mutations in rpoA and rpoC are necessary and sufficient to mitigate impaired fitness due to rifampicin resistance-conferring mutations in rpoB in that species as well [67]. Taken together, the available evidence strongly suggests that the notion of drug-resistance being invariably linked to a fitness burden in the absence of drugs is too simplistic. Because of the occurrence of low/no-cost drug resistance mutations and the potential for compensatory evolution, drug-resistant bacteria are unlikely to disappear, even if the drug pressure is removed. In fact, over time, more opportunity for compensatory evolution arises, and circulating MDR and XDR strains might gradually become more and more transmissible.
The broader role of epistasis
The interaction between a drug resistance-conferring mutation and a compensatory mutation is an example of epistasis, which is defined as the effect of the interaction between two or more mutations on an organism’s phenotype [68]. Similarly, different drug resistance-conferring mutations can also interact epistatically. Studies in E. coli, P. aeruginosa and Streptococcus pneumoniae found that generally, the observed cumulative fitness cost of carrying multiple drug resistance-conferring mutations was below the expected sum of the fitness cost associated with each individual mutation; an observation referred to as “positive” or “antagonistic” epistasis [69–72]. Moreover, in some cases, strains resistant to two drugs had a higher fitness than at least one of the corresponding single drug-resistant mutants. In other words, the acquisition of one or more additional resistance determinants may sometimes ameliorate the fitness-cost caused by a preceding resistance mutation; a phenomenon termed “sign epistasis” [71, 73]. Although the interaction between different drug resistance-conferring mutations has not yet been studied in M. tuberculosis, a similar phenomenon in MDR-TB would represent a worst case scenario, as MDR strains could improve their fitness by acquiring mutations conferring resistance to additional drugs.
Epistatic interactions may also determine the order in which drug resistance-conferring mutations and associated compensatory mutations are acquired over time [74]. The nonrandom nature of mutational pathways to drug resistance has been demonstrated in several bacterial species [75–77]. Whilst non-bacterial factors will play a key role in the acquisition of the different drug resistance mutations (Figure 2), epistatic interactions may determine the optimal mutational pathway. Epistatic interactions may also occur between drug resistance mutations and pre-existing genomic differences, which characterize a given strain genetic background [78]. Phylogenetic analyses of human disease-associated strains of the M. tuberculosis complex identified six main phylogenetic lineages [23], and there is mounting evidence that the specific strain genetic background as defined by these lineages plays a role in the emergence and spread of MDR-TB [13]. For example, strains harbouring identical rifampicin resistance mutations but belonging to different lineages of M. tuberculosis showed different levels of fitness-cost [50]. Moreover, different phylogenetic lineages of M. tuberculosis have been associated with different drug resistance mutations [56, 79, 80], and the level of resistance conferred by specific isoniazid resistance-conferring mutations has been shown to differ depending on the M. tuberculosis lineage [80]. As mentioned above, the “Beijing family” of M. tuberculosis has often been associated with drug resistance [20]. The association between specific M. tuberculosis lineages and drug resistance could result from an increased ability to tolerate a larger range of resistance mutations, either due to a higher baseline fitness, or more favourable epistatic interactions between the strain genetic background and drug resistance mutations. This idea is supported by the presence of various lineage-specific non-synonymous nucleotide substitutions in genes associated with drug-resistance (Figure 4) [25, 81]. Thus it is possible that the pre-existence of non-resistance mutations in drug resistance-associated genes could modulate the fitness effects of subsequently acquired resistance-conferring mutations in these genes.
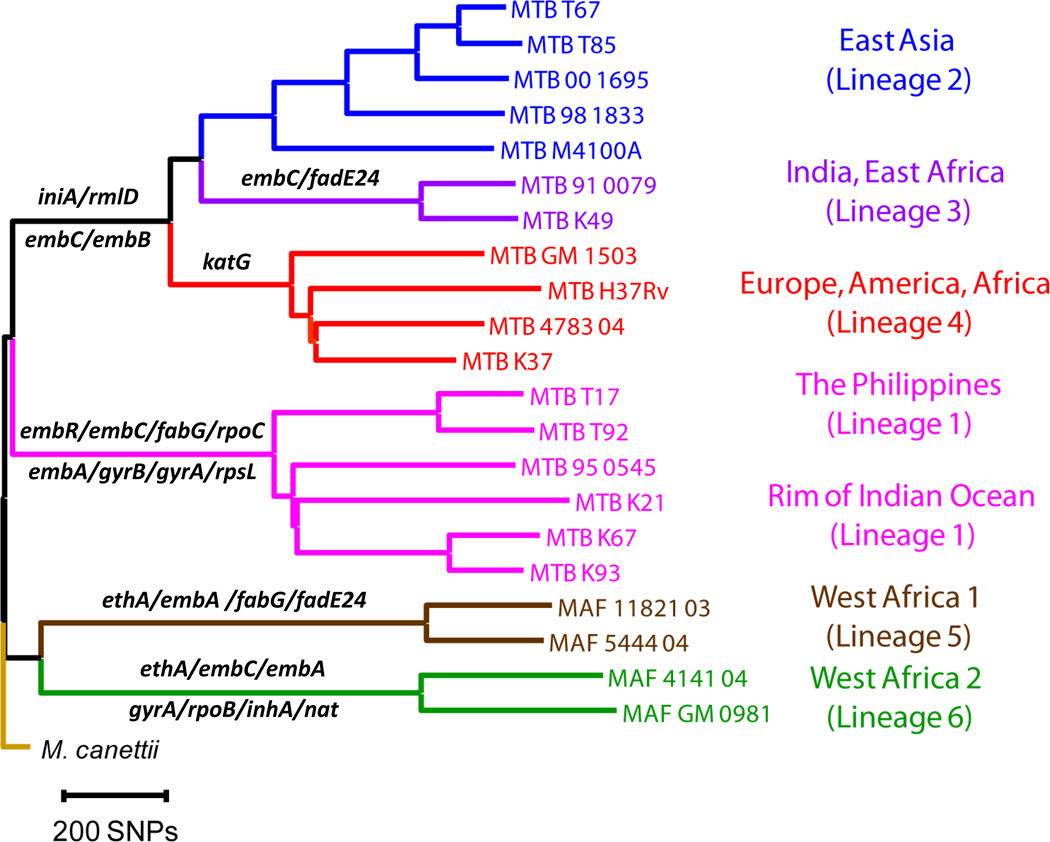
Figure 4. Drug resistance-associated genes containing M. tuberculosis lineage-specific mutations.
Phylogenetic tree of the six main lineages of M. tuberculosis associated with human TB, based on 21 whole genome sequences [81]. Genes indicated are associated with drug resistance (Table 1) and harbour lineage-defining, non-synonymous substitutions. This creates the potential for epistatic interactions between the genetic background of a given strain and specific drug resistance-conferring mutations.
Taken together, current evidence suggests that epistatic interactions between the strain genetic background, drug resistance-conferring mutations, and compensatory mutations may play a role in defining evolutionary trajectories towards multidrug resistance in TB. The genetic landscape of an MDR strain differs from a drug-sensitive strain by at least the presence of resistance mutations. Being forced to manage the potential fitness-cost associated with drug resistance mutations (e.g., through the acquisition of compensatory mutations), certain selective pressures imposed onto MDR strain populations will differ from those acting on drug-sensitive strains. Moreover, treatment regimens for drug-sensitive and MDR-TB differ and are variably effective, and are therefore likely to differentially affect bacterial transmission. Thus, altogether, we expect evolutionary trajectories to differ considerably between drug-sensitive and MDR strains. This view is supported by the observation that the population structures of drug-sensitive and MDR strains of M. tuberculosis in the same geographic regions can differ substantially [82, 83].
Clinical implications
Because in M. tuberculosis drug resistance is encoded on the chromosome [22, 25], rapid detection of resistance by molecular methods is possible. These techniques overcome some of the limitations of the classical techniques of phenotypic drug susceptibility testing (DST), which may require standard mycobacterial culture for up to several months [84]. Hence, developing improved methods of molecular DST in TB is currently high on the agenda, and recently, important progress has been made [85]. However, for many anti-TB drugs, at least a proportion of drug resistance-conferring mutations remain unknown. Hence, current molecular DST is still not 100% sensitive, making culture-based DST indispensable. With the advent of next-generation sequencing technologies, efforts are underway to identify these unknown resistance determinants in M. tuberculosis, which then could be incorporated into more sensitive diagnostic tools for drug-resistant TB. Once more sensitive molecular diagnostics become available, drug-resistant profiles could be determined at treatment onset, and drug regimens could be tailored to the individual patient needs. Ultimately, this would help curb the transmission of drug-resistant strains. However, in high-incidence areas that lack appropriate resources for DST, WHO recommends the use of standard treatment regimens (Box 1) [3]. As a result, these standard regimens are often administered empirically, (i.e. without knowing whether a given patient might harbour an M. tuberculosis strain resistant to one or more of the drugs included in the standard regimen), raising the possibility of treating patients with drug-resistant TB with suboptimal or ineffective drugs (Box 1). Even though the standard WHO treatment protocols are highly efficacious for the treatment of drug-susceptible TB [3], in regions with a high burden of drug-resistant TB, empiric regimens without proper DST can contribute to the amplification of drug resistance [2]. The long prevailing dogma of drug-resistant strains being less transmissible than drug-sensitive strains created the belief that by preventing the acquisition of drug resistance by ensuring treatment adherence, drug resistance could be controlled [15, 16, 59]. However, the presence of low-cost resistance mutations, compensatory mutations, and the potential for positive epistasis between drug-resistance-conferring mutations suggests that MDR bacteria can evolve in a range of different ways to alleviate the detrimental effects of drug resistance mutations. Indeed, in the case of M. tuberculosis, the most recent global surveillance data revealed the highest rates of MDR-TB ever recorded, as well as increasing trends in MDR-TB rates in certain regions of Eastern Europe, Central Asia, and parts of Africa, although MDR-TB rates have remained constant or have decreased in many other areas [7]. Hence, preventing the acquisition of resistance through the use of standard regimens will not suffice to curb MDR-TB epidemics globally. Rather, increased efforts to directly target circulating MDR strains should be considered. Moreover, new drugs are urgently needed to control M. tuberculosis strains which are becoming resistant to an ever increasing number of compounds [86]. These new drugs will have to be deployed rationally to minimize the likelihood of resistance arising. An increased knowledge of the compensatory mechanisms and epistatic interactions among drug resistance-conferring mutations might be able to facilitate defining the ideal treatment regimens. One possibility would be to combine drugs in which the respective drug resistance-conferring mutations interact negatively, thereby lowering the fitness of strains carrying both drug resistance determinants. As discussed here, the evolution of MDR-TB is highly heterogeneous, which is likely to pose a major challenge to the elaboration of a universally effective anti-TB regimen in the future. For example, the high HIV prevalence among TB cases, such as observed in Southern Africa, complicates TB treatment because of the interactions between anti-TB drugs and antiretrovirals [86]. Finally, given the global phylogeography of M. tuberculosis, and the heterogeneous distribution of strains with different genetic backgrounds, ideal drug regimens might have to be tailored to different geographical settings as well as to individual patients.
Concluding remarks
The emergence of MDR-TB is highly heterogeneous, involving a multitude of non-bacterial as well as bacterial factors (Figure 2). Epistatic interactions between drug resistance-conferring mutations, different strain genetic backgrounds, and compensatory mutations can influence the evolutionary trajectories of drug-resistant bacteria, including M. tuberculosis. These factors could impact the effectiveness of current as well as future treatment regimens against MDR-TB. However, as yet, chemotherapy of TB and MDR-TB has been based on globally standardized strategies (Box 1). Building on the evidence discussed here, future TB control might benefit from new strategies that target individual patients and the diverse populations of MDR strains circulating in different parts of the world. Moreover, much of our understanding of the importance of persistence, hypermutation, compensatory evolution, and epistasis in drug resistance evolution stems from research in other organisms. Hence more work is needed to explore these factors in M. tuberculosis, and determine whether they have any clinical relevance for the emergence and spread of MDR- and XDR-TB. Historically, actions against drug-resistant TB have been taken long after the damage was done [15]. We believe that understanding the bacterial factors driving the evolution of drug-resistant M. tuberculosis will help anticipate its future trajectories and curb the progression of an ever deadlier disease.
Acknowledgements
We thank the other members of our group for the stimulating discussions and comments on the manuscript. Work in our laboratory is supported by the Swiss National Science Foundation (grant number PP00A-119205) and the National Institutes of Health (AI090928 and HHSN266200700022C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. Global tuberculosis control - surveillance, planning, financing. Geneva: World Health Organization; 2012. [Google Scholar]
- 2.Gandhi NR, et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet. 2010;375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 3.Caminero JA, et al. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis. 2010;10:621–629. doi: 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
- 4.Velayati AA, et al. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest. 2009;136:420–425. doi: 10.1378/chest.08-2427. [DOI] [PubMed] [Google Scholar]
- 5.Udwadia ZF, et al. Totally drug-resistant tuberculosis in India. Clin Infect Dis. 2012;54:579–581. doi: 10.1093/cid/cir889. [DOI] [PubMed] [Google Scholar]
- 6.Alanis AJ. Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res. 2005;36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Zignol M, et al. Surveillance of anti-tuberculosis drug resistance in the world: an updated analysis, 2007–2010. Bull World Health Organ. 2012;90:111–119D. doi: 10.2471/BLT.11.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells CD, et al. HIV infection and multidrug-resistant tuberculosis: the perfect storm. J Infect Dis. 2007;196(Suppl 1):S86–S107. doi: 10.1086/518665. [DOI] [PubMed] [Google Scholar]
- 9.Dalton T, et al. Prevalence of and risk factors for resistance to second-line drugs in people with multidrug-resistant tuberculosis in eight countries: a prospective cohort study. Lancet. 2012 doi: 10.1016/S0140-6736(12)60734-X. Published online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan G, et al. Mycobacterium tuberculosis growth at the cavity surface: a microenvironment with failed immunity. Infect Immun. 2003;71:7099–7108. doi: 10.1128/IAI.71.12.7099-7108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner M, et al. Effects of rifampin and multidrug resistance gene polymorphism on concentrations of moxifloxacin. Antimicrob Agents Chemother. 2007;51:2861–2866. doi: 10.1128/AAC.01621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coscolla M, Gagneux S. Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech. 2010;7:e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrell S, Gagneux S. Strain diversity, epistasis and the evolution of drug resistance in Mycobacterium tuberculosis. Clin Microbiol Infect. 2011;17:815–820. doi: 10.1111/j.1469-0691.2011.03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin BR, et al. The population genetics of antibiotic resistance. Clin Infect Dis. 1997;24(Suppl 1):S9–S16. doi: 10.1093/clinids/24.supplement_1.s9. [DOI] [PubMed] [Google Scholar]
- 15.Keshavjee S, Farmer PE. Tuberculosis, drug resistance, and the history of modern medicine. N Engl J Med. 2012;367:931–936. doi: 10.1056/NEJMra1205429. [DOI] [PubMed] [Google Scholar]
- 16.Dye C, Espinal MA. Will tuberculosis become resistant to all antibiotics? Proc R Soc Lond B Biol Sci. 2001;268:45–52. doi: 10.1098/rspb.2000.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dye C, et al. Erasing the world's slow stain: strategies to beat multidrug-resistant tuberculosis. Science. 2002;295:2042–2046. doi: 10.1126/science.1063814. [DOI] [PubMed] [Google Scholar]
- 18.Cohen T, Murray M. Modeling epidemics of multidrug-resistant M. tuberculosis of heterogeneous fitness. Nat Med. 2004;10:1117–1121. doi: 10.1038/nm1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blower SM, Chou T. Modeling the emergence of the 'hot zones': tuberculosis and the amplification dynamics of drug resistance. Nat Med. 2004;10:1111–1116. doi: 10.1038/nm1102. [DOI] [PubMed] [Google Scholar]
- 20.Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13:1456–1466. [PubMed] [Google Scholar]
- 21.Nguyen L, Pieters J. Mycobacterial subversion of chemotherapeutic reagents and host defense tactics: challenges in tuberculosis drug development. Annu Rev Pharmacol Toxicol. 2009;49:427–453. doi: 10.1146/annurev-pharmtox-061008-103123. [DOI] [PubMed] [Google Scholar]
- 22.Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 23.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 24.Namouchi A, et al. After the bottleneck: Genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res. 2012;22:721–734. doi: 10.1101/gr.129544.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandgren A, et al. Tuberculosis Drug Resistance Mutation Database. PLoS Med. 2009;6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.David HL. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl Microbiol. 1970;20:810–814. doi: 10.1128/am.20.5.810-814.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillespie SH. Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob Agents Chemother. 2002;46:267–274. doi: 10.1128/AAC.46.2.267-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Post FA, et al. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J Infect Dis. 2004;190:99–106. doi: 10.1086/421501. [DOI] [PubMed] [Google Scholar]
- 30.Pasipanodya JG, Gumbo T. A new evolutionary and pharmacokinetic-pharmacodynamic scenario for rapid emergence of resistance to single and multiple anti-tuberculosis drugs. Curr Opin Pharmacol. 2011;11:457–463. doi: 10.1016/j.coph.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perlman DC, et al. The clinical pharmacokinetics of rifampin and ethambutol in HIV-infected persons with tuberculosis. Clin Infect Dis. 2005;41:1638–1647. doi: 10.1086/498024. [DOI] [PubMed] [Google Scholar]
- 32.Muller B, et al. InhA promoter mutations: a gateway to extensively drug-resistant tuberculosis in South Africa? Int J Tuberc Lung Dis. 2011;15:344–351. [PubMed] [Google Scholar]
- 33.Louw GE, et al. Rifampicin reduces susceptibility to ofloxacin in rifampicin-resistant Mycobacterium tuberculosis through efflux. Am J Respir Crit Care Med. 2011;184:269–276. doi: 10.1164/rccm.201011-1924OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balaban NQ. Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr Opin Genet Dev. 2011;21:768–775. doi: 10.1016/j.gde.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Dhar N, McKinney JD. Mycobacterium tuberculosis persistence mutants identified by screening in isoniazid-treated mice. Proc Natl Acad Sci U S A. 2010;107:12275–12280. doi: 10.1073/pnas.1003219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams KN, et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balaban NQ, et al. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 38.Aldridge BB, et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ford CB, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43:482–6. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolivet-Gougeon A, et al. Bacterial hypermutation: clinical implications. J Med Microbiol. 2011;60:563–573. doi: 10.1099/jmm.0.024083-0. [DOI] [PubMed] [Google Scholar]
- 41.Rad ME, et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg Infect Dis. 2003;9:838–845. doi: 10.3201/eid0907.020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werngren J, Hoffner SE. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J Clin Microbiol. 2003;41:1520–1524. doi: 10.1128/JCM.41.4.1520-1524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Steenwinkel JE, et al. Drug susceptibility of Mycobacterium tuberculosis Beijing genotype and association with MDR TB. Emerg Infect Dis. 2012;18:660–663. doi: 10.3201/eid1804.110912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boshoff HI, et al. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 45.Ysern P, et al. Induction of SOS genes in Escherichia coli and mutagenesis in Salmonella typhimurium by fluoroquinolones. Mutagenesis. 1990;5:63–66. doi: 10.1093/mutage/5.1.63. [DOI] [PubMed] [Google Scholar]
- 46.Ren L, et al. Escherichia coli cells exposed to streptomycin display a mutator phenotype. J Bacteriol. 1999;181:1043–1044. doi: 10.1128/jb.181.3.1043-1044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takiff H, Guerrero E. Current prospects for the fluoroquinolones as first-line tuberculosis therapy. Antimicrob Agents Chemother. 2011;55:5421–5429. doi: 10.1128/AAC.00695-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barnett M, et al. Tubercle bacilli resistant to isoniazid: Virulence and response to treatment with isoniazid in guinea-pigs. Brit. J. Exp. Path. 1953;34:568–581. [PMC free article] [PubMed] [Google Scholar]
- 49.Andersson DI, Levin BR. The biological cost of antibiotic resistance. Curr Opin Microbiol. 1999;2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 50.Gagneux S, et al. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- 51.Billington OJ, et al. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1999;43:1866–1869. doi: 10.1128/aac.43.8.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies AP, et al. Comparison of fitness of two isolates of Mycobacterium tuberculosis one of which had developed multi-drug resistance during the course of treatment. J Infect. 2000;41:184–187. doi: 10.1053/jinf.2000.0711. [DOI] [PubMed] [Google Scholar]
- 53.Pym AS, et al. Effect of katG Mutations on the Virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun. 2002;70:4955–4960. doi: 10.1128/IAI.70.9.4955-4960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sander P, et al. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob Agents Chemother. 2002;46:1204–1211. doi: 10.1128/AAC.46.5.1204-1211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bottger EC, et al. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat Med. 1998;4:1343–1344. doi: 10.1038/3906. [DOI] [PubMed] [Google Scholar]
- 56.Gagneux S, et al. Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog. 2006;2:e61. doi: 10.1371/journal.ppat.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Doorn HR, et al. Public health impact of isoniazid-resistant Mycobacterium tuberculosis strains with a mutation at amino-acid position 315 of katG: a decade of experience in The Netherlands. Clin Microbiol Infect. 2006;12:769–775. doi: 10.1111/j.1469-0691.2006.01495.x. [DOI] [PubMed] [Google Scholar]
- 58.van Soolingen D, et al. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in the Netherlands. J Infect Dis. 2000;182:1788–1790. doi: 10.1086/317598. [DOI] [PubMed] [Google Scholar]
- 59.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 60.Levin BR, et al. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maisnier-Patin S, Andersson DI. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res Microbiol. 2004;155:360–369. doi: 10.1016/j.resmic.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 62.Hazbon MH, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50:2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sherman DR, et al. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 64.Shcherbakov D, et al. Directed mutagenesis of Mycobacterium smegmatis 16S rRNA to reconstruct the in-vivo evolution of aminoglycoside resistance in Mycobacterium tuberculosis. Mol Microbiol. 2010;77:830–840. doi: 10.1111/j.1365-2958.2010.07218.x. [DOI] [PubMed] [Google Scholar]
- 65.Comas I, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Casali N, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–745. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brandis G, et al. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol Microbiol. 2012;85:142–51. doi: 10.1111/j.1365-2958.2012.08099.x. [DOI] [PubMed] [Google Scholar]
- 68.Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ward H, et al. The cost of multiple drug resistance in Pseudomonas aeruginosa. J Evol Biol. 2009;22:997–1003. doi: 10.1111/j.1420-9101.2009.01712.x. [DOI] [PubMed] [Google Scholar]
- 70.Hall AR, MacLean RC. Epistasis buffers the fitness effects of rifampicin-resistance mutations in Pseudomonas aeruginosa. Evolution. 2011;65:2370–2379. doi: 10.1111/j.1558-5646.2011.01302.x. [DOI] [PubMed] [Google Scholar]
- 71.Trindade S, et al. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genetics. 2009;5:e1000578. doi: 10.1371/journal.pgen.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rozen DE, et al. Fitness costs of fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2007;51:412–416. doi: 10.1128/AAC.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinreich DM, et al. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- 74.Salverda ML, et al. Initial mutations direct alternative pathways of protein evolution. PLoS Genetics. 2011;7:e1001321. doi: 10.1371/journal.pgen.1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bergval I, et al. Pre-existing isoniazid resistance, but not the genotype of Mycobacterium tuberculosis drives rifampicin resistance codon preference in vitro. PLoS ONE. 2012;7:e29108. doi: 10.1371/journal.pone.0029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elena SF, Lenski RE. Epistasis between new mutations and genetic background and a test of genetic canalization. Evolution. 2001;55:1746–1752. doi: 10.1111/j.0014-3820.2001.tb00824.x. [DOI] [PubMed] [Google Scholar]
- 79.Baker L, et al. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg Infect Dis. 2004;10:1568–1577. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fenner L, et al. Effect of mutation and genetic background on drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2012;56:3047–3053. doi: 10.1128/AAC.06460-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Niemann S, et al. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J Clin Microbiol. 2010;48:3544–3550. doi: 10.1128/JCM.00715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chihota VN, et al. Population structure of multi- and extensively drug-resistant Mycobacterium tuberculosis strains in South Africa. J Clin Microbiol. 2012;50:995–1002. doi: 10.1128/JCM.05832-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Deun A, et al. Diagnosis of drug-resistant tuberculosis: reliability and rapidity of detection. Int J Tuberc Lung Dis. 2010;14:131–140. [PubMed] [Google Scholar]
- 85.Boehme CC, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zumla A, et al. Advancing the development of tuberculosis therapy. Nat Rev Drug Discov. 2012;11:171–172. doi: 10.1038/nrd3694. [DOI] [PubMed] [Google Scholar]
- 87.McNerney R, et al. Tuberculosis diagnostics and biomarkers: needs, challenges, recent advances, and opportunities. J Infect Dis. 2012;205(Suppl 2):S147–S158. doi: 10.1093/infdis/jir860. [DOI] [PubMed] [Google Scholar]
- 88.Goldfeld AE, et al. Association of an HLA-DQ allele with clinical tuberculosis. JAMA. 1998;279:226–228. doi: 10.1001/jama.279.3.226. [DOI] [PubMed] [Google Scholar]
- 89.Comas I, Gagneux S. A role for systems epidemiology in tuberculosis research. Trends Microbiol. 2011;19:492–500. doi: 10.1016/j.tim.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gagneux S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]