Abstract
Eukaryotic cells transcribe a vast number of noncoding RNA species. Among them, long noncoding RNAs (lncRNAs) have been widely implicated in the regulation of gene transcription. However, examples of post-transcriptional gene regulation by lncRNAs are emerging. For example, through extended base-pairing, lncRNAs can stabilize or promote the translation of target mRNAs, while partial base-pairing facilitates mRNA decay or inhibits target mRNA translation. In the absence of complementarity, lncRNAs can suppress pre-mRNA splicing and translation by acting as decoys of RNA-binding proteins or microRNAs, and can compete for microRNA-mediated inhibition leading to increased expression of the mRNA. Through these regulatory mechanisms, lncRNAs can elicit differentiation, proliferation, and cytoprotective programs, underscoring the rising recognition of lncRNA roles in human disease. In this review, we summarize the mechanism of post-transcriptional gene regulation by lncRNAs.
Introduction
The majority of RNAs transcribed in mammalian cells do not contain protein-coding sequences.1–3 Although many transcripts in this group are eventually processed into small mature RNAs (e.g., microRNAs, piRNAs, tiRNAs, and snoRNAs), a subset of them produce long noncoding (lnc)RNAs, with lengths of over 200 nucleotides. LncRNAs are transcribed by RNA polymerase II, even though many lncRNA genes contain histone modification signatures distinct from those of protein-coding genes (H3K4me3, H3K36me).4,5 After transcription, most lncRNAs are processed like protein-coding RNAs, including 5’-end capping, 3’-end polyadenylation, splicing of introns, and intracellular transport. Many lncRNAs have small open-reading frames, but they are not predicted to codify for proteins.6–8 However, recent RNA-seq analysis identified many lncRNAs associated with ribosomes, suggesting that they could have protein-coding potential and may play additional cytoplasmic roles in mRNA metabolism.9
LncRNAs as transcriptional regulators
Functionally, lncRNAs are best known for their roles as regulators of transcription. Over thirty years ago, Paul and Duerksen10 reported the surprising discovery that chromatin is purified with twice as much as RNA as DNA, suggesting that RNA may regulate chromatin structure and gene transcription. Many subsequent studies have shown that some lncRNAs are associated with chromatin modification enzymes and mediate gene activation or silencing.3 For instance, during X chromosome dosage compensation in mammals, the lncRNA XIST is expressed from one X chromosome in female cells and inactivates the other X chromosome by recruiting PRC2 (Polycomb repressive complex 2).11 In plants, the seasonal timing of flowering (vernalization) is mediated by COLDAIR, a cold-inducible intronic lncRNA which silences FLC, a gene that regulates flowering.12 In mammalian cells, the lncRNA HOTAIR associates with PRC2 and modulates H3K27me3 distribution in genomic targets.13,14 In addition, two p53-regulated lncRNAs, lincRNA-p21 and PANDA, repress target gene transcription by interacting with DNA-binding proteins hnRNP K and NF-YA, respectively.15,16 Together with other examples, the role of lncRNAs as regulators of gene transcription is well established. However, their involvement in other modes of gene regulation remains relatively unknown.
LncRNAs as post-transcriptional regulators
Recently, a small number of lncRNAs have been reported to regulate gene expression post-transcriptionally in a variety of ways. For example, the lncRNA MALAT1 (metastasis-associated long adenocarcinoma transcript 1) was implicated in pre-mRNA splicing by influencing the distribution of SR proteins.17 The cytoplasmic 1/2-sbsRNAs (half-Staufen 1-binding site long noncoding RNAs) promoted mRNA decay by partial base-pairing with specific target mRNAs and recruiting the protein Staufen 1;18 by contrast, longer base-pairing of β amyloid-cleaving enzyme (BACE)1 mRNA and BACE1-AS (antisense) protected BACE1 mRNA from RNase cleavage, resulting in mRNA stabilization.19 A global function of the lncRNA BC1 in translation repression was linked to its interaction with the eukaryotic translation initiation factor eIF4A and with the poly(A)-binding protein (PABP),20 while lincRNA-p21 was recently shown to repress the translation of mRNAs encoding β-catenin and JunB by partial base-pairing and recruitment of translation repressor proteins.21 In sum, besides the well-established function of lncRNAs in transcriptional and epigenetic gene regulation, lncRNAs also possess the potential to promote and inhibit the post-transcriptional processes of mRNA splicing, degradation, and translation. In this review, we will discuss these and other emerging examples of lncRNAs with post-transcriptional functions.
Post-transcriptional control of lncRNAs
As knowledge of the post-transcriptional roles of lncRNAs rises, our understanding of the mechanisms that control lncRNA abundance are also expanding. Besides the transcriptional regulation of lncRNAs, shared with that of host genes (as for intronic lncRNAs) or controlled independently (as for intergenic lncRNAs –lincRNAs), evidence is mounting that lncRNAs are also regulated post-transcriptionally. Global measurement of the half-lives of lncRNA in mouse neuronal cells by the Mattick laboratory revealed that some lncRNAs are unstable (http://stability.matticklab.com).22 In general, intronic lncRNAs are less stable than intergenic and antisense lncRNAs, whereas spliced lncRNAs are more stable than unspliced transcripts. In addition, cytoplasmic lncRNAs are more stable than nuclear lncRNAs, as exemplified by the extremely labile lncRNA NEAT1, involved in paraspeckle assembly. Very recently, a triple helix was found in the 3’ end of long noncoding RNAs MALAT1 and multiple endocrine neoplasia β (MENβ) which protected the ends of lncRNAs from 3’→5’exonucleolytic cleavage, and stabilized these transcripts.23,24 This triple-helical structure resembled the Kaposi’s sarcoma-associated Herpesvirus (KSHV) expression and nuclear retention element (ENE) present in the KSHV PAN (polyadenylated nuclear) lncRNA.25 The dynamic nature of lncRNA turnover emphasizes the complexity of regulating RNA metabolism, sometimes elicited by other RNAs which are themselves subject to post-transcriptional control.
Although the transcriptome-wide survey of lncRNA stability revealed a rich regulation of lncRNA turnover, the mechanisms that change the stability of lncRNAs in cells are unknown. A recent report linked the stability of lincRNA-p21 to its interaction with the RNA-binding proteins HuR and Ago2 [Argonaute, a component of RNA-induced silencing complex (RISC)], and the microRNA let-7b in human cervical carcinoma cells.21 In this model system, silencing HuR or Ago2 increased lincRNA-p21 stability, whereas overexpressing let-7b promoted lincRNA-p21 decay, uncovering a cooperative mechanism of lincRNA-p21 decay by HuR and RISC. However, most aspects of the degradation of lncRNAs are not known at present, including how decapping and deadenylation contribute to lncRNA decay. Molecular insight into lncRNA turnover will be essential in order to understand how lncRNA abundance is controlled.
LncRNAs and pre-mRNA splicing
The Mattick group identified many lncRNAs residing in the nucleus (191 lncRNAs) and many in the cytoplasm (499 lncRNAs).22 The nuclear lncRNAs may be implicated in post-transcriptional regulatory steps such as pre-mRNA splicing, mRNA capping, polyadenylation and export to the cytoplasm. In particular, alternative splicing of pre-mRNAs is a key mechanism to achieve protein diversification in higher eukaryotes26–28 and a process through which lncRNAs can profoundly affect gene expression patterns.
MALAT1
Prasanth and colleagues recently uncovered a role of the nuclear lncRNA MALAT1 in alternative splicing.17 MALAT1 interacts with and influences the distribution of serine/arginine (SR) splicing factors in nuclear speckle domains in order to direct alternative splicing in a cell type-specific manner.17 Accordingly, MALAT1 depletion or SR protein overexpression affects alternative splicing in similar sets of pre-mRNAs, suggesting that they share pathways for control of alternative splicing. Interestingly, lowering MALAT1 levels increased the abundance and phosphorylation of SR proteins, while it inhibited their accumulation in nuclear speckles. These findings suggest that MALAT1 may modulate SR protein concentration in nuclear domains for optimal alternative splicing.
Although MALAT1 is expressed in various tissues, it is particularly abundant in the nervous system.29 In neurons, the concentration of MALAT1 in nuclear speckles declines after inhibition of RNA polymerase II activity. In this cell type, depleting MALAT1 disrupts the localization of SR proteins in transcriptionally active sites, lowering the transcription of mRNAs encoding proteins with roles in nuclear processing and in synapse function. In hippocampal neurons, silencing MALAT1 reduces synaptic density whereas overexpressing MALAT1 increases it. These results illustrate a function for a lncRNA, MALAT1, in synapse assembly through transcription-coupled alternative splicing.
Sno-lncRNAs
Recently, the Chen and Carmichael groups identified sno-lncRNAs, a class of nuclear-enriched intron-derived lncRNAs containing snoRNA sequences on both ends, named sno-lncRNAs,307 which lack 5’caps and 3’poly(A) tails. After synthesis, sno-lncRNAs accumulate in nuclear foci distinct from nucleoli or Cajal bodies; these sno-lncRNAs associate with Fox family splicing regulators such as Fox2 and modulate splicing in pluripotent cells. Interestingly, the genomic region encoding sno-lncRNAs (15q11-q13) is deleted in Prader-Willi Syndrome (PWS), implicating this class of splicing regulatory lncRNAs in the molecular pathogenesis of PWS.
LncRNAs and mRNA turnover
Gene expression is also robustly regulated via processes that affect mRNA half-life. Although RNA-binding proteins and microRNAs are major factors affecting the stability of mRNAs, lncRNAs are increasingly recognized as a prominent class of molecules that interact with mRNAs and affect their half-lives.
Global mRNA stability
In a recent study, arthropod-born flaviviruses were reported to generate a small ‘subgenomic’ flavivirus RNA (sfRNA) from the 3’ end of the viral genome. Interestingly, the sfRNA interacted with the cellular exonuclease XRN1 and repressed its activity; this inhibitory effect led to the accumulation of stable, uncapped cellular mRNAs, as well as to increased levels of mRNA decay intermediates typically degraded by XRN1.31
½-sbsRNAs
LncRNAs exported to the cytoplasm are able to interact with other RNA species like mRNAs, siRNA, miRNAs, and antisense RNAs. The Maquat laboratory reported that Staufen 1 binds double-stranded (ds)RNAs and promotes their decay, even though most Staufen 1 target mRNAs did not have distinct dsRNA structures and instead contained Alu elements.32,33 Recently, Gong and Maquat18 identified a group of lncRNAs bearing repeats of the Alu element, which formed imperfect base-pairing with Staufen 1-target mRNAs. This group of transcripts, termed ½-sbsRNAs, constitutes a novel class of trans-acting lncRNAs that activate the decay of specific target mRNAs.
BACE1-AS
In contrast to promoting mRNA decay, perfect base-pairing of lncRNA with an mRNA can protect mRNA from degradation. Wahlestedt and colleagues identified a role for a conserved noncoding antisense (AS) transcript for β-secretase-1 (BACE1), an enzyme with a key function in Alzheimer’s disease.19 Depletion of BACE1-antisense transcript (BACE1-AS) reduced BACE1 mRNA and protein levels in neuronal cell culture and in mouse brain. Introduction of BACE1-AS resulted in protection of BACE1 mRNA from degradation by RNase in vitro, whereas silencing of BACE1-AS promoted degradation of BACE1 mRNA in cultured cells. These results suggest that besides promoting target mRNA decay, extended base-pairing can also protect mRNA from degradation. Wahlestedt and colleagues further proposed that BACE1 mRNA and BACE1-AS may form part of similar regulatory circuits, based on the observation that their levels are elevated in neuronal cells upon treatment with Aβ(1–42), in transgenic mice expressing Aβ(1–42), and in Alzheimer`s disease patients. These results implicate a lncRNA that governs mRNA decay, BACE1-AS, in neurodegeneration.
Wahlestedt and colleagues uncovered another mechanism of the control of BACE1 mRNA stability by BACE1-AS.34 BACE1-AS forms perfect base-pairing with BACE1 mRNA exon 6, which bears a miR-485-5p target site. In human embryonic kidney (HEK 293T-C3) cells, overexpression of miR-485-5p reduced the steady-state level of BACE1 mRNA and this effect was rescued by introducing BACE1-AS. Accordingly, the authors proposed that miR-485-5p and BACE1-AS compete for binding with the same region of BACE1 mRNA. They also observed that BACE1-AS and miR-485-5p levels are dysregulated in Alzheimer's disease patients compared to control individuals, further highlighting the variety of potential regulatory interactions between endogenous antisense transcripts and miRNAs upon target mRNAs.
gadd7
Very recently, Liu and coworkers showed that the growth arrest and DNA-damage-inducible lncRNA gadd7 increases in response to irradiation of Chinese hamster ovary (CHO) cells with ultraviolet light and associates with the RNA-binding protein TDP-43 (TAR DNA-binding protein-43). Since TDP-43 functions as a stabilizing factor for cdk6 mRNA, its mobilization from the cdk6 mRNA to the gadd7 lncRNA resulted in increased degradation of cdk6 mRNA, reduced Cdk6 levels, and cell cycle arrest.35
LncRNAs and mRNA translation
Even though lncRNAs are not predicted to engage in translation, recent RNA-seq analysis identified many lncRNAs associated with ribosomes.9 This observation supports the idea that lncRNAs could have additional cytoplasmic functions in mRNA translation.
LincRNA-p21
LincRNA-p21 was recently shown to co-distribute with ribosomes and to represses the translation of target mRNAs.21 Transcripts CTNNB1 and JUNB mRNAs (encoding β-catenin and JunB, respectively), base-pair imperfectly with lincRNA-p21 in several places throughout the coding regions and untranslated regions (UTRs). The complex [lincRNA-p21–mRNA] further interacts with translation repressor Rck and Fmrp, suggesting that lincRNA-p21 can repress the translation of target mRNAs by operating via multiple mechanisms.
AS Uchl1
Very recently, antisense (AS) Uchl1 was identified as a nuclear-enriched lncRNA complementary to the mRNA that encodes mouse ubiquitin carboxyterminal hydrolase L1 (Uchl1), an enzyme with roles in brain development and neurodegeneration.36 Through a SINEB2 sequence and a 73-nt region of overlapping complementarity with the 5’ end of Uchl1 mRNA, AS Uchl1 enhanced the formation of active polysomes on Uchl1 mRNA and hence its translation. This mechanism is activated when mTORC1 is inhibited, since treatment with rapamycin triggered the export of AS Uchl1 from the nucleus to the cytoplasm, in turn increasing Uchl1 mRNA translation. AS Uchl1 constitutes the first example of a lncRNA promoting target mRNA translation by sequence complementarity.
BC1
Besides modulating the translation of specific targets, lncRNAs can also modulate the general translation machinery. The small lncRNA BC1 RNA (152 nt long), expressed in neurons and germ cells, inhibits the assembly of the translation initiation complex.37 A 3’ segment of the BC1 RNA interacts with eIF4A and PABP and represses general translation in Xenopus oocyte extracts.20 Introduction of excess eIF4A and PABP rescued the repression of translation by BC1 RNA, demonstrating that translation repression by BC1 occurs through eIF4A and PABP. The sequestration of translation initiation factors by BC1 is reminiscent of the function of MALAT1 as a decoy for SR proteins to influence splicing.
Linc-MD1
In some cases, lncRNAs modulate gene expression by acting upon miRNAs. A muscle-specific lncRNA, linc-MD1, controls muscle differentiation by competing with mRNAs for binding to shared target miRNAs.38 As proposed by Pandolfi and colleagues, the cytoplasmic availability of microRNAs can be titrated by modulating the abundance of RNA species containing specific target sites.394 In this manner, lncRNAs with specific miRNA target sites can act as ‘competing endogenous’ (ce)RNAs to modulate post-transcriptional gene expression. In agreement with this hypothesis, depletion or overexpression of linc-MD1 retarded or accelerated muscle differentiation, respectively. Further, linc-MD1 positively regulated the translation of MAML1 and MEF2C mRNAs by sequestering miR-133 and miR-135 in mouse and human myoblasts.38 Interestingly, miR-206 and miR-133b arise from an intron and an exon of linc-MD1, respectively, highlighting additional layers of mRNA regulation by lncRNAs and miRNAs.38
H19
The long-studied lncRNA H19 also contains microRNAs and is also dually implicated in the control of mRNA translation. Reik and colleagues identified HuR as an RNA-binding protein that interacts with H19 and potently represses the biogenesis of miRNAs present in H19.405 HuR represses the synthesis of the placenta-specific microRNA miR-675, contained within the first exon of H19, at the level of Drosha-mediated processing. In turn, miR-675 inhibits proliferation of many cells including embryonic stem cells, trophoblastic stem cells, mouse embryo fibroblasts, and mouse myoblasts (C2C12). Among other targets, miR-675 reduces the translation of the insulin growth factor receptor 1 (Igf1r) without affecting Igf1r mRNA levels. Thus, H19 limits placenta growth in part by regulating miR-675 production in an HuR- and Dicer-dependent manner.
LncRNAs and model organisms
To understand the biological context of the processes in which lncRNAs regulate gene expression post-transcriptionally, there is pressing need for model organisms where we can study their roles in growth, differentiation and development. Recently, the phenotype of Malat1 knock-out mice was studied simultaneously by three groups.416–43 Even though Malat1 is among the most abundant and conserved lncRNAs, it was unexpectedly found to be dispensable for pre- or post-natal mouse development. Indeed, the absence of Malat1 did not lead to gross phenotypic or histological abnormalities compared to wild type mouse, although small number of neighboring genes were dysregulated in adult Malat1-null mice.41–43
In contrast to MALAT1, suppression of neuronal lncRNA function in zebrafish resulted in abnormal neural development.8 Bartel and colleagues characterized 550 zebrafish lncRNAs by analyzing a combination of chromatin marks, poly(A)-site maps, and RNA-seq data; among these, only 29 lncRNAs shared sequence similarity and genomic location with putative mammalian orthologs. Morpholino-mediated suppression of two particular lncRNAs in zebrafish embryos (cyrano and megamind), showed strong defects in embryonic development, a defect that was rescued by introducing human or mouse orthologs.8 This study provides a roadmap for identifying and analyzing lncRNAs in model organisms.
Concluding remarks and perspectives
An expanding body of evidence reveals that lncRNAs control gene expression on multiple levels via a number of complex mechanisms. In addition to their well-established influence as regulators of transcription, lncRNAs are also effective modulators of pre-mRNA splicing, mRNA decay, and translation. In light of these functions, lncRNAs should be included as bona fide post-transcriptional regulators of gene expression alongside two well-established families of factors, RNA-binding proteins and microRNAs. Moreover, many more examples are expected to come to light of lncRNAs eliciting post-transcriptional actions by competing or cooperating with RNA-binding proteins, microRNAs, and perhaps even other lncRNAs.
LncRNAs constitute a unique family of post-transcriptional regulators, since their function is not mainly dependent on sequence (as is the case for microRNAs) or structure (as for RNA-binding proteins). Instead, lncRNAs, particularly larger lncRNAs, can function both by sequence homology/complementarity with other nucleic acids, and by structure, forming molecular frames and scaffolds for assembly of macromolecular complexes. Some of these functions have already been tied to transcriptional processes such as the recruitment of transcriptional activators and repressor, and of chromatin remodeling factors.3 In a similar manner, assembly of post-transcriptional processing centers may also be facilitated by lncRNAs. For example, they may exert this effect in the nucleus by helping assemble components of the splicing apparatus, and in the cytoplasm by assisting with the building and/or function of processing bodies, stress granules, the exosome complex, or cytoskeletal mRNP (messenger ribonucleoprotein) motors.
In closing, the analysis of lncRNAs remains challenging, since lncRNAs are a heterogeneous class of transcripts, are incompletely annotated, exist in the nucleus and the cytoplasm, can function through their sequence and structure, and associate with DNA, RNA, and protein. However, the growing recognition that lncRNAs are implicated in development, phsyiology, and disease44 warrants a great deal of future attention to this RNA family. Given the rich, versatile, and sometimes unexpected functions of lncRNAs identified thus far, future investigation of lncRNA actions promises to be an exciting journey.
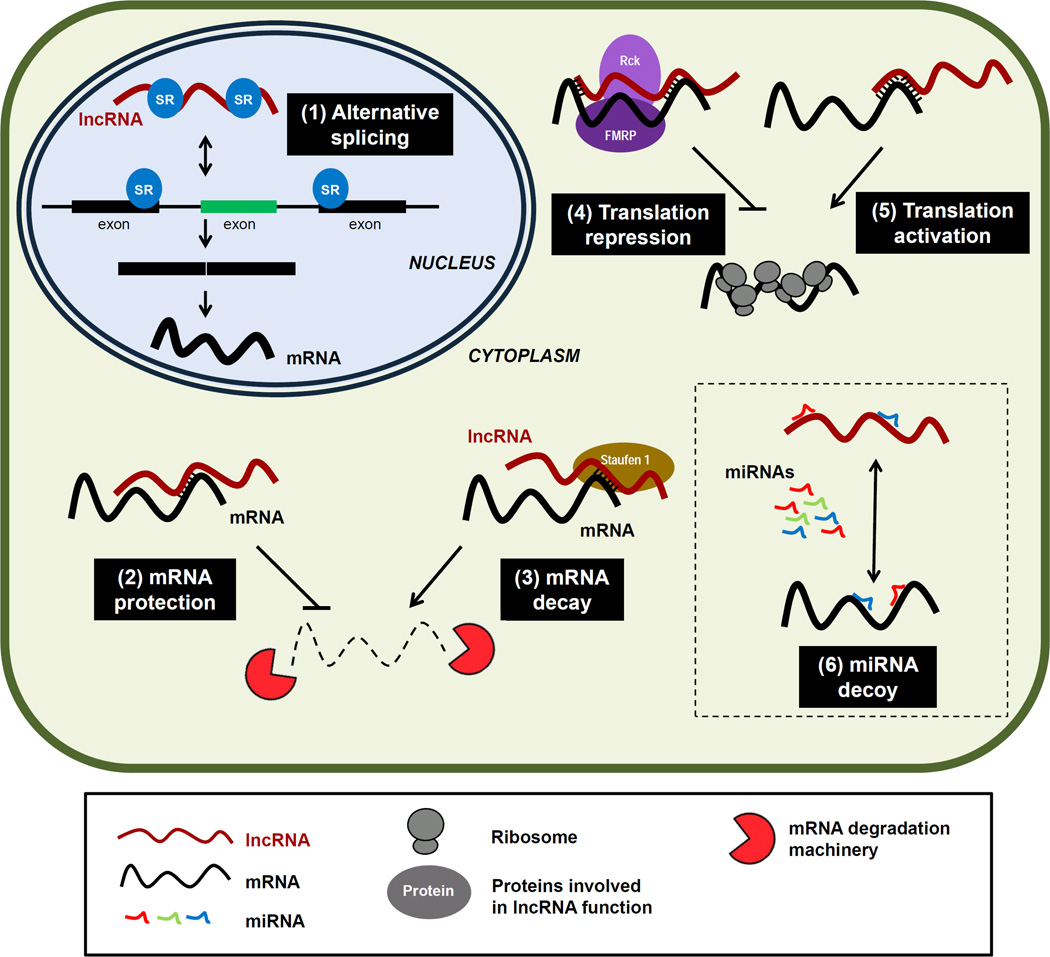
Figure 1.
Levels of post-transcriptional gene regulation by lncRNAs. The major post-transcriptional processes influenced by lncRNAs are indicated in black boxes. (1) Splicing of pre-mRNAs, which is modulated by lncRNAs that compete for binding for splicing regulatory proteins (e.g., SR, Fox). (2) Protection from mRNA decay, as exemplified by lncRNA BACE1-AS, which forms a hybrid and hence prevents the decay of BACE1 mRNA. (3) Acceleration of mRNA decay, as reported for ½-sbsRNAs, which function jointly with Staufen 1 to promote the decay of Alu-containing mRNAs. (4) Repression of mRNA translation, as demonstrated for lincRNA-p21 via interaction with partially complementary mRNAs and recruitment of translation repressors Rck and Fmrp. (5) Activation of mRNA translation, as illustrated by the interaction of AS Uchl1 via a SINEB2 sequence and a segment fully complementary with the 5’ end of Uchl1 mRNA; this association helps to recruit ribosomes to Uchl1 mRNA and enhances its translation. (6) Functional association with microRNAs, as shown for linc-MD1, which ‘sponges’ microRNAs, for H19, which hosts microRNAs, and for BACE1-AS, which promotes BACE1 mRNA translation by competing with a microRNA. See text for further details.
Table 1.
LncRNAs controlling gene expression post-transcriptionally. Listed are the post-transcriptional processes affected by lncRNAs (column 1), the lncRNAs reported in each regulatory process (column 2), the target molecules (RNAs and proteins) through which the lncRNAs elicit a post-transcriptional influence (column 3), and additional details of the post-transcriptional consequences of lncRNA activity.
| Post-transcriptional processes |
LncRNA | Target | Comments | References |
|---|---|---|---|---|
| Splicing | MALAT1 | Pre-mRNAs regulated by SR proteins | Modulation of splicing patterns | [17] |
| sno-lncRNAs | Pre-mRNAs regulated by Fox proteins | Modulation of splicing patterns | [30] | |
| mRNA Turnover | ½-sbsRNAs | mRNAs bearing Alu sequences, Staufen-1 targets | Increase in mRNA decay | [18] |
| BACE1-AS | BACE1 mRNA | Increase in mRNA stabilization | [19] | |
| Translation | lincRNA-p21 | JUNB and CTNNB mRNAs | Inhibition translation (specific mRNAs) | [21] |
| AS Uchl1 | Uchl1 mRNA | Promotion of translation (specific mRNA) | [36] | |
| BC1 | eIF4A and PABP | Modulation of translation (global) | [20] | |
| miRNA Function | Linc-MD1 | Decoy for miR-133 and miR-135 | Increased translation of MAML1 and MEF2C mRNAs | [38] |
| H19 | Gives rise to miR-675 | Decreased expression of IGF1R mRNA (miR-675 target) | [40] | |
| BACE1-AS | BACE1 mRNA | Competition with miR-485-5p for binding to BACE1 mRNA | [34] | |
HIGHLIGHTS.
Long noncoding RNAs are emerging as key post-transcriptional gene regulatory factors
LncRNAs can control splicing by sequestering splicing regulatory proteins
LncRNAs can modulate target mRNA turnover via partial or extended complementarity
LncRNAs can affect translation by interacting with target mRNAs, recruiting proteins
LncRNAs can function by competing or cooperating with microRNAs
ACKNOWLEDGMENT
JHY, KA, and MG were supported by the NIA-IRP, NIH.
Abbreviations
- lncRNA
long noncoding RNA; lncRNA
- lincRNA
large intergenic noncoding RNA
- RBP
RNA-binding protein
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 2.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. Highresolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 8.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul J, Duerksen JD. Chromatin-associated RNA content of heterochromatin and euchromatin. Mol. Cell. Biochem. 1975;9:9–16. doi: 10.1007/BF01731728. [DOI] [PubMed] [Google Scholar]
- 11.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 12.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 13.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclearretained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed- forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Iacoangeli A, Lin D, Williams K, Denman RB, Hellen CU, et al. Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 2005;171:811–821. doi: 10.1083/jcb.200506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilusz JE, Jnbaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3'-end sequences of MALAT1 and MENâ noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 2012 doi: 10.1073/pnas.1217338109. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. Conservation of a triplehelix- forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012;2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Hallegger M, Llorian M, Smith CW. Alternative splicing: global insights. FEBS. J. 2010;277:856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 28.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO. J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, et al. Long Noncoding RNAs with snoRNA Ends. 2012 doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 31.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 33.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. EMBO. J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO. J. 2012 doi: 10.1038/emboj.2012.292. 2012 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. New Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012 doi: 10.1038/nature11508. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 37.Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu. Rev. Cell. Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA Malat1 Is Dispensable for Mouse Development but Its Transcription Plays a cis-Regulatory Role in the Adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eißmann M, Gutschner T, Hämmerle M, Günther S, Caudron-Herger M, Groß M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8) doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]