Abstract
The chimeric proteins, silk-elastin-like protein polymers (SELPs), consist of repeating units of silk and elastin to retain the mechanical strength of silk, while incorporating the dynamic environmental sensitivity of elastin. A retinal-modified silk-elastin-like protein polymer was prepared, modified and studied for photodynamic responses. The protein was designed, cloned, expressed and purified with lysine present in the elastin repeats. The purified protein was then chemically modified with the bio-compatible moiety retinal via the lysine side chains. Structural changes with the polymer were assessed before and after retinal modification using Fourier Transform Infrared Spectroscopy and Circular Dichroism Spectroscopy. Optical studies and spectral analysis were performed before and after retinal-modification. The random coil fraction of the protein increased after retinal modification while the β-sheet fraction significantly decreased. Birefringence of the modified protein was induced when irradiated with a linearly polarized 488 nm laser light. Retinal modification of this protein offers a useful strategy for potential use in biosensors, controlled drug delivery and other areas of biomedical engineering.
Keywords: Silk, elastin, retinal, light, photoresponsive
INTRODUCTION
Protein-based biomaterials such as silks,1 elastins,2 collagens,3,4 and resilins5 are studied for drug delivery and tissue engineering because of their biocompatibility, controllable degradation and minimal cytotoxicity.6 Recombinant DNA technologies allow the synthesis of biopolymers with precise control of primary sequence and chain length, as well as placement of chemical handles to permit functionalization to enhance interactions with cells and surfaces. This versatility and control of protein polymers provides remarkable design options to extend biological and mechanical properties. Silk-elastin-like protein polymers (SELPs), consisting of repeating units of silk and elastin have been utilized to exploit the mechanical strength of silk with the environmental sensitivity of elastin.7 Some SELPs have been fabricated into nanoparticles, hydrogels, microdiameter fibers and nanofibrous scaffolds, displaying useful properties for drug delivery and tissue engineering.3,7-14 The structure of SELPs can be modified at the single amino acid residue level to allow the introduction of functional motifs to control self-assembly and sensitivity to external stimuli.9 For example, in our previous studies a two-step self-assembly process of SELPs was reported relative to the ratio of silk-to-elastin domains, indicating the potential tunable control of structural transitions.7
The small-size of the repetitive sequence in the elastin primary structure, such as VPGXG (X = V, I, A) allows for the introduction of alternative amino acid residues such as K, Y and E for facile chemical modifications of the polypeptides.3 Azobenzene derivatives, a group of photosensitive molecules, were successfully introduced onto genetically engineered elastin molecules, and the transition temperature (Tt) of these modified elastins was controlled by ultraviolet irradiation due to the trans-cis isomerization of the azobenzene groups.15 Azo-modified silk films, synthesized via diazonium coupling between tyrosine residues of silk and azobenzene derivatives, also demonstrated nonlinear optical properties, such as optically induced birefringence, holographic recording and optically induced surface relief gratings.16 The studies of these materials related to optical holography, reaction efficiencies and ultrafast dynamics, are all areas of interest in the optics and materials communities. While azo materials do possess attractive optical properties, their questionable biocompatibility motivated us to consider the use of retinal, which, as the basis of the visual system, represents a truly biocompatible photosensitizer. Retinal also has more complex photoisomerization pathsways, and so is more amenable to engineering optimization.
Retinal is a promising candidate for holographic memory applications triggered by all-trans to cis isomerization of a protein-bound retinal protonated Schiff base (RPSB) chromophore.17,18 Recently new retinal nano-ceramic thin films have been fabricated for photonic applications, and the Schiff base in the retinal films had a substantial effect on optical properties, suggesting a good candidate for holographic storage.18 In the present study, silk-elastin-like polymers were synthesized and modified with retinal protonated Schiff Base, to generate an material for light-induced dynamic changes. This new silk-elastin-like polymer, termed PS2E8K, includes the introduction of lysine. The structure and optical properties of the protein were analyzed by circular dichroism (CD) spectroscopy, Fourier Transform Infrared (FTIR) and optical polarization.
RESULTS AND DISCUSSION
SELP Synthesis and Purification
Seamless cloning strategies were used for the biosynthesis of the SELP 7 and PS2E8K with a silk-to-elastin ratio at 2: 8 was generated with lysine (K) in the second amino acid position of the fifth elastin block. This chemical handle provided for further chemical cross-linking via –NH2 groups. The purity and molecular weight of SELPs was confirmed by SDS-PAGE analysis (Figure 1A). The yield of purified PS2E8K was about 50 mg per liter. The molecular weight of PS2E8K was 38-49 kDa (Figure 1A), estimated from SDS-PAGE, and determined to be 44.5 kDa by MALDI-TOF mass spectrometry (Figure 1B).
Figure 1.
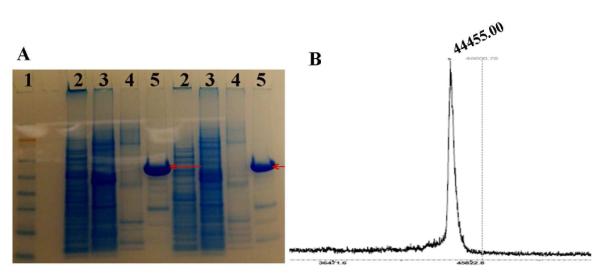
Characterization of purified silk-elastin-like polymer PS2E8K. A. Protein purification determination by 4-12% SDS-PAGE. Lane 1, blue plus2 pre-stained standard (Invitrogen) used as size markers (188, 98, 62, 49, 38, 28, 17, 14, 6, 3 kDa); Lane 2-4, collected fractions from Ni-NTA columns after washing buffer; Lane 5, collected fractions from Ni-NTA columns with elution buffer. B. MALDI-TOF mass spectrum of PS2E8K.
Chemical Modification
The formation of a Schiff base between the lysine and retinal was conducted as shown in Figure 2. The amine groups of lysine side chains act as nucleophilic agents and attack the aldehyde carbons of retinal molecules to form carbon nitrogen double bonds under basic environment. Once the photosensitive molecules were integrated into PS2E8Y, optical properties of the modified biopolymer were investigated (Figure 2). Organic solvents can induce β-sheets to form with silk proteins and result in insolubility,1,5 therefore, we selected dimethyl sulfoxide (DMSO) versus methanol in these reactions to avoid this complication. The silk-elastin copolymer had good solubility in DMSO (Figure 2, insert). The retinal-modified silk-elastin products appear red-orange color. This finding suggests that the retinal Schiff base was protonated, and the positive charge delocalization along the conjugated carbon chain of retinal was stabilized by one or more interactions, such as hydrogen bonding or electrostatic repulsion, probably attributed from the appropriate folding of the copolymer. The charge delocalization caused red-shifts of retinal modified silk-elastin copolymer, but the hydrogen of the protonated Schiff base may not be distinguished by NMR spectra.17,27
Figure 2.
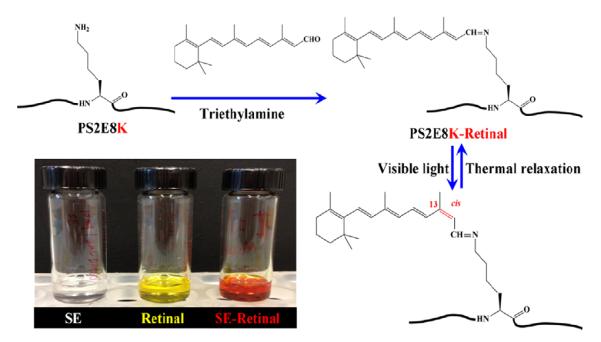
Schematic for retinal modification of the lysine residues in the silk-elastin copolymer PS2E8K. Insert: Chemical reactions for PS2E8K before and after retinal modification (Left: PS2E8K in DMSO solution; Middle: retinal molecules in DMSO solution; Right: retinal-modified PS2E8K in DMSO solution).
1H-NMR analysis of silk-elastin-like polymers before and after retinal modification
Comparative studies of silk-elastin-like polymers before and after retinal modification were performed by 1H-NMR (Figure 3).
Figure 3.
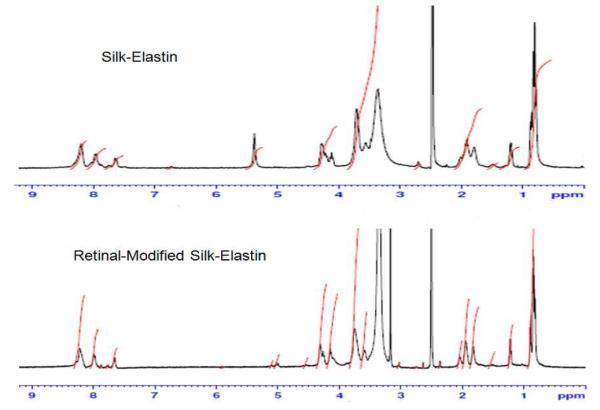
1H-NMR analysis of silk-elastin-like polymers before (upper panel) and after (lower panel) retinal modification.
Silk-elastin-like polymer PS2E8K (500 MHz, DMSO-d6): δH/ppm 0.83, 0.89 (s, Valγ), 1.22, 1.23 (m, Alaβ), 1.51 (br, Lysγ), 1.81 (br, Lysβ/Lysδ), 1.90, 1.92 (br, Proγ/Valβ), 2.01 (m, Proβ), 3.57 (br, Proδ), 3.72 (br, Serβ/Glyα), 4.12 (br, Glyα), 4.21 (br, Alaα/Valα/Lysα), 4.28 (br, Serα/Hisα/Proα), 7,63 (br, Lys ε (Lys-NH2)), 7.98, 8.20 (br, HN/Hisδ, ε).
Retinal-modified silk-elastin-like polymer PS2E8K (500 MHz, DMSO-d6): δH/ppm 0.84, 0.89 (s, Valγ ), 1.22, 1.23 (m, Alaβ), 1.52 (br, Lysγ), 1.83 (br, Lysβ/Lysδ), 1.95 (br, Proγ/Valβ), 2.04 (m, Proβ), 3.17 (s, Hisβ), 3.60 (br, Proδ), 3.75 (br, Serβ/Glyα), 4.15 (br, Glyα), 4.26 (br, Alaα/Valα/Lysα), 4.31 (br, Serα/Hisα/Proα), 7,65 (br, Lys ε (Lys-NH2)), 7.98, 8.21 (br, HN/Hisδ, ε).
The chemical shift of lys-NH2 is around 7.65 ppm, and 3.57% of lys-NH2 dropped to 2.21% after retinal modification according to peak area integral analysis, implying that about 38.1% of lysine residues were involved in the Schiff base reaction. The peak of 2.51 ppm is assigned for DMSO-d6 solvent, and 3.36 ppm is the peak of H O in CD3SO.28,29
Structural characterization
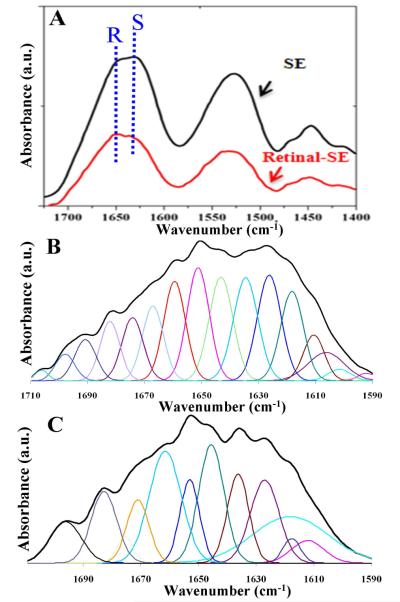
Lyophilized powders or dried films of the recombinant protein and retinal-modified PS2E8K were examined by attenuated total reflectance (ATR) FTIR spectroscopy (Figure 4A), which includes an expansion of the Amide I region for secondary structure analysis. The broad nature of the Amide I bands (centered around 1650 cm−1) indicated a range of heterogeneous conformations for both the unmodified and retinal-modified silk-elastin copolymer PS2E8K. Peak deconvolution suggested potential contributions from all known secondary structures (Figure 4B,C). Secondary structure changes were further analyzed for the polymer PS2E8K before and after retinal-modification (Table 1). Random coil content increased from 31% to 43% while the beta-sheet content decreased from 31% to 20% before and after retinal-modification, indicating that the modification induced conformational changes. To confirm the structural changes of the retinal-modified PS2E8K, secondary structure was also assessed by CD spectroscopy (Figure 5). CD spectra of both the unmodified and retinal-modified PS2E8K showed an increase in signal at 210 nm, suggestive of induction of a type II beta-turn conformation, similar to elastin-like polymers as observed previously.7 The positive ellipticity at 190 nm and negative ellipticity at 220 nm indicated the induction of a beta-sheet conformation in both polymers, a feature usually observed for silk polymers.6 Compared with unmodified PS2E8K, the negative signal near 195 nm suggested more disordered structure in the retinal-modified PS2E8K, consistent with the FTIR spectra.
Figure 4.
Structure characterization of unmodified and retinal-modified silk-elastin polymer PS2E8K by FTIR. A. FTIR spectra. R represents random coil, S for beta-sheet structure. B & C. Selected FTIR absorbance spectra of unmodified (B) and retinal-modified (C) silk-elastin copolymer in the amide I’ regions after Fourier self-deconvolution. The heavy line represents the deduced absorbance band. The light lines represent the contributions to the amide I’ band.
Table 1.
Secondary structure of unmodified and retinal- modified silk-elastin polymer PS2E8K by FTIR.
| Beta- Sheets |
Random Coils |
Alpha Helices |
Turns | |
|---|---|---|---|---|
| PS2E8K | 31% | 31% | 13% | 25% |
| Retinal-modified PS2E8K | 20% | 43% | 10% | 26% |
Figure 5.
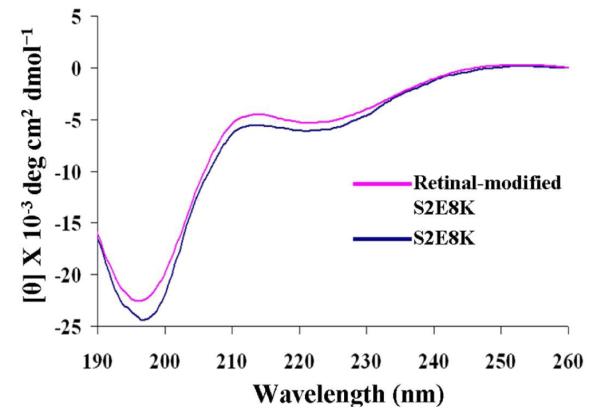
Far-UV CD spectra of unmodified and retinal-modified silk-elastin copolymer PS2E8K. All spectra were recorded at room temperature (25°C) using a 0.5 mm path-length quartz cell with protein concentrations of 0.5 mg/ml.
Optical polarization and spectral analysis
Thin films of approximately 10 μm thick retinal-modified PS2E8K were prepared and dried on glass slides (Figure 6A). As reported previously and shown in Figure 6B,16 the probe was split into p and s polarization by means of a Wollaston prism after passing through the all-trans retinal-modified PS2E8K film. The setup was slightly different from that described in reference 15 in that the incident 632.8 nm probe beam was p polarized instead of circularly polarized. This change enabled more accurate measurement of the birefringence to be made since the birefringence in the PS2E8k film was a factor of 10 lower than that observed for azo-modified silk. The smaller effect is more easily seen in an instrument where a null result leads to a zero output: p polarized light whose polarization is unaltered by the film is completely blocked by an orthogonally oriented polarizer. The intensity of the transmitted s polarized beams was monitored using photodiodes and compared to that resulting from the use of a known azo-silk standard. Figure 6C shows the birefringence versus exposure time of the all-trans retinal-modified silk-elastin copolymer when illuminated with 1 Wcm−2 of 488 nm wavelength argon laser light. The first part of the figure is the response to linearly polarized light and the second part is the erasure response to circularly polarized light. The retinal-modified films exhibited the optically in-duced effects similar to those found in other polymers incorporating azobenzene moieties, such as azobenzene modified Bombyx mori silk.16 We hypothesize that initially, the all-trans retinal molecules retained their low energy state, the trans state. After the retinal-modified sample was irradiated by the light of proper wavelength with a polarization component parallel to the trans structure, the retinal molecule was excited to the cis state, while the excess trans isomers perpendicular to the polarization build up in a form of spatial hole-burning. This optically induced anisotropy in the all-trans retinal distribution leads to birefringence.16 Therefore, the detected bire-fringence of the retinal-modified silk-elastin copolymer was caused by the anisotropic structure of the sample which was induced by the trans-cis isomerization of retinal molecules. The double exponential data fits for birefringence retardation amplitude and time constants for azo-silk and retinal-PS2E8K are shown in Table 2.
Figure 6.
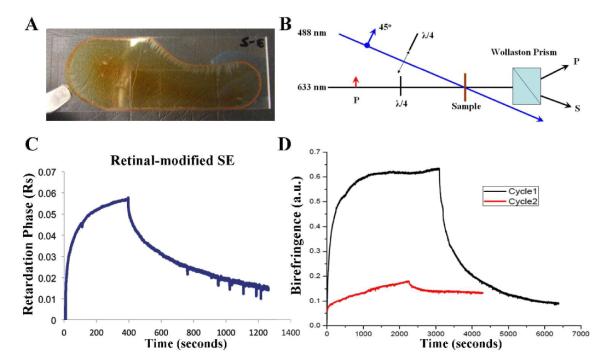
Optical polarization of retinal-modified polymer films. A. Retinal-modified PS2E8K film dried onto a glass slide. B. Birefringence measurement setup. The birefringence written using a 488-nm argon ion laser beam polarized at 45°
Table 2.
The double exponential data fits for azo-silk and retinal-PS2E8K.
| Steady State Phase Retardation (radians) |
Time Constant 1 (seconds) |
Time Constant 2 (seconds) |
|
|---|---|---|---|
| Azo-silk | 1.38 | 6.2 | 173 |
|
Retinal-
PS2E8K |
0.058 | 15.3 | 124 |
We also noticed that the optical behavior of retinal-silk-elastin was different from that of azo dye modified polymers. The optical effects in azo polymers are usually simply and repetitively reversible by switching the polarization of the incident light, since there is only one cis state,16 while the retinal molecule has several different cis states, such as 11-cis, 9-cis, and 13-cis. Retinal in protein environment would give trans to 13-cis isomerization, while retinal protonated Schiff base in solution would prefer trans to 11-cis transition,17 and which cis form generated in this situation needs to be determined in the near future work. After one cycle of writing and erasure, the optical sensitivity of the retinal-modified silk-elastin films was substantially reduced. (Figure 6D) to the horizontal. The birefringence was probed with a p polarized He-Ne laser. In the absence of birefringence, the powers in the P and S polarizations after the Wollaston prism were equal. In the presence of birefringence, the powers became unbalanced, and the degree of imbalance indicated the extent of birefringence. The birefringence was erased by inserting a quarter-wave plate into the writing beam to render it circularly polarized. C. The birefringence retardation versus exposure time of the retinal-modified silk-elastin copolymer PS2E8K, first with linearly polarized light to induce birefringence, then with circularly polarized light for erasure. The intensity of the actinic light was 1 Wcm−2 at a wavelength of 488 nm. D. Bire-fringence retardation in PS2E8K for the same experimental conditions as C, showing the first cycle of recording and erasing, and the second cycle in the same spot of recording and erasing. This behavior contrasts to that of azo-silk in which the second cycle would be the same as the first.
Therefore, investigating the proportion of the various isomers in a retinal-modified silk-elastin film after light irradiation would be of interest, as well as detection of volume changes due to isomerization. 30-32
In total, the studies reported here demonstrate that this new SELP with retinal modifications provides a useful platform to study optically-indicated changes in material structure and morphology.33,34 The tunable features of the proteins, the SELPs, combined with the interesting polarization associated changes in structure, suggest future directions towards novel biomaterials for a range of potential utilities.35,36 This may include selective control of multimodal drug delivery, medical materials that can change properties upon irradiation inputs, and many related themes.
CONCLUSIONS
A recombinant silk-elastin-like polymer, PS2E8K, was generated and characterized as a soluble protein, with the lysine residues as chemical handles for reaction with retinal. The material features of both the unmodified and retinal-modified PS2E8K were investigated for structural and optical properties. The findings suggest this retinal-modified silk-elastin-like polymer with efficient light-induced processes could be further studied for self-assembly, drug delivery, and biomedical applications.
Supplementary Material
ACKNOWLEDGMENT
Support from the NIH EB014283 and EB002520 is gratefully acknowledged, as is support from the Air Force Office of Scientific Research.
Footnotes
The authors declare no competing financial interest.
SUPPORTING INFORMATION Please see Supporting Information for experimental details. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Omenetto FG, Kaplan DL. Science. 2010;329:528. doi: 10.1126/science.1188936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Vasconcelos A, Gomes AC, Cavaco-Paulo A. Acta Biomater. 2012;8:3049. doi: 10.1016/j.actbio.2012.04.035. [DOI] [PubMed] [Google Scholar]
- (3).Bracalello A, Santopietro V, Vassalli M, Marletta G, Del Gaudio R, Bochicchio B, Pepe A. Biomacromolecules. 2011;12:2957. doi: 10.1021/bm2005388. [DOI] [PubMed] [Google Scholar]
- (4).An B, Desrochers TM, Qin G, Xia X, Thiagarajan G, Brodsky B, Kaplan DL. Biomaterials. 2013;34:402. doi: 10.1016/j.biomaterials.2012.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Qin G, Hu X, Cebe P, Kaplan DL. Nat Commun. 2012;3:1003. doi: 10.1038/ncomms2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Chow D, Nunalee ML, Lim DW, Simnick AJ, Chilkoti A. Mater Sci Eng R Rep. 2008;62:125. doi: 10.1016/j.mser.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Xia XX, Xu Q, Hu X, Qin G, Kaplan DL. Biomacromolecules. 2011;12:3844. doi: 10.1021/bm201165h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Qiu W, Cappello J, Wu X. Appl Phys Lett. 2011;98:263702. doi: 10.1063/1.3604786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chang J, Peng XF, Hijji K, Cappello J, Ghandehari H, Solares SD, Seog J. J Am Chem Soc. 2011;133:1745. doi: 10.1021/ja110191f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Qiu W, Huang Y, Teng W, Cohn CM, Cappello J, Wu X. Biomacromolecules. 2010;11:3219. doi: 10.1021/bm100469w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gustafson JA, Price RA, Greish K, Cappello J, Ghandehari H. Mol Pharm. 2010;7:1050. doi: 10.1021/mp100161u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Greish K, Araki K, Li D, O’Malley BW, Jr., Dandu R, Frandsen J, Cappello J, Ghandehari H. Biomacromolecules. 2009;10:2183. doi: 10.1021/bm900356j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Haider M, Leung V, Ferrari F, Crissman J, Powell J, Cappello J, Ghandehari H. Mol Pharm. 2005;2:139. doi: 10.1021/mp049906s. [DOI] [PubMed] [Google Scholar]
- (14).Megeed Z, Cappello J, Ghandehari H. Adv Drug Deliv Rev. 2002;54:1075. doi: 10.1016/s0169-409x(02)00063-7. [DOI] [PubMed] [Google Scholar]
- (15).Alonso M, Reboto V, Guiscardo L, Mate V, Rodriguez-Cabello JC. Macromolecules. 2001;34:8072. [Google Scholar]
- (16).Cronin-Golomb M, Murphy AR, Mondia JP, Kaplan DL, Omenetto FG. J Polym Sci Pol Phys. 2012;50:257. [Google Scholar]
- (17).Sovdat T, Bassolino G, Liebel M, Schnedermann C, Fletcher SP, Kukura P. J Am Chem Soc. 2012;134:8318. doi: 10.1021/ja3007929. [DOI] [PubMed] [Google Scholar]
- (18).Wu PF, Bhamidipati M, Coles M, Rao DVGLN. Chem Phys Lett. 2004;400:506. [Google Scholar]
- (19).Qin G, Lapidot S, Numata K, Hu X, Meirovitch S, Dekel M, Podoler I, Shoseyov O, Kaplan DL. Biomacromolecules. 2009;10:3227. doi: 10.1021/bm900735g. [DOI] [PubMed] [Google Scholar]
- (20).Qin G, Rivkin A, Lapidot S, Hu X, Preis I, Arinus SB, Dgany O, Shoseyov O, Kaplan DL. Biomaterials. 2011;32:9231. doi: 10.1016/j.biomaterials.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Murphy AR, Kaplan DL. J Mater Chem. 2009;19:6443. doi: 10.1039/b905802h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Serban MA, Panilaitis B, Kaplan DL. J Biomed Mater Res A. 2011;98:567. doi: 10.1002/jbm.a.33149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Serban MA, Kaplan DL. Biomacromolecules. 2010;11:3406. doi: 10.1021/bm100925s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Murphy AR, St John P, Kaplan DL. Biomaterials. 2008;29:2829. doi: 10.1016/j.biomaterials.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Xu L, Tremblay ML, Meng Q, Liu XQ, Rainey JK. Biomol NMR Assign. 2012;6:147. doi: 10.1007/s12104-011-9344-z. [DOI] [PubMed] [Google Scholar]
- (26).Yazawa K, Yamaguchi E, Knight D, Asakura T. Biopolymers. 2012;97:347. doi: 10.1002/bip.21718. [DOI] [PubMed] [Google Scholar]
- (27).Han M, Smith SO. Biochemistry. 1995;34:1425. doi: 10.1021/bi00004a037. [DOI] [PubMed] [Google Scholar]
- (28).Asakura T, Nishi H, Nagano A, Yoshida A, Nakazawa Y, Kamiya M, Demura M. Biomacromolecules. 2011;12:3910. doi: 10.1021/bm2011196. [DOI] [PubMed] [Google Scholar]
- (29).Creager MS, Izdebski T, Brooks AE, Lewis RV. Comp Biochem Physiol A Mol Integr Physiol. 2011;159:219. doi: 10.1016/j.cbpa.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Omenetto F, Kaplan D. Sci Am. 2010;303:76. [PubMed] [Google Scholar]
- (31).Tao H, Chieffo LR, Brenckle MA, Siebert SM, Liu M, Strikwerda AC, Fan K, Kaplan DL, Zhang X, Averitt RD, Omenetto FG. Adv Mater. 2011;23:3197. doi: 10.1002/adma.201100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tao H, Kaplan DL, Omenetto FG. Adv Mater. 2012;24:2824. doi: 10.1002/adma.201104477. [DOI] [PubMed] [Google Scholar]
- (33).Tao H, Amsden JJ, Strikwerda AC, Fan K, Kaplan DL, Zhang X, Averitt RD, Omenetto FG. Adv Mater. 2010;22:3527. doi: 10.1002/adma.201000412. [DOI] [PubMed] [Google Scholar]
- (34).Tao H, Brenckle MA, Yang M, Zhang J, Liu M, Siebert SM, Averitt RD, Mannoor MS, McAlpine MC, Rogers JA, Kaplan DL, Omenetto FG. Adv Mater. 2012;24:1067. doi: 10.1002/adma.201103814. [DOI] [PubMed] [Google Scholar]
- (35).Omenetto FG, Kaplan DL. Biomaterials. 2010;31:6119. doi: 10.1016/j.biomaterials.2010.05.003. [DOI] [PubMed] [Google Scholar]
- (36).Omenetto FG, Kaplan DL. Nat Mater. 2012;11:273. doi: 10.1038/nmat3291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.